Open Access Article
Open Access ArticleAntioxidant activity of Schiff base ligands using the DPPH scavenging assay: an updated review
Md. Sohel Rana a,
Noor Mohammad Azbar Rayhana,
Md. Shahadat Hossain Emon
a,
Noor Mohammad Azbar Rayhana,
Md. Shahadat Hossain Emon a,
Md. Tanvir Islam
a,
Md. Tanvir Islam a,
Khandaker Rathry
a,
Khandaker Rathry a,
Md. Mahadi Hasana,
Md. Munna Islam Mansura,
Bishal Chakrabarty Srijona,
Md Shohidul Islamb,
Anik Raya,
Md. Abdur Rakib
a,
Md. Mahadi Hasana,
Md. Munna Islam Mansura,
Bishal Chakrabarty Srijona,
Md Shohidul Islamb,
Anik Raya,
Md. Abdur Rakib a,
Azharul Islamc,
Md. Kudrat-E-Zahana,
Md. Faruk Hossena and
Md. Ali Asraf
a,
Azharul Islamc,
Md. Kudrat-E-Zahana,
Md. Faruk Hossena and
Md. Ali Asraf *a
*a
aDepartment of Chemistry, University of Rajshahi, Rajshahi-6205, Bangladesh
bDepartment of Pharmacy, University of Rajshahi, Rajshahi-6205, Bangladesh
cDepartment of Biochemistry and Molecular Biology, University of Rajshahi, Rajshahi-6205, Bangladesh
First published on 21st October 2024
Abstract
Schiff base ligands, formed from primary amines and carbonyl compounds, are potential antioxidants because they scavenge 2,2-diphenyl-1-picrylhydrazyl (DPPH) radicals via hydrogen atom transfer (HAT) and single electron transfer (SET) routes. This review aims to help design, synthesize, and discuss the antioxidant activity of Schiff base ligands based on their structure. This study critically discussed the solvent effect and the structural changes of Schiff base ligands responsible for DPPH scavenging activity, such as proton donating, electron-donating, and electron-withdrawing substituents, conjugation and ring structure. The ligands with electron-donating substituent groups in the phenolic ring demonstrated greater activity by readily stabilizing the radical and some of them showed higher activity than the standard. The activity also depends on the solvent used; the activity increases in those solvents that promote the proton and electron donation of the Schiff base. Schiff bases are most important due to their versatile applications, which can be explained by their antioxidant activity. The data led to the conclusion that the Schiff base ligand will serve as a source of synthetic antioxidants. There should be lots of scope for research on the antioxidant activity of Schiff bases. This review will assist researchers in studying Schiff base-based antioxidants and their applications. All the data analyzed in this paper was found from in vitro tests; for more clearance supplementary tests and in vivo investigations are crucial.
1. Introduction
The concept of Schiff's base originates from the name of the German scientist Hugo Schiff, who in 1864 was the first to elucidate the chemicals formed when primary amines react with carbonyl compounds. These chemicals (Fig. 1) are called Schiff bases after Hugo Schiff and are well-known for their strong coordination capabilities.1,2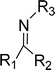 | ||
| Fig. 1 The general structure of Schiff bases, where R1, R2 and R3 are alkyl or aryl groups. R1 or/and R2 may also be hydrogen atoms.1,2 | ||
From the literature, Schiff bases have been synthesized using lots of methods but now the green synthesis methods are the main focus of researchers. Traditionally, Schiff bases are made by refluxing (heating) a mixture of an amine and an aldehyde in organic solvents like alcohol or acetic acid for several hours. This method requires an acid catalyst and often uses techniques to remove water formed during the reaction, which improves yield.3 In the microwave irradiation method, the reaction mixture was radiated at (180–600 W) for about several minutes which offers a faster and cleaner alternative. It generates Schiff bases quickly and with minimal byproducts, making it a more efficient method.4 Recently, researchers have been exploring greener and more sustainable approaches. One method uses natural acids from fruit juices as catalysts, allowing reactions to occur at room temperature with good yields. Additionally, ultrasonic waves can be used to accelerate reactions and achieve higher yields under milder conditions.5 Grinding the starting materials together is another simple and green technique that doesn't require harsh chemicals. Finally, water itself can be used as a solvent for Schiff base synthesis. This method is cost-effective, non-toxic, and allows for easy product separation. Interestingly, water can significantly increase reaction speed compared to traditional methods.6 These advancements provide researchers with a variety of options for synthesizing Schiff bases, catering to factors like speed, efficiency, and environmental impact.7 For electron or proton donating capacity, Schiff bases exhibit versatile applications. In medicine, they act as metal chelators, potentially combating diseases.8 They can also be used as catalysts for speeding up chemical reactions.9 Their ability to bind with metals makes them valuable in material science for energy storage,10 chemo-sensing,11 bio-sensing,12 biomedical,13 nanotechnology applications14 and creating new compounds with desired properties. Additionally, Schiff bases play a role in the dye industry,15 contributing vibrant colors to textiles and other materials. Their biological activity makes them promising candidates for developing new drugs and pesticides.16 Some Schiff bases have remarkable antibacterial,17 antifungal18 and anticancer activities.19 Schiff bases possess antioxidant properties that enable them to stabilize free radicals via the donation of protons and electrons.20
Our bodies constantly produce free radicals as a byproduct of normal functions and exposure to outside elements. These free radicals form through various reactions within cells, both those driven by enzymes (like respiration, immune cell activity, and hormone production) and those that happen spontaneously (like oxygen interacting with molecules or due to radiation). Internally, some major sources of free radicals including our powerhouses like mitochondria, and enzymes like xanthine oxidase. Other internal contributors are inflammation, immune cell activity (phagocytosis), pathways involved in certain fats (arachidonate pathways), exercise, and situations where oxygen supply is cut off and then restored (ischemia/reperfusion injury). Externally, we encounter free radicals from cigarette smoke, pollutants in the air and environment, radiation exposure, certain medications and pesticides, industrial solvents, and even ozone in the atmosphere.21,22 Oxygen utilization in cells may result in the formation of free radicals. These free radicals damage cells in various ways, including breaking down fats, fragmenting DNA, and altering proteins. This damage, known as oxidative stress, contributes not only to the toxicity of outside chemicals but also to many diseases. Examples include injuries, organ failure, inflammation, and even conditions like cancer and dementia. Interestingly, iron buildup seems to play a role in worsening these effects, particularly in brain diseases like amyotrophic lateral sclerosis and multiple sclerosis. This suggests that iron accumulation may be a contributing factor, potentially causing inflammation and harming blood flow in the brain.23 Antioxidants protect cells from damage caused by free radicals.24 These kinds of substances are also employed as food preservatives that prevent oxidation.25 Antioxidants are often used as catalysts in antibiotics for their anti-inflammatory, antifungal, antibacterial, and antiviral properties, as well as in the industrial sector for their anticorrosive effects.26 Antioxidants include a variety of compounds, such as beta-carotene, lycopene, vitamins A, C, and E, as well as other naturally occurring and synthetic compounds.24 Even though antioxidants are produced by our bodies, we can get a lot more of them from food. Colorful fruits, vegetables, berries, green leaves, carrots, nuts and seeds, like almonds and sunflower seeds, brown rice, quinoa, and fatty fish, such as salmon and tuna are the natural sources of antioxidants.27,28 The natural antioxidants are important in their application, but they are limited. The use of synthetic antioxidants has become prevalent due to their superior efficacy and cost-effectiveness compared to natural antioxidants.26 For specific radicals, specific antioxidants are needed. That's why research focuses on synthetic antioxidants.25 Schiff bases would be a source of synthetic antioxidants because they showed antioxidant activity.29,30 Further investigation is required about the antioxidant efficacy of the mechanism of Schiff bases in both human physiological systems and industrial applications. This study centers its attention on the examination of the activity, mechanism, and impact of the replaced group on the activity.
2. Antioxidant and free radicals
Free radicals refer to atoms, molecules, or ions containing unpaired electrons. They are highly unstable and active in chemical reactions with other molecules. Leonor Michaelis's research in the 1930s sparked an interest in free radicals within cells. He observed a two-step electron loss during oxidation, suggesting a free radical intermediate (semiquinone) existed. While this theory wasn't entirely accurate, it fueled further investigation. With the development of better detection methods like electron spin resonance (ESR) spectroscopy in the 1950s, scientists confirmed the presence of free radicals in various enzyme reactions, including those involving vitamin B2. This led to the recognition of free radicals in a wider range of biological processes beyond just oxidation. Though Michaelis's initial theory had limitations, it paved the way for understanding the prevalence of free radicals. These reactive molecules are constantly produced in cells as byproducts of normal metabolism, a concept solidified by research following his observations. The discovery of mechanisms behind oxygen toxicity and ionizing radiation further supported the idea of continuous free radical formation within cells.31,32Reactive oxygen species (ROS) consist of three chemical species found in the Fenton/Haber Weiss pathway: the superoxide radical (O2˙−), hydrogen peroxide (H2O2), and the hydroxyl radical (HO˙). These species are formed when oxygen is partially reduced. The reduction of molecular oxygen by four electrons results in the production of water without the synthesis of ROS. However, when molecular oxygen is reduced by just one electron, it leads to the formation of the O2˙−, H2O2, and HO˙ radicals. O2˙− and HO˙ are classified as free radicals due to the presence of unpaired electrons in their outer orbitals. In contrast, H2O2 does not possess unpaired electrons and is hence not considered a radical. Oxygen derivatives that are not radical in nature include H2O2, ozone (O3), and singlet oxygen (1O2). Reactive nitrogen species (RNS) are radicals that are based on nitrogen. These radicals include three chemical species from the Beckman–Radi–Freeman pathway: nitrogen dioxide 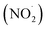 , nitric oxide (NO˙), and peroxynitrite (ONOO˙). The radicals that are part of this group including oxygen radical
, nitric oxide (NO˙), and peroxynitrite (ONOO˙). The radicals that are part of this group including oxygen radical 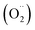 , O2˙−, ˙OH, peroxyl radical (ROO˙−), alkoxy radical (RO˙),
, O2˙−, ˙OH, peroxyl radical (ROO˙−), alkoxy radical (RO˙), 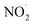 , and NO˙. The non-radical entities including hypobromous acid (HOBr), O3, H2O2, hypochlorous acid (HOCl), 1O2, nitrosyl cation (NO+), nitrous acid (HNO2), nitroxyl anion (NO−), dinitrogen tetroxide (N2O4), dinitrogen trioxide (N2O3), nitronium (nitryl) cation (NO2+), organic peroxides (ROOH), peroxynitrite (ONOO−), and aldehydes (HCOR). These non-radical entities are not free radicals themselves, but they may easily trigger free radical reactions that occurs in living organisms. Free radicals exhibit varying degrees of reactivity, with ROS being ranked in decreasing order as follows: HO˙ > O2˙− > H2O2. The chemical reactivity of free radicals is directly linked to their capacity to harm biological molecules. The reactivity of HO˙ is higher than others and exhibits fast reactivity with almost all chemical species, while H2O2, NO˙, and O2˙− react swiftly with just a limited number. Other species, such as
, and NO˙. The non-radical entities including hypobromous acid (HOBr), O3, H2O2, hypochlorous acid (HOCl), 1O2, nitrosyl cation (NO+), nitrous acid (HNO2), nitroxyl anion (NO−), dinitrogen tetroxide (N2O4), dinitrogen trioxide (N2O3), nitronium (nitryl) cation (NO2+), organic peroxides (ROOH), peroxynitrite (ONOO−), and aldehydes (HCOR). These non-radical entities are not free radicals themselves, but they may easily trigger free radical reactions that occurs in living organisms. Free radicals exhibit varying degrees of reactivity, with ROS being ranked in decreasing order as follows: HO˙ > O2˙− > H2O2. The chemical reactivity of free radicals is directly linked to their capacity to harm biological molecules. The reactivity of HO˙ is higher than others and exhibits fast reactivity with almost all chemical species, while H2O2, NO˙, and O2˙− react swiftly with just a limited number. Other species, such as 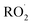 ,
, 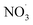 , RO˙, HOCl,
, RO˙, HOCl, 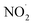 , ONOO−,
, ONOO−, 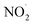 , and O3, exhibit moderate levels of reactivity. RO˙ exhibits higher reactivity compared to ROO˙, which are the primary byproducts of lipid peroxidation. Concerning RNS, the reactivity of
, and O3, exhibit moderate levels of reactivity. RO˙ exhibits higher reactivity compared to ROO˙, which are the primary byproducts of lipid peroxidation. Concerning RNS, the reactivity of 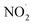 lies between that of NO˙ and ONOO−. NO˙ has a very limited chemical reactivity and as a result, its toxicity is not significant. Nevertheless, when reacting with O2˙−, it generates a profoundly harmful compound called ONOO−, which can cause damage to lipids, proteins, and DNA. The term ROS encompasses both radicals and non-radical species. Nevertheless, oxygen-derived radicals are the primary cause of damage to biological systems. The relative abundance of free radicals ranked in decreasing order is as follows: ROS > RNS > RSS (reactive sulfur species).33
lies between that of NO˙ and ONOO−. NO˙ has a very limited chemical reactivity and as a result, its toxicity is not significant. Nevertheless, when reacting with O2˙−, it generates a profoundly harmful compound called ONOO−, which can cause damage to lipids, proteins, and DNA. The term ROS encompasses both radicals and non-radical species. Nevertheless, oxygen-derived radicals are the primary cause of damage to biological systems. The relative abundance of free radicals ranked in decreasing order is as follows: ROS > RNS > RSS (reactive sulfur species).33
Free radicals exhibit various reaction mechanisms, including reducing radicals, oxidizing radicals (a), hydrogen abstraction (b), addition reactions (c), self-annihilation reactions (d), and disproportionation (e) (Fig. 2). These mechanisms allow them to interact with surrounding molecules. These interactions result in the generation of RNS, ROS, and RSS which have been associated with several serious illnesses.34
 | ||
| Fig. 2 Mechanism of reactions of free radicals.34 | ||
Free radicals can alter DNA and potentially cause illness. DNA damage is defined as any alteration in the structure of DNA that modifies its coding characteristics and/or disrupts cellular functions.
