Open Access Article
Open Access ArticleBiomimetic synthesis of RPL14B-based CdSe quantum dots for the detection of heavy metal copper ions
Lipeng Zhong†
 ab,
Wenyue Liu†
ab,
Wenyue Liu† b,
Zhixiong Xie‡
*b and
Jiye Liu‡*b
b,
Zhixiong Xie‡
*b and
Jiye Liu‡*b
aDepartment of Clinical Laboratory, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang, Jiangxi 330006, China
bHubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan 430072, China. E-mail: zxxie@whu.edu.cn; 2017102040034@whu.edu.cn; jiyeliu-doc@whu.edu.cn
First published on 24th May 2024
Abstract
In the present study, an Escherichia coli-expressed yeast ribosomal protein was used as a template for synthesizing RPL14B-based CdSe quantum dots in vitro via the quasi-biosynthesis strategy at low temperature. The synthetic bionic RPL14B-based CdSe quantum dots were characterized using TEM, HRTEM, and EDX spectra, and the results showed that the synthesized quantum dots were CdSe quantum dots with a crystal face spacing of 0.21 and 0.18 nm. The biomimetic method-synthesized quantum dots exhibited the characteristics of a uniform particle size, good dispersion, and strong photobleaching resistance. Moreover, the fluorescence of the RPL14b-based CdSe quantum dots could be specifically quenched using Cu2+ in a linear range of 0.2–10 μM. Finally, these RPL14b-based CdSe quantum dots can be used for the specific detection of heavy metal copper ions in addition to other applications in biological analyses.
Introduction
Copper is a heavy metal and one of the essential trace elements for the human body.1–3 Copper deficiency can lead to certain dysfunctions in the human body; however, its excessive intake can pose a serious threat to health.4 At present, the methods commonly used for copper detection include inductively coupled mass spectrometry, electrochemical luminescence, atomic absorption spectrometry, and emission spectrometry.5–7 These detection methods have some disadvantages, including difficult operation and weak anti-interference ability. Therefore, it is important to develop a method with high sensitivity, strong specificity, and simple operation for detecting copper.With the rapid development of nanotechnology, nanofluorescent probes with high sensitivity, short detection time, simple operation, and good selectivity have been favored.8,9 In this context, semiconductor quantum dots (QDs) have attracted much attention because of their unique size-dependent optical properties, narrow spectral luminescence, strong fluorescence intensity, and excellent photobleaching resistance. At present, semiconductor QDs and carbon dots have been used for detecting several heavy metal ions, such as mercury, iron, chromium, copper, and lead.10–12 QD synthesis methods can be divided into chemical synthesis, microbial synthesis, and quasi-biological synthesis.13–15 In previous studies, a method of QD synthesis in Saccharomyces cerevisiae cells was proposed using the temporal–spatial coupling strategy.16 Moreover, in other studies on CdSe QD synthesis in S. cerevisiae, proteins wrapped on the surface of CdSe QDs were identified and the ribosomal protein RPL14B was determined to participate in CdSe QD synthesis (data not published). Moreover, it has been shown that embedding fluorescent quantum clusters in proteins can improve their water solubility, stability, and target specificity with reduced cost compared with chemically synthesized QDs.17 In the current study, the yeast ribosomal protein RPL14B expressed in Escherichia coli was used as a template for synthesizing bionic composite QDs. The in vitro bionic CdSe QDs wrapped in RPL14B were used to detect copper ions, suggesting broad application prospects.
