Open Access Article
Open Access ArticleSynthesis and mutagenic risk of avanafil's potential genotoxic impurities†
Yunkai Sun *ab,
Xiaoxia Wubc,
Pei Zuoc,
Zhao Liu*c,
Xuepei Miaoa,
Jian Liua and
Hairuo Wen*d
*ab,
Xiaoxia Wubc,
Pei Zuoc,
Zhao Liu*c,
Xuepei Miaoa,
Jian Liua and
Hairuo Wen*d
aSchool of Chemical Engineering and Materials, Changzhou Institute of Technology, Changzhou 213022, China. E-mail: sunyunkai1983@163.com
bSchool of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China
cHarvest Pharmaceutical Co., Ltd, Changsha 410000, China. E-mail: liuz@harvest-pharm.com
dNational Center for Safety Evaluation of Drugs, National Institutes for Food and Drug Control, Key Laboratory of Beijing for Nonclinical Safety Evaluation Research of Drugs, Beijing, 100176, P. R. China. E-mail: wenhairuo@nifdc.org.cn
First published on 8th July 2024
Abstract
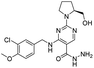
In the technical route for the synthesis of avanafil, 1-ethyl-(3-dimethylaminopropyl)carbamyldiimide hydrochloride (EDCI) and 1-hydroxybenzotriazole (HOBT) are used as reactive acid–amine binding agents. HOBT contains trace amounts of hydrazine residue, and there is a risk of introducing potentially mutagenic impurities with hydrazide-containing structures. The potentially genotoxic impurities E (Imp-E) and F (Imp-F) of avanafil with altering hydrazide-structure were synthesized by chemical method; subsequently, the impurities were evaluated and classified according to ICH M7 guidelines. Two complementary quantitative structure–activity relationship (QSAR) evaluation systems (Derek and Sarah) based on expert rules and statistics were used to preliminarily predict the genotoxicity of Imp-E and Imp-F, and the prediction result of E was suspected to be positive. In the Ames test of Imp-E and Imp-F, in the dose range of 62.5–1000 μg per plate, with or without the presence of metabolic activation system S9, the number of revertant colonies did not exceed 2 times the number of colonies in the solvent control group and did not show a dose–response relationship, and the test results were negative. Imp-E and Imp-F were determined to be negative for genotoxicity, which could be controlled as class 5 in ICH M7, that is, non mutagenic impurity.
1. Introduction
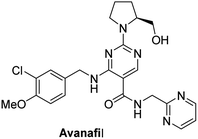
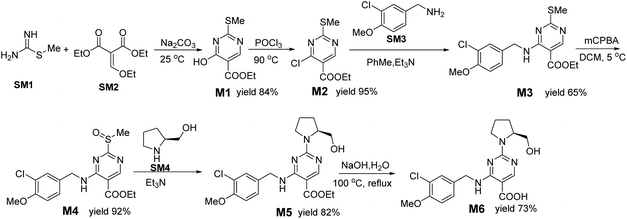
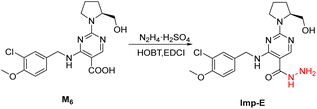
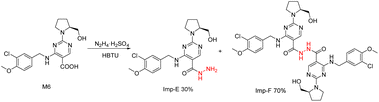
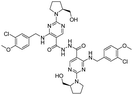
Avanafil is a novel potent phosphodiesterase-5 (PDE-5) inhibitor (Fig. 1) with rapid onset and few adverse reactions and is used for the treatment of hypertension, hyperlipidemia and hyperglycemia.1,2 As the fourth-generation drug for the treatment of erectile dysfunction, its safety has been widely concerned. Limit control of drug impurity is an important part of drug quality standard research.3,4 The process route of drug synthesis involves chemical reagents such as reactants, solvents and catalysts, as well as chemical reactions and degradation. Impurities are unavoidable in raw materials and preparations. In recent years, increasing attention has been paid to the genotoxicity risk of drug impurities, and the European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) have successively promulgated the guiding principles for the control of genotoxic impurities (GTIs).5,6 In 2014, ICH officially issued the M7 guidelines,7 which set out general requirements for the control of DNA-active (mutagenic) drug impurities in order to control the evaluation and control of DNA reactive (mutagenic) drug impurities with potential carcinogenic risks and have been used in the drug registration of member states.Genotoxic impurities refer to DNA reactive substances that could directly cause DNA damage at low levels, leading to DNA mutation and even carcinogenesis.8,9 Hydrazine has been identified as a mutagenic and carcinogenic compound,10–15 and the hydrazine group is also an alert structure of typical genotoxic carcinogens in compounds.16–18 In the synthesis of avanafil, EDCI and HOBT were used as acid–amine condensation agents, while hydrazine was used in the upper source process of HOBT, resulting