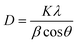Open Access Article
Open Access ArticleModulation of miR-205 expression using a Cheiranthus cheiri phyto-nano hybrid as a potential therapeutic agent against breast cancer
Fatima Razzaq†
a,
Samiah Shahid† *ab and
Wajeehah Shahid
*ab and
Wajeehah Shahid c
c
aInstitute of Molecular Biology and Biotechnology (IMBB), The University of Lahore, Lahore, Pakistan. E-mail: samiah.shahid@yahoo.com
bResearch Centre for Health Sciences (RCHS), The University of Lahore, Lahore, Pakistan
cDepartment of Physics, The University of Lahore, Lahore, Pakistan
First published on 21st November 2024
Abstract
Breast cancer is the fifth major cause of fatalities associated with cancer worldwide and in Pakistan, 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 066 female breast cancer cases were recorded in 2018. This study was designed to understand extracts of Cheiranthus cheiri (C. cheiri) and to evaluate the epigenetic modulation of microRNA expression for breast cancer therapy using a selected phyto-nanohybrid treatment. The phytochemical screening revealed the presence of potential phytochemicals and antioxidant scavenging activity in the C. cheiri extracts with a DPPH (2-diphenyl-1-picryl-hydroxyl) assay giving an IC50 value of 20.63 μg mL−1. GC-MS (gas chromatography-mass spectroscopy) analysis of the C. cheiri n-hexane extract detected 42 phytocompounds. Titanium oxide (TiO2) nanoparticles were synthesized and characterized using XRD (X-ray diffraction), SEM (scanning electron microscopy) and EDX (energy dispersive X-ray spectrometry) to confirm the synthesis of anatase (tetragonal) TiO2. The prepared nanoparticles were conjugated with the selected plant i.e., C. cheiri. The resulting phyto-nanohybrid was used for the subsequent treatment of breast cancer induced in a female rat model. The treatment groups were as follows: doxorubicin as the standard treatment, C. cheiri, TiO2 and the phyto-nano hybrid treatment. After 8 weeks of treatment, the groups induced to exhibit breast cancer (with and without treatment) and the control groups were dissected and analysed for histopathological, haematological and microRNA expression. Histopathological examination revealed chronic inflammation in the dilated ducts and tumour embolus formation, thus confirming the presence of breast cancer in the DMBA-induced female rat model. MicroRNA expression analysis showed a statistically significant decrease in levels of miR-205 in the plasma of the breast cancer rat model compared to the control (p < 0.05). After treatment with the phyto-nano hybrid, a statistically significant increase in the expression of miR-205 was observed in the rat models induced to exhibit breast cancer compared to the rat model without any treatment (p < 0.05). The downregulation of miR-205 in the plasma of the breast cancer exhibiting model, as compared to the control, and its upregulation after treatment with the selected phyto-nano hybrid indicated its diagnostic and prognostic significance. It is concluded that the phyto-nano hybrid used in this study is effective against breast cancer induced female rat model. All the results support the finding that the selected phyto-nano hybrid has great potential as a possible therapeutic agent for the treatment of breast cancer.
066 female breast cancer cases were recorded in 2018. This study was designed to understand extracts of Cheiranthus cheiri (C. cheiri) and to evaluate the epigenetic modulation of microRNA expression for breast cancer therapy using a selected phyto-nanohybrid treatment. The phytochemical screening revealed the presence of potential phytochemicals and antioxidant scavenging activity in the C. cheiri extracts with a DPPH (2-diphenyl-1-picryl-hydroxyl) assay giving an IC50 value of 20.63 μg mL−1. GC-MS (gas chromatography-mass spectroscopy) analysis of the C. cheiri n-hexane extract detected 42 phytocompounds. Titanium oxide (TiO2) nanoparticles were synthesized and characterized using XRD (X-ray diffraction), SEM (scanning electron microscopy) and EDX (energy dispersive X-ray spectrometry) to confirm the synthesis of anatase (tetragonal) TiO2. The prepared nanoparticles were conjugated with the selected plant i.e., C. cheiri. The resulting phyto-nanohybrid was used for the subsequent treatment of breast cancer induced in a female rat model. The treatment groups were as follows: doxorubicin as the standard treatment, C. cheiri, TiO2 and the phyto-nano hybrid treatment. After 8 weeks of treatment, the groups induced to exhibit breast cancer (with and without treatment) and the control groups were dissected and analysed for histopathological, haematological and microRNA expression. Histopathological examination revealed chronic inflammation in the dilated ducts and tumour embolus formation, thus confirming the presence of breast cancer in the DMBA-induced female rat model. MicroRNA expression analysis showed a statistically significant decrease in levels of miR-205 in the plasma of the breast cancer rat model compared to the control (p < 0.05). After treatment with the phyto-nano hybrid, a statistically significant increase in the expression of miR-205 was observed in the rat models induced to exhibit breast cancer compared to the rat model without any treatment (p < 0.05). The downregulation of miR-205 in the plasma of the breast cancer exhibiting model, as compared to the control, and its upregulation after treatment with the selected phyto-nano hybrid indicated its diagnostic and prognostic significance. It is concluded that the phyto-nano hybrid used in this study is effective against breast cancer induced female rat model. All the results support the finding that the selected phyto-nano hybrid has great potential as a possible therapeutic agent for the treatment of breast cancer.
1 Introduction
Breast cancer is a leading cause of death nationally in the 21st century, after cardiovascular diseases and skin cancer, in the United States.1 During 2020, 19 million new cases of breast cancer were recorded in all ages of Pakistani females, and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 838 people died of breast cancer.2 Breast cancer rates are high in Asia and particularly in Pakistan. In 2020, one woman in 9 became a breast cancer patient at some stage of their life.3 Studies have reported 45 genes in the human breast cancer panel and the most common hereditary genes are BRCA1 and BRCA2, whilst some other genes, such as PALB2, PTEN and TP53, are linked with an increased risk of breast cancer.4 Multiple therapies exist for breast cancer; however, breast cancer can develop resistance to chemotherapy, radiation, and hormonal treatments. These conventional treatments for breast cancer are linked to severe side effects, the development of drug resistance, and the possibility of cancer recurrence. Many research findings indicate that medicinal plants have the potential to have therapeutic benefits.5
838 people died of breast cancer.2 Breast cancer rates are high in Asia and particularly in Pakistan. In 2020, one woman in 9 became a breast cancer patient at some stage of their life.3 Studies have reported 45 genes in the human breast cancer panel and the most common hereditary genes are BRCA1 and BRCA2, whilst some other genes, such as PALB2, PTEN and TP53, are linked with an increased risk of breast cancer.4 Multiple therapies exist for breast cancer; however, breast cancer can develop resistance to chemotherapy, radiation, and hormonal treatments. These conventional treatments for breast cancer are linked to severe side effects, the development of drug resistance, and the possibility of cancer recurrence. Many research findings indicate that medicinal plants have the potential to have therapeutic benefits.5