Experimental
Materials
IPTG, reduced glutathione (GSH), NaBH4, Na2SeO3, CdCl2 (Sigma Corporation USA); plasmid extraction kit (Tiangen Biochemical Technology Co., Ltd); E. coli DH5α, E. coli BL21(DE3) and pET26b plasmids (lab-preserved). Xho I and Nde I enzymes (New England Biolabs). Culture medium. CaCl2, FeCl3, KCl, MgCl2, BaCl2, CuCl2 (Sinopsin Group Chemical Reagent Co., Ltd). All chemicals used are analytical grade. Ultrapure water is used for all aqueous solutions.Recombinant expression of RPL14B by E. coli
E.coli DH5α and E.coli BL21(DE3) were used to construct heterogeneously expressing strains of ribosomal protein. The pET26b-rpl14b vector is constructed by E.coli DH5α, while RPL14B22 is expressed by E.coli BL21(DE3). PCR amplification was utilized to extract the DNA fragment of RPL14B. After doubly digesting the PCR product and pET26b vector with Xho I and Nde I, the purified enzyme-digested product was linked for an overnight period at 16 °C. After that, the conjugate product was introduced into E.coli DH5α, positive clones were chosen, plasmids were removed and submitted to be sequenced, and the appropriate vector—dubbed pET26b-rpl14b—was chosen. For the purpose of the next studies, the vector was moved to the BL21/pET26b-rpl14b strain of E.coli BL21(DE3). After being reconstituted in 500 mL of LB medium at 37 °C and 200 rpm, the activated BL21/pET26b-rpl14b strain was introduced. To increase protein expression at low temperature, IPTG was administered at a final concentration of 0.1 mM, and the bacteria were centrifuged and collected. Rinse the bacteria three times with binding buffer. The precipitate was then eliminated by centrifuging the cells after they had been re-suspended in soluble binding buffer and subjected to ultrasonic treatment.Quasi-biosynthetic CdSe quantum dots
Na2SeO3 was selected as Se source, CdCl2 as Cd source, GSH as reducing agent and stabilizer, and the buffer solution was BR (in 100 mL triacid mixture, in which phosphoric acid, acetic acid and boric acid, the concentration is 0.04 mol L−1, 60 mL 0.2 mol L−1 NaOH is added, pH = 7.9). The 0.3 mM Na2SeO3, 0.4 mM CdCl2, 1.6 mM GSH or RPL14B proteins were each 10 mL quickly added into the 20 mL BR buffer reaction system, and the reaction temperature was set at 40 °C. Samples with different reaction time (0–30 min) were taken every 5 min for fluorescence intensity detection, and products with the optimal reaction time of 30 min were selected for sample collection. All the reaction processes of synthesizing quantum dots are carried out under the protection of high purity argon.Determination of fluorescence intensity and absorption spectra of CdSe quantum dots
Fluorescence intensity was measured using Fluorolog-3 fluorometers with excitation wavelengths of 400 nm and detection emission spectra of 420–700 nm. The absorption spectrum was measured by UV-2550 UV-visible spectrophotometer.Purification and characterization of RPL14B-based CdSe quantum dots
The CdSe quantum dots synthesized in vitro and quasi-biological systems were first filtered by 100 K MWCO ultrafiltration tube, and the filter liquid was concentrated by 10 K MWCO ultrafiltration tube. Then, CdSe quantum dots were washed with 1 × PBS at least 3 times to obtain purified CdSe quantum dots under ultrafiltration conditions of 4 °C, 3500 g, 20 min. Finally, the CdSe quantum dots were diluted to a suitable concentration for characterization. The purified CdSe quantum dots were diluted to a suitable concentration (about 10 times), then the 3 μL sample was dropped onto the ultra-thin carbon film and dried for at least 3 h, and then characterized by transmission electron microscopy (TEM), high resolution transmission electron microscopy (HRTEM), and energy dispersive X-ray spectroscopy (EDX).18 TEM and HRTEM images were obtained using JEM-1400plus and JEM-2100(HR) microscopes, respectively, and EDX data were obtained using JEM-2100(HR) microscopes equipped with GENESIS XM.Detection of photobleach resistance of RPL14B-based CdSe quantum dots