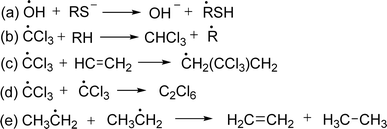
ROS and RNS induce DNA and strand breaks DNA by different mechanisms, such as base alterations. Oxidative DNA damage, specifically 8-oxo-dG, could potentially promote cancer development through two mechanisms: gene expression alteration or mutation initiation.35 The mechanism of hydroxyl radical is discussed here. Hydroxyl radicals inhibit the double bonds of DNA bases and abstract an H-atom from thymine and carbon atom of 2′-deoxyribose followed by the addition reactions which yield OH-adduct radicals of bases or abstraction reactions that generate allyl radical of thymine and sugar radicals. Further reaction of base and sugar radicals give various modified sugars and bases, base-free sites, strand breaks, and DNA-protein cross-links. For example, hydroxyl radical adds to the thymine and cytosine giving rise to C5–OH− and C6–OH− adduct radicals whereas the hydroxyl radical reacts with purines to yield C4–OH−, C5–OH−, and C8–OH− adduct radicals. Oxidation of C5–OH− adduct radicals of cytosine and thymine followed by the addition of OH− give cytosine glycol and thymine glycol respectively. On the other side, C4–OH− and C5–OH− adduct radicals of purines undergo dehydration and lead to the formation of oxidizing purine radicals whereas the C8–OH− adduct radicals give 8-hydroxypurines and formamidopyrimidines from oxidation and reduction respectively. The mechanism of the formation of guanine products is discussed in the following figure (Fig. 3).36
 | ||
| Fig. 3 Mechanism of binding of free radicals with nitrogen base of DNA.36 | ||
Free radicals damage several cellular and plasma proteins like immunoglobulin G (IgG) and the alpha-1-proteinase inhibitor. The addition of free radical-altered IgG prolongs bovine serum albumin-induced allergic inflammation according to rat air pouch model. Moreover, the radical nitric oxide (NO) is a cytotoxic effector molecule in defense against tumor cells, parasitic fungi, protozoa, helminthes and mycobacteria. It is a potent vasodilator in vitro, contributing to the cardinal signs of inflammation, heat, redness and swelling.37 Antioxidants are stable molecules that donate electrons to extremely reactive free radicals, neutralizing them and reducing their damage. Antioxidants can protect the human body against the detrimental effects of free radicals, ROS, and RNS. They impede the advancement of numerous chronic disorders as well as lipid peroxidation.21 Antioxidants can interact with free radicals through two main methods: HAT or SET mechanism (Fig. 4). In some cases, both HAT and SET mechanisms can work together. The HAT reaction involves the simultaneous transfer of a proton and an electron in a single kinetic step. Within HAT processes, a free radical eliminates a single hydrogen atom from an antioxidant, causing the antioxidant to transform into a radical. The BDE plays a crucial role in assessing the antioxidant activity in this pathway. The lower the BDE of the hydrogen-donating group in the potential antioxidant, the more readily the process of free radical inactivation will occur. The SET reaction involves the transfer of a single electron from the nucleophile to the substrate, resulting in the formation of a radical intermediate. The subsequent fate of this intermediate might lead to several outcomes. In the context of SET mechanisms, the role of the antioxidant is to donate an electron, resulting in the formation of a radical cation. The IP of the antioxidant is the primary energy component that determines the effectiveness of the antioxidant action in this pathway. As the ionization potential decreases, the process of electron abstraction becomes easier. Differentiating between HAT and SET reactions can be highly challenging. Typically, these two reactions occur at the same time, and the reaction's mechanism is influenced by the structure of antioxidant and solubility, as well as the partition coefficient and solvent polarity. 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) ABTS and oxygen radical absorbance capacity (ORAC) are two examples of HAT-based assays. Some examples of assays that are based on the use of SET include DPPH and ferric reducing antioxidant power (FRAP).38–41
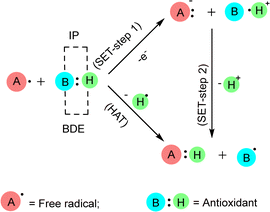 | ||
| Fig. 4 Interaction of antioxidants with free radicals by SET and HAT. When measuring the antioxidant effect in SET mechanism, the IP (ionization potential) of the antioxidant is the most crucial energetic component. In the HAT mechanism, the BDE (bond dissociation enthalpy) of the antioxidant is crucial for assessing its effectiveness.38 | ||
3. Schiff base as antioxidant
Antioxidants are natural compounds that can stop other molecules from oxidizing. They do this by breaking the chain reactions and giving up their electrons to feed the free radicals. Antioxidants are known as electron and proton generators.42 In general, they can help the body's defense system against free radicals obtained through dietary intake.26 The main antioxidant chemicals include vitamin C, which helps in lowering serum uric acid levels, and vitamin E, which is a significant barrier against lipid peroxidation and ROS across membranes, carotenoids, specifically xanthophylls, have been implicated in the mitigation of cardiovascular disease risk, and polyphenols such as flavonoids, flavonones, flavonols, phenolic acids, lignans, etc. show activities against severe diseases.27 The majority of these requisite antioxidants are found in natural sources.28 Furthermore, for low prices and large possibilities, researchers focused on synthetic ones.26 Butylated hydroxyanisole (BHA), butylated hydroxytoluene (BHT), propyl gallate (PG), and tertiary butylhydroquinone (TBHQ) are some of the synthetic antioxidants that are frequently used. But they don't show any specific medicinal activity. In higher concentrations, they can instead induce cytotoxicity and apoptosis. For example, BHT has been shown to induce DNA damage and apoptosis in several cell lines due to the production of ROS, and constant consumption of BHT induces the development of inflammation and liver damage.43,44 Adverse toxicity of BHA could cause mitochondrial malfunctions, endoplasmic reticulum stress, and abnormal calcium levels in the testis.45,46 TBHQ has dual nature, where lower concentrations may offer protective effects against oxidative stress, while higher concentrations can induce cytotoxicity and apoptosis in thymocytes.47 PG induces apoptosis and DNA damage in lung cancer cells, with mitochondrial impairment being a key factor in its toxic effects. It may lead to abnormal implantation and placental development in early pregnancy.48,49 It has become imperative to identify substitutes for synthetic antioxidants. Schiff bases will be one of the potential sources of antioxidants due to their proton and electron donating ability. According to the literature, certain Schiff bases are nontoxic and have strong, similar antioxidant activity to conventional antioxidants.30 Schiff bases have some crucial properties that give them more advantages to show antioxidant activity such as structural diversity and chelating ability.50,51 The extensive availability of aldehydes, ketones, and amines allows the production of Schiff bases that can fine-tune the antioxidant activity, solubility, and other characteristics.30,52A study by Kareem et al. examined the antioxidant properties of Schiff base derivatives. They discovered that the imine group (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) is a potential free radical scavenger.53 The RN
N–) is a potential free radical scavenger.53 The RN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–R′ group is generated through the condensation reaction of aldehydes and primary amines; both R and R′ are substituent function groups that are attached to the nuclei. Subsequent research has shown that the quantity of –OH groups in diverse compounds and their locations had a major influence in improving antioxidant activity.26,54 Experimentally, Choon Young Lee et al. demonstrated that electron-donating groups (EDGs) such as –OCH3 enhance antioxidant activity whereas electron-withdrawing groups (EWGs), e.g., –NO2, diminish antioxidant activity.55 N and O donor atoms containing chelating ligands are important categories of ligands because of the many ways in which they may attach to metal ions and their capacity to exhibit strong antioxidant activity.56 A further investigation found that amino acids containing sulfur had significant antioxidant properties via the process of chelation.57 While certain antioxidants contain a –COOH or –OH functional group, also an oxo group exhibited the capacity to coordinate with diverse be ascribed to the capacity of Schiff base ligands to form coordination complexes with metal ions, leading to the formation of compounds that effectively scavenge ROS.26 There are many in vitro assays for assessing the antioxidant activity of various Schiff bases such as DPPH radical assay,58 FRAP,59 cupric reducing antioxidant capacity (CUPRAC),58 nitric oxide radical scavenging assay,60 superoxide anion radical scavenging assay,60 H2O2 scavenging activity,60 ABTS assay58 and so on. Beena et al. proposed that Schiff bases remove DPPH free radicals as antioxidants through electron and proton transport mechanisms. In the electron transfer mechanism, the Schiff bases lose the proton and prepare ions, then the ions transfer the electrons to the DPPH radical to convert it into a charged species, which neutralized by the lost proton results, converts the Schiff base to a radical. The Schiff base contains a labile proton and stabilizes the radical in one step by transferring the proton directly (Fig. 5). The electron-donating functional group that stabilized Schiff base free radicals had a higher antioxidant activity than the phenolic benzine electron-removing substituent.61
CH–R′ group is generated through the condensation reaction of aldehydes and primary amines; both R and R′ are substituent function groups that are attached to the nuclei. Subsequent research has shown that the quantity of –OH groups in diverse compounds and their locations had a major influence in improving antioxidant activity.26,54 Experimentally, Choon Young Lee et al. demonstrated that electron-donating groups (EDGs) such as –OCH3 enhance antioxidant activity whereas electron-withdrawing groups (EWGs), e.g., –NO2, diminish antioxidant activity.55 N and O donor atoms containing chelating ligands are important categories of ligands because of the many ways in which they may attach to metal ions and their capacity to exhibit strong antioxidant activity.56 A further investigation found that amino acids containing sulfur had significant antioxidant properties via the process of chelation.57 While certain antioxidants contain a –COOH or –OH functional group, also an oxo group exhibited the capacity to coordinate with diverse be ascribed to the capacity of Schiff base ligands to form coordination complexes with metal ions, leading to the formation of compounds that effectively scavenge ROS.26 There are many in vitro assays for assessing the antioxidant activity of various Schiff bases such as DPPH radical assay,58 FRAP,59 cupric reducing antioxidant capacity (CUPRAC),58 nitric oxide radical scavenging assay,60 superoxide anion radical scavenging assay,60 H2O2 scavenging activity,60 ABTS assay58 and so on. Beena et al. proposed that Schiff bases remove DPPH free radicals as antioxidants through electron and proton transport mechanisms. In the electron transfer mechanism, the Schiff bases lose the proton and prepare ions, then the ions transfer the electrons to the DPPH radical to convert it into a charged species, which neutralized by the lost proton results, converts the Schiff base to a radical. The Schiff base contains a labile proton and stabilizes the radical in one step by transferring the proton directly (Fig. 5). The electron-donating functional group that stabilized Schiff base free radicals had a higher antioxidant activity than the phenolic benzine electron-removing substituent.61
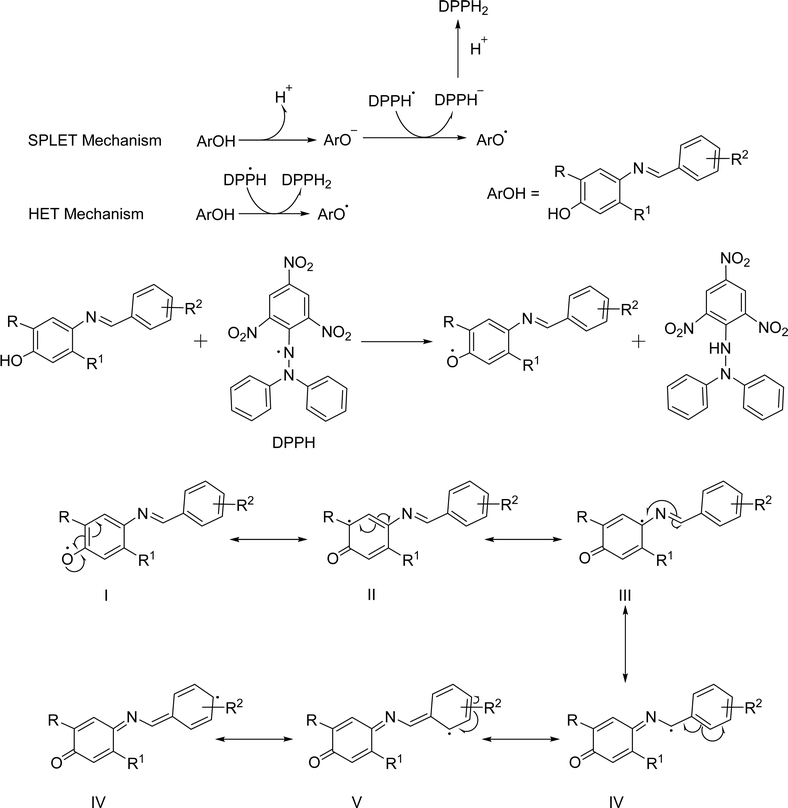 | ||
| Fig. 5 Proposed mechanism of reaction of Schiff bases with DPPH.61 | ||
4. DPPH antioxidant assay
There are various assays for determining the antioxidant activities of Schiff base ligands.26 Among them, the DPPH scavenging assay came out as a promising technique because of its simplicity, rapidity and affordability and it also gives precise information on the test system's total antioxidant capacity.62 The test is based on measuring antioxidants' scavenging capability towards DPPH free radicals. The odd electron of the nitrogen atom in DPPH is reduced by obtaining a hydrogen atom from antioxidants and the associated hydrazine.63 DPPH is one of the few safe organic nitrogen radical tests that can be bought in stores. At first, it was tracked using ESR spectroscopy, which relied on the fact that the signal strength of the DPPH˙ radical was inversely linked to the quantity of antioxidants and the reaction time. Recently, the discoloration test has been used to measure this reaction. The simplest technique involves mixing a prospective Schiff base ligand with DPPH solution and evaluating absorbance at 515–528 nm after a certain period64 in different solvents, like DMSO, ethanol, methanol or buffered methanol, depending on how well the Schiff base ligands dissolve because activity depending on the solvent. During this method, the room should be kept dark or with dim lighting.655. Classifications of Schiff bases
Schiff bases are classified based on the starting material to focus on certain functional groups that are responsible for their antioxidant activity and discuss the effect of substituted groups in the activity of the ligands.5.1 Isoniazid based Schiff base ligands
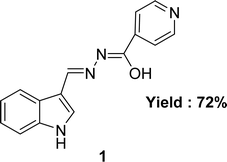
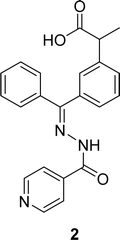
The aroyl-hydrazone Schiff base ligand 1 (Fig. 6) was prepared from isonicotinic hydrazide and indole-3-carboxaldehyde and FTIR, UV-visible, EPR, 13C and 1H NMR spectroscopy and single crystal X-ray studies were used to fully characterize ligand 1 by Masrat Bashir and his colleagues in 2023. Ligand showed DPPH radical scavenging activity with an IC50 of 36.09 μM, whereas conventional ascorbic acid had higher activity with a lower IC50 value of 19.95 μM (Table 1). Ligand 1 exhibited cell viability closer to DMSO (100.572–101.583%) against lung cancer cell line A549 (109.957–118.376%) and breast cancer cell line MDA-MB-231 (100.209–102.999%), respectively, leading to poor cytotoxic activity.66In 2018, Naima Rehman et al. mentioned the synthesis of Schiff base ligand (Z)-2-(3-((2-benzoylhydrazineylidene)(phenyl)methyl)phenyl)propanoic acid 2 (Fig. 7) from ketoprofen and isoniazid. The obtained ligand was characterized by FT-IR, X-ray crystallographic, and DSC studies. From the DPPH scavenging method, they found that the ligand showed better antioxidant activity with IC50 = 6.12 ppm compared to the starting material ketoprofen an anti-inflammatory drug with IC50 = 18.85 ppm (Table 1).29 Compared to ketoprofen an anti-inflammatory drug the ligand showed better activity because the ligand can donate electrons easily from its keto structure and donate protons easily from its enol structure.
5.2 Nicotinic acid hydrazide based Schiff base ligands
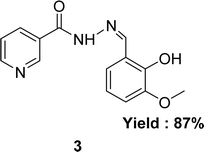
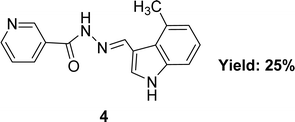
In 2020, nicotinic acid hydrazide and o-vanillin were used to synthesize nicotinic acid (2-hydroxy-3-methoxybenzylidene)hydrazide 3 (Fig. 8) Schiff base ligand by Neetu Patel et al. FT-IR, elemental analysis and 1H NMR were used to characterize Schiff base ligand. The IC50 value for the Schiff base ligand in a DPPH radical scavenging experiment was 729.258 μg mL−1. Lower DPPH scavenging activity of 3 compared to positive control ascorbic acid (730.120 μg mL−1) was found (Table 2).67 Due to the presence of phenolic and enolic hydroxyl groups which may easily donate a proton to stabilize free radicals and show better antioxidant activity.Ohyla A. EL-Gammal and his colleagues synthesized (E)-N′-((4-methyl-1H-indol-3-yl)methylene)nicotinohydrazide 4 (Fig. 9) from nicotinohydrazide and 4-methyl-1H-indol-3-carbaldehyde in 2019 and found 25% yield. The ligand was characterized using IR, 1H NMR, UV-visible and EPR. After that, it was evaluated the antioxidant activity using the DPPH antioxidant assay and found to have an IC50 value of 3.82 μg mL−1 which is respectively better than that of the standard ascorbic acid (IC50 = 144.56 μg mL−1) (Table 2).68 Electron-donating groups in the imidazole ring increase electron density near N atoms, making them more likely to give hydrogen to the free radical, while the keto–enol group ligand donates a proton to the free radical and outperforms ascorbic acid.