The bioactive chemicals present in medicinal plants have proven to be valuable for their therapeutic properties and their potential in pharmacological treatments for malignant tumours. Flavonoids and polyphenols that have been derived from plants are synthetic antioxidants which frequently act as chemotherapeutic agents. In addition, antioxidants protect cells from reactive oxygen species and limit the oxidative damage to an organism's cells, thus effectively reducing hazardous degenerative disorders.4
C. cheiri is a medicinal plant belongs to the Brassicae family and is a tonic, diuretic, aphrodisiac and expectorant that is useful in dry bronchitis, paralysis and cancer.6 Bioactive components of plant phenols, cardiac steroids, isothiocyanates and flavonoids have high anti-cancer, anti-inflammatory and antioxidant activity.7 The essential oils of C. cheiri were identified by GC-MS analysis, and were found to be γ-tocopherol, β-tocopherol, campesterol, γ-sitosterol, estragole, carnosic acid, gamma muurolene, β-sitosterol and delta cadinol. Different studies have identified plant components as being modulators of miRNA expression in different types of cancer. An effective approach while dealing with miRNA regulation involves utilizing a natural plant source as it will be cost-effective and safe.8
MicroRNAs can be used as non-invasive diagnostic tools and prognostic biomarkers for cancer. Traditional targeted therapies can cause adverse side effects, drug resistance and can be ineffective for the treatment of metastasis so there is a need to explore some new strategies, such as microRNA based therapies, which can evaluate alterations in oncogenic microRNA expression.9 Because of their ability to regulate microRNAs, plant-based products or their synthetic analogues have recently gained attention as a potential cancer prevention treatments. The modification of specific miRNA responses was aided by plant compounds such as artemisinin, curcumin, isoflavones, diindolylmethane (DIM), indole-3-carbinol (I3C), and epigallocatechin gallate (EGCG). The modified miRNA expression boosts the sensitivity of the cancer cells to conventional medicines, hence limiting tumour growth. It is reported that changing miRNA regulation could provide a new approach for developing an effective cancer therapy method.10
Understanding the potential of plant products and miRNAs for the breast cancer prevention and treatment in low-income populations is of paramount importance, given that evidence suggests that this illness is a result of health inequality. Therefore breast cancer can be prevented and treated with the use of natural medicines or their derivatives, either in isolation or in conjunction with traditional medicine.11 In this research the role of plant-mediated regulation of miRNAs in breast cancer prevention and treatment was evaluated. Understanding the anti-breast-cancer drug response mechanism can be aided by discussing the interactions with miRNA and their targeted genes. By altering miRNA expression and tumour suppressor genes, and by targeting oncogenes, natural medicines selectively inhibit malignant cells.12
MicroRNA expression can be modulated by nanoparticles and they represent an effective approach to drug delivery for the treatment of many disorders. Nanotechnology may have remarkable potential to prevent breast cancer recurrence. Nanoparticle drug delivery systems are currently emerging and nanoparticles are a renewable material and chemotherapeutic drug in breast cancer models. Different nanoparticles, such as zinc-oxide and selenium nanoparticles, prevent the growth of mammary tumours10 whilst some other metal-based or magnetic nanoparticles are used for metastatic breast cancer detection and protection.13 It is reported that metallic nanoparticles have antibacterial, antioxidant and anticancer effects. Metallic nanoparticles such as titanium oxide, in combination with plant extracts, show anti-breast cancer activity and reduce tumour volume by destroying free radicals.14
Studies on the anticancer effects of many plants and nanoparticles have been conducted. These new therapy options appear to be more effective, less expensive, safer, and more widely applicable than their predecessors.15 These compounds could potentially be harnessed to develop therapeutic strategies that complement conventional remedies in an effort to improve cancer prevention and treatment. The potential for transforming phytochemicals into commercially available pharmaceuticals with cancer prevention and treatment capabilities is substantial.16
2 Methodology
2.1. Plant collection
The C. cheiri was collected from various areas in Punjab, Pakistan, it was validated by a taxonomist and submitted with voucher number IMBB.UOL.931-7 to the herbarium bank of the university.2.2. Extraction and fractionation
For preparing the plant extracts, its aerial parts were shade dried for 10 days, crushed into powder using an electromechanical grinder and stored in sterilized zipper plastic bags for further processing. The plant powder (400 grams) was soaked in 600 mL of ethanol and hexane solvents each for two weeks and kept at room temperature. After two weeks, Whatman filter paper was utilized for the filtration of the aqueous extract. Moreover, the aqueous extract was evaporated through rotary evaporation and the dried extract was lyophilized for research purposes.2.3. Phytochemical analysis
The preliminary phytochemical screening of the n-hexane and ethanol extracts of C. cheiri was performed using standard procedures.122.4. Total phenolic content determination (TPC)
A stock solution was prepared by dissolving the dried plant extracts obtained from ethanol and n-hexane in a minute quantity of dimethyl sulphoxide (DMSO) to make dilutions at 1 mg mL−1 with distilled water. The standard gallic acid (0–6 μg mL−1) was taken as a reference. Then, 0.5 mL of each extract was dissolved in 2.25 mL of Folin reagent (10%) and mixed thoroughly. Sodium carbonate solution (7.5%, 2.5 mL) was added and the volume was raised up to 10 mL with distilled water. Then this solution was kept at 37 °C for half an hour. Using a spectrophotometer, the absorbance was measured at 760 nm. Based on the calibration curve, the TPC was determined and it is equivalent to a mg of gallic acid.172.5. Total flavonoid content determination (TFC)
A stock solution was prepared by dissolving the dried plant extracts obtained from ethanol and n-hexane in a minute quantity of dimethyl sulphoxide (DMSO) to make dilutions at 1 mg mL−1 with distilled water. As the standard, the drug catechin was used and 0–100 μg mL−1 was dissolved in methanol. 0.5 mL of sample was dissolved in 0.15 mL of aluminum chloride solution (10%), 0.15 mL of potassium acetate (0.1 mM) and the volume was raised to 5 mL. The mixture was incubated at room temperature for 40 minutes and the volume was made up to 10 mL with distilled water. Then this solution was kept at 37 °C for half an hour. Using a spectrophotometer, the absorbance was measured at 415 nm. Based on the calibration curve, the TFC was determined and it is equivalent to a mg of catechin.172.6. Determination of antioxidant potential
2.7. Gas chromatography-mass spectrometry (GC-MS) analysis