Detection of photobleach resistance: the fluorescence spectra of RPL14B-based CdSe quantum dots are determined continuously for 3 times with Fluorolog-3 spectrometer to detect the change of fluorescence intensity with excitation wavelengths of 400 nm and detection emission spectra of 420–700 nm.RPL14B-based CdSe quantum dots detects Cu2+
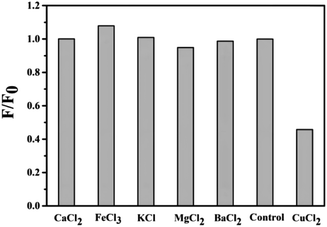
RPL14B-based CdSe quantum dots were synthesized by quasi-biological method, and the specificity of RPL14B-based CdSe quantum dots as fluorescent probes was determined with different solutions as follows: CaCl2, FeCl3, KCl, MgCl2, BaCl2, CuCl2. The detection method is as follows: adding the prepared RPL14B-based CdSe quantum dots into different above solutions, mixing them with slight shock, and using a fluorometer to detect the effect of the prepared RPL14B-based CdSe quantum dots on the fluorescence intensity of RPL14B-based CdSe quantum dots.19 Cu2+ solution with 0.2–10 μM concentration was added into the prepared RPL14B-based CdSe quantum dot solution, mixed well, and the fluorescence intensity of the mixed solution was detected.Results and discussion
Recombinant expression and purification of RPL14B
Previous studies found that ribosomal protein RPL14B was involved in the synthesis of CdSe (unpublished data). In this study, strain BL21/pET26b-rpl14b was constructed, and RPL14B protein containing 6× His label was obtained through induced expression. High-purity RPL14B recombinant protein was purified by Ni-NTA affinity chromatography (Fig. 1).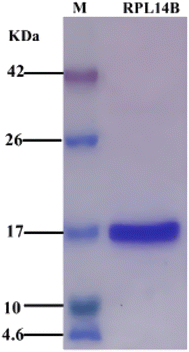 | ||
| Fig. 1 SDS-PAGE of recombinant expression and purification of RPL14B protein. M: protein maker, corresponding band size marked on the left; RPL14B lane: purified RPL14B recombinant protein. | ||
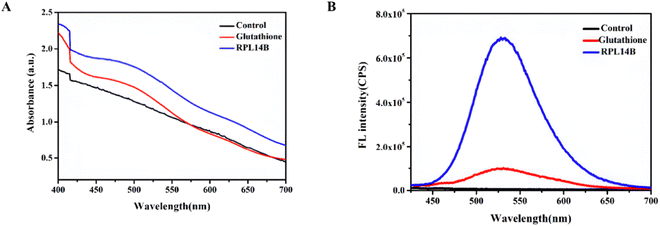
Some studies have reported that NaBH4 can prepare NaHSe as a selenium precursor by reducing selenium powder or selenium compounds. Therefore, synthetic CdSe with 4 mM Na2SeO3 as Se source, excess NaBH4 as reducing agent, 0.5 mM CdCl2 as Cd source, and buffer solution as BR (pH = 7.9) was obtained. The experimental results show that no absorption and emission peaks can be observed without the addition of templates, but the absorption peaks (Fig. 2A) and emission peaks of CdSe quantum dots can be clearly observed in the reaction system with the addition of RPL14B protein (Fig. 2B). By replacing GSH with RPL14B protein, the absorption peaks and emission peaks of CdSe quantum dots can be clearly observed. The above results clearly indicate that RPL14B protein can play a role in controlling the growth of quantum dots, that is, CdSe quantum dots can be synthesized as a template alone.
Characterization of RPL14B-based QDs developed via quasi-biosynthesis
The results of the in vitro synthesis of CdSe QDs indicated that selenium precursors can be prepared by reducing Na2SeO3 with NaBH4. Therefore, GSH was selected as the reducing agent to prepare the selenium precursor. The mechanism of the GSH reduction of SeO32− is as follows:20| SeO32− + 2H3O+ + 4RSH → RS–Se–SR + RS–SR + 5H2O |
| RS–Se–SR → RS–SR + Se0 |
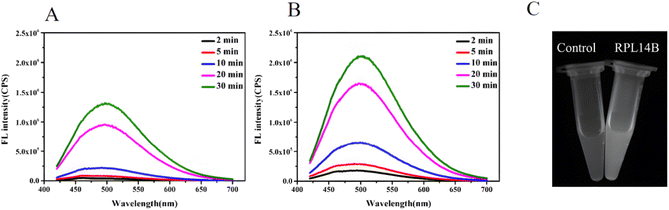
The above reaction shows that GSH can reduce SeO32− to the complex RS–Se–SR and that RS–Se–SR can participate in the combination of CdSe QDs as a precursor of selenium. To improve the quasi-biosynthesis system and assess the role of the ribosomal protein RPL14B in the system, a quasi-biosynthesis system of CdSe QDs was established. The in vitro synthesis results showed that CdSe QDs could be synthesized at 40 °C. Therefore, 40 °C was selected as the optimal synthesis temperature for CdSe QDs in the quasi-biosynthesis system. The RPL14B protein was then added to the system (experimental group), whereas it was omitted in the control group. Compared with the control group, the fluorescence intensity of CdSe QDs in the RPL14B protein addition group was significantly increased (Fig. 3A and B).