From the data, it would be concluded that, during the synthesis of nicotinic acid hydrazide based on Schiff base ligands, researchers should focus on the electron-pushing substituted indole-based aldehydes to get better activity.
5.3 Benzoic acid hydrazide-based Schiff base ligands
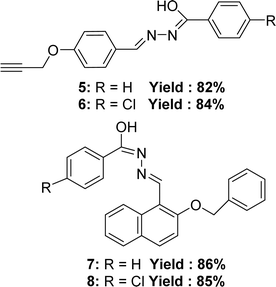
In 2021, Manju Yadav et al. synthesized four Schiff bases (Z)-N-methyl-2-((2-phenethoxynaphthalen-1-yl)methylene)hydrazinecarbothioamide 5 (yield: 82%), (Z)-2-((2-(benzyloxy)naphthalen-1-yl)methylene)-N-methylhydrazine carbothioamide 6 (yield: 84%), (E)-2-(4-(but-3-yn-1-yl)benzylidene)-N-methylhydrazine carbothioamide 7 (yield: 86%), (Z)-2-(3,5-dichloro-2-phenethoxybenzylidene)-N-methylhydrazinecarbothioamide 8 (yield: 85%). 5 was synthesized from 4-prop-2-ynyloxy-benzaldehyde and benzoic acid hydrazide; 6 was synthesized from 4-prop-2-ynyloxy-benzaldehyde and 4-chloro-benzoic acid hydrazide; 7 was synthesized from 2-benzyloxy-naphthalene-1-carbaldehyde and benzoic acid hydrazide; 8 (Fig. 10) was synthesized from 2-benzyloxy-naphthalene-1-carbaldehyde and 4-chloro-benzoic acid hydrazide and compounds were characterized using FT-IR, 1H and 13C NMR, mass spectrometry, elemental analysis, conductivity, powder XRD studies. From the DPPH assay, 6 (IC50 = 4.36 μM) exhibited higher activity antioxidant activity compared to 5 (IC50 = 5.38 μM) due to the presence of a chlorine group containing lone electron pairs. Chlorine also resonates, and the resonance effect may contribute to electron density in the benzene ring. Furthermore, it was also observed that 8 (IC50 = 4.78 μM) has a higher activity than 7 (IC50 = 6.18 μM) also due to the presence of the chlorine group (Table 3). In terms of antioxidant activity, the ligands demonstrated the following order: 6 > 8 > 5 > 7.695.4 Hydroxy naphthaldehyde-based ligands
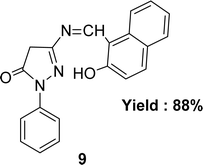
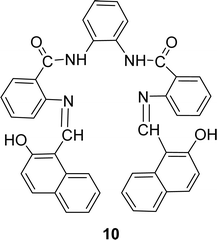
Nazar M. Abdalsahib and his team reported a synthetic route for the Schiff base ligand 5-[(2-hydroxy-naphthalen-1-ylmethylene)-amino]-2-phenyl-2,4-dihydro-pyrazol-3-one 9 (Fig. 11) from 3-amino-1-phenyl-2-pyrazoline-5-one and 2-hydroxynaphthaldehyde and they characterized the ligand by mass, 1H and 13C NMR, FTIR, and UV-visible analysis in 2023. They found much lower antioxidant activity of 9 (IC50 = 589.6 μg mL−1) than standard ascorbic acid (IC50 = 36.3 μg mL−1) from the DPPH test (Table 4).70Schiff base ligand N,N′(1,2-phenylene)bis(2-(((Z)-(2-hydroxynaphthalen-1-yl)methylene)amino)benzamide) 10 (Fig. 12) was synthesized using 2-hydroxy-1-naphthaldehyde and FTIR, 1H NMR, microanalysis and UV-visible analysis to characterize the ligand by H. A. El-Boraey et al. in 2020. In contrast to standard ascorbic acid (IC50 = 28.21 μg mL−1), the free radical DPPH scavenging activity of 10 was determined to be 253.15 μg mL−1 (Table 4).71
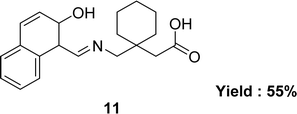
In 2015, the team of Muhammad Ikram described the synthesis of Schiff base ligand, [1-({[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]amino}methyl)cyclohexyl]acetic acid 11 (Fig. 13), from the condensation reaction of [1-(aminomethyl)cyclohexyl]acetic acid and 2-hydroxynaphthaldehyde. The ligand 11 was characterized by using various spectroscopic techniques such as elemental analyses, IR spectra, 1H and 13C NMR, UV-visible spectra, and single-crystal diffraction. The antioxidant activity was significantly less than the standard reference gallic acid (IC50 = 23.46 ± 0.43 μM) (Table 4).72
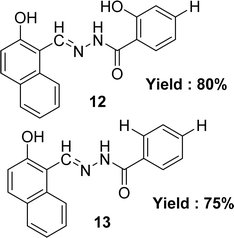
Lijun Liu et al. noted the preparation of two Schiff base ligands (E)-2-hydroxy-N′-[(2-hydroxynaphthalen-1-yl)methylene]benzohydrazide 12 and (E)-4-hydroxy-N′-[(2-hydroxynaphthalen-1-yl)methylene]benzohydrazide 13 (Fig. 14) from 2-hydroxybenzohydrazide and 4-hydroxybenzohydrazide respectively with 2-hydroxynaphthaldehyde in 2013. They were also characterized the synthesized by UV-visible, elemental analysis, 1H NMR, IR and fluorescence spectra methods. The IC50 value of ligands was given as 13.79 μM and 23.52 μM for 12 and 13 respectively which is higher than the standard ascorbic acid with IC50 value of 0.037 μM in the DPPH method (Table 4).73
All this naphthaldehyde-based ligand showed very negligible antioxidant activity, so researchers should avoid hydroxy naphthaldehyde as the starting material in synthesizing Schiff base to get better antioxidant activity.
5.5 Aminophenol and substituted aminophenol-based Schiff base ligands
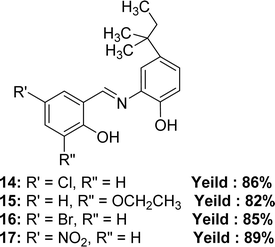
In 2023, Pinki Barwa et al. mentioned the synthetic route of 4-chloro-2-(((2-hydroxy-5-(tert-pentyl)phenyl)imino)methyl)phenol 14, 3-ethoxy-2-(((2-hydroxy-5-(tert-pentyl)phenyl)imino)methyl)phenol 15, 4-bromo-2-(((2-hydroxy-5-(tert-pentyl)phenyl)imino)methyl)phenol 16, 4-nitro-2-(((2-hydroxy-5-(tert-pentyl)phenyl)imino)methyl)phenol 17 (Fig. 15). They used 2-amino-4-tert-amylphenol with 5-chlorosalicylaldehyde for 14, 3-ethoxysalicylaldehyde for 15, 5-bromosalicylaldehyde for 16, and 5-nitrosalicylaldehyde for 17. MS, FT-IR, 1H and 13C-NMR, SEM, powder XRD to characterize the Schiff base ligands. The antioxidant activity of the ligands and standard ascorbic acid from DPPH assay followed the trend: ascorbic acid (IC50 = 1.49 ± 0.012 μM) > 15 (IC50 = 4.39 ± 0.026 μM) > 16 (IC50 = 5.48 ± 0.025 μM) > 14 (IC50 = 6.00 ± 0.018 μM) > 17 (IC50 = 7.90 ± 0.022 μM) (Table 5).74 The ligand 17 has low activity because of the (–I) action of the NO2 group which makes the Schiff base radicals less stable after scavenging DPPH radicals.30| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference. | ||
| 14 (6.00 ± 0.018 μM), 15 (4.39 ± 0.026 μM), 16 (5.48 ± 0.025 μM), 17 (7.90 ± 0.022 μM) | Ascorbic acid (1.49 ± 0.012 μM) | 74 |
| 18 1.85 μg mL−1 (methanol), 2.94 μg mL−1 (chloroform), 23.2 μg mL−1 (acetonitrile), 33.3 μg mL−1 (acetone), 150 μg mL−1 (ethyl acetate) | BHA 3.46 μg mL−1 (methanol), 5.34 μg mL−1 (chloroform), 41.9 μg mL−1 (acetonitrile), 56.0 μg mL−1 (acetone), 170 μg mL−1 (ethyl acetate) | 30 |
| 19 4.08 μg mL−1 (methanol), 26.01 μg mL−1 (chloroform), 43.2 μg mL−1 (acetonitrile), 90.3 μg mL−1 (acetone), 206 μg mL−1 (ethyl acetate) | ||
| 20 1.60 μg mL−1 (methanol), 2.77 μg mL−1 (chloroform), 18.5 μg mL−1 (acetonitrile), 26.0 μg mL−1 (acetone) 134 μg mL−1 (ethyl acetate) | ||
| 21 1.66 μg mL−1 (methanol), 2.80 μg mL−1 (chloroform), 26.0 μg mL−1 (acetonitrile), 32.0 μg mL−1 (acetone), 123 μg mL−1 (ethyl acetate) | ||
| 22 2.25 μg mL−1 (methanol), 4.15 μg mL−1 (chloroform), 42.1 μg mL−1 (acetonitrile), 52.9 μg mL−1 (acetone), 150 μg mL−1 (ethyl acetate) | ||
| 23 1.50 μg mL−1 (methanol), 2.31 μg mL−1 (chloroform), 22.0 μg mL−1 (acetonitrile), 26.1 μg mL−1 (acetone), 120 μg mL−1 (ethyl acetate) | ||
| 24 (32.0 μM), 25 (45.5 μM) | BHA (44.2 μM) | 75 |
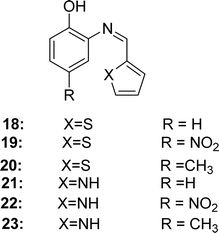
In 2018, Angamaly Antony Shanty and his colleagues synthesized the heterocyclic imine-based reagent from thiophene-2-carboxaldehyde, pyrrole-2-carboxaldehyde, 2-aminophenol, 2-amino-4-nitrophenol, 2-amino-4-methylphenol by condensation reaction. It had been found the IC50 values of 18 (in methanol 1.85 μg mL−1, in chloroform 2.94 μg mL−1, in acetonitrile 23.2 μg mL−1, in acetone 33.3 μg mL−1, in ethyl acetate 150 μg mL−1), 19 (in methanol 4.08 μg mL−1, in chloroform 26.01 μg mL−1, in acetonitrile 43.2 μg mL−1, in acetone 90.3 μg mL−1, in ethyl acetate 206 μg mL−1), 20 (in methanol 1.60 μg mL−1, in chloroform 2.77 μg mL−1, in acetonitrile 18.5 μg mL−1, in acetone 26.0 μg mL−1, in ethyl acetate 134 μg mL−1), 21 (in methanol 1.66 μg mL−1, in chloroform 2.80 μg mL−1, in acetonitrile 26.0 μg mL−1, in acetone 32.0 μg mL−1, in ethyl acetate 123 μg mL−1), 22 (in methanol 2.25 μg mL−1, in chloroform 4.15 μg mL−1, in acetonitrile 42.1 μg mL−1, in acetone 52.9 μg mL−1, in ethyl acetate 150 μg mL−1), 23 (in methanol 1.50 μg mL−1, in chloroform 2.31 μg mL−1, in acetonitrile 22.0 μg mL−1, in acetone 26.1 μg mL−1, in ethyl acetate 120 μg mL−1) and standard BHA (in methanol 3.46 μg mL−1, in chloroform 5.34 μg mL−1, in acetonitrile 41.9 μg mL−1, in acetone 56.0 μg mL−1, in ethyl acetate 170 μg mL−1) by using DPPH antioxidant assay/method (Fig. 16). The synthesized compounds showed the following order of antioxidant activity: 23 > 20 > 21 > 18 > 22 > 19 (Table 5). Compounds 20 and 23 containing electron-donating methyl substituents at the para position of the phenol ring showed greater activity than 18 and 21 (without substituents). However, compounds 19 and 22 containing an electron-withdrawing NO2 group at the phenol ring exhibited modest activity owing to their (−I) action, which destabilizes the radical. In the cytotoxicity study, all Schiff bases 18–23 showed 100% cell viability at four different concentrations (10 μM, 50 μM, 100 μM, and 150 μM) against normal 3T3LI cells.30 This means that ligands 18–23 are not toxic, since compounds showing 80%, 80–60%, 60–40%, and below 40% cell viability are thought to have non-, weak-, moderate-, and strong cytotoxicity, respectively.76
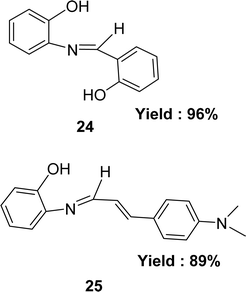
Preparation of Schiff bases 2-{[(2-hydroxyphenyl)methylidene]amino}phenol 24 and 2-{[3-4-(dimethylamino)phenyl-2-propenylidene]amino}phenol 25 (Fig. 17) from 2-aminophenol with 2-hydroxybenzaldehyde and 4-dimethylaminocinnamaldehyde respectively were mentioned by the team of Muhammad Aslam in 2013. They characterized the ligands by IR, 1H NMR, MS, elemental analysis and crystallography. The ligands were found to exhibit antioxidant activity with IC50 value of 32.0 μM and 45.5 μM for 24 and 25 respectively in the DPPH assay. Standard BHA (IC50 = 44.2 μM) showed better antioxidant activity than 25 but less than that of 24 (Table 5). Multiple phenolic rings in ligand 24 increased its activity making it more active than ligand 25 which contains a single one.75
Because of the acidic phenolic –OH, aminophenol-based ligands had an activity that was similar to or sometimes greater than that of conventional antioxidants because they stabilized the radicals by giving them protons. Adding an electron-pushing group to them also made their activity better because it helped stabilize the resonance structure of Schiff base-free radicals.