GC-MS was conducted on an Agilent USA Technologies GC system on a GC-7890B/MS-5977 A model to identify the bioactive chemicals in the n-hexane extract of the entire C. cheiri plant. The inlet temperature was held at 280 °C by using helium as a carrier gas while maintaining a 1 mL min−1 flow rate. A 1 μL sample volume was injected into the column (DB 5 MS, 3 cm length, and 0.25 mm diameter by 250 mm thickness). However, the carrier gas that was utilized here was helium. The oven temperature was set to be 50 °C for 1 minute, 25–120 °C for 5 minutes, while a temperature of 250 °C was maintained for the injector port and detector. The runtime was held at 51.133 minutes. However, a calibration was carried out with a comparison of the mass spectrum (molecular fingerprint) of the separated molecule by way of the mass to charge ratio. Finally, the mass spectrum of each chemical was compared using the PubChem libraries.212.8. Synthesis and characterization of TiO2 nanoparticles
Titanium dioxide nanoparticles were synthesized via a hydrothermal method.22 Titanium dioxide with a molecular weight of 79.88 and sodium hydroxide with a molecular weight of 40 were used. Here, the molar ratio of TiO2 is 1 M and the mass of the NaOH is calculated by taking 1 M. First, the calculated mass of 1 g of NaOH is taken in a beaker containing 25 mL of distilled water. It was stirred for 30 minutes to obtain a clear solution. Then, the calculated mass of 2 g of TiO2 is added to the above solution. The mixture was stirred vigorously and continuously for 24 hours. The mixture was transferred to an alumina crucible and kept undisturbed for the setting of the sol–gel. Then, it was dried at 50 °C. Finally, it was placed in a furnace at 400 °C for 2 hours for the calcination of the sample. The sample was taken and made into a very fine powder by grinding it with a mortar and pestle for 1 hour to obtain the TiO2 nanopowder. Further, XRD, SEM analysis and EDX were used to characterize the produced nanoparticles.232.9. Synthesis of the phyto-nano hybrid
C. cheiri solution was prepared by dissolving 3 mg of lyophilized C. cheiri in 3 mL of DMSO. The C. cheiri solution was gradually added to the functionalized TiO2 NPs with stirring with a magnetic stirrer at 800 rpm for 30 min. The C. cheiri-loaded TiO2 NPs were centrifuged at 6000 rpm for 10 min. The pellet was separated from the supernatant, washed using deionized water, dried at room temperature, and kept at −20 °C.92.10. Animal model
A sample size of 30 female Wistar rats was chosen using the G*Power 3 program.22 For the experiment, 40–50 days old Wistar rats were used. Before the induction of DMBA, the animals underwent a two-week acclimatization period. The study was carried out strictly following the guidelines of the institutional animal ethical committee for handling experimental animals and was approved by the Bioethical, Biosafety and Biosecurity Committee (BBBC) of the Institute of Molecular Biology and Biotechnology at the University of Lahore with Ref No: CRiMM/23/Research/33.| Groups | Treatment | Number of rats | Induction |
|---|---|---|---|
| a Doxorubicin (20 mg per kg bodyweight) for 28 days, C. cheiri for 28 days (100 mg per kg bodyweight), TiO2 for 28 days (50 mg per kg bodyweight), phyto-nano hybrid (50 mg per kg bodyweight). | |||
| A (Placebo) | Normal saline | 5 | Control |
| B (Induced) | DMBA + olive oil + normal saline | 5 | Induced female rat |
| C (T1) | Doxorubicin | 5 | Standard drug |
| D (T2) | C. cheiri | 5 | C. cheiri |
| E (T3) | TiO2 nanoparticles | 5 | TiO2 nanoparticles |
| F (T4) | Phyto-nano hybrid | 5 | C. cheiri + TiO2 nanoparticles hybrid |
2.11. Tumor volume
The tumor volume was calculated using the method suggested by Geran.24 A Vernier caliper was used to perform the measurement.2.12. Blood sample collection
Using 27 gauge needles, 3 mL of blood was drawn from the heart of the rat by an experienced lab assistant and collected in EDTA vials (1.5 mL for EDTA and 1.5 mL for serum tubes). Blood samples were sent immediately for hematological analysis in an ice box.2.13. MicroRNA extraction and quantification
Plasma was isolated using a two-step centrifugation method and stored at −20 °C for further processing. After that, 200 μL of plasma was added to 600 μL of TRIzol with 10 minutes of incubation on ice, followed by 10 minutes of centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C. With an addition of 200 μL of chloroform and manual vibration 10 times, a further 10 minutes of incubation on ice followed along with 10 minutes of centrifugation at 12
000g and 4 °C. With an addition of 200 μL of chloroform and manual vibration 10 times, a further 10 minutes of incubation on ice followed along with 10 minutes of centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C. After discarding the supernatant, 1 mL of freshly prepared 75% ethanol was added in every tube, followed by 15 minutes of centrifugation at 12
000g and 4 °C. After discarding the supernatant, 1 mL of freshly prepared 75% ethanol was added in every tube, followed by 15 minutes of centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C. Next, the ethanol was discarded and the cap of the centrifuge tube was removed to carry out natural air-drying. Finally, 30 μL of RNase free water was added for dissolving the RNA. The optical density (OD) values and the purity of the RNA were measured.25
000g and 4 °C. Next, the ethanol was discarded and the cap of the centrifuge tube was removed to carry out natural air-drying. Finally, 30 μL of RNase free water was added for dissolving the RNA. The optical density (OD) values and the purity of the RNA were measured.25
2.14. Primer designing
To quantify the selected miRNAs, RNA primers were used and designed according to the protocol in ref. 26 (Table 2).| Primer name | Mature sequence (5′–3′) |
|---|---|
| miR-205 | CCTTAAAAGGCTAAGATTTTA |
| miR-16 | CGCAGTAGCAGCACGTA |
2.15. Reverse transcription
Further processing of the mixture was carried out for the synthesis of cDNA using the Thermo Scientific™ RevertAid™ first strand cDNA synthesis kit (cat no. K1622). Moreover, the recommended protocol used, 1 μL of random hexamer and 5 μg total of RNA, and DEPC water was added until the total volume was 12 μL. Additionally, the mixture was incubated at 65 °C for 5 minutes and it was chilled on ice. M-MuLV Revert Aid Reverse Transcriptase enzyme (200 U), dNTPs mix (10 mM), and 1× RT buffer were added and DEPC water was added until the total volume reached 20 μL. Lastly, the solution was gently mixed while executing reverse transcription with the help of incubation at 70 °C for five minutes, 42 °C for sixty minutes and 25 °C for five minutes.272.16. Real-time PCR
For the quantitative PCR analysis, Maxima SYBR Green/ROX qPCR Master Mix (2X) (Thermo Scientific (Pvt.) Ltd, USA) (cat no. K0221) in a StepOne Plus system (Thermo Fisher Scientific) was used. The qRT-PCR reaction setups were prepared in 0.2 mL sterile tubes by mixing 10 μL of 2X Maxima SYBR Green/ROX qPCR Master Mix fluorescein, 250 nM of each primer set in 1 μL of 10× diluted cDNA with a total volume of 20 μL. Therefore, the cyclic conditions set for the STEP ONE PLUS Real-Time System were: annealing at primer precise temperatures between 55–60 °C for 60 s with extension at 72 °C for 30 s and 95 °C for 10 minutes followed by 40 heating cycles at 95 °C for 15 s. Hence, all the qRT-PCR experiments were carried out in replicates with a quantification basis of the threshold cycle estimation (ΔCt value). Normalization of the raw data was done through subtraction of the Ct value of the normalizer mir-16 from the Ct value of the target microRNA and target gene prior to the analysis.273 Results