To verify the above experimental results, the two samples with a reaction time of 30 min were subjected to ultraviolet light. The results showed that the RPL14B protein addition group emitted stronger fluorescence than the control group under ultraviolet irradiation (Fig. 3C).
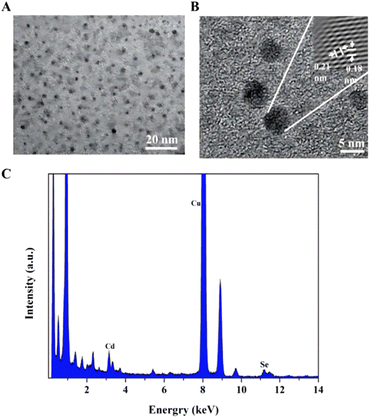
After adding the RPL14B protein to the quasi-biosynthesis system, the synthetic QDs were purified and characterized. The results are shown in Fig. 4. TEM revealed that the ribosomal protein RPL14B-associated QDs had a uniform particle size and good dispersion (Fig. 4A). HRTEM showed that the crystal plane spacing of the synthesized quantum dots was 0.21 and 0.18 nm (Fig. 4B). Finally, the EDX energy spectrum suggested that the synthesized QDs comprised Cd and Se elements (Fig. 4C), indicating that the nanoparticles synthesized using the quasi-biosynthesis system were CdSe QDs.
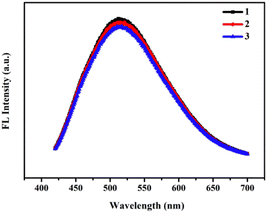
To evaluate the optical properties of RPL14B-based CdSe QDs, the photobleachability of the synthetic QDs synthesized with the RPL14B protein in the quasi-biosynthesis system was investigated. The fluorescence properties of the QDs were measured using a fluorometer for three consecutive excitation times (Fig. 5). The results showed that the fluorescence intensity of the QDs remained stable and that the fluorescence emission peak did not shift, indicating excellent light drift-resistant properties.
The above experimental results confirmed that the quasi-biosynthesis system used in this study was optimally processed at the constant temperature of 40 °C, in line with the current green synthesis concept. Subsequent experiments revealed that CdSe QDs with higher fluorescence intensity can be obtained by adding RPL14B to the system. QDs with high fluorescence intensity and light bleaching resistance have broad application prospects in detection copper ion.
RPL14B-based CdSe QDs detected Cu2+
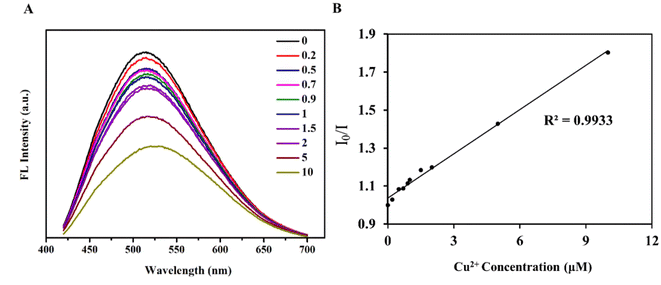
For detecting ions, in addition to the sensitivity of detection, the specificity of the detection probe is particularly critical. To eliminate the influence of anions in the experiment, a salt solution of the same anion (Cl−) was selected to detect the influence of cations on the RPL14B-based CdSe QDs. Based on this consideration, the effects of CaCl2, FeCl3, KCl, MgCl2, BaCl2, and CuCl2 solutions on the fluorescence intensity of the QDs were detected, and the results are shown in Fig. 6. The experimental results revealed that Cu2+ significantly quenched the fluorescence of RPL14B-based CdSe QDs and Ca2+, Fe3+, K+, Mg2+, and Ba2+ had little effect on the fluorescence. Moreover, the results showed that the QDs could be used as fluorescent probes for detecting Cu2+ with excellent selectivity.Then, the influence of Cu2+ on the fluorescence quenching of RPL14B-based CdSe quantum dots was studied by measuring the fluorescence intensity (Fig. 7). The results show that the concentration of Cu2+ can be detected from 0.2 μM to 10 μM by using RPL14B-based CdSe quantum dots as fluorescent probes. The fluorescence quenching data showed a linear relationship (R2 = 0.9933), which was obtained using the Stern–Volmer equation,21 as shown below:
| I0/I = 1 + KSV[Q] |
Based on literature and the experimental results, Cu2+ interacted with QDs through the coating layer and quenched their fluorescence.22,23 Thus, the QDs synthesized using the quasi-biological synthesis system can be used for Cu2+ detection. Copper ions can form selenides with selenium, producing the associated redshift and fluorescence quenching. A weak redshift was observed in the presence of high copper ion concentration, indicating a possible displacement between copper and cadmium ions (Fig. 7A). In addition, the RPL14B protein contains a large number of amide, hydroxyl, and carboxyl groups. The transfer of electrons from excited CdSe QDs to Cu2+ may have also caused fluorescence quenching.
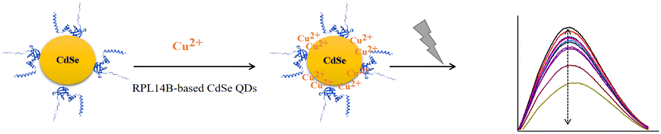
The possible mechanism of fluorescence quenching was evaluated (Fig. 8). The results revealed that the fluorescence quenching mechanism of copper ions can be explained by the presence of the RPL14B shell. As per this mechanism, Cu2+ first binds to the RPL14B scaffold, thereby transferring electrons from excited CdSe QDs to Cu2+ and reducing Cu2+ to Cu+. With the increase in copper ion concentration, substitution between the copper and cadmium ions occur, which produces CuSe and results in the redshift and fluorescence quenching phenomena. These results indicated that CdSe QDs synthesized via the quasi-biological system with the RPL14B protein can be used to develop an effective method for detecting Cu2+.
Conclusion
The specific interaction between quantum dots and some molecules can cause fluorescence quenching. Different types of quantum dots have been widely used in copper ion detection, and have good detection limits, which are suitable for a variety of scenarios (Table 1).The conditions required for the quasi-biosynthesis of CdSe QDs established in the current study were significantly mild, with the synthesis temperature of 40 °C. Moreover, the bionic synthesis of CdSe QDs was regulated by adding ribosomal proteins. The RPL14B-based CdSe QDs were sensitive to Cu2+. This property was utilized to evaluate a method for detecting Cu2+ via the QDs as fluorescent probes. The results showed that the QD probe had the characteristics of light bleaching resistance, strong specificity, simple operation, and short detection time.| Types of quantum dots | Materials | Synthesis principle | LOD (μM) | Linea concentration Range(μM) |
|---|---|---|---|---|
| Carbon dot | Carbon dots composite with PCL/PLA24 | In situ electrospinning technology | 0.83 | 0.5–5 |
| Carbon dot | The CD/SiO2/AuNC ratiometric fluorescent nanocomposites25 | Condensation reaction | 0.013 | 0.025–4 |
| Carbon dot | Nitrogen-doped carbon quantum dots26 | Hydrothermal method | 0.071 | 0.3–0.7 |
| Carbon dot | Dual function polyamine-functionalized carbon dots27 | Plant-dependent thermolysis procedure | 0.02 | 0.07–60 |
| Other quantum dots | N-doped molybdenum oxide QDs28 | Hydrothermal method | 0.78 | 1–100 |
| Other quantum dots | N,S,P-CDs and NAC-AuNCs29 | Acid–base reaction heat method | 0.5 | 0.65–26.58 |
| Other quantum dots | AuAgNCs and WS2-QDs30 | Hydrothermal method | 0.12 | 0.3–3 |