5.6 Salicylaldehyde and substituted salicylaldehyde based ligands
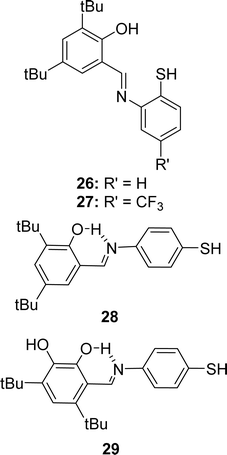
In 2023, Ivan V. Smolyaninov et al. described the synthesizing methods of Schiff bases, i.e., 2-(2-mercaptophenylimino)methyl-4,6-di-tert-butylphenol 26, 2,4-di-tert-butyl-6-(5-(trifluoromethyl)-2,3-dihydrobenzo[d]thiazol-2-yl)phenol 27, 2,4-di-tert-butyl-6-((4-mercaptophenylimino)methyl)phenol 28, and 4,6-di-tertbutyl-3-((4-mercaptophenylimino)methyl)catechol 29 (Fig. 18) from 3,5-di-tert-butyl-2-hydroxybenzaldehyde, 2-amino-4-(trifluoromethyl)benzenethiol hydrochloride, appropriate hydrochloride, triethylamine, and solution of aldehyde. The Schiff bases were characterized by 1H and 13C NMR spectroscopy, IR, and HR-MS. Using the DPPH antioxidant assay, they found 29 (10.0 ± 0.5 μM) showed higher antioxidant activity than standard trolox (12.0 ± 0.5 μM) and other ligands 26 (42.1 ± 1.9 μM), 27 (43.5 ± 1.5 μM), and 28 (30.0 ± 1.3 μM) (Table 6). A significant dependency of the IC50 values (antioxidant activity) was found on hydroxyl groups in the ligands. Ligand 29 (IC50 = 3.53–11.03 μM) is highly cytotoxic and 28 (IC50 = 25.50–43.19 μM) is poor cytotoxic compared to cisplatin (IC50 = 9.02–15.2 μM) against MCF-7, HCT-116, and A-549 cell lines, but both are less cytotoxic compared to doxorubicin (IC50 = 0.13–0.62 μM).77| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference; NG = not given. | ||
| 26 (42.1 ± 1.9 μM), 27 (43.5 ± 1.5 μM), 28 (30.0 ± 1.3 μM), 29 (10.0 ± 0.5 μM) | Trolox (12.0 ± 0.5 μM) | 77 |
| 30 (6.13 μM), 31 (5.76 μM), 32 (4.98 μM), 33 (7.09 μM) | Ascorbic acid (1.95 μM) | 78 |
| 34 (8.24 ± 0.03 μM), 35 (9.33 ± 0.06 μM), 36 (8.60 ± 0.07 μM), 37 (7.35 ± 0.01 μM) | Ascorbic acid (1.95 ± 0.02 μM) | 79 |
| 38 (0.531 mg mL−1) | NG | 80 |
| 39 (62 μM) | Ascorbic acid (52 μM) | 81 |
| 40 (1437.63 μM) | Ascorbic acid (13.58 μM) | 82 |
| 41 (0.18 mg mL−1) | Ascorbic acid (0.14 mg mL−1), quercetin (0.13 mg mL−1) | 83 |
| 42 (4.51 mg mL−1) | NG | 84 |
| 43 (132.60 nmol mL−1), 44 (51.80 nmol mL−1), 45 (482.20 nmol mL−1) | Trolox (18.83 nmol mL−1), BHA (25.08 nmol mL−1), BHT (33.81 nmol mL−1) | 85 |
| 46 (198.37 ppm), 47 (300 ppm), 48 (108.65 ppm) | NG | 86 |
| 49 (204 μg mL−1), 50 (224 μg mL−1) | Ascorbic acid (46 μg mL−1) | 87 |
| 51 (136 μM) | Ascorbic acid (46.8 μM) | 88 |
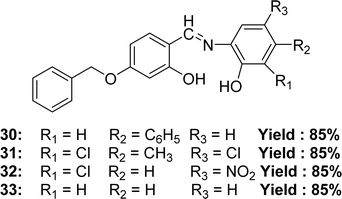
In 2022, Jai Devi et al. mentioned the synthesis of 4-((4-(benzyloxy)-2-hydroxybenzylidene)amino)-[1,1′-biphenyl]-3-ol 30, 6-((4-(benzyloxy)-2-hydroxybenzylidene)amino)-4,6-dichloro-3-methyl-phenol 31, 2-((4-(benzyloxy)-2-hydroxybenzylidene)amino)-6-chloro-4-nitro-phenol 32, 5-(benzyloxy)-2-(((2-hydroxyphenyl)imino)methyl)phenol 33 (Fig. 19) from 4-(benzyloxy)-2-hydroxybenzaldehyde with various aminophenol derivatives by condensation reaction. Characterization of the ligands had been studied by element analysis, FT-IR, conductivity, mass spectrometry, 13C-NMR, 1H-NMR spectroscopy. It had been found that the IC50 values of 30, 31, 32 and 33 ligands were 6.13 μM, 5.76 μM, 4.98 μM and 7.09 μM respectively by using the DPPH antioxidant assay whereas the standard ascorbic acid had an IC50 value of 1.95 μM. The increasing order of IC50 values for Schiff base ligands (32 < 31 < 30 < 33) shows how electron-withdrawing or electron-donating groups affect their strength (Table 6).78 From the previous and lateral study, we know that the presence of an electron-donating substituent of phenolic ring increased the activity but an electron-withdrawing substituent decreased the activity. This is an exceptional case, the presence of two electron-withdrawing groups in phenolic ring increased the acidity of phenolic ring through resonance structure in a result the Schiff base can easily provide a proton to stabilize the radicals.
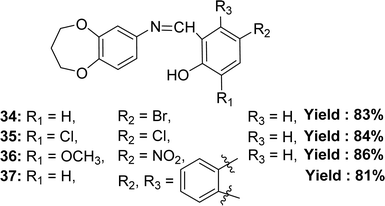
Binesh Kumar and his colleagues elaborated the preparation of 4-bromo-2-(((3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)imino)methyl)phenol 34, 2,4-dichloro-6-(((3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)imino)methyl)phenol 35, 2-(((3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)imino)methyl)-6-methoxy-4-nitrophenol 36 and 1-(((3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)imino)methyl)naphthalen-2-ol 37 (Fig. 20) from 3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-amine(3,4-dihydro-2H-1,5-benzodioxepin-7-amine), 5-bromosalicylaldehyde, 3,5-dichlorosalicylaldehyde, 3-methoxy-5-nitrosalicylaldehyde, and 2-hydroxy-1-naphthaldehyde in 2023. They characterized the synthesized ligand by ESR, FTIR, mass spectrometry, NMR, UV-visible, XRD, and SEM. The ligands and standard (ascorbic acid) exhibited DPPH radical scavenging activity in the following order: ascorbic acid (IC50 = 1.95 ± 0.02 μM) > 37 (IC50 = 7.35 ± 0.01 μM) > 34 (IC50 = 8.24 ± 0.03 μM) > 36 (IC50 = 8.60 ± 0.07 μM) > 35 (IC50 = 9.33 ± 0.06 μM) (Table 6). The ligand 37 is more effective at antioxidant activity because it doesn't have any electron-withdrawing groups on its aromatic ring. On the other hand, the ligand 34 is only moderately effective because it has a bromo group, and the ligand 36 has methoxy and nitro groups. The 35 ligand has the least antioxidant effect because it has chloro groups attached to the aromatic ring that draws electrons. So, the antioxidant action depends on how well the linked group in the molecule can donate electrons. Ligand 36 (33.11% cytotoxicity) and ligand 37 (42.57% cytotoxicity) were less cytotoxic than DMSO (79.87% cytotoxicity) at 1000 μg mL−1 on Vero cell lines.79
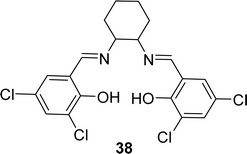
A novel tetra-dentate ligand named 6,6′-((1E,1′E)-(cyclohexane-1,2-diylbis(azanylylidene))bis(methanylylidene))bis(2,4-dichlorophenol) 38 (Fig. 21) was synthesized in 2022 by J. Priya et al. where the starting compounds were 3,5-dichlorosalicylaldehyde and trans-1,2-diaminocyclohexane. The ligand was characterized by element analysis, IR, 1H NMR spectroscopy, electronic spectra and ESI-MS studies. According to the DPPH radical scavenging method, the IC50 value of 38 was 0.531 mg mL−1 (Table 6).80
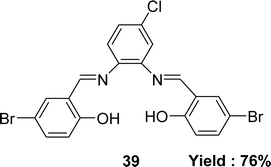
In 2022, Laila H. Abdel-Rahman et al. mentioned the synthesis of the Schiff base ligand 2,2′-{(4-chloro-1,2-phenylene)bis(nitrilo(E)methylylidene)}bis(4-bromophenol) 39 (Fig. 22) from the starting materials 4-chloro-o-phynelendiamine and 5-bromo-salicyaldehyde. The synthesized ligand was characterized by FT-IR, 1H NMR, 13C NMR, electronic spectra, X-ray diffraction and elemental analysis. It had been notified that the IC50 value of the ligand was 62 μM by using the DPPH antioxidant method whereas the ascorbic acid had an IC50 value of 52 μM (Table 6). The ligand 39 had a significant effect on normal HEK-293 cell (IC50 = 83 μM) compared to anticancer drug vinblastine (IC50 = 95 μM).81
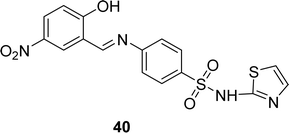
Schiff base ligand 40 (Fig. 23) was synthesized from sulfathiazole and 5-nitro salicylaldehyde by I. Rama and his colleague in 2020. Ligand 40 was characterized using FT-IR, 1H, 13C NMR, UV-visible, and EI-mass analysis. It showed free radical scavenging activities in DPPH antioxidant experiments, with an IC50 value of 1437.63 μM. The antioxidant activity of the ligand was not significant compared to standard ascorbic acid (13.58 μM) (Table 6).82
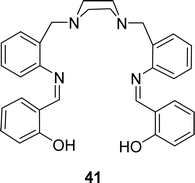
H. Keypour and his team described the synthesis of 2,2′-(1Z,1′Z)-(2,2′-(piperazine-1,4-diylbis(methylene))bis(2,1-phenylene)bis(azan-1-yl-1-ylidene)bis(methan-1-yl-1-ylidene)diphenol) 41 (Fig. 24) in 2020. They characterized the ligands by FT-IR, molar conductivity, elemental analysis, MS, 1H and 13C NMR. Antioxidant activity of the ligand (IC50 = 0.18 mg mL−1) was insignificant compared to conventional ascorbic acid (IC50 = 0.14 mg mL−1) and quercetin (IC50 = 0.13 mg mL−1) in DPPH assay (Table 6). From the cytotoxic study, ligand 41 showed cytotoxic activity with IC50 value of 27.66 μM and 188.12 μM respectively against human gastric (AGS) and lung (A549) cancer cell lines.83
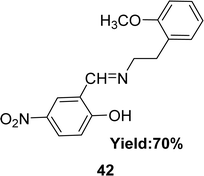
In 2020, a Schiff base ligand 42 (Fig. 25) was synthesized from 2-methoxyphenylethylamine and 5-nitro-2-hydroxybenzaldehyde by Imène Bougossa et al. 42 was characterized using IR, 1H, 13C NMR, elemental analysis and UV-visible spectroscopy. Using DPPH method the free radical scavenging capacity of 42 has been found IC50 value of 4.51 mg mL−1 (Table 6).84
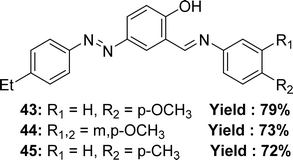
In 2018, Ayşe İnan and her team explained the synthesis Schiff base ligand, 4-((E)-(4-ethylphenyl)diazenyl)-2-((E)-((4-methoxyphenyl)imino)methyl)phenol 43, 2-((E)-((3,4-dimethoxyphenyl)imino)methyl)-4-((E)-(4-ethylphenyl)diazenyl)phenol 44, 4-((E)-(4-ethylphenyl)diazenyl)-2-((E)-(p-tolylimino)methyl)phenol 45 (Fig. 26) which was derived via refluxing the mixture of (E)-5-((4-ethylphenyl)diazenyl)-2-hydroxybenzaldehyde, 4-methoxyaniline, 3,4-dimethoxyanline and p-toluidine. The synthesized ligands had been characterized via elemental analyses, IR, UV-visible spectroscopy and 1H and 13C NMR spectra, X-ray diffraction studies. It had been informed that the IC50 value of 43 was 132.60 nmol mL−1, 44 was 51.80 nmol mL−1 and 45 was 482.20 nmol mL−1 and trolox 18.83 nmol mL−1, BHA 25.08 nmol mL−1, BHT 33.81 nmol mL−1 by DPPH antioxidant method (Table 6).85 More methoxy substituted ligand 44 showed better activity than others.
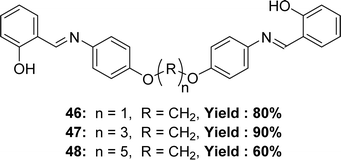
Three new Schiff base ligands 2-((4-(1-(4-(2-hydroxybenzylideneamino)phenoxy)methoxy)phenylimino)methyl)phenol 46, 2-((4-(3-(4-(2-hydroxybenzylideneamino)phenoxy)propoxy)phenylimino)methyl)phenol 47, 2-((4-(5-(4-(2-hydroxybenzylideneamino) phenoxy) pentyloxy) phenylimino) methyl) phenol 48 (Fig. 27) were synthesized through the reaction of salicylaldehyde with semi-aromatic diamines by Bushra Iftikhar et al. in 2017 and FT-IR, 1H and 13C NMR, UV-visible spectroscopic techniques were used to characterize the synthesized ligands. They found IC50 values of compounds 46, 47, 48 were 198.37 ppm, 300 ppm and 108.65 ppm respectively, in DPPH free radical scavenging method (Table 6).86
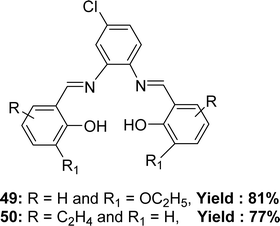
The preparation of Schiff base ligands 49 from 4-chloro-o-phenylenediamine and 3-ethoxy salicylaldehyde and 50 (Fig. 28) from 4-chloro-o-phenylenediamine and 2-hydroxy-1-naphthaldehyde were reported by Krishnan Sampath in 2015. Infrared spectra, electronic spectra, NMR spectra, and mass spectral analysis have characterized the ligands. The ligands showed lower antioxidant activity with IC50 = 204 and 224 μg mL−1 for 49 and 50 respectively in comparison to the activity of standard ascorbic acid (IC50 = 46 μg mL−1) (Table 6).87
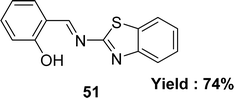
Subbaiyan Sathiyaraj and his colleagues mentioned the synthesis of Schiff base ligand 2-(benzothiazol-2-yliminomethyl)-phenol 51 (Fig. 29) from the reactions of salicylaldehyde and 2-amino benzothiazole in 2013. The ligand was characterized by single crystal X-ray diffraction, 1H and 13C NMR, elemental analysis, UV-visible and IR. The DPPH free radical scavenging abilities of the ligand 51 (IC50 = 136 μM) were less than those observed for ascorbic acid (IC50 = 46.8 μM) (Table 6).88
Except for ligand 29, none of the ligands showed better activity than standard. Also, ligands 39 (Fig. 22) and 41 (Fig. 24) were closer to the standard ascorbic acid. In conclusion, the salicylaldehyde ligand did not show any significant antioxidant activity. Further research should be advised to clear it.
5.7 Furyl acrolein based Schiff base ligands
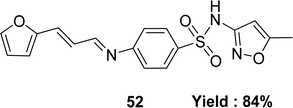
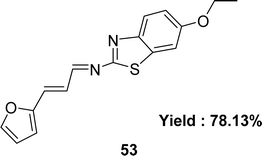
Form the condensation reaction of 3-(2-furyl)acrolein and sulfamethoxazole (SMX), Schiff base ligand 4-(((1E,2E)-3-(furan-2-yl)allylidene)amino)-N-(5-methylisoxazol-3-yl) benzene-sulfonamide 52 (Fig. 30) was produced by Jibril I. Al-Hawarin et al. in 2023 and HR-ESI-MS, FTIR, 1H and 13C NMR was used to characterize the ligand. The DPPH radical scavenging technique determined antioxidant activity the ligand showed moderate activity, with a low IC50 value of 0.7372 μg mL−1 (Table 7).89Abdel-Aziz Abu-Yamin et al. elucidated the synthesis of (1E,2E)-N-(6-ethoxybenzo[d]thiazol-2-yl)-3-(furan-2-yl)prop-2-en-1-imine 53 (Fig. 31) with the general formula C16H14N2O2S in 2022. The Schiff base was prepared through the thermal condensation of 3-(2-furyl)acrolein with 2-amino-6-ethoxybenzothiazole. The ligand had been characterized by elemental analysis, 1H and 13C NMR, FTIR, UV-visible spectroscopy and mass spectrometry. The DPPH radical scavenging activity of ligand 53 was IC50 = 478.52 ± 13.84 μg mL−1 which showed very weak activity compared to standard ascorbic acid and BHT, which were IC50 = 10.56 ± 0.78 and 5.78 ± 0.27 μg mL−1 respectively (Table 7).90
The furyl acrolein-based Schiff base ligand with sulfamethoxazole had considerable antioxidant activity compared to ascorbic acid but the one with ethoxybenzothiazole had very poor antioxidant activity. Furyl acrolein-based Schiff base ligand synthesis should prioritize sulfamethoxazole-based ligand.