3.1. Qualitative phytochemical analysis
The qualitative phytochemical screening revealed the presence of phenol flavonoids, alkaloids, tannins, steroids and terpenoids in the n-hexane and ethanol extracts of C. cheiri (Table 3).| Sr. no. | Phytochemical constituents | Ethanol extract of C. cheiri | n-Hexane extract of C. cheiri |
|---|---|---|---|
| a (+) Indicates the presence and (−) indicates the absence of phytochemical compounds. | |||
| 1 | Flavonoids | + | + |
| 2 | Phenols | + | + |
| 3 | Alkaloids | + | + |
| 4 | Tannins | + | + |
| 5 | Steroids | + | + |
| 6 | Terpenoids | + | − |
| 7 | Saponins | − | + |
3.2. Phytochemical evaluation of plant extracts
3.3. GC-MS analysis of Cheiranthus cheiri
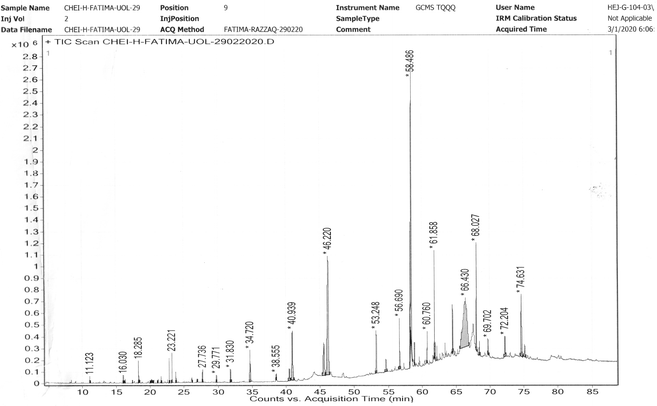
A reliable GC-MS technique for the identification of the constituents of volatile matter, such as long obtained branched hydrocarbons, esters, acids and alcohols was used. On the basis of the antioxidant activities, the n-hexane extract was selected for GC-MS analysis. The analysis of the GC-MS for n-hexane extract of C. cheiri yielded 48 prominent peaks with concentration, molecular weight, molecular formula, and retention time (Table 4). The GC-MS analysis of the C. cheiri n-hexane extract showed 48 prominent peaks with diverse retention time and area sums ranging from 11.123 to 75.105 minutes and 0.05 to 19.83, respectively. Heptacosane had the longest retention time of 75.105 minutes and, while cyclohexene, 4-methylene-1-(1-methylethyl) had the shortest retention time within the range of 11.123 minutes. Z-(13,14-Epoxy)tetradec-11-nen-1-ol acetate was found to be present in the highest concentration (19.83) in the C. cheiri n-hexane extracts (Fig. 2).| No. of peaks | List of compounds | Molecular weight | Formula | Retention time | Area sum% | Class of phytochemical |
|---|---|---|---|---|---|---|
| 1 | Cyclohexene, 4-methylene-1-(1-methylethyl)- | 136 | C10H16 | 11.123 | 0.14 | Terpenoid |
| 2 | 3-Cyclohexene-1-ol, 4-methyl-1-(1-methylethyl)-, (R-) | 154 | C10H18O | 16.03 | 0.16 | Terpineol |
| 3 | Octanoic acid, ethyl ester | 172 | C10H20O2 | 16.194 | 0.05 | Fatty acid ester |
| 4 | Dodecane | 170 | C12H26 | 16.294 | 0.09 | Alkane |
| 5 | Benzaldehyde, 4-(1-methylethyl)- | 148 | C1010H12O | 17.388 | 0.06 | Benzaldehyde |
| 6 | Thymol | 150 | C10H14O | 18.285 | 0.47 | Phenol |
| 7 | 1,3-Benzodioxole, 5-(1-propenyl),-(Z)- | 162 | C10HO2 | 18.385 | 0.09 | Benzodioxole |
| 8 | Propanoic acid, 2-methyl-, 3-hydroxy-2,4,4-trimethylpentyl ester | 216 | C12H24O3 | 20.028 | 0.05 | Carboxylic acid |
| 9 | Copaene | 204 | C15H24 | 20.231 | 0.07 | Sesquiterpene |
| 10 | Benzene, 1,2-dimethoxy-4-(2-propenyl)- | 178 | C11H14O2 | 20.484 | 0.05 | Dimethoxybenzene |
| 11 | Bicyclo[7.2.0]undec-4-ene, 4,11,11-trimethyl-8-methylene-, [1R-(1R*,4Z,9S*)]- | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 411 411 |
C15H24 | 21.15 | 0.08 | Terpenoid |
| 12 | Octanoic acid, 2-ethoxyethyl ester | 216 | C12H24O3 | 21.632 | 0.13 | Benzoic acid ester |
| 13 | 1,3-Benzodioxole, 4-methoxy-6-(2-propenyl)- | 192 | CH12O3 | 22.8 | 0.56 | Benzodioxole |
| 14 | Asarone | 208 | C12H16O3 | 23.221 | 0.63 | Phenylpropanoid |
| 15 | Dodecanoic acid, ethyl ester | 228 | C14H28O2 | 23.805 | 0.22 | Fatty acid ester |
| 16 | Methyl tetradecanoate | 242 | C15H30O2 | 26.179 | 0.1 | Fatty acid methyl ester |
| 17 | Tetradecanoic acid | 228 | C14H28O2 | 26.923 | 0.1 | Long chain fatty acid |
| 18 | Tetradecanoic acid, ethyl ester | 256 | C16H32O2 | 27.736 | 0.38 | Fatty acid ester |
| 19 | 3-Tetradecanone | 212 | C14H28O | 29.771 | 0.3 | Alkane |
| 20 | Hexadecanoic acid, methyl ester | 270 | C17H34O2 | 31.83 | 0.64 | Fatty acid methyl ester |
| 21 | Hexadecanoic acid, ethyl ester | 284 | C18H36O2 | 34.72 | 1.91 | Fatty acid ethyl ester |
| 22 | 3-Hexadecanone | 240 | C16H32O | 38.555 | 0.37 | Ketone |
| 23 | 9,12-Octadecadienoic acid, methyl ester | 294 | C19H34O2 | 40.49 | 1 | Linoleic acid |
| 24 | 9,12-Octadecadienoyl chloride, (Z,Z)- | 298 | C18H31ClO | 40.939 | 4.81 | Alkyl chloride |
| 25 | 9,12,15-Octadecatrienoic acid, methyl ester, (Z,Z,Z)- | 292 | C19H32O2 | 45.557 | 3.82 | Fatty acid methyl ester |
| 26 | 9,12-Octadecadienoic acid, ethyl ester | 308 | C20H36O2 | 46.22 | 16.49 | Fatty acid ethyl ester |
| 27 | Ethyl 9,12,15-octadecatrienoate | 306 | C20H34O2 | 53.248 | 2.5 | Fatty acyls, linoleic acid |
| 28 | Stearic acid, 2-hydroxy-1-methylpropyl ester | 356 | C22H44O3 | 54.659 | 0.75 | Fatty acid ester |
| 29 | cis-11-Eicocenoic acid | 310 | C20H38O2 | 56.69 | 2.23 | Fatty acyl |
| 30 | Methyl 9-eicosenoate | 324 | C21H40O2 | 58.28 | 4.55 | Ester |
| 31 | cis-11-Eicocenoic acid, methyl ester | 324 | C21H40O2 | 58.486 | 17.77 | Fatty acid ester |
| 32 | 9,12-Octadecadienoic acid (Z,Z)-, 2,3-dihydroxypropyl ester | 354 | C21H38O4 | 58.896 | 0.57 | Monoacylglycerol |
| 33 | Butyl 9,12,15-octadecatrienoate | 334 | C22H38O2 | 60.76 | 0.87 | Fatty acyl |
| 34 | Stearic acid, 2-hydroxy-1-methylpropyl ester | 356 | C22H44O3 | 61.858 | 2.96 | Fatty acid ester |
| 35 | Ethyl 13-docosenoate | 366 | C24H46O2 | 61.943 | 0.43 | Fatty acid ethyl ester |
| 36 | cis-13-Eicosenoic acid | 310 | C20H38O2 | 62.189 | 0.34 | Fatty acyl |
| 37 | 9,12,15-Octadecatrienoic acid, 2,3-dihydroxypropyl ester, (Z,Z,Z)- | 352 | C21H36O4 | 64.516 | 1.35 | Glycerophospholipid |
| 38 | Z-(13,14-Epoxy) tetradec-11-nen-1-ol acetate | 268 | C16H28O3 | 66.43 | 19.83 | Fatty acyl |
| 39 | cis-13-Docosenoyl chloride | 356 | C22H41ClO | 68.087 | 4.49 | Fatty acyl chloride |
| 40 | (E)-13-Docosenoic acid | 338 | C22H42O2 | 68.455 | 0.55 | Fatty acyl |
| 41 | Dodecanoic acid, 1,2,3-propanetriyl ester | 638 | C39H74O6 | 69.702 | 0.76 | Triacylglycerol |
| 42 | γ-Tocopherol | 416 | C28H48O2 | 72.204 | 1.45 | Phenol |
| 43 | β-Tocopherol | 416 | C28H48O2 | 74.631 | 4.89 | Phenol lipid |
| 44 | Heptacosane | 380 | C27H56 | 75.105 | 0.94 | Alkane |
| 45 | Cholestane-3,5-diol,5-acetate, (3β,5α)- | 446 | C29H50O3 | 76.12 | 1.23 | Diterpene |
| 46 | Campesterol | 400 | C28H48O | 76.34 | 4.33 | Phytosterol |
| 47 | γ-Sitosterol | 414 | C29H50O | 76.48 | 4.28 | Stigmastane |
3.4. XRD analysis
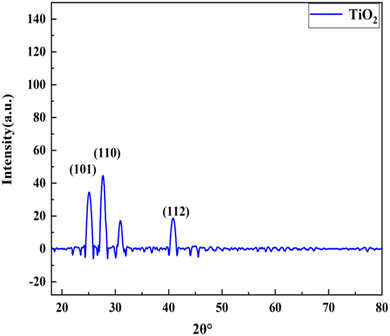
The TiO2 nanoparticle crystal structure was studied using XRD. Many diffraction peaks are present in the TiO2 nanoparticle XRD patterns, as illustrated in Fig. 3. The scanning range goes from 2θ to 80°. Sharp peaks were located at 2θ = 25.12°, 27.68°, and 39.62° and coincide with hkl values of (101), (110), and (112), respectively, which correlates with the tetragonal phase and matches JCPDs (card no 84-128) as well as JCPDs card no 01-073-1764 (Table 5). The findings reveal that TiO2 has an anatase (tetragonal) crystalline shape with parameters of a = b = 3.789603 and c = 9.516015, as shown in Fig. 3. We can determine the average length of the crystalline nanoparticles using the Debye–Sheerer equation.| 2θ | FWHM (°) | d-spacing | Cross ponding plane (hkl) | Crystallite size (nm) |
|---|---|---|---|---|
| 25.12448 | 0.6396 | 1.329 | (101) | 13.29 |
| 27.6830 | 2.9862 | 2.862 | (110) | 28.62 |
| 39.6279 | 3.6258 | 2.435 | (112) | 24.32 |
β represents the full width at half-maximum (FWHM) of the relevant peak, λ is the wavelength and 2θ is the Bragg diffraction angle, while D is utilized to indicate the crystal form of the nano powder.