| CdSe quantum dots | DES-CdSe QDs31 | Aqueous phase synthesis | 0.0053 | 0.001–0.06 |
| CdSe quantum dots | CdSe QDs32 | Biosynthesized by B. Licheniformis | 0.91 | 0–20 |
| CdSe quantum dots | Polymer-capped CdSe/ZnS quantum dots33 | High-temperature injection method and the SILAR method | 0.00694 | 0.02–0.7 |
Author contributions
Jiye Liu, Lipeng Zhong and Zhixiong Xie designed all experiments, discussed the data and wrote the manuscript. Wenyue Liu performed the copper ion quenching test, Lipeng Zhong performed protein expression, Jiye Liu performed all other experiments. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
Part of this article has applied for the invention patent of the People's Republic of China: Preparation method of single selenium-containing protein of S. cerevisiae (202110178408.8).Acknowledgements
This work was supported by the Natural Science Foundation of Jiangxi Province (20224BAB216083); Young Talent Cultivation Fund of the First Affiliated Hospital of Nanchang University (YFYPY202207); Science and Technology Project of Jiangxi Provincial Health Commission (202210350); Science and Technology Project of Jiangxi Administration of Traditional Chinese Medicine (2022A363); Research Training Project of Nanchang University by 2022 years; National Natural Science Foundation of China (31570090 and 31800028) and the National Infrastructure of Natural Resources for Science and Technology Program of China (NIMR-2020-8).References
- K. Jomova, M. Makova, S. Y. Alomar, S. H. Alwasel, E. Nepovimova, K. Kuca, C. J. Rhodes and M. Valko, Chem.–Biol. Interact., 2022, 367, 110173 CrossRef CAS PubMed.
- A. Punia, Environ. Sci. Pollut. Res. Int., 2021, 28, 4056–4072 CrossRef CAS PubMed.
- E. V. Soares, K. Hebbelinck and H. M. Soares, Can. J. Microbiol., 2003, 49, 336–343 CrossRef CAS PubMed.
- J. F. Collins, J. R. Prohaska and M. D. Knutson, Nutr. Rev., 2010, 68, 133–147 CrossRef PubMed.
- A. F. Robson, P. Lockett, L. Tetlow and C. Chaloner, Nutr. Rev., 2020, 57, 246–248 CAS.
- J. Wang, M. Rao, C. Ye, Y. Qiu, W. Su, S. R. Zheng, J. Fan, S. L. Cai and W. G. Zhang, RSC Adv., 2020, 10, 4621–4629 RSC.
- T. U. Gosterisli, I. Z. Kublay, S. Oflu, Y. Kilinc, E. S. Kocoglu, B. T. Zaman, S. Keyf and S. Bakirdere, Food Chem., 2022, 377, 132057 CrossRef CAS PubMed.
- P. Li, X. Duan, Z. Chen, Y. Liu, T. Xie, L. Fang, X. Li, M. Yin and B. Tang, Chem. Commun., 2011, 47, 7755–7757 RSC.
- J. Liu, K. Ning, Y. Fu, Y. Sun and J. Liang, Spectrochim. Acta, Part A, 2023, 294, 122553 CrossRef CAS PubMed.
- S. D. Torres Landa, N. K. Reddy Bogireddy, I. Kaur, V. Batra and V. Agarwal, iScience, 2022, 25, 103816 CrossRef CAS PubMed.
- M. Batool, H. M. Junaid, S. Tabassum, F. Kanwal, K. Abid, Z. Fatima and A. T. Shah, Crit. Rev. Anal. Chem., 2022, 52, 756–767 CrossRef CAS PubMed.
- H. M. Junaid, A. R. Solangi and M. Batool, Analyst, 2021, 146, 2463–2474 RSC.
- J. Liu, D. Zheng, L. Zhong, A. Gong, S. Wu and Z. Xie, Biochem. Biophys. Res. Commun., 2021, 544, 60–64 CrossRef CAS PubMed.
- Y. Li, R. Cui, P. Zhang, B. B. Chen, Z. Q. Tian, L. Li, B. Hu, D. W. Pang and Z. X. Xie, ACS Nano, 2013, 7, 2240–2248 CrossRef CAS PubMed.