5.8 Triazole-based Schiff base ligands
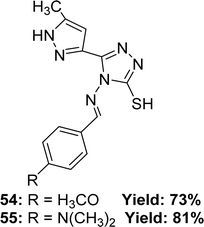
Renjith Raveendran Pillai and his colleagues mentioned the synthesis process of ligand 4-((4-methoxybenzylidene)amino)-5-(5-methyl-1H-pyrazole-3-yl)-4H-1,2,4-triazole-3-thiol 54 and 4-((4-(dimethylamino)benzylidene)amino)-5-(5-methyl-1H-pyrazol-3-yl)-4H-1,2,4-triazole-3-thiol 55 (Fig. 32) from 4-amino-5-(5-methyl-1H-pyrazol-3-yl)-4H-1,2,4-triazole-3-thiol and appropriate aromatic aldehydes in 2019. They found 73% yield of ligand 54 and 81% of 55. Which was characterized using 1H and 13C NMR, FTIR and ESI-MS spectroscopic methods. The antioxidant activity of the two ligands was evaluated using a DPPH antioxidant assay and found an IC50 value of 5.65 ± 0.38 μg mL−1 for ligand 54 and 1.39 ± 0.11 μg mL−1 for 55, whereas the IC50 value of standard BHA was 4.20 ± 0.02 μg mL−1 (Table 8).91 The 55-ligand showed higher activity than 54 and BHA due to the presence of a dimethyl amine substituted group, which offers a large delocalization area of the excited electron as a result of a stable Schiff base radical after scavenging radicals.| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference. | ||
| 54 (5.65 ± 0.38 μg mL−1), 55 (1.39 ± 0.11 μg mL−1) | BHA (4.20 ± 0.02 μg mL−1) | 91 |
| 56 (1.35 μg mL−1, 5.62 μM) | BHT (0.67 μg mL−1) (3.04 μM) | 92 |
| 57 (5809.65 ± 1639.21 μg mL−1), 58 (1782.51 ± 56.09 μg mL−1), 59 (1977.04 ± 19.76 μg mL−1), 60 (2705.19 ± 109.05 μg mL−1) | BHT (315.75 ± 3.66 μg mL−1) | 93 |
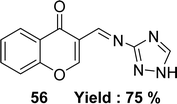
Schiff base 3-(((1H-1,2,4-triazol-3-yl)imino) methyl)-4H-chromen-4-one 56 (Fig. 33) was synthesized from the starting material 3-formylchromone, 3-amino-1,2,4-triazole by Mayuri Bheemarasetti et al. in 2018. FT-IR analysis, 1H and 13C NMR, elemental analysis, UV-visible spectrophotometer, ESI-MS were used to characterize the synthesized ligand. It had been informed that the IC50 value of ligand 56 was 1.35 μg mL−1 (5.62 μM) which showed lower activity than BHT (IC50 = 0.67 μg mL−1 (3.04 μM)) (Table 8).92
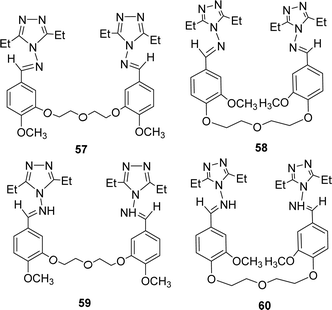
Bahar Bilgin Sokmen and his team mentioned the synthesis of methoxy substituted four novel bis triazole-Schiff bases 57, 58 and 59, 60 (Fig. 34) from the reaction of 4-amino-3,5-diethyl-4H-1,2,4-triazole and various bis-aldehydes in 2013. They were characterized by 1H and 13C-NMR. The antioxidant activity of ligands was noted with IC50 values of ligands 57, 58 and 59, 60 respectively 5809.65 ± 1639.21, 1782.51 ± 56.09, 1977.04 ± 19.76 and 2705.19 ± 109.05 μg mL−1. BHT (315.75 ± 3.66 μg mL−1) showed better antioxidant activity than the two ligands (Table 8). They can be arranged as BHT > 58 > 59 > 60 > 57 in terms of antioxidant activity.93
From the data of all triazole-based ligands 54 and 55 (Fig. 32) showed better activity than others due to the presence of electron-donating substituted group in the pyrazole ring.86
5.9 Thiosemicarbazide based Schiff base ligands
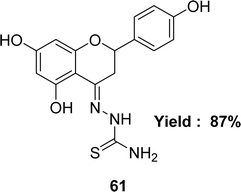
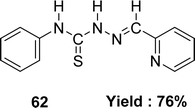
Katarzyna Brodowska et al. synthesized a Schiff base ligand, namely 61 (Fig. 35) from naringenin and thiosemicarbazide and characterized the ligand by IR, UV-visible and 1H NMR spectroscopy in 2015. They also demonstrated the antioxidant activity of the average inhibitory ratio of the free ligand, whose IC50 value was 0.43 ± 0.00 mM in DMSO and methanol-mixed solvent (Table 9).94Diana-Carolina Ilies and her colleagues reported the synthesis of the Schiff base ligand named 2-formylpyridine-N(4)-phenylthiosemicarbazone 62 (Fig. 36) from the reaction of 4-phenylthiosemicarbazide and 2-formylpyridine in 2015. The ligand underwent characterization using elemental analysis, FTIR, NMR (1H and 13C) spectroscopy and X-ray diffraction. The author noted that the ligand 62 exhibited antioxidant action with an IC50 value of 157.55 μM. On the contrary, the IC50 values of standard BHT and BHA were >250 μM and 34.49 μM respectively (Table 9). The thiosemicarbazone ligand exhibited more activity than the BHT standard but lower activity than the BHA based on its IC50 value.95
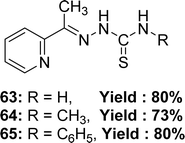
Rajendran Manikandan et al. synthesized Schiff base ligands: 2-acetylpyridine-thiosemicarbazone 63 from 2-acetylpyridine and thiosemicarbazide, 2-acetylpyridine-4-methyl-thiosemicarbazone 64 from 2-acetylpyridine and 4-methyl-thiosemicarbazide, 2-acetylpyridine-4-phenyl-thiosemicarbazone 65 (Fig. 37) from 2-acetylpyridine and 4-phenyl-thiosemicarbazide and they characterized the ligands by single-crystal X-ray diffraction studies. The author reported the antioxidant activity of ligands with IC50 values of 63.03 ± 0.2, 53.45 ± 0.3 and 65.56 ± 0.2 μM for 63, 64, and 65, respectively. The ligands exhibited enhanced antioxidant activity compared to the conventional BHT (IC50 = 86.53 ± 0.6 μM) and vitamin C (IC50 = 147.20 ± 0.8 μM) (Table 9). The inclusion of electron-donating methyl groups enhances the antioxidant activity of 64.96
The electron-donating substituent of the ligands increased the electron density in the aromatic ring of the ligand which helped to build a stable resonance structure for the Schiff base radical after scavenging the free radical and enhanced the antioxidant activity of all thiosemicarbazide-based ligands; therefore, researchers should concentrate on it while synthesizing Schiff base to get better antioxidant activity.
5.10 Carbohydrazide-based Schiff base ligands
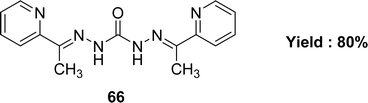
The preparation of Schiff base 66 (Fig. 38) from carbohydrazide and 2-acetylpyridine was mentioned by G. M. Abu El-Reash et al. in 2014. The Schiff base ligand was characterized by infrared spectra, electronic spectra, NMR spectra. The antioxidant activity of the Schiff base ligand 66 (IC50 = 0.964 mg mL−1) was less than that of conventional ascorbic acid (Table 10). Ligand 66 was also screened for antitumor activity using in vitro Ehrlich ascites assay and showed high cytotoxic activity with 61.41% inhibition.975.11 Anili-nohydrazide based Schiff base ligands
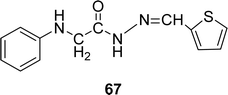
Schiff base ligand (Z)-2-(phenylamino)-N′-(thiophen-2-ylmethylene)acetohydrazide 67 (Fig. 39) from anili-nohydrazide and 2-thiophene carbaldehyde was synthesized and 1H and 13C NMR, elemental analysis mass and IR spectra were used to characterize them by the team of S. A. Aly et al. in 2020. They found the IC50 = 0.514 μg mL−1 of 67 for DPPH-free radical scavenging (Table 11). The Schiff base ligand has a lower scavenging ability compared to the positive standards BHA and TBHQ.985.12 Isatin-based Schiff base ligands
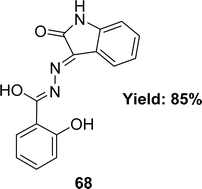
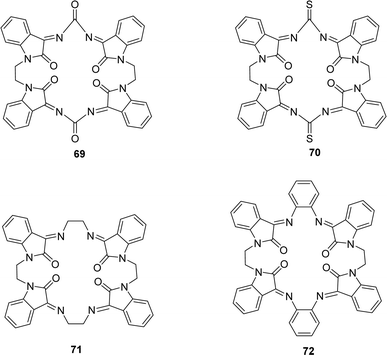
Waqas Jamil and his colleagues synthesized 2-hydroxy salicylhydrazide isatin hydrazone 68 (Fig. 40) in 2019 and achieved 85% yield. They characterized 68 using 1H NMR, UV-Vis, IR spectroscopy and elemental analysis. After that, it was evaluated for antioxidant activity using the DPPH antioxidant assay and found to have an IC50 value of 54.14 ± 2.43 μM whereas the standard BHA was measured at 44.7 ± 1.21 μM (Table 12).99A. G. Bharathi Dileepan et al. synthesized four isatin-based macrocyclic Schiff base ligands, C38H24N8O6 69, C38H24N8O4S2 70, C40H32N8O4 71 and C48H32N8O4 72 (Fig. 41) by nucleophilic addition of a Mannich base ligand in 2018. The structure of the ligand was completely defined by using elemental analysis, infrared, Raman, NMR, and ESI-MS spectroscopy. They used DPPH antioxidant assay and found IC50 values of compounds 69, 70, 71, and 72 were 23.59 μM, 25.79 μM, 30.45 μM, and 28.47 μM respectively (Table 12). The following order of 69 > 70 > 72 > 71 of antioxidant activity reveals that among all the ligands, 69 showed a higher antioxidant property. The oxygen atom of ligand 69 stabilizes free radicals such as DPPH,˙OH, and NO˙ radicals better than other ligands.100
5.13 Isovanillin-based Schiff base ligands
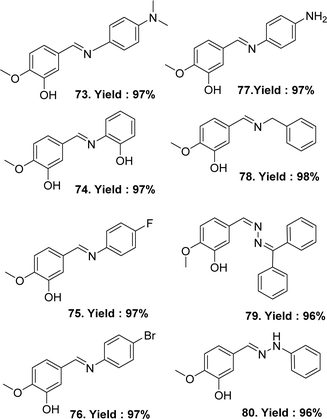
In 2023, Sertan Aytac et al. synthesised the following ligands: (E)-5-((4-(dimethylamino)phenyl)imino)methyl-2-methoxyphenol 73, (E)-5-(((2-hydroxyphenyl)imino)methyl)-2-methoxyphenol 74, (E)-5-(((4-fluorophenyl)imino)methyl)-2-methoxyphenol 75, (E)-5-(((4-bromophenyl)imino)methyl)-2-methoxyphenol 76, (E/Z)-5-(((4-aminophenyl)imino)methyl)-2-methoxyphenol 77, (E)-5-((benzylimino)methyl)-2-methoxyphenol 78, (E)-5-(((diphenylmethylene)hydrazineylidene)methyl)-2-methoxyphenol 79, (E)-2-methoxy-5-((2-phenylhydrazineylidene)methyl)phenol 80 (Fig. 42) from Isovanillin, N1-dimethylbenzene-1,4-diamine, 2-aminophenol, 4-fluoroaniline, 4-bromoaniline, p-phenylenediamine, benzlyamine, benzophenone hydrazone and phenylhydrazine. All the Schiff base ligands were characterized by HRMS (ESI), FTIR, 1H and 13C-NMR spectroscopy. After a comparative study of the DPPH radical scavenging activity of the ligand and standard, their increasing order was: 78 (IC50 = 99.01 μg mL−1) < 76 (IC50 = 87.72 μg mL−1) < 75 (IC50 = 57.75 μg mL−1) < 79 (IC50 = 30.13 μg mL−1) < 73 (IC50 = 16.90 μg mL−1) < 80 (IC50 = 14.74 μg mL−1) < 77 (IC50 = 13.86 μg mL−1) < BHT (IC50 = 13.32 μg mL−1) < trolox (IC50 = 12.15 μg mL−1) ≈ 74 (IC50 = 12.15 μg mL−1) < BHA (IC50 = 11.55 μg mL−1) < α-tocopherol (IC50 = 10.04 μg mL−1). The values exhibited that the DPPH radical activity of Schiff bases 73–80 was close to the standards. The most effective DPPH radical scavenging value was found for 74 (IC50 = 12.15 μg mL−1), which was similar to trolox (IC50 = 12.15 μg mL−1) and lower than BHA (IC50 = 11.55 μg mL−1) and α-tocopherol (IC50 = 10.04 μg mL−1) (Table 13).101| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference. | ||
| 73 (16.90 μg mL−1), 74 (12.15 μg mL−1), 75 (57.75 μg mL−1), 76 (87.72 μg mL−1), 77 (13.86 μg mL−1), 78 (99.01 μg mL−1), 79 (30.13 μg mL−1), 80 (14.74 μg mL−1) | Torolox (12.15 μg mL−1), BHA (11.55 μg mL−1), α-tocopherol (10.04 μg mL−1), BHT (13.32 μg mL−1) | 101 |
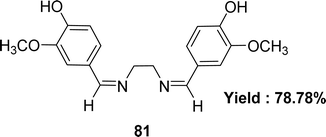
| 81 (5.59 ± 1.16 μg mL−1) | Gallic acid (2.02 ± 0.42 μg mL−1), ascorbic acid (1.22 ± 0.84 μg mL−1) | 102 |
Schiff base ligand 4,4-{ethane-1,2-diylbis[nitrilo(Z)methylylidene]}bis(2-methoxyphenol) 81 (Fig. 43) using ethane-1.2-diamine and 4-hydroxy-3-methoxy benzaldehyde was synthesized and characterized using elemental analysis, FTIR and UV-visible spectroscopic methods by Racheal O. Awolope and her team members in 2023. The ligand 81 exhibited DPPH scavenging action (IC50 = 5.59 ± 1.16 μg mL−1) which was compared with standards gallic acid (IC50 = 2.02 ± 0.42 μg mL−1) and ascorbic acid (IC50 = 1.22 ± 0.84 μg mL−1) (Table 13). The ligand has a lower DPPH scavenging ability than gallic acid and ascorbic acid.102
The study on Schiff base ligands derived from isovanillin showed that ligands with a greater quantity of phenolic groups showed increased antioxidant activity. This may be attributed to their capacity to give protons to free radicals, facilitated by the acidic nature of phenol, which increases the likelihood of proton donation.