in residual hydrazine in HOBT.19 Hydrazine reacts with intermediate 6 (M6) to introduce potentially mutagenic impurities with warning structures into avanafil. In our synthesis of avanafil, HOBT containing trace hydrazine leads to the creation of impurities (Imp-E) with hydrazine groups. Genotoxic impurities have a higher risk of carcinogenesis than common impurities20,21 and generally have low acceptable limits, posing great challenges to synthesis control processes and detection. Further mutagenic evaluation of the potentially genotoxic impurity of avanafil was performed in accordance with the ICH M7 guidelines: that is, the potential genotoxic impurities E (Imp-E) and impurities F (Imp-F) are synthesized and confirmed firstly, and then the compound toxicity prediction softwares of Derek Nexus and Sarah Nexus22–26 were used to predict the risk and possible warning structure of mutation resulting from mutation as the toxicity endpoint. Finally, the bacterial reverse mutation test (Ames test)27–30 was conducted to provide guidance and a basis for the classification and control of related substances in avanafil.
2. Experimental
2.1 Chemicals and materials
1-Hydroxybenzotriazole (HOBT Guangdong Wengjiang Chemical Reagent Co., LTD.); diethyl ethoxymethylenemalonate and L-prolinol (Qifei Pharmaceutical); S-methyl isothiourea semisulfate and 3-chloro-4-methoxybenzylamine hydrochloride (Emeishan Hongsheng Pharmaceutical); M-chloroperoxybenzoic acid (Baokang Pharmaceutical Chemical Co., Ltd.); sodium azide (Henan Standard Material Research and Development Center); O-benzotriazole-tetramethylurea hexafluorophosphate (HBTU), N,N-diisopropylethylamine (DIEA), 1-ethyl-(3-dimethylaminopropyl) carbamyldiimide hydrochloride (EDCI), hydrazine sulfate, L-histidine, D-biotin, D-glucose 6-phosphate sodium salt, β-NADP-Na2, ampicillin, tetracycline, crystal purple, mitomycin C, 2-aminoanthracene, 2-aminofluorene, dioxone (Aladdin); agar plate (Bio-high Technology); metabolic activation system S9 (Bioplus Biotech). Citric acid monohydrate, disodium hydrogen phosphate, sodium dihydrogen phosphate, sodium ammonium hydrogen phosphate, D-(+)-glucose, magnesium chloride, potassium chloride, dipotassium phosphate, magnesium sulfate, sodium chloride, anhydrous sodium carbonate, phosphorus oxychloride, methanol, triethylamine (Nanjing Chemical Reagent Co., Ltd.).2.2 Instruments
Instrument: Bruker Avance III 400 NMR instrument (solvent: DMSO-d6, internal standard: TMS); high resolution mass spectrometer (UPLC1290/TOF MS 6230); high pressure preparation liquid chromatograph (Bonna-Agela CHEETAH II); ultra-performance liquid chromatography (Agilent 1260 UHPLC system); WTD electronic balance; SGW-3 automatic polarimeter; N-1300 rotary evaporation instrument; SHZ-D vacuum pump; DZF-6051 vacuum drying oven; DHG-9140 electric blast drying oven; SPX-70BE biochemical incubator; NMSP-600 horizontal rotating oscillator; DW-86W28 ultra-low temperature storage box; HYC-3105 medical cooler.2.3 UPLC and chromatographic conditions
The UPLC method was developed for the analysis of the related impurities on an Agilent 1260 UHPLC system.31 The chromatographic analysis was carried out on a Waters ACQUITY HSS C18 with 50 mm × 2.1 mm and 1.8 μm particle size column. The mobile phase was composed of 20 mM ammonium formate aqueous solution; pH was adjusted to 5.00 ± 0.05 with dilute formic acid (mobile phase-A) and acetonitrile (mobile phase-B). The gradient program is set to (Tmin/mobile phase-A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B): T0/90
B): T0/90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, T1/90
10, T1/90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, T8/20
10, T8/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80, T10/90
80, T10/90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, T11/10
10, T11/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90. It was degassed and filtered through 0.22 μm filters under vacuum before injecting into the system. The mobile phases were set with a flow rate of 0.3 mL min−1, and the column temperature was kept at 35 °C. The analytes were detected in a photodiode array detector at 239 nm.