3.5. SEM analysis
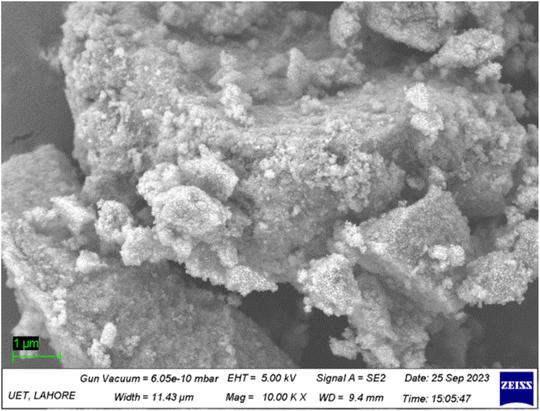
The creation of TiO2 nanoparticles was confirmed through SEM analysis, which is illustrated in Fig. 4, and the shape and structure is demonstrated on the micro-meter scale. The scale bar in the SEM micrograph represents 1 μm. The SEM image clearly exhibits that the morphology of the TiO2 nanoparticles is a spherical agglomeration and that they are homogenously distributed through the entire surface. The structure of the prepared nanoparticles of TiO2 accumulates within the aqueous suspension (Fig. 4). The formation of an agglomeration appears to be due to the fragility of the TiO2 nanoparticles and their small nanoparticular shape. Thus, the nanoparticles have the tendency to bind together until the particles become slightly more stable.3.6. EDX analysis
The energy dispersive X-ray spectroscopy technique was applied to identify the chemical composition of the prepared nanoparticles and to obtain information about the elemental composition of every single element in the samples. Fig. 5 shows the EDX chemical composition analysis of the specimen. The spectrum indicates the existence of pure titanium and oxygen peaks in the specimen being examined. The weight percentages and atomic percentages of Ti and O are 19.91% and 64.41%, and 7.6% and 73.35%, respectively, as given in the inset table. Some impurity peaks are seen in the spectrum and the atomic and weight percentages of Rb, Cl, and Cu are 0.13% and 0.46%, 0.06% and 0.63%, and 0.89% and 0.19%, respectively. The existence of pure oxygen and titanium in the EDX analysis confirmed the creation of pure TiO2 nanoparticles.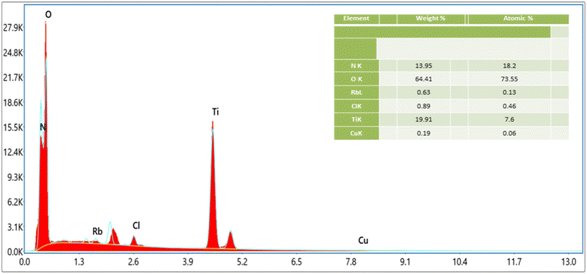 | ||
| Fig. 5 EDX spectrum of the TiO2 nanoparticles. Peaks representing pure titanium and oxygen are present in the specimen. | ||
3.7. Animal body weight study
The body weight of the rats in the DMBA treated group was found to increase with an increase in time duration of the experiment. However, after treatment with the whole plant extract nanoparticles and the standard drug doxorubicin, a substantial reduction (p < 0.05) in the bodyweight was observed compared to the group that received DMBA alone.3.8. Complete blood count
A significant difference was observed between the control, DMBA-induced and treatment groups. The difference between the breast cancer and the treatment groups was significant in terms of the Hb, HCT, and MCV parameters (p < 0.05). Also, there was a moderate difference between the DMBA and treatment groups in the MPV and MCH parameters (p < 0.05). Meanwhile, there was a minimal difference between the groups in terms of the MCHC parameter and the WBC and RBC counts. There was no significant difference found in the platelet, lymphocyte, monocyte, and basophil counts between these groups (p > 0.05) (Table 6).| Hematological parameters | Normal range | Control | DMBA treated | T1 (Dox) | T2 (C. cheiri) | T3 (TiO2) | T4 (phyto-nano hybrid) |
|---|---|---|---|---|---|---|---|
| a The values are the mean ± SD (standard deviation). p-Values are significant at <0.05. | |||||||
| Hb (g dL−1) | 11.7–14 | 14.9 ± 0.01 | 19.1 ± 0.01 | 17.1 ± 0.02 | 18.3 ± 0.05 | 15.2 ± 0.01 | 12.9 ± 0.01 |
| RBC (× 106/μL) | 5.1–6.6 | 7.61 ± 0.01 | 10.2 ± 0.005 | 9.1 ± 0.05 | 9.5 ± 0.05 | 8.4 ± 0.05 | 8.61 ± 0.01 |
| HCT (%) | 30 – 45 | 42.07 ± 0.06 | 59.1 ± 0.05 | 55 ± 0.050 | 57.1 ± 0.05 | 53 ± 0.06 | 43.08 ± 0.06 |
| MCV (fL) | 60–72 | 55.03 ± 0.05 | 56.9 ± 0.05 | 57 ± 0.06 | 57 ± 0.05 | 55 ± 0.05 | 56.12 ± 0.05 |
| MCH (pg) | 20–24 | 19.06 ± 0.05 | 18.9 ± 0.05 | 18 ± 0.07 | 18.1 ± 0.06 | 19 ± 0.05 | 20.08 ± 0.05 |
| MCHC (g dL−1) | 32–35 | 34 ± 0.04 | 33 ± 0.04 | 32 ± 0.05 | 32.5 ± 0.01 | 31 ± 0.06 | 31 ± 0.04 |
| PLT (× 103/μL) | 160–462 | 983 ± 0.05 | 1099 ± 0.50 | 999 ± 0.1 | 1090 ± 0.5 | 995 ± 0.01 | 999 ± 0.05 |
| WBC (× 103/μL) | 3.6–7.2 | 18.17 ± 0.01 | 13.7 ± 0.01 | 14.5 ± 0.01 | 14 ± 0.07 | 15 ± 0.01 | 16.42 ± 0.01 |
| Neutrophils (%) | 27–74 | 10.1 ± 0.05 | 17.07 ± 0.06 | 16 ± 0.01 | 16.1 ± 0.05 | 15 ± 0.02 | 9.21 ± 0.05 |
| Lymphocytes (%) | 16–70 | 9.1 ± 0.01 | 8.3 ± 0.05 | 8.5 ± 0.05 | 8.5 ± 0.03 | 8.8 ± 0.01 | 9.1 ± 0.01 |
| Monocytes (%) | 0–3 | 2 ± 0.05 | 2 ± 0.05 | 2.2 ± 0.01 | 2.5 ± 0.2 | 2.9 ± 0.05 | 2 ± 0.05 |
| Eosinophil (%) | 0–3 | 2.06 ± 0.1 | 2.03 ± 0.07 | 2.21 ± 0.01 | 2.4 ± 0.05 | 2 ± 0.01 | 2.06 ± 0.1 |
3.9. Histopathology of the mammary glands
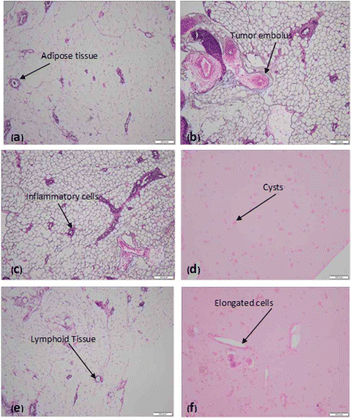
Histological examination of a given section of the mammary glands reveals a skin covered piece of tissue. The epidermis is unremarkable, the underlying dermis reveals benign skin appendages and the deep dermis reveals fibro collagenous tissue and mature adipose tissue. No granulomas are seen and there is no evidence of malignancy (Fig. 6a). Histological examination of a given section of the mammary glands reveals features of non-invasive cystic carcinoma and formation of tumour embolus, composed of malignant epithelial cells with hyper chromatic nuclei, a high N/C ratio and scanty clear to eosinophilia cytoplasm and inconspicuous nucleoli. The intervening stroma reveals fibrosis, congested blood vessels and adipose tissue infiltrated by mild chronic inflammatory cell infiltrate (Fig. 6b). Histopathology reveals chronic inflammation in the dilated ducts (Fig. 6c). Histopathology also reveals cystic changes (Fig. 6d) and mature adipose tissue along with collection of lymphoid tissue. Inflammatory cells are also seen. No evidence of granulomatous or a neoplastic process was found (Fig. 6e). The mature adipose tissue and a collection of lymphoid tissue have been seen. Inflammatory cells are also seen (Fig. 6f).3.10. miR-205 expression in breast cancer rats
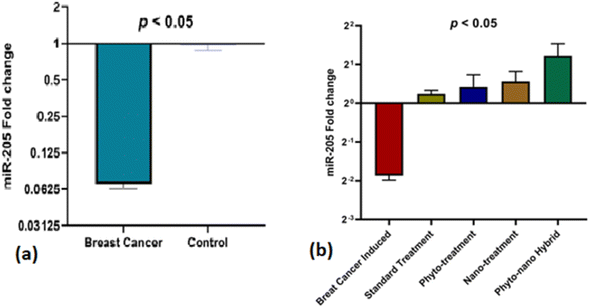
Fold change expression of miR-205 in the DMBA-induced plasma of the rats was downregulated as compared to the control. But after treatment with C. cheiri, TiO2, doxorubicin and the phyto-nano hybrid, a significant difference was observed. Fold change expression in the plasma of the doxorubicin treated rats was slightly upregulated. In the nanoparticles and plant treatment group, microRNA expression was upregulated and it is highly upregulated with the treatment of the phyto-nano hybrid (Fig. 7).MicroRNA expression analysis showed a statistically significant decrease in levels of miR-205 in the plasma of a breast cancer rat model compared to the control (p < 0.05). After treatment with the phyto-nano hybrid, a statistically significant increase in the expression of miR-205 was observed in the breast cancer induced rat models compared to the rat model without any treatment (p < 0.05).
4 Discussion
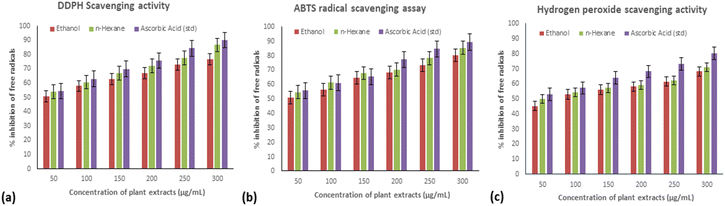