5.14 Pyrazole-based Schiff base ligands
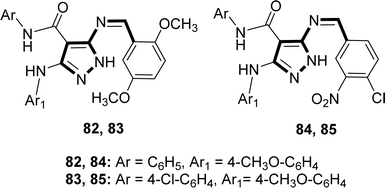
Hamad M. Alkahtani and his colleagues synthesized the Schiff base ligands 82, 83, 84 and 85 (Fig. 44) from the reaction of 5-aminopyrazole with 2,5-dimethoxybenzaldehyde or 4-chloro-3-nitrobenzaldehyde where 5-aminopyrazoles were synthesized using multistep reactions and characterized by 1H-NMR in 2023. The ligands 82–85 showed DPPH radical scavenging action with IC50 values of 82 (16.22 ± 0.04 μg mL−1), 83 (13.20 ± 0.03 μg mL−1), 84 (15.21 ± 0.03 μg mL−1), and 85 (16.13 ± 0.04 μg mL−1) which was lower than conventional ascorbic acid (4.05 ± 0.01 μg mL−1) with higher activity (Table 14).103| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference. | ||
| 82 (16.22 ± 0.04 μg mL−1), 83 (13.20 ± 0.03 μg mL−1), 84 (15.21 ± 0.03 μg mL−1), 85 (16.13 ± 0.04 μg mL−1) | Ascorbic acid (4.05 ± 0.01 μg mL−1) | 103 |
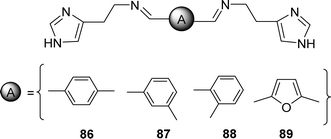
| 86 (508.66 ± 0.66 μM), 87 (414.49 ± 1.56 μM), 88 (>1000 μM), 89 (112.23 ± 1.11 μM) | BHT (232.11 ± 3.01 μM), BHA (61.72 ± 0.85 μM), α-tocopherol (56.86 ± 0.77 μM) | 104 |
In 2022, Nebih Lolak et al. reported the antioxidant properties of bis-histamine Schiff bases (86, 87, 88 and 89) (Fig. 45) by the DPPH free radical scavenging method and found the IC50 values of 86, 87, 88 and 89 ligands were 508.66 ± 0.66, 414.49 ± 1.56, >1000 and 112.23 ± 1.11 μM respectively by using the DPPH antioxidant assay which was compared with standard BHT (IC50 = 232.11 ± 3.01 μM), BHA (IC50 = 61.72 ± 0.85 μM) and α-tocopherol (IC50 = 56.86 ± 0.77 μM) (Table 14). Only the 89 ligand showed better activity than the standard BHT in the presence of a furan ring.104
5.15 Antipyrine-based Schiff base ligands
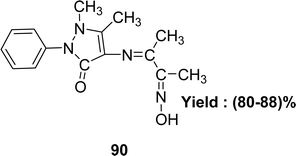
The synthesis of C15H18N4O2 90 (Fig. 46) utilizing 4-aminoantipyrine and 2,3-butanedione monoxime was described by the team of Safaa S. Hassan and IR, elemental analysis, UV-visible and 1H-NMR spectra were used to characterize the ligand in 2020. The DPPH experiment revealed that the antioxidant activity of the ligand was lower with an IC50 value of 70 μg mL−1 than standard ascorbic acid (Table 15). The Schiff base 90 showed high cytotoxic activity against the liver cancer cell line HEPG2 (IC50 = 18.2 μg mL−1).105| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference; NG = not given. | ||
| 90 (70 μg mL−1) | Ascorbic acid | 105 |
| 91 (48.49 ± 0.13 μg mL−1), 92 (91.83 ± 0.28 μg mL−1), 93 (616.36 ± 2.04 μg mL−1), 94 (724.79 ± 2.06 μg mL−1), 95 (591.62 ± 3.16 μg mL−1), 96 (593.66 ± 3.54 μg mL−1) | BHT (11.81 ± 0.42 μg mL−1), BHA (5.19 ± 0.08 μg mL−1), trolox (5.34 ± 0.06 μg mL−1) | 106 |
| 97 (>200 μM), 98 (NG), 99 (>200 μM), 100 (28.33 ± 4.35 μM), 101 (>200 μM), 102 (>200 μM), 103 (>200 μM), 104 (129.4 ± 18.7 μM), 105 (18.9 ± 2.4 μM), 106 (15.7 ± 3.2 μM), 107 (>200 μM), 108 (>200 μM) | Ascorbic acid (14.5 ± 2.2 μM), quercetin (7.3 ± 1.0 μM), caffeic acid (16.2 ± 2.4 μM) | 107 |
| 109 (134.80 μM) | Ascorbic acid (46.81 μM) | 108 |
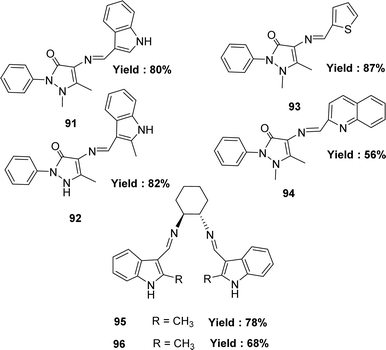
In 2020, Hakan Kizilkaya and her colleagues reported the preparation of Schiff base ligands such as 4-(1H-Indol-3-ylmethyleneamino)-1,5-dimethyl-2-phenyl-1H-pyrazol-3(2H)-one 91 from 4-amino-1,5-dimethyl-2-phenyl-1H-pyrazol-3(2H)-one and indole-3-carbaldehyde; 1,5-dimethyl-4-[(2-methyl-1H-indol-3-yl)methyleneamino]-2-phenylpyrazol-3-one 92 from 4-aminopyridine and methylindole-3-caroboxaldehyde, ligand (E)-1,5-dimethyl-2-phenyl-4-(thiophen-2-ylmethyleneamino)-1H-pyrazol-3(2H)-one 93 from 4-aminopyridine, thiophene-2-carbaldehyde, and ligand (E)-1,5-dimethyl-2-phenyl-4-(quinolin-2-ylmethyleneamino)-1H-pyrazol-3(2H)-one 94 from 4-aminopyridine and quinoline-2-carbaldehyde they also synthesize the ligand (1S,2S,N1,N2)-N1,N2-bis((1H-indol-3-yl)methylene) cyclohexane-1,2-diamine 95 ligand from indole-3-carbaldehyde and trans-cyclohexane-1,2-diamine and (1S,2S,N1,N2)-N1,N2-bis((2-methyl-1H-indol-3-yl)methylene)cyclohexane-1,2-diamine 96 (Fig. 47) ligand from 2-methyl-indole-3-carbaldehyde and (1S,2S)-cyclohexane-1,2-diamine and all the Schiff base ligands were characterized by FTIR, 1H and 13C NMR. The antioxidant activity of synthesized compounds was investigated using 1,1-diphenyl-2-picryl-hydrazyl free radical (DPPH˙) scavenging, and the values of 91–96 were found by IC50 values such as 48.49 ± 0.13, 91.83 ± 0.28, 616.36 ± 2.04, 724.79 ± 2.06, 591.62 ± 3.16 and 593.66 ± 3.54 μg mL−1 respectively. They reported that compound 91 exhibited the most activity (IC50 = 48.49 ± 0.13 μg mL−1). But in comparison with standard BHT (IC50 = 11.81 ± 0.42 μg mL−1), BHA (IC50 = 5.19 ± 0.08 μg mL−1), and trolox (IC50 = 5.34 ± 0.06 μg mL−1), all the ligands showed very lower antioxidant activity (Table 15).106
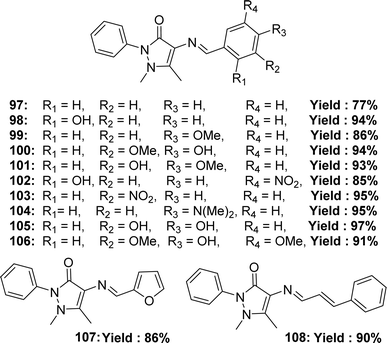
Rommy Terana and his colleagues synthesized 12 ligands 97–108 (Fig. 48) from 4-amino antipyrine in 2019 and characterized them using 1H and 13C NMR, IR, HRMS spectroscopic methods. They reported the yield % of the 97–108 ligands was 77–97% respectively. The antioxidant activity of the ligands was evaluated using DPPH antioxidant assay and found to have IC50 values of the ligands 97, 99, 101, 102, 103, 107, and 108 >200 μM and of the ligands 100, 104, 105, 106 were 28.33 ± 4.35, 129.4 ± 18.7, 18.9 ± 2.4 and 15.7 ± 3.2 μM. The experimental data of the ligands was compared with standard ascorbic acid (IC50 = 14.5 ± 2.2 μM), quercetin (IC50 = 7.3 ± 1.0 μM), caffeic acid (IC50 = 16.2 ± 2.4 μM) and found none of the ligands showed better activity than that of the standard ascorbic acid, caffeic acid, quercetin except 106 that showed higher activity than caffeic acid (Table 15). The presence of hydroxyl groups in the para positions of aromatic rings (100, 105, and 106) usually results in better radical scavenging activity than other positions or electron-donating groups, except for 104, which contains a diethylamino group which stabilizes the radical by resonance structure. Ligands 97–106 exhibited significant cytotoxicity, resulting in <40% cell survival against murine RAW 264.7 and ATCC TIB71 mammal macrophage cells.76,107
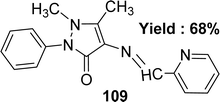
Subbaiyan Sathiyaraj et al. elucidated the preparation of a Schiff base ligand of the molecular formula C17H16N4O 109 (Fig. 49). The ligand was characterized by elemental analysis, UV-visible, FTIR, 1H and 13C NMR, and X-ray crystallography. The ligand exhibited antioxidant activity with an IC50 value of 134.80 μM, whereas the standard ascorbic acid showed 46.81 μM (Table 15). Thus, the antioxidant activity of the ligand was lower than the standard one. Additionally, ligand 109 did not give any significant cytotoxic activity against HeLa, MCF-7, and Hep-2 cell lines with IC50 values > 100 μM.108
Antipyrine is not a good choice for the synthesis of Schiff base ligands to get antioxidant activity because most of the antipyrine-based Schiff base ligands did not show significant activity compared to the well-known standard.
5.16 Pyrimidine-based Schiff base ligands
Wail Al Zoubi et al. synthesized 6-[(4-dimethylamino-benzylidene)amino]-pyridine-2,4-diol 110 (Fig. 50) and found 79.5% yield from 4-dimethyl aminobenzaldehyde and 4-amino-2,6-dihydroxy pyrimidine in 2019. The ligand was characterized using 1H and 13C NMR, FTIR, UV-visible spectra, and elemental microanalysis. After that, it was evaluated for antioxidant activity using a DPPH antioxidant assay and found to have an IC50 value of 11.4 M (Table 16). DPPH radical-scavenging activity is influenced by the presence of –OH groups.1095.17 Pyridine-based Schiff base ligands
The synthesis of Schiff base ligands 111–126 (Fig. 51) from the starting material 3-amino-6-methyl-4-phenylpyridin-2(1H)-one and aromatic aldehydes was mentioned by Ivan V. Kulakov et al. in 2018. The obtained ligands were characterized by IR and NMR spectroscopy, elemental analysis, and single-crystal X-ray diffraction. The IC50 (DPPH) values of compounds 115 and 116 were found to be 25.4 μM and 17.6 μM, respectively, and compounds 117, 120, 124 and 126 could not be measured. The ligand 116 showed higher activity than the standard ascorbic acid (IC50 = 19.4 μM) (Table 17).110Amani S. Alturiqi et al. reported that the Schiff base ligand 127 (Fig. 52) was synthesized from the reaction of 4-oxo-4H-chromene-3-carbaldehyde and 2-aminopyridine-3-thiol and they characterized the ligand using 1H NMR, mass spectra, IR, XRD, and UV-visible spectroscopy in 2017. Ligand showed stronger antioxidant activity (IC50 = 78.92 μg mL−1) compared to standard vitamin C (IC50 = 66.96 μg mL−1). Ligand 127 is non-toxic against LCLC-103H (IC50 = 145.368 μM), A427 (IC50 = 172.354 μM), 5637 (IC50 = 152.258 μM), and SISO (IC50 = 163.357 μM) cell lines compared to doxorubicin (IC50 = 0.138–0.252 μM) (Table 17).111
5.18 Phenyl thiazole-based Schiff base ligands
Ligands 128–130 (Fig. 53) from phenyl thiazole and benzaldehyde/benzaldehyde derivatives and 131, 132 ligands from p-nitrobenzadehyde and acetophenonethiazole/p-nitroacetophenone thiazole were synthesized by Fitsum Lemilemu and his colleagues in 2021. FTIR, UV-visible, 1H and 13C NMR were used to characterize the ligands. Ligands 128 (IC50 = 3.60 μg mL−1) and 130 (IC50 = 3.65 μg mL−1) have higher antioxidant activity than standard ascorbic acid (IC50 = 3.98 μg mL−1), moderately potent ligand 129 (IC50 = 5.56 μg mL−1), and less potent ligands 131 and 132 (IC50 = 5.36 μg mL−1 and 5.14 μg mL−1 respectively) (Table 18). The ligands 128 and 130 are notable for their potent ability to scavenge free radicals. Ligand 128 may have significant antioxidant activity owing to the presence of multiple –OH substituents, which can stabilize the resonance structure during radical scavenging. The author found that the compound quantum mechanical descriptors of the ligands support this experimental finding.1125.19 Amino-thiadiazol-based Schiff base ligands
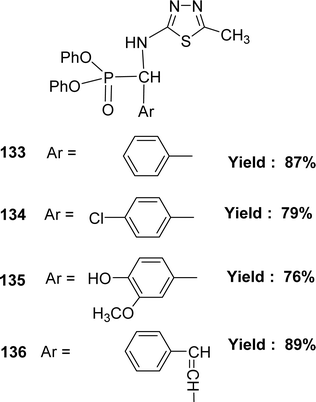
In 2018, the team of Mohamed M. Azaam described the synthesis of Schiff base ligand diphenyl(5-methyl-1,3,4-thiadiazol-2-ylamino)(phenyl)methylphosphonate 133, diphenyl(4-chlorophenyl)(5-methyl-1,3,4-thiadiazol-2-ylamino)methylphosphonate 134, diphenyl(4-hydroxy-3-methoxyphenyl)(5-methyl-1,3,4-thiadiazol-2-ylamino)methylphosphonate 135 and diphenyl1-(5-methyl-1,3,4-thiadiazol-2-ylamino)-3-phenylallyl phosphonate 136 (Fig. 54) from the starting material 2-amino-5-methyl1,3,4-thiadiazole with various aldehydes. The Schiff base ligand was characterized by FT-IR analysis, ESI-MS, 1H NMR, and elemental analysis. It had been notified that the IC50 values of ligands 133–136 were 198.6, 159.6, 129.6 and 204 μg mL−1 respectively. The compounds were arranged following their antioxidant activities as follows: 135 > 134 > 133 > 136 (Table 19).113 The multiple –OH group may enhance the activity of ligand 135.5.20 Ethylenediamine-based Schiff base ligand
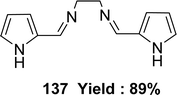
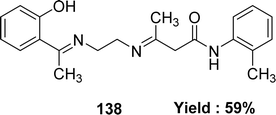
In 2023, the ligand [(N1Z,N2Z)-N1,N2-bis((1H-pyrrol-2-yl)methylene)ethane-1,2-diamine] 137 (Fig. 55) was synthesized by Ibtissam Elaaraj et al. via condensation of 1H-pyrrole-2-carbaldehyde with ethylenediamine. The ligand was characterized by mass spectrometry, UV-visible, FT-IR, and 1H, 13C NMR. Using the DPPH radical test, ligand 137 showed lesser DPPH scavenging activity (IC50 = 1.065 ± 0.44 mg mL−1) than normal ascorbic acid (IC50 = 0.133 ± 0.03 mg mL−1) (Table 20).114K. Subin Kumar and his colleagues explained the synthesis of a novel mixed Schiff base ligand, 3-((2-((1-(2-hydroxyphenyl)ethylidene)amino)ethyl)imino)-N-(o-tolyl)butanamide 138 (Fig. 56) which was derived via refluxing the mixture of o-hydroxyacetophenone, 2-methylacetoacetanilide and 1,2-ethylenediamine by the one-pot method in 2022. They characterized the synthesized ligands via element analysis, IR, 1H and 13C NMR, UV-visible, and high-resolution mass spectrometry. From the result of DPPH antioxidant assay the ligand 138 with an IC50 value of 18.4 μM has shown better antioxidant activity than conventional vitamin C with an IC50 value of 32.5 μM (Table 20). The ligand 138 showed weak cytotoxic activity against Dalton's Lymphoma Ascites (36% cytotoxicity).76,115