90. It was degassed and filtered through 0.22 μm filters under vacuum before injecting into the system. The mobile phases were set with a flow rate of 0.3 mL min−1, and the column temperature was kept at 35 °C. The analytes were detected in a photodiode array detector at 239 nm.
2.4 High resolution mass spectrometer (HRMS)
HRMS analyses were performed on a UPLC1290/TOF MS 6230 system. Analysis of all compounds was performed on Waters C18 columns. The operating parameters were as follows: a nebulizer gas pressure of 7.5 bar, dry gas flow rate of 5.0 L min−1, capillary voltage of 3.5 kV, ion flight time of 0.5 s, and transfer capillary temperature of 300 °C. All data is collected and processed by Agilent's data analysis software, Mass Hunter.2.5 Synthesis of target compounds
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B), T0/70
B), T0/70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, T70/60
30, T70/60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, T200/50
40, T200/50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45, the flow rate was 10 mL min−1, and the detection wavelength was maintained at 254 nm. 450 mg of solid was dissolved in about 2 mL solution with DMSO, and the column liquid with Imp-F was collected and treated to obtain 65 mg yellow solid with a purity of 95%. mp 161.5–163.3 °C (see Fig. S7 and S8†). 1H NMR (400 MHz, DMSO-d6) δ: 9.95 (s, 2H), 9.00 (s, 2H), 8.50 (s, 2H), 7.40 (s, 2H), 7.28 (d, J = 8.2 Hz, 2H), 7.08 (d, J = 8.4 Hz, 2H), 4.89 (s, 1H), 4.69 (s, 1H), 4.51 (s, 4H), 4.09 (s, 2H), 3.81 (s, 6H), 3.61 (s, 2H), 3.49 (s, 4H), 1.95 (m, 8H), 1.23 (s, 2H); 13C NMR (100 MHz, DMSO-d6) δ: 167.16, 153.89, 133.48, 129.83, 129.59, 128.20, 121.19, 113.15, 59.79, 59.36, 56.52, 47.85, 42.75, 28.01, 22.95.
45, the flow rate was 10 mL min−1, and the detection wavelength was maintained at 254 nm. 450 mg of solid was dissolved in about 2 mL solution with DMSO, and the column liquid with Imp-F was collected and treated to obtain 65 mg yellow solid with a purity of 95%. mp 161.5–163.3 °C (see Fig. S7 and S8†). 1H NMR (400 MHz, DMSO-d6) δ: 9.95 (s, 2H), 9.00 (s, 2H), 8.50 (s, 2H), 7.40 (s, 2H), 7.28 (d, J = 8.2 Hz, 2H), 7.08 (d, J = 8.4 Hz, 2H), 4.89 (s, 1H), 4.69 (s, 1H), 4.51 (s, 4H), 4.09 (s, 2H), 3.81 (s, 6H), 3.61 (s, 2H), 3.49 (s, 4H), 1.95 (m, 8H), 1.23 (s, 2H); 13C NMR (100 MHz, DMSO-d6) δ: 167.16, 153.89, 133.48, 129.83, 129.59, 128.20, 121.19, 113.15, 59.79, 59.36, 56.52, 47.85, 42.75, 28.01, 22.95.