Breast cancer is a primary cause of death for women worldwide due to its aggressive invasion process, its early metastasis resistance to chemotherapeutic treatments, and its high mortality rates. Because traditional chemotherapeutic medicines influence the entire body system via the blood, there can be several systematic adverse effects, including tissue damage and gastrointestinal discomfort. Plants have been used as treatments for thousands of years across various civilizations.20 Thus, apart from treatment with chemotherapy and radiotherapy, there should be some natural way to treat breast cancer. The discovery and development of innovative pharmaceutical medications derived from medical plants has a substantial impact on human well-being by treating a variety of ailments. Folk medicine uses C. cheiri for several therapeutic applications. The growth of pathogenic bacteria is inhibited by secondary metabolites, such as flavonoids, alkaloids, phenolic compounds, and steroids.24 This current phytochemical screening of the n-hexane and ethanolic extracts of the whole C. cheiri plant exposed the presence of flavonoids, tannins, terpenoids, steroids, glycosides, and phenolic compounds. The secondary metabolites were shown to have pharmacological characteristics. For example, tannins are anti-cancer and anti-inflammatory chemicals; flavonoids are anti-inflammatory and antioxidant compounds; and saponins are antibacterial substances which decrease blood cholesterol.28 Therefore, the total phenolic content of the ethanolic and hexane C. cheiri extracts was estimated to be 478.80 mg gallic acid equivalent per gram and 765.65 mg gallic acid equivalent per gram, correspondingly. The overall phenolic contents of the same extracts were calculated to be 53.34 mg catechin equivalent per gram and 59.19 mg catechin equivalent per gram, respectively. According to earlier studies, medicinal plants contain a high concentration of phenolics as well as flavonoids. Hence, it was found that extracts of C. cheiri comprised more total phenols than flavonoids.29Plants are capable of preventing oxidative damage with growth and thus can moderate stress. Enzymatic and non-enzymatic mechanisms protect tissues from activated oxygen species, which can be caused by environmental stressors such as cold, dryness, and air pollution.30 The results of this study support the use of DPPH, NO, and H2O2 models to investigate the oxidant scavenging capability of C. cheiri. The ethanolic extract showed positive antioxidant activity in both the NO model, with an IC50 of 15.70 ± 0.024 μg mL−1, and in the DPPH model, with an IC50 of 17.98 ± 0.04 μg mL−1, values which are comparable to the corresponding standard, ascorbic acid. In comparison, the n-hexane extract exhibited the lowest antioxidant capacities in NO (IC50 24.08 ± 0.024) and DPPH (IC50 30.80 ± 0.024). At the same percentage, the H2O2 model demonstrated modest activity, reflected in an IC50 value of 57.59 ± 0.01 μg mL−1. The differences in the antioxidant assay results demonstrate that the best antioxidant activities depend upon their comparative efficacy. The presence of the NO, DPPH and H2O2 radicals may account for the observed disparities.31 Therefore, biological antioxidant assays were performed to evaluate the effectiveness of the biologically active compounds present in the ethanol and n-hexane extracts. This was done by comparison of the outcomes of the DPPH, NO, and H2O2 free radical scavenging activity assays based on the nature of compounds present in the extracts.32
Medicinal chemistry involves using convenient analytical techniques such as GCMS for detecting complex mixtures of phytocompounds based on solvent polarity.33 GCMS is utilized for the identification and analysis of the chemicals found in plant samples. In our study, 42 phytocompounds were identified in the n-hexane extract through GCMS analysis. This technique is vital for phytochemical research along with chemotaxonomy investigations for medicinal plants with biologically active constituents. Thus, the treatment of breast cancer with herbal medication is a better approach, but for the delivery of these phytocompounds direct to their target, nanoparticles are the best drug delivery system.34
The work of different researchers on nanotechnology compounds with anti-cancer properties has led to advancements in the pharmacotherapeutic field. Nanotechnology is a rapidly growing research field in both the global and national context.35 A previous study reported TiO2 nanoparticles36 and here the X-ray diffraction (XRD) pattern revealed the presence of TiO2 nanoparticles. The morphology revealed in the SEM images clearly shows that the TiO2 nanoparticles are a spherical agglomeration and are homogenously distributed over the entire surface. The FWHM showed that the TiO2 nanoparticles were spherical in shape with sizes ranging from 13.29 to 24.32 nm. The EDX profile of the TiO2 nanoparticles showed a strong Ti peak. The presence of the O peaks along with the Ti signals suggests that the TiO2 nanoparticles were protected with phenolate ions. The spectrum data of XRD pattern of the synthesized TiO2 nanoparticles revealed the occurrence of four major peaks. The peaks appeared with 2θ values that correspond to the (111), (200), (220), and (311) planes of the face-centered cubic (fcc) phase, which are comparable with the standard JCPDS 89-3722. The peak positions shed light on the translational proportion, namely the size and shape of the unit cell, while the peak intensities give details about the electron density inside the unit cell where the atoms are located. The width of the (111) peak was utilized to calculate the average crystallite size using the Scherrer equation.37,38 Our study confirmed that the scanning range goes from 2θ to 80°. Sharp peaks were located at 2θ = 25.12°,27.68°, and 39.62° and coincide with hkl values of (101), (110), and (112), respectively, which correlates with the tetragonal phase and matches with JCPDs card no 84-1285 as well as JCPDs card no 01-073-1764. The findings reveal that the TiO2 has the anatase (tetragonal) crystalline shape with parameters of a = b = 3.789603 and c = 9.516015. We can determine the average length of the crystalline nanoparticles using the Debye–Sheerer equation. Several studies have shown that plants and nanoparticles can modulate microRNA expression, which in turn can inhibit the proliferation of breast cancer.35 Studies have reported that changes in the regulation of microRNAs performs a significant part in the progress, advancement, and promotion of breast cancer, therefore, these microRNAs are essential in controlling the disease. Different microRNAs have been identified which are dysregulated in breast cancer. And these microRNAs play a significant regulation role for several molecular procedures such as invasion, proliferation, metastasis, self-renewal, caspase-mediated cell death, and epithelial–mesenchymal transition. MicroRNAs have the potential to act as both oncogenic and tumor suppressors.39