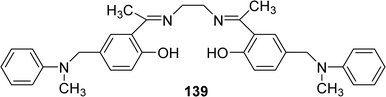
In 2021, Nadjah Maghraoui et al. synthesized the Schiff base ligand 139 (Fig. 57). The ligand showed DPPH radical scavenging activity with IC50 values of 373.0 μg mL−1 (Table 20).116
5.21 o-Phenylenediamine-based Schiff base ligand
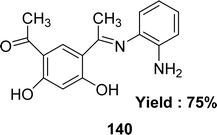
In the year of 2015, Magdy Shebl et al. synthesized ligand 140 (Fig. 58) from 4,6-diacetylresorcinol and o-phenylenediamine. The ligand was characterized by elemental analysis, IR, 1H and 13C NMR, UV-visible and mass spectra. The antioxidant activity of the ligand was tested against the 1,1-diphenyl-2-picrylhydrazyl radical (DPPH) and it exhibited antioxidant activity with an IC50 value of 118.1 ± 3.51 μM which was compared with the standard ascorbic acid. The standard IC50 value of ascorbic acid was 126.4 ± 11.3 μM greater than the IC50 value of the ligand (Table 21).117 The ligand has two phenolic –OH groups, and because of its acidic properties, protons may be transferred to the radical to stabilize it and create a stable free radical.1185.22 2-Aminobenzimidazole-based Schiff base ligand
Nadia El-wakiel and her team members elucidated the preparation of 4-[(1H-benzoimidazol-2-ylimino)-methyl]-benzene-1,3-diol 141 (Fig. 59) Schiff base ligand, using a condensation reaction between 2-aminobenzimidazole and 2,4-dihydroxy benzaldehyde in 2015. The Schiff base ligand was identified by elemental analyses, IR, EI-mass, UV-visible and ESR spectral studies. The ligand exhibited lower antioxidant activity with an IC50 value of 77.85 μg mL−1 in comparison to the standard ascorbic acid (IC50 = 67.44 μg mL−1) (Table 22). The ligand 141 has moderate cytotoxic activity against liver carcinoma cell HEPG2 (IC50 = 9.03 μg mL−1).1195.23 6-tert-Butyl 3-ethyl 2-amino-4,5-dihydrothieno[2,3-c] pyridine-3,6(7H)-dicarboxylate-based ligand
Synthesis of ((E)-6-tert-butyl 3-ethyl 2-(2-hydroxybenzylideneamino)-4,5-dihydrothieno[2,3-c]pyridine-3,6(7H)-dicarboxylate) 142 (Fig. 60) from 6-tert-butyl 3-ethyl 2-amino-4,5-dihydrothieno[2,3-c]pyridine-3,6(7H)-dicarboxylate and 2-hydroxybenzaldehyde by condensation reaction was reported by Nevin Turan and her colleagues in 2022. They evaluated the antioxidant activity using DPPH antioxidant assay and found that the Schiff base ligand has lower antioxidant activity with IC50 value 57.9 ± 4.4 μg mL−1 compared to standard BHT (IC50 = 24.3 ± 2.8 μg mL−1), BHA (IC50 = 25.8 ± 2.3 μg mL−1), α-tocopherol (IC50 = 26.6 ± 3.8 μg mL−1) and ascorbic acid (IC50 = 38.4 ± 3.5 μg mL−1) (Table 23).120| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference. | ||
| 142 (57.9 ± 4.4 μg mL−1) | Ascorbic acid (38.4 ± 3.5 μg mL−1), BHT (24.3 ± 2.8 μg mL−1), BHA (25.8 ± 2.3 μg mL−1), α-tocopherol (26.6 ± 3.8 μg mL−1) | 120 |
| 143 (15.5 ± 7.8 μg mL−1) | BHT (24.6 ± 4.4 μg mL−1), BHA (37.9 ± 11.4 μg mL−1), α-tocopherol (43.7 ± 9.4 μg mL−1), ascorbic acid (52.4 ± 5.7 μg mL−1) | 121 |
Ercan Bursal and his colleagues reported the synthesis of the Schiff base ligand ((E)-6-tert-butyl 3-ethyl 2-(2-hydroxy-3-methoxybenzylideneamino)-4,5-dihydrothieno[2,3-c]pyridine-3,6(7H)-dicarboxylate) 143 (Fig. 61) in 2021. Characterization of the ligand was performed using FT-IR, 1H and 13C NMR, UV-visible, mass spectrometry, and elemental analysis. The free radical scavenging antioxidant effects of the Schiff base were determined by the DPPH method and the IC50 values were found as follows: 15.5 ± 7.8 μg mL−1 for BHT, 24.6 ± 4.4 μg mL−1 for BHA, 37.9 ± 11.4 μg mL−1 for α-tocopherol, 43.7 ± 9.4 μg mL−1 for the ligand, and 52.4 ± 5.7 μg mL−1 for ascorbic acid. The IC50 values indicate that the ligand exhibits moderate antioxidant activity compared to well-known antioxidants like BHT, BHA, and α-tocopherol, but slightly higher than ascorbic acid (Table 23).121
6-tert-Butyl 3-ethyl 2-amino-4,5-dihydrothieno[2,3-c]pyridine-3,6(7H)-dicarboxylate based ligand showed closer activity compared to standard so this kind of ligand can be further studied for different biological activities.
5.24 Other types of Schiff base ligand
In 2021, May Juda. Kareem et al. reported a synthetic scheme synthesized a novel Schiff base ligand (3Z,NZ)-3-(2-(1H-indol-3yl)ethylimino)-N-(4-((Z)-((Z)-3(ethylimino)-1,5-dimethyl-2-phenyl-2,3-dihydro-1H-pyrazole-4-ylimino)methyl)benzylidene)-1,5-dimethyl-2-phenyl-2,3-dihydro-1H-pyrazol-4-amine,3-methyl-1H-indole 144 (Fig. 62) from 3-[2-(1H-indol-3-yl)-ethylimino]-1.5-dimethyl-2-phenyl-2.3-dihydro-1H-pyrazol-4-ylamine and benzene-1.4-dicarbaldehyde. Characterization of 144 employed elemental analysis, FTIR, 1H and 13C NMR, UV-visible, and mass spectrometry. Free radical scavenging activity of the compounds 144 has been studied using DPPH method. The IC50 values revealed significant activity in various solvents 3.02 μg mL−1 (methanol); 6.85 μg mL−1 (chloroform); 43.3 μg mL−1 (acetonitrile); 86.2 μg mL−1 (acetone); 206 μg mL−1 (ethyl acetate) for 144, depending on the solvent used. The IC50 values indicate significant activity for 144, with the activity being dependent on the solvent used (Table 24).53| Compound (IC50 value) | Standard (IC50 value) | Ref. |
|---|---|---|
| a Ref. = reference; NG = not given. | ||
| 144 3.02 μg mL−1 (methanol), 6.85 μg mL−1 (chloroform), 43.3 μg mL−1 (acetonitrile), 86.2 μg mL−1 (acetone), 206 μg mL−1 (ethyl acetate) | NG | 53 |
| 145 (6.12 ± 0.04 μM mL−1), 146 (4.91 ± 0.03 μM mL−1), 147 (5.29 ± 0.06 μM mL−1), 148 (4.83 ± 0.01 μM mL−1), 149 (4.61 ± 0.02 μM mL−1), 150 (5.86 ± 0.03 μM mL−1), 151 (4.45 ± 0.06 μM mL−1), 152 (5.18 ± 0.00 μM mL−1), 153 (4.48 ± 0.06 μM mL−1), 154 (1.01 ± 0.04 μM mL−1), 155 (0.22 ± 0.00 μM mL−1), 156 (0.15 ± 0.01 μM mL−1), 1c57 (0.42 ± 0.01 μM mL−1), 158 (0.36 ± 0.02 μM mL−1), 159 (0.19 ± 0.00 μM mL−1), 160 (0.51 ± 0.03 μM mL−1), 161 (0.67 ± 0.02 μM mL−1), 162 (0.29 ± 0.00 μM mL−1) | Trolox (0.04 ± 0.00 μM mL−1) | 122 |
| 163 (5.74 μg mL−1), 164 (0.8 μg mL−1) | Ascorbic acid (1.34 μg mL−1) | 123 |
| 165 (290 ± 9 μM), 166 (390 ± 5 μM), 167 (360 ± 5 μM), 168 (290 ± 6 μM), 169 (260 ± 5 μM), 170 (380 ± 2 μM), 171 (200 ± 0 μM), 172 (210 ± 5 μM), 173 (220 ± 6 μM) | 3,4,5-Trihydroxybenzoic acid (420 ± 3 μM), 5-(1,2-dihydroxyethyl)-3,4-dihydroxyfuran-2(5H)-one (350 ± 5 μM), 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one (240 ± 3 μM) | 124 |
Arif Mermer and his colleagues synthesized 18 ligands 145–162 (Fig. 63) in 2019 and they characterized the compounds using 1H and 13C NMR, FTIR and MS spectra. They achieved 81–96% yield. The antioxidant activity of the ligands was evaluated using a DPPH antioxidant assay and found to have IC50 values of the ligands 145 (6.12 ± 0.04 μM mL−1), 146 (4.91 ± 0.03 μM mL−1), 147 (5.29 ± 0.06 μM mL−1), 148 (4.83 ± 0.01 μM mL−1), 149 (4.61 ± 0.02 μM mL−1), 150 (5.86 ± 0.03 μM mL−1), 151 (4.45 ± 0.06 μM mL−1), 152 (5.18 ± 0.00 μM mL−1), 153 (4.48 ± 0.06 μM mL−1), 154 (1.01 ± 0.04 μM mL−1), 155 (0.22 ± 0.00 μM mL−1), 156 (0.15 ± 0.01 μM mL−1), 157 (0.42 ± 0.01 μM mL−1), 158 (0.36 ± 0.02 μM mL−1), 159 (0.19 ± 0.00 μM mL−1), 160 (0.51 ± 0.03 μM mL−1), 161 (0.67 ± 0.02 μM mL−1), 162 (0.29 ± 0.00 μM mL−1) and standard trolox (0.04 ± 0.00 μM mL−1). So none of the ligands shows better activity than that of the standard trolox (Table 24).122
The synthesis of novel Schiff base ligands 163 and 164 (Fig. 64) which was derived via refluxing the mixture of ethylbenzoate, hydrazine hydrate and ethylsalicylate that was characterized via UV-visible spectroscopy, IR spectra, 1H NMR spectra and mass spectra reported by Mohammad Nasir Uddin et al. in 2018. It had been informed that the IC50 value of 163, 164 and ascorbic acid were 5.74, 0.8 and 1.34 μg mL−1 from DPPH free radical scavenging assay. 164 ligand showed higher antioxidant activity than the standard ascorbic acid (Table 24).123 The presence of the naphthol ring, hydroxyl group, conjugated double bond and azo group which can stabilize the resonance structure of Schiff base-free radicals after the radical scavenging reaction may enhance the activity of ligand 164.
Bamidele Joseph Okoli and his colleagues reported the preparation of Schiff base ligands 3-(((4-nitrophenyl)methylidene)amino)-1H-pyrazol-5-ol 165, 3-(((2-hydroxyphenyl)methylidene)amino)-1H-pyrazol-5-ol 166, 3-((phenylmethylidene)amino)-1H-pyrazol-5-ol 167, 4-(((4-nitrophenyl)methylidene)amino)phenol 168, 4-(2-hydroxybenzylideneamino)phenol 169, 4-(benzylidene amino)phenol 170, 4-(4-nitrobenzylideneamino)-3-hydroxynaphthalene-1-sulphonic acid 171, 4-(2-hydroxybenzylideneamino)-3-hydroxynaphthalene-1-sulphonic acid 172 and 4-(benzylidene amino)-3-hydroxynaphthalene-1-sulphonic acid 173 (Fig. 65) in 2018. The ligands were characterized by TGA, XRD, 1H and 13C NMR, elemental analyses, and FT-IR. From DPPH scavenging assay the IC50 value of the ligands and controls were 165 (290 ± 9 μM), 166 (390 ± 5 μM), 167 (360 ± 5 μM), 168 (290 ± 6 μM), 169 (260 ± 5 μM), 170 (380 ± 2 μM), 171 (200 ± 0 μM), 172 (210 ± 5 μM), 173 (220 ± 6 μM), 3,4,5-trihydroxybenzoic acid (420 ± 3 μM), 5-(1,2-dihydroxyethyl)-3,4-dihydroxyfuran-2(5H)-one (350 ± 5 μM), 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one (240 ± 3 μM) respectively (Table 24). Introducing a para or ortho substituent to the phenolic ring of Schiff bases enhanced their scavenging capability. The presence of a nitro group at the para position on other sides of the phenolic ring of the Schiff base enhanced its activity. This is due to the nitro group reducing the electron density in the ring, so facilitating the donation of acidic protons towards the quenching of the radicals. The extra hydroxyl group on Schiff base 169 helps to stabilize the phenoxy ion compared to Schiff bases 168 and 170. This changes the balance to make the more stable bi-phenoxy ion, which makes it easier to donate acidic protons. Considering that phenolic hydrogen is not very acidic, it will not be as likely to donate a proton into the medium as p-nitro Schiff base derivatives 165 and 171. Adding p-nitro or o-hydroxy to a Schiff base makes it better at removing radicals than Schiff bases that haven't been modified (167, 170, and 173).124
6. Conclusions
Over the last decade, Schiff bases, with well-established chemical structures, have been widely used due to their ability to interact with biological systems. They have applications in biological systems, medicine, pharmacy, chemical analysis, and new technologies, among other fields. Their antioxidant action may further enhance these various uses. Schiff bases have exhibited antioxidant activity that is sometimes greater than, or comparable to, conventional antioxidants, due to their structural characteristics and their capacity to donate protons and electrons. Thus, Schiff bases should be rapidly developed into novel, ecologically sound techniques in medicinal chemistry for the benefit of both individuals and ecosystems.In recent years, the use of bioinorganic chemistry in pharmaceutical research has significantly increased. The development of Schiff bases as antioxidants in clinical trials is expected to enhance the recognition of pharmaceutical industry and facilitate further research in this promising and innovative field. This review aims to summarize the antioxidant activity of Schiff base ligands and the effects of substituent groups.