| Sample | Solvent | Concentration of solution (g per 100 mL) | [α]Dt |
|---|---|---|---|
| a [α]Dt: t = 20 °C; D = 589 nm. | |||
| SM4 | DMF | 0.2612 | +9.7 |
| M5 | DMF | 0.2518 | −49.5 |
| M6 | DMF | 0.2534 | −92.4 |
| Imp-E | DMF | 0.2512 | −108.3 |
| Imp-F | DMF | 0.2534 | −110.5 |
2.6 QASR evaluation model for genotoxicity
Derek Nexus (Derek Nexus version 6.2.1, knowledge base 2022 2.0) provided by Lhasa Limited (Leeds, UK) is a computerized expert system that uses a knowledge-based expert system approach to predict the toxicological hazard of molecules. Thus, it contains expert knowledge rules derived from the known relationship of a given substructure and a toxicological effect of the molecule, and it applies these rules to predict potential toxicological effects of substances for which no experimental data usually exist.Sarah Nexus (Sarah Nexus version 3.2.0, model 2022.2), provided by Lhasa Limited (Leeds, UK), is a statistics-based system. Structures submitted for processing are fragmented, and these fragments are reviewed for activity versus inactivity. The model then arranges those fragments of interest into a network of hypotheses (or nodes) and relevant hypotheses are used to inform an overall prediction of toxicity. Sarah Nexus predicts activity or inactivity in the Ames test and provides information on the coverage of a query compound.
Nexus version: Nexus 2.5.2 is a software integration platform. In this study, the solidified ICH M7 batch classification model in the Nexus system was used to predict and automatically classify the genotoxicity of impurities.
2.7 Ames test
The test articles were impurities Imp-E and Imp-F. Positive controls for the Ames test were sodium azide (CAS no. 26628-22-8), mitomycin C (CAS no. 50-07-7), 2-aminofluorene (CAS no. 153-78-6), 2-anthramine (CAS no. 613-13-8) and dioxone (CAS no. 140-56-7), respectively. DMSO, which was used as a solvent for the test chemicals and positive controls, served as the vehicle control.Salmonella typhimurium histidine-dependent strains TA98, TA100, TA1535, TA102 and TA97a were obtained from Moltox Company. The strains were freeze-dried and stored in the refrigerator at 2–8 °C. The strains were inoculated in nutrient broth medium of 20 mL and cultured at 37 °C (120 times per min) for 11 h. The metabolic activation system (rat liver microsomes, S9) was purchased from IPHASE. S9 mixture (10% S9 v/v) was prepared in a glucose-6-phosphate shortly before use and kept on crushed ice to keep it cold. The top layer medium 2.0 mL containing 0.5 mmol per L histidine and 0.5 mmol per L biotin was divided into a sterilized EP tube and kept warm in a water bath at 50 °C. Then, 0.1 mL of the bacterial suspension, 0.1 mL test substance solution, and 10% (v/v) of liver S9 mixture or phosphate buffer were added. The mixture solution was thoroughly mixed, quickly poured over the surface of an agar plate, and incubated in a 37 °C incubator for 48 h, and the number of reverted colonies per plate was counted. Three parallel plates were made for each dose, and a blank control group, solvent control group and positive control group were set up.
2.8 Statistics
The number of revertant colonies was counted manually, and the average plate counts under each concentration gradient were taken. SPSS 27.0 software was used for statistical analysis of the data. The quantitative statistical description was![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, and the multiple change of reversed colony number was calculated by comparing with spontaneous control.
± s, and the multiple change of reversed colony number was calculated by comparing with spontaneous control.