There has been prior reporting of miR-205 downregulation in the triple-negative patient population, and its role as a breast tumor suppressor has also been documented.40 Although miR-205 has been demonstrated to be either increased or downregulated in breast cancer compared with normal tissue, its breast cancer stem cell expression remains unknown.41 A study found that miR-205, which directly targets and downregulates erbb receptors, is more abundantly expressed in human breast cancer stem cells than in more differentiated tumor cells.42 Although it was previously known that miR-205 specifically targets ERBB3, research has shown that it also regulates ERBB2 and EGFR. While it appears that ERBB2 is a direct target of this microRNA, p63, which has already been established to have the potential to regulate EGFR transcription, it is responsible for mediating the regulation of EGFR.43 A study found that breast cancers are resistant to Lapatinib and other breast cancer treatments because they express less ERBB receptor family members. This suggests that miR-205 may be an important target to improve outcomes in patients with breast cancer who overexpress Her2 and if these cells survived then the tumor will proliferate.42
MicroRNA profiling and the molecular features of tumor cells has become research of interest in recent years.44 The primary objective of our research was to evaluate epigenetic modulation in microRNA expression by phytocompounds, nanoparticles and a phyto-nano hybrid. An in vivo study was conducted and cancer was induced using DMBA (7,12-dimethylbenzanthracene). After the confirmation of the cancer by histopathology and hematology techniques, treatment was started with plants, nanoparticles and a phyto-nano hybrid. After the treatment, blood was collected and microRNA expression was quantified using real time PCR. We analyzed the data using a real-time RT-PCR technique with SYBR Green. Our study confirmed that miR-205 was downregulated in the breast cancer sample as compared to the control. It was slightly upregulated in treatment with the standard drug doxorubicin. However, microRNA expression was upregulated in treatment with plant and nanoparticles and it was significantly upregulated in treatment with the phyto-nanohybrid. Thus, our study concluded that the phyto-nano hybrid could be a better approach to treat breast cancer and other diseases.
5 Conclusions
Breast cancer affects a large number of women worldwide, yet treating it remains difficult. The C. cheiri plant and TiO2 nanoparticles have shown anti-tumorigenic properties, but the therapeutic impact of a phyto-nano hybrid on microRNA expression has never been investigated. The present work explored the ability of a chemically modified phyto-nano hybrid to inhibit tumorigenesis as depicted by expression of microRNA-205 in breast cancer. MicroRNA expression was downregulated in breast cancer when compared with a control, but after treatment with C. cheiri, TiO2 and a phyto-nano hybrid, microRNA expression was upregulated significantly. The significant change in microRNA expression before and after treatment demonstrated the therapeutic potential of the tested phyto-nano hybrid against breast cancer.Data availability
The data is accessible in the supplementary file provided.Author contribution
Samiah Shahid conceptualized and designed the research. Fatima Razzaq, Samiah Shahid and Wajeehah Shahid performed the experimental work, analysed data, and drafted the manuscript. All authors critically reviewed and approved the final draft of the manuscript.Conflicts of interest
The authors have no conflicts of interests.References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2018, 68(6), 394–424 CrossRef PubMed.
- L. Torre, F. Islami, R. L. Siegel, E. Ward and A. Jemal, Global cancer in women: burden and trends, Cancer Epidemiol., Biomarkers Prev., 2017, 26(4), 444–457 CrossRef.
- D. M. Anwa, M. El-Sayed, A. Reda, J. Y. Fang, S. N. Khattab and A. O. Elzoghby, Recent advances in herbal combination nanomedicine for cancer: delivery technology and therapeutic outcomes, Expert Opin. Drug Delivery, 2021, 2(11), 1609–1625 CrossRef PubMed.
- M. A. Taylor, K. Sossey-Alaoui, C. L. Thompson, D. Danielpour and W. P. Schiemann, TGF-β upregulates miR-181a expression to promote breast cancer metastasis, J. Clin. Invest., 2013, 123(1), 150–163 CrossRef CAS PubMed.
- F. Ahmed, B. Ijaz, Z. Ahmad, N. Farooq, M. B. Sarwar and T. Husnain, Modification of miRNA Expression through plant extracts and compounds against breast cancer: mechanism and translational significance, Phytomedicine, 2020, 68, 153168 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method, Methods, 2001, 25(4), 402–408 CrossRef CAS PubMed.
- G. Mosleh, P. Badr, A. Azadi, Z. Abolhassanzadeh and S. V. Hosseini, Wallflower (Erysimum cheiri (L.) Crantz) from past to future, Res. J. Pharmacogn., 2019, 6(2), 85–95 CAS.
- B. Singh, A. M. Ronghe, A. Chatterjee, N. K. Bhat and H. K. Bhat, MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis, Carcinogenesis, 2013, 34(5), 1165–1172 CrossRef CAS PubMed.
- P. Sadhukhan P, M. Kundu, S. Chatterjee, N. Ghosh, P. Manna and J. Das J, Targeted delivery of quercetin via pH-responsive zinc oxide nanoparticles for breast cancer therapy, Mater. Sci. Eng., C, 2019, 100, 129–140 CrossRef PubMed.
- K. K. Vekariya, J. Kaur and K. Tikoo, ERα signaling imparts chemotherapeutic selectivity to selenium nanoparticles in breast cancer, Nanomed.: Nanotechnol. Biol. Med., 2012, 8(7), 1125–1132 CrossRef CAS PubMed.
- H. Jiang, J. Sun, F. Liu, X. Wu and Z. Wen, An immune-related long noncoding RNA pair as a new biomarker to predict the prognosis of patients in breast cancer, Front. Genet., 2022, 13, 895200 CrossRef CAS.
- M. M. Haque, N. Sultana, S. M. T. Abedin and S. E. Kabir, Phytochemical screening and determination of minerals and heavy metals in the flowers of Nyctanthes arbor-tristis L, Bangladesh J. Sci. Ind. Res., 2019, 54(4), 321–328 CrossRef CAS.
- J. Ye, J. Shi, M. Zhang, Y. Zhang, J. Tao and S. Jiang S, Novel Alhagi maurorum leaves mediated synthesis of titanium nanoparticles for human breast carcinoma applications: a preclinical trial study, Arch. Med. Sci., 2021, 28, 143345 Search PubMed.
- N. A. Al-Shabib, F. M. Husain, F. A. Qais, N. Ahma, A. Khan and A. A. Alyousef AA, Phyto-mediated synthesis of porous titanium dioxide nanoparticles from Withania somnifera root extract: broad-spectrum attenuation of biofilm and cytotoxic properties against HepG2 cell lines, Front. Microbiol., 2020, 11, 1680 CrossRef PubMed.