- G. Li, X. Fei, H. Liu, J. Gao, J. Nie, Y. Wang, Z. Tian, C. He, J. L. Wang, C. Ji, D. Oron and G. Yang, ACS Nano, 2020, 14, 4196–4205 CrossRef CAS PubMed.
- Y. Yu, X. Chen, Y. Wei, J. H. Liu, S. H. Yu and X. J. Huang, Small, 2012, 8, 3274–3281 CrossRef CAS PubMed.
- J. H. Liu, J. B. Fan, Z. Gu, J. Cui, X. B. Xu, Z. W. Liang, S. L. Luo and M. Q. Zhu, Langmuir ACS J. Colloid. Surface., 2008, 24, 5241–5244 CrossRef CAS PubMed.
- R. Zhang, M. Shao, X. Han, C. Wang, Y. Li, B. Hu, D. Pang and Z. Xie, Int. J. Nanomed., 2017, 12, 3865–3879 CrossRef CAS PubMed.
- V. Duran-Toro, A. Gran-Scheuch, N. Ordenes-Aenishanslins, J. P. Monras, L. A. Saona, F. A. Venegas, T. G. Chasteen, D. Bravo and J. M. Perez-Donoso, Anal. Biochem., 2014, 450, 30–36 CrossRef CAS PubMed.
- M. Bjornstedt, S. Kumar and A. Holmgren, J. Biol. Chem., 1992, 267, 8030–8034 CrossRef CAS.
- T. Htun, J. Fluoresc., 2004, 14, 217–222 CrossRef CAS PubMed.
- K. M. Omer, Anal. Bioanal. Chem., 2018, 410, 6331–6336 CrossRef CAS PubMed.
- J. Yao, K. Zhang, H. Zhu, F. Ma, M. Sun, H. Yu, J. Sun and S. Wang, Anal. Chem., 2013, 85, 6461–6468 CrossRef CAS PubMed.
- X. Cui, Y. Zhang, Z. Chen, H. Xiao, R. Xiong and C. Huang, Int. J. Biol. Macromol., 2023, 252, 126431 CrossRef CAS PubMed.
- H. Liu, L. Jia, Y. Wang, M. Wang, Z. Gao and X. Ren, Anal. Bioanal. Chem., 2019, 411, 2531–2543 CrossRef CAS PubMed.
- Y. Liu, M. Zhao and Q. Zhu, J. Fluoresc., 2023, 33, 2391–2401 CrossRef CAS PubMed.
- H. R. H. Ali, A. I. Hassan, Y. F. Hassan and M. M. El-Wekil, Anal. Bioanal. Chem., 2020, 412, 1353–1363 CrossRef CAS.
- T. Ma, M. Liu, J. Sun, J. Wu, Z. Zhao, J. Bai, Y. Fang and X. Jin, Analytical Methods : Advancing Methods and Applications, 2023, 15, 6239–6244 Search PubMed.
- W. Dong, R. Wang, X. Gong, W. Liang, L. Fan, S. Song and C. Dong, Mikrochim. Acta, 2020, 187, 299 CrossRef CAS.
- Z. Wang, R. Liu, Z. Fu, X. Yi, Y. Hu, C. Liu, D. Pan and Z. Wu, Analytical Methods : Advancing Methods and Applications, 2023, 15, 2505–2511 Search PubMed.
- S. Sadeghi and A. Davami, Mikrochim. Acta, 2020, 187, 147 CrossRef CAS PubMed.
- Z. Y. Yan, Q. Q. Du, D. Y. Wan, H. Lv, Z. R. Cao and S. M. Wu, Enzyme Microb. Technol., 2017, 107, 41–48 CrossRef CAS PubMed.
- Q. Zhou, Z. Li, Q. Wang, L. Peng, S. Luo and F. L. Gu, Analytical Methods : Advancing Methods and Applications, 2021, 13, 2305–2312 Search PubMed.
Footnotes |
| † Lipeng Zhong and Wenyue Liu are co-first author. |
| ‡ Zhixiong Xie and Jiye Liu are co-corresponding authors. |
| This journal is © The Royal Society of Chemistry 2024 |