3. Results and discussion
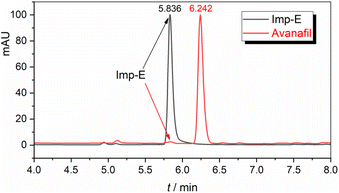
3.1 Discovery and structural resolution of Imp-E
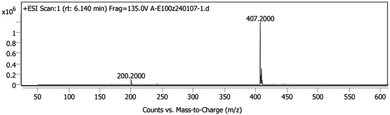
The synthesized avanafil samples were analyzed using the newly developed method described in Section 2.3, and the results showed that the Imp-E with the retention time (5.836 min) was detected (Fig. 5). The structure of Imp-E is determined by mass spectrometry (MS) and nuclear magnetic resonance spectrometry (NMR). The HRMS spectrum (Fig. 6) shows that Imp-E has only one high molecular ion peak (m/z 407.2000). The formation of Imp-E is caused by a small amount of hydrazine in HOBT. Although Imp-F is not found in avanafil, the confirmation of the structure of Imp-E makes us realize that Imp-F with mutagenic alert structure needs to be considered as a potential genotoxic impurity.3.2 QSAR evaluation of Imp-E and Imp-F
Imp-E and Imp-F had mutagenic alert structures, and the ICH classification model was used for prediction, as shown in Table 2. Derek Nexus considered that Imp-E was suspiciously positive for mutagenic risk due to the presence of hydrazine sulfate, hydrazine hydrate, and isoniazid, which could increase the mutation rate of TA1535. However, for Imp-F, the predicted result is inactive. The prediction results of Sarah Nexus were based on the search of compound structure decomposition, and the probability of mutagenic risk between all decomposed structures and substances with the same structure was estimated. Sarah Nexus' prediction results of impurity E and impurity F were negative. The results of computer simulation toxicity assessment of impurity E by QSAR methods based on different principles are inconsistent, and the predicted results between Imp-E and Imp-F (with a similar structure) are also inconsistent.3.3 Genotype identification of the detected strains
As shown in Table 3, the rfa membrane mutation of the strain increased the permeability of macromolecular compounds, making it easier for macromolecular compounds to enter the bacteria. UvrB-strains have defective excision repair functions and cannot excise DNA adducts, making the strains susceptible to mutagenicity and bacterial toxicity of a large number of mutants. The bacteria containing plasmid pkm101 had a higher number of spontaneous recovery colonies, and plasmid pkm101 had muc+ gene, which was involved in DNA damage-induced DNA repair pathway, and increased the susceptibility to mutability while giving bacteria resistance to bacterial toxicity. The strains with the R factor were resistant to ampicillin. Strains containing pAQ1 had tetracycline resistance. The different strains have different sensitivities to mutants. TA98 and TA100 are sensitive to most mutants, and the preliminary screening test can only detect these two strains. In the formal experiment, TA98 and TA97a detected frameshift mutation agents, TA1535 and TA100 detected base-displacement mutagenic agents, and oxidizing mutants, DNA crosslinkers, and hydrazine could be detected by strains TA102 containing AT-site mutations.| Bacterial strain | His− | Rfa | R | pAQ1 | UvrB− repair-deficiency |
|---|---|---|---|---|---|
| TA98 | + | + | + | − | − |
| TA97a | + | + | + | − | − |
| TA100 | + | + | + | − | − |
| TA102 | + | + | + | + | + |
| TA1535 | + | + | − | − | − |
3.4 Ames test for Imp-E and Imp-F
Spectral grade DMSO is selected as the solvent to dissolve Imp-E and Imp-F because it does not react with the subject matter and is not toxic to the strain and S9. Cytotoxicity and the solubility of the substance should be considered when determining the maximum concentration for the Ames test. For compounds with good solubility and no or little cytotoxicity, the maximum dose can be 5 mg per plate.Preliminary data showed that Imp-E and Imp-F were toxic in the dose groups of 5000, 2500 and 1600 μg per plate. In the formal test, 1000, 500, 250, 125, 62.5 μg per plate doses were applied. As shown in Tables 4 and 5, the number of revertants in the solvent control group was within the normal range, and the number of revertants in the different positive control groups was more than 2 times that of the solvent control group. Imp-E and Imp-F at the dose of 1000, 500, 250, 125, 62.5 μg per plate had no difference in the number of revertants induced by TA97a, TA98, TA100, TA102 and TA1535 compared with the solvent control group and showed a dose-independent effect. There was also no reproducible positive response for a single test point. These results suggested that hydrazine with large steric acyl substitution reduces the genotoxicity of hydrazine, and that hydrazine compounds containing 1 or 2 acyl substitutions have no mutagenic effect and cannot cause base substitution and shift-type mutation. Imp-F showed a similar result. These indicated that both Imp-E and Imp-F were not mutagenic to Salmonella typhimurium.