- P. Banthia, L. Gambhir, D. Daga, A. Sharma, N. Kapoor and R. D. Agarwal, Phytogenic synthesis of metallic nanoparticles: application for breast cancer nanomedicine, Vegetos, 2023, 1(1), 10–19 CrossRef.
- S. Behboodi, F. Baghbani-Arani, S. Abdalan and S. A. Sadat Shandiz, Green Engineered Biomolecule-Capped Silver Nanoparticles Fabricated from Cichorium intybus Extract: In Vitro Assessment on Apoptosis Properties Toward Human Breast Cancer (MCF-7) Cells, Biol. Trace Elem. Res., 2019, 187(2), 392–402 CrossRef CAS.
- M. A. Mir, K. Parihar, U. Tabasum and E. Kumari, Estimation of alkaloid, saponin and flavonoid, content in various extracts of Crocus sativa, J. Med. Plants Stud., 2016, 4(5), 171–174 Search PubMed.
- V. L. Singleton, R. Orthofer and R. M. Lamuela-Raventos, Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent, in Methods in Enzymology, Elsevier, 2001, pp. 152–78 Search PubMed.
- S. Sultana and S. Mir, Chemical constituents from the roots of Trichodesma Indicum (L.), Eur. J. Pharm. Med. Res., 2022, 1(9), 450–456 Search PubMed.
- M. Ebrahimzadeh, F. Pourmorad and S. Hafeezi, Antioxidant Activities of Iranian Corn Silk, Turk. J. Biol., 2008, 1(1), 43–49 Search PubMed.
- H. Jaberian, K. Piri and J. Nazari, Phytochemical composition and in vitro antimicrobial and antioxidant activities of some medicinal plants, Food Chem., 2013, 1(1), 237–244 CrossRef PubMed.
- M. Hossain and S. Islam, Synthesis of Carbon Nanoparticles from Kerosene and their Characterization by SEM/EDX, XRD and FTIR, Am. J. Nanosci. Nanotechnol., 2013, 1, 1–52 CrossRef.
- I. Romero, D. L. Garcia-Gonzalez, R. Aparicio-Ruiz and M. T. Morales, Validation of SPME–GCMS method for the analysis of virgin olive oil volatiles responsible for sensory defects, Talanta, 2015,(134), 394–401 CrossRef CAS PubMed.
- F. Faul, E. Erdfelder, A. Buchner and A. G. Lang, Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses, Behav. Res. Methods, 2009, 41(4), 1149–1160 CrossRef PubMed.
- D. S. Metibemu, O. A. Akinloye, A. J. Akamo, J. O. Okoye, D. A. Ojo and E. Morifi, Carotenoid isolates of Spondias mombin demonstrate anticancer effects in DMBA-induced breast cancer in Wistar rats through X-linked inhibitor of apoptosis protein (XIAP) antagonism and anti-inflammation, J. Food Biochem., 2020, 44, 12 CrossRef PubMed.
- A. E. Simoes, D. M. Pereira, J. D. Amaral, A. F. Nunes, S. E. Gomes, P. M. Rodrigues, R. D'Hooge, C. J. Steer, S. N. Thibodeau and P. M. Borralho, Efficient recovery of proteins from multiple source samples after trizol® or trizol® LS RNA extraction and long-term storage, BMC Genomics, 2013, 14, 1–5 CrossRef.
- U. Baykal, Development of a sensitive primer extension method for direct detection and quantification of miRNAs from plants, PLoS One, 2024, 15(3), e0230251 CrossRef.
- H. Jonckheere, J. Anne and E. DeClercq, The HIV-1 reverse transcription (RT) process as target for RT inhibitors, Med. Res. Rev., 2000, 20(2), 129–154 CrossRef CAS.
- T. S. Richard, A. H. N. Kamdje and F. Mukhtar, Medicinal plants in breast cancer therapy, J. Med. Plants, 2015, 1(1), 19–23 Search PubMed.
- E. Lambertini, R. Piva, M. T. H. Khan, I. Lampronti, N. Bianchi and M. Borgatti, Effects of extracts from Bangladeshi medicinal plants on in vitro proliferation of human breast cancer cell lines and expression of estrogen receptor α gene, Int. J. Oncol., 2004, 24(2), 419–423 Search PubMed.
- V. S. Slambrouck, A. Daniel, C. Hooten, S. Brock, A. Jenkins and M. Ogasawara, Effects of crude aqueous medicinal plant extracts on growth and invasion of breast cancer cells, Oncol. Rep., 2007, 17(6), 1487–1492 Search PubMed.
- M. M. Mainasara, M. F. A. Bakar and A. C. Linatoc, Malaysian medicinal plants potential for breast cancer therapy, Asian J. Pharm. Clin. Res., 2018, 101–117 CrossRef.
- M. I. Khan, A. Bouyahya, N. E. L. Hachlafi, N. E. Menyiy, M. Akram and S. Sultana, Anticancer properties of medicinal plants and their bioactive compounds against breast cancer: a review on recent investigations, Environ. Sci. Pollut. Res., 2022, 29(17), 24411–24444 CrossRef CAS PubMed.
- S. S. Kamble and R. N. Gacche, Evaluation of anti-breast cancer, anti-angiogenic and antioxidant properties of selected medicinal plants, Eur. J. Integr. Med., 2019, 25, 13–19 CrossRef.
- Z. O. Omogbadegun, Medicinal plants-based foods for breast cancer treatment: an ethnobotanical survey and digitization, Int. J. Med. Aromat. Plants., 2013, 1(8), 137–163 Search PubMed.
- R. Mund, N. Panda, S. Nimesh and A. Biswas, Novel titanium oxide nanoparticles for effective delivery of paclitaxel to human breast cancer cells, J. Nanopart. Res., 2014, 22(12), 2739 CrossRef.
- K. Murugan, D. Dinesh, K. Kavithaa, M. Paulpandi, T. Ponraj and M. S. Alsalhi, Hydrothermal synthesis of titanium dioxide nanoparticles: mosquitocidal potential and anticancer activity on human breast cancer cells (MCF-7), Parasitol. Res., 2016, 115(3), 1085–1096 CrossRef PubMed.
- N. Lagopati, P. V. Kitsiou, A. L. Kontos, P. Venieratos, E. Kotsopoulou and A. G. Kontos, Photo-induced treatment of breast epithelial cancer cells using nanostructured titanium dioxide solution, J. Photochem. Photobiol., A, 2010, 15(2), 215–223 CrossRef.
- N. Chauhan, A. Dhasmana, M. Jaggi and M. M. Yallapu, miR-205: a potential biomedicine for cancer therapy, Cells, 2020, 25(9), 1957 CrossRef PubMed.
- H. Wu and Y. Y. Mo, Targeting miR-205 in breast cancer, Expert Opin. Ther. Targets, 2009, 13(12), 1439–1448 CrossRef CAS PubMed.
- I. Plantamura, A. Cataldo, G. Cosentino and M. V. Iorio, miR-205 in Breast Cancer: State of the Art, Int. J. Mol. Sci., 2021, 22(1), 27 CrossRef CAS PubMed.
- A. De Cola, S. Volpe, M. C. Budani, M. Ferracin, R. Lattanzio and A. Turdo, et al., miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance, Cell Death Dis., 2015, 6(7), e1823 CrossRef CAS PubMed.
- Y. Hu, Y. Qiu, E. Yague, W. Ji, J. Liu and J. Zhang, miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer, Cell Death Dis., 2016,(6), e2291 CrossRef CAS.
- H. Wu, S. Zhu and Y. Y. Mo, Suppression of cell growth and invasion by miR-205 in breast cancer, Cell Res., 2009, 19(4), 439–448 CrossRef CAS PubMed.
Footnote |
| † The first two authors, Fatima Razzaq and Samiah Shahid, have contributed equally to this study and will be considered as first authors. |
| This journal is © The Royal Society of Chemistry 2024 |