Isoniazid-based, nicotinic acid hydrazide-based, and other Schiff base ligands can donate electrons and protons to stabilize free radicals in the presence of keto–enol, phenolic, and enolic hydroxyl groups. Electron-donating groups in the imidazole ring enhance this ability by increasing electron density near nitrogen atoms. Chlorine-substituted benzoic acid hydrazide-based Schiff base ligands show higher activity due to the resonance effect of chlorine. Aminophenol-based ligands with multiple phenolic rings exhibit higher activity, which is enhanced by electron-donating substituents like methyl groups and reduced by electron-withdrawing groups such as –NO2. The activity of salicylaldehyde-based ligands depends on hydroxyl groups, and larger substituted Schiff bases show significant activity. Electron-withdrawing groups like nitro, chloro, bromo, and methoxy generally decrease activity, with chloro having the most significant effect. However, methoxy-substituted ligands on the opposite side of the phenolic ring show better activity. Two electron-withdrawing groups in the phenolic ring can increase acidity, aiding proton donation. Electron-donating groups like dimethylamine in triazole-based ligands enhance activity. Methyl groups also enhance the activity of thiosemicarbazide-based ligands. Isatin-based ligands exhibit activity due to the keto group. Schiff base ligands derived from isovanillin, with more phenolic groups, show increased activity due to their proton-donating capacity. Hydroxyl groups in the para positions, as opposed to other positions on aromatic rings in antipyrine-based ligands, result in better radical-scavenging activity due to greater resonance stability after neutralizing free radicals. The activity of pyrimidine-based ligands is attributed to –OH groups. O-Phenylenediamine-based ligands, with two phenolic –OH groups, transfer protons to stabilize radicals. Naphthol rings, hydroxyl groups, conjugated double bonds, and azo groups enhance the activity of ligands by stabilizing the resonance structure of Schiff base radicals.
There is some solvent effect on the antioxidant activity of Schiff bases, which varies with the dielectric constant of the solvent. In polar solvents like methanol, ethanol, ethyl acetate, and acetone, their antioxidant activity is enhanced due to the higher ionizing capacity and dielectric constant compared to non-polar solvents like chloroform. The antioxidant efficacy in polar protic solvents is influenced by the hydrogen bond strength between the Schiff base and the solvent. Polar protic solvents, such as methanol and ethanol, facilitate the sequential proton loss electron transfer (SPLET) and electron transfer-proton transfer (ET-PT) processes, transforming a neutral substrate into charged species and stabilizing the transition state in polar media. In HAT, the solvent effect is determined by solvation and hydrogen bonding in the transition state. In polar aprotic solvents, compounds perform better in acetonitrile than in acetone and ethyl acetate due to the lower electronegativity of nitrogen atoms. The intermolecular hydrogen bond between the phenol moiety and acetone or ethyl acetate is stronger than the bond with acetonitrile, enhancing hydrogen reactivity in acetonitrile.
Thus, Schiff bases represent a potential source of antioxidants for combating diseases associated with oxidative stress. Further studies on Schiff bases with superior DPPH-scavenging activity are necessary to advance the field. This review will assist researchers in designing Schiff bases with improved activity. The knowledge presented here could help researchers explore the potential applications of Schiff-base-derived antioxidants in various fields.
7. Future aspects of Schiff base ligands
The field of Schiff base ligands is rapidly expanding, offering numerous opportunities for future research, including the study of antioxidant activity and mechanisms of action. While the DPPH scavenging assay is essential for evaluating antioxidant potential, gaining a deeper understanding of specific mechanisms, such as electron transfer, hydrogen atom transfer, or metal chelation, will provide valuable insights for developing more effective and selective antioxidants.From the present study, the Schiff base ligands 2–4, 18, 20–24, 55, 63–65, 128, 130, 138, 140, 143, 164, and 171–173 exhibited higher antioxidant activity than the conventional standard antioxidant. Additionally, the ligands 29, 39, 54, 68, 74, 77, 80, 127, 141, 165, 168, and 169 showed comparable activity to the conventional antioxidant in the in vitro DPPH scavenging assay. Therefore, further in vivo and clinical trials are necessary to develop more potent conventional antioxidants. A toxicity study is also important for assessing the safety of Schiff base antioxidants. In this study, ligands 29, 39, 41, 66, 90, and 97–106 were found to be highly toxic, while ligand 141 was moderately toxic. However, the in vitro analysis revealed that ligands 1, 28, 36, 37, 109, and 138 were poor toxic, while ligands 18–23, and 127 showed no toxicity. Among the studied ligands, the toxicity of 2–17, 24–27, 30–35, 38, 40, 42–65, 67–89, 91–96, 107, 108, 110–126, 128–137, 139, 140, and 142–173 was not reported. Therefore, more toxicological studies are needed to develop non-toxic antioxidants.
Researchers should also investigate the mechanisms of action of these Schiff base ligands in different solvents, as solvents can significantly affect their antioxidant properties. The study found that not all synthesized compounds exhibited effective antioxidant properties. Therefore, researchers should focus on developing new compounds with tailored antioxidant properties by exploring various structural modifications, incorporating natural products or bioactive molecules, and utilizing computational modeling approaches. This may lead to the identification of novel lead compounds for applications in fields such as food preservation, medicinal research, and cosmetics.
Furthermore, Schiff base ligands hold great potential in various industries. In the food and beverage sector, these compounds can act as preservatives to enhance product shelf-life and quality. In the pharmaceutical sector, Schiff base ligands could be explored as potential therapeutic agents for diseases associated with oxidative stress. Additionally, their use in skincare formulations could protect against environmental damage and promote healthy aging.
In summary, the future of Schiff base ligand research is promising, with many opportunities to further elucidate their antioxidant mechanisms, develop innovative molecules, and explore their applications across multiple industries. By considering these future prospects, researchers may develop effective antioxidant solutions to meet the growing demand.
Abbreviations
| DPPH | 2,2-diphenyl-1-picrylhydroxyl |
| HAT | hydrogen atom transfer |
| SET | single electron transfer |
| ROS | reactive oxygen species |
| RNS | reactive nitrogen species |
| BHA | butylated hydroxyanisole |
| BHT | butylated hydroxytoluene |
| TBHQ | tertiary butylhydroquinone |
| DNA | deoxyribonucleic acid |
| ESR | electron spin resonance |
| IgG | immunoglobulin G |
| RSS | reactive sulphur species |
| IP | ionization potential |
| BDE | bond dissociation enthalpy |
| ABTS | 2,2′-azino-bis-(3-ethylbenzothiozoline-6- sulphonic acid) |
| ORAC | oxygen radical absorbance capacity |
| FRAP | ferric reducing antioxidant power |
| EDGs | electron donating groups |
| CUPRAC | cupric reducing antioxidant capacity |
| FTIR | Fourier transform infrared |
| UV-Visible | ultraviolet visible spectroscopy |
| EPR | electron paramagnetic resonance |
| DSC | differential scanning calorimetry |
| NMR | nuclear magnetic resonance spectroscopy |
| XRD | X-ray diffraction |
| MS | mass spectrometry |
| SEM | scanning electron microscopy |
| ESI-MS | electrospray ionization mass spectrometry |
| HR-MS | high resolution mass spectrometry |
| HR-ESI-MS | high resolution electrospray ionization mass spectrometry |
| EI-mass | electron ionization mass spectrometry |
| DMSO | dimethyl sulfoxide |
| A549 | adenocarcinoma of the human lung type II alveolar cells |
| MDA-MB-231 | breast cancer cells from metastatic site |
| 3T3LI | mouse embryo fibroblast cells |
| MCF-7 | breast cancer cells |
| HCT-116 | colorectal cancer cells |
| HEK-293 | human embryonic kidney cells |
| AGS | gastric adenocarcinoma cells |
| HEPG2 | hepatoma cells |
| RAW 264.7 | mouse leukemic monocyte macrophage cells |
| ATCC TIB71 | normal human fibroblast cells |
| HeLa | human cervical cancer cells |
| Hep-2 | laryngeal carcinoma cells |
| LCLC-103H | lung cancer cells |
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Md. Sohel Rana: conceptualization, investigation, formal analysis, writing – original draft preparation, writing – reviewing and editing, supervision; Noor Mohammad Azbar Rayhan: writing – original draft preparation; Md. Shahadat Hossain Emon: writing – original draft preparation; Md. Tanvir Islam: writing – original draft preparation; Khandaker Rathry: writing – original draft preparation; Md. Mahadi Hasan: writing – original draft preparation; Md. Munna Islam Mansur: writing – original draft preparation; Bishal Chakrabarty Srijon: writing – original draft preparation; Md Shohidul Islam: writing – original draft preparation, writing – reviewing and editing; Anik Ray: writing – original draft preparation; Md. Abdur Rakib: writing – original draft preparation; Azharul Islam: writing – original draft preparation; Md. Kudrat-E-Zahan: writing – reviewing and editing; Md. Faruk Hossen: writing – reviewing and editing; Md. Ali Asraf: writing – reviewing and editing, supervision.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors are grateful to the inorganic research laboratory (Organometallic Chemistry), Department of Chemistry, University of Rajshahi, for their technical support. The authors would like to acknowledge the assistance of QuillBot in language editing and grammatical corrections.References
- E. Raczuk, B. Dmochowska, J. Samaszko-Fiertek and J. Madaj, Molecules, 2022, 27, 787 CrossRef.
- D. Iacopetta, J. Ceramella, A. Catalano, C. Saturnino, M. G. Bonomo, C. Franchini and M. S. Sinicropi, Appl. Sci., 2021, 11, 1877 CrossRef.
- M. Alias, K. Huda and C. Shakir, J. Assoc. Arab Univ. Basic Appl. Sci., 2014, 15, 28–34 Search PubMed.
- A. Jain, S. De and P. Barman, Res. Chem. Intermed., 2022, 48, 2199–2251 CrossRef CAS.
- L. Ma'rufah, A. Hanapi, R. Ningsih and A. G. Fasya, Advances in Social Science, Education and Humanities Research, Atlantis Press, 2021, pp. 297–301 Search PubMed.
- H. Sachdeva, R. Saroj, S. Khaturia and D. Dwivedi, Green Process. Synth., 2012, 1, 469–477 CAS.
- I. R. Pathan and M. K. Patel, Inorg. Chem. Commun., 2023, 158, 111464 CrossRef CAS.
- M. T. Kaczmarek, M. Zabiszak, M. Nowak and R. Jastrzab, Coord. Chem. Rev., 2018, 370, 42–54 CrossRef CAS.
- M. Kaur, S. Kumar, S. A. Younis, M. Yusuf, J. Lee, S. Weon, K.-H. Kim and A. K. Malik, Chem. Eng. J., 2021, 423, 130230 CrossRef CAS.
- P.-P. Zhao, P. Lu, Z.-Y. Zhao, S.-W. Chen, X.-Y. Li, C. Deng and Y.-Z. Wang, Chem. Eng. J., 2022, 437, 135461 CrossRef CAS.
- F. Ndidi Ejiah, M. Olarewaju Rofiu, O. Adekunbi Oloba-Whenu, T. Mojisola Fasina, F. N. Ejiah, M. O. Rofiu, O. A. Oloba-Whenu, T. M. Fasina, F. Ndidi Ejiah, M. Olarewaju Rofiu, O. Adekunbi Oloba-Whenu, T. Mojisola Fasina, F. N. Ejiah, M. O. Rofiu, O. A. Oloba-Whenu and T. M. Fasina, Mater. Adv., 2023, 4, 2308–2321 RSC.
- Z.-Y. Li, H.-K. Su, K. Zhou, B.-Z. Yang, T. Xiao, X.-Q. Sun, J. Jiang and L. Wang, Dyes Pigm., 2018, 149, 921–926 CrossRef CAS.
- M. N. Uddin, S. S. Ahmed and S. M. R. Alam, J. Coord. Chem., 2020, 73, 3109–3149 CrossRef CAS.
- X. Zhong, Z. Li, R. Shi, L. Yan, Y. Zhu and H. Li, ACS Appl. Nano Mater., 2022, 5, 13998–14020 CrossRef CAS.
- V. Pawariya, S. De and J. Dutta, Carbohydr. Polym., 2024, 323, 121395 CrossRef CAS.
- L. M. Aroua, S. K. Alhag, L. A. Al-Shuraym, S. Messaoudi, J. A. Mahyoub, M. Y. Alfaifi and W. M. Al-Otaibi, Saudi J. Biol. Sci., 2023, 30, 103598 CrossRef CAS PubMed.
- M. Li, H. Wang, X. Chen, S. Jin, W. Chen, Y. Meng, Y. Liu, Y. Guo, W. Jiang, X. Xu and B. Wang, Chem. Eng. J., 2020, 382, 122973 CrossRef CAS.
- L. Wei, W. Tan, G. Wang, Q. Li, F. Dong and Z. Guo, Carbohydr. Polym., 2019, 226, 115256 CrossRef.
- S. Shekhar, A. M. Khan, S. Sharma, B. Sharma and A. Sarkar, Emergent Mater., 2022, 5, 279–293 CrossRef CAS.
- W. Al Zoubi, A. A. S. Al-Hamdani and M. Kaseem, Appl. Organomet. Chem., 2016, 30, 810–817 CrossRef CAS.
- V. Lobo, A. Patil, A. Phatak and N. Chandra, Pharmacogn. Rev., 2010, 4, 118–126 CrossRef CAS PubMed.
- S. Di Meo, T. T. Reed, P. Venditti and V. M. Victor, Oxid. Med. Cell. Longevity, 2016, 2016, 1–44 Search PubMed.
- P. Chaudhary, P. Janmeda, A. O. Docea, B. Yeskaliyeva, A. F. Abdull Razis, B. Modu, D. Calina and J. Sharifi-Rad, Front. Chem., 2023, 11, 1158198 CrossRef CAS.
- Y. Wang, L. L. Pruteanu, D. S. Bailey, A. C. Grădinaru, L. Jäntschi, Y. Wang, L. L. Pruteanu, D. S. Bailey, A. C. Grădinaru and L. Jäntschi, Antioxidants, 2023, 12, 860 CrossRef.
- D. R. Berdahl, R. I. Nahas and J. P. Barren, Oxid. Foods Beverages Antioxid. Appl., 2010, pp. 272–320 Search PubMed.
- H. M. A. El-Lateef, T. El-Dabea, M. M. Khalaf and A. M. Abu-Dief, Antioxidants, 2023, 12, 213 CrossRef CAS PubMed.
- M. Oroian and I. Escriche, Food Res. Int., 2015, 74, 10–36 CrossRef CAS PubMed.
- S. C. Lourenço, M. Moldão-Martins and V. D. Alves, Molecules, 2019, 24, 4132 CrossRef.
- N. Rehman, M. Khalid, M. H. Bhatti, U. Yunus, A. A. C. Braga, F. Ahmed, S. M. A. Mashhadi and M. N. Tahir, Turk. J. Chem., 2018, 42, 639–651 CAS.
- A. A. Shanty and P. V. Mohanan, Spectrochim. Acta, Part A, 2018, 192, 181–187 CrossRef CAS PubMed.
- S. Di Meo and P. Venditti, Oxid. Med. Cell. Longevity, 2020, 2020, 1–32 CrossRef.
- E. G. Rozantsev and D. V. Loshadkin, Des. Monomers Polym., 2001, 4, 281–300 CrossRef CAS.
- G. Martemucci, C. Costagliola, M. Mariano, L. D'andrea, P. Napolitano and A. G. D'Alessandro, Oxygen, 2022, 2, 48–78 CrossRef CAS.
- M. Carocho and I. C. F. R. Ferreira, Food Chem. Toxicol., 2013, 51, 15–25 CrossRef CAS.
- C. M. C. Andrés, J. M. P. D. La Lastra, C. A. Juan, F. J. Plou and E. Pérez-Lebeña, Int. J. Mol. Sci., 2023, 24, 15240 CrossRef.
- J. Lunec, K. A. Holloway, M. S. Cooke, S. Faux, H. R. Griffiths and M. D. Evans, Free Radical Biol. Med., 2002