| Group μg per plate | S9 | Number of revertants | ||||
|---|---|---|---|---|---|---|
| TA98 | TA97a | TA100 | TA102 | TA1535 | ||
| a Dioxone, 50 μg per plate.b 2-Aminofluorene, 20 μg per plate.c Sodium azide, 1 μg per plate.d Mitomycin C, 1 μg per plate.e 2-Aminoanthracene, 20 μg per plate. | ||||||
| 1000 | − | 33 ± 4 | 129 ± 17 | 122 ± 11 | 210 ± 14 | 41 ± 6 |
| + | 36 ± 4 | 172 ± 8 | 159 ± 32 | 266 ± 12 | 57 ± 2 | |
| 500 | − | 43 ± 5 | 91 ± 5 | 124 ± 12 | 274 ± 8 | 41 ± 6 |
| + | 37 ± 2 | 185 ± 9 | 159 ± 32 | 241 ± 25 | 53 ± 3 | |
| 250 | − | 41 ± 3 | 152 ± 34 | 174 ± 13 | 223 ± 12 | 55 ± 2 |
| + | 42 ± 3 | 178 ± 14 | 124 ± 13 | 223 ± 12 | 54 ± 2 | |
| 125 | − | 39 ± 4 | 144 ± 12 | 143 ± 15 | 218 ± 14 | 61 ± 3 |
| + | 34 ± 3 | 154 ± 7 | 158 ± 30 | 262 ± 7 | 61 ± 3 | |
| 62.5 | − | 34 ± 4 | 153 ± 19 | 143 ± 22 | 264 ± 14 | 56 ± 2 |
| + | 30 ± 2 | 194 ± 16 | 153 ± 25 | 270 ± 5 | 57 ± 2 | |
| Positive control | − | 1111 ± 117a | 1872 ± 104a | 1697 ± 54c | 2043 ± 185d | 1190 ± 40c |
| + | 1171 ± 78b | 1920 ± 120b | 2053 ± 65b | 2121 ± 21e | 1323 ± 69b | |
| Solvent control | − | 27 ± 4 | 137 ± 24 | 177 ± 4 | 196 ± 12 | 45 ± 5 |
| + | 35 ± 7 | 164 ± 14 | 212 ± 31 | 212 ± 17 | 47 ± 4 | |
| Blank control | − | 29 ± 1 | 152 ± 10 | 181 ± 15 | 252 ± 2 | 50 ± 5 |
| + | 47 ± 4 | 192 ± 5 | 183 ± 10 | 277 ± 4 | 45 ± 7 | |
| Group μg per plate | S9 | Number of revertants | ||||
|---|---|---|---|---|---|---|
| TA98 | TA97a | TA100 | TA102 | TA1535 | ||
| a Dioxone, 50 μg per plate.b 2-Aminofluorene, 20 μg per plate.c Sodium azide, 1 μg per plate.d Mitomycin C, 1 μg per plate.e 2-Aminoanthracene, 20 μg per plate. | ||||||
| 1000 | − | 16 ± 3 | 57 ± 6 | 174 ± 12 | 201 ± 7 | 41 ± 2 |
| + | 31 ± 5 | 38 ± 7 | 184 ± 12 | 172 ± 22 | 33 ± 2 | |
| 500 | − | 26 ± 4 | 129 ± 26 | 131 ± 11 | 235 ± 13 | 38 ± 4 |
| + | 30 ± 3 | 137 ± 12 | 181 ± 10 | 208 ± 11 | 35 ± 7 | |
| 250 | − | 34 ± 3 | 107 ± 13 | 167 ± 19 | 262 ± 8 | 25 ± 4 |
| + | 38 ± 2 | 161 ± 31 | 189 ± 2 | 226 ± 14 | 23 ± 2 | |
| 125 | − | 28 ± 9 | 98 ± 5 | 169 ± 5 | 257 ± 15 | 37 ± 2 |
| + | 28 ± 8 | 146 ± 32 | 192 ± 9 | 209 ± 22 | 35 ± 5 | |
| 62.5 | − | 31 ± 6 | 142 ± 6 | 151 ± 15 | 274 ± 6 | 33 ± 2 |
| + | 36 ± 2 | 177 ± 9 | 184 ± 7 | 273 ± 2 | 32 ± 2 | |
| Positive control | − | 1111 ± 117a | 1872 ± 104a | 1697 ± 54c | 2043 ± 185d | 1333 ± 53c |
| + | 1171 ± 78b | 1920 ± 120b | 2053 ± 65b | 2121 ± 21e | 1534 ± 77b | |
| Solvent control | − | 32 ± 1 | 137 ± 24 | 177 ± 4 | 242 ± 10 | 43 ± 5 |
| + | 35 ± 4 | 164 ± 14 | 212 ± 31 | 227 ± 10 | 47 ± 3 | |
| Blank control | − | 29 ± 1 | 152 ± 10 | 181 ± 15 | 252 ± 2 | 38 ± 1 |
| + | 47 ± 4 | 192 ± 5 | 183 ± 10 | 277 ± 4 | 48 ± 4 | |
4. Conclusions
In this study, the potential mutagenic impurities Imp-E and Imp-F of avanafil were synthesized using the design method. QSAR genotoxicity assessment and Ames test were performed for Imp-E and Imp-F according to ICH M7 requirements. The QSAR evaluation showed that Derek's prediction of Imp-E was positive and that of Imp-F was inactive. Sarah's prediction results for Imp-E and Imp-F were negative, and the prediction results could not accurately distinguish the toxicity differences of compounds having structural differences. The results of the Ames test showed that the genotoxicity of Imp-E and Imp-F in 62.5–1000 μg per plate was negative with or without the presence of S9. In summary, the results eliminate concerns about the genotoxicity of Imp-E and Imp-F based on the altered structure of the hydrazine group. Imp-E and Imp-F could be classified as class 5 impurities, that is, ordinary impurities. This study provides theoretical guidance and an experimental basis for the classification and classification control of potential genotoxic impurities Imp-E and Imp-F in avanafil.Data availability
All relevant data are included in the manuscript, along with additional information.Author contributions
Yunkai Sun: writing-original draft, supervision, funding acquisition; Xiaoxia Wu: investigation, experiment, data curation; Pei Zuo: design of Ames test; Zhao Liu: experimental design, supervision and revise the paper; Xuepei Miao: instruction of the experiment; Jian Liu: revise the paper. Hairuo Wen: QSAR evaluation, revise the paper. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Hunan Province (No. 2022JJ80096), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (23KJA150003) and Fund of CIT (YN22039).Notes and references
- M. Kumar, A. D. Pathade, V. B. Gupta, S. V. Gupta, S. Goyal, D. Rath, M. Thakre, J. Sanmukhani and R. Mittal, Int. J. Urol., 2022, 29(4), 351–359 CrossRef CAS PubMed.
- J. Li, L. Peng, D. Cao, L. J. He, Y. X. Li and Q. Wei, J. Mens Health, 2019, 13(5), 1557–9883 CAS.
- R. K. Chawla, S. Panda, K. Umasankar, S. P. Panda and D. Damayanthi, Curr. Pharm. Anal., 2020, 16(7), 801–805 CrossRef CAS.
- D. Pokar, N. Rajput and P. Sengupta, Int. J. Pharm., 2020, 576, 119018 CrossRef CAS.
- D. K. Singh, A. Sahu, S. Kumar and S. Singh, TrAC, Trends Anal. Chem., 2018, 101, 85–107 CrossRef CAS.
- R. Holm and D. P. Elder, Eur. J. Pharm. Sci., 2015, 87, 118–135 CrossRef PubMed.
- European Medcines Agency (EMA), Guideline on the Limits of Genotoxic Impurities [EB/OL] (2006-06-28) [2020-02-22], https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-limits-genotoxic-impurities_en.pdf Search PubMed.
- European Medcines Agency (EMA), Questions & answers on the CHMP Guideline on the Limits of Genotoxic Impurities [EB/OL] (2010-09-23) [2020-02-22], https://www.ema.europa.eu/en/documents/scientific-guideline/questions-answers-guideline-limits-genotoxic-impurities_en.Pdf Search PubMed.
- International Conference on Harmonisation (ICH) M7 (R2), Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk Impurities [EB/OL], (2023-07), https://www.fda.gov/media/170461/download Search PubMed.
- M. E. Crosby, R. Ciurlionis, T. G. Brayman, A. Kondratiuk and J. J. Nicolette, Environ. Mol. Mutagen., 2022, 63(7), 336–350 CrossRef CAS PubMed.
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, Some Industrial Chemicals, International Agency for Research on Cancer, Lyon (FR), 2018, PMID: 29863829 Search PubMed.
- B. Z. Levi, J. C. Kuhn and S. Ulitzur, Mutat. Res. Lett., 1986, 173(4), 233–237 CrossRef CAS PubMed.
- J. Nicolette, J. Murray, P. Sonders, J. Murray, P. Sonders, A. Kondratiuk and M. Crosby, Environ. Mol. Mutagen., 2021, 62(1), 4–17 CrossRef CAS PubMed.
- L. Malcamor and A. A. Stark, Appl. Environ. Microbiol., 1982, 44(4), 801–808 CrossRef CAS PubMed.
- A. V. Wright and L. Tikkanen, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1980, 78(1), 17–23 Search PubMed.
- T. Maeda, A. Shibai, N. Yokoi, Y. Tarusawa, M. Kawada, H. Kotani and C. Furusawa, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2021, 823, 111–759 CrossRef.
- P. Silvio, T. Maurizio, R. Patrizia, P. Mauro, T. Marisa and M. B. Carlo, Carcinogenesis, 1981, 2(12), 1317–1326 CrossRef.
- S. A. Glover, Aust. J. Chem., 2023, 76(1), 1–24 CrossRef CAS.
- D. S. Treitler, A. S. Marriott, J. Chadwick and E. Quirk, Org. Process Res. Dev., 2019, 23(11), 2562–2566 CrossRef CAS.
- M. A. Sandhu, A. A. Saeed, M. S. Khilji, A. L. Anwaar, S. Z. Malik and N. Khalid, J. Toxicol. Sci., 2013, 38(2), 237–244 CrossRef CAS PubMed.
- G. Susanne, O. Ulrich, E. Azeddine, S. Muthusamy, R. Sangana, H. J. Martus, T. Bedman and A. Hartmann, Regul. Toxicol. Pharmacol., 2022, 134, 105245 CrossRef.
- J. Yin, H. Wen and H. Chen, J. Appl. Toxicol., 2023, 43(2), 230–241 CrossRef CAS.
- A. B. Raies and V. B. Bajic, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2016, 6(2), 147–172 CAS.
- M. Honma, A. Kitazawa, T. Kasamatsu and K. I. Sugiyama, Gene Environ., 2020, 42(1), 32–37 CrossRef CAS.
- T. Kasamatsu, A. Kitazawa and S. Tajima, Gene Environ., 2021, 43(1), 1–17 CrossRef.
- A. Sutter, A. Amberg, S. Boyet, F. J. Contrera, L. L. Custer and L. K. Dobo, Regul. Toxicol. Pharmacol., 2013, 67(1), 39–52 CrossRef CAS.
- M. S. Diehl, S. L. Willaby and R. D. Snyder, Environ. Mol. Mutagen., 2015, 36(1), 72–77 CrossRef.
- M. O. Kenyon, J. R. Cheung, K. L. Dobo and W. K. Warren, Regul. Toxicol. Pharmacol., 2007, 48(1), 75–86 CrossRef CAS PubMed.
- K. P. Cross and D. M. DeMarini, Mutat. Res., 2023, 827, 111838 CrossRef CAS PubMed.
- B. C. Fischer, Y. Musengi, J. Konig, B. Sachse, S. Hessel-Pras, B. Schafer, C. Kneuer and K. Herrmann, Mutagenesis, 2024, 39(1), 32–42 CrossRef PubMed.
- Y. K. Sun, X. X. Wu, M. Cao, X. Q. Xue, D. f. Jiang and Z. Liu, J. Mol. Struct., 2024, 1295, 136601 CrossRef CAS.
- Y. Koichiro, M. Kenji and O. Kenii, et al., US Pat., US20030229095A1, 2003.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02345e |
| This journal is © The Royal Society of Chemistry 2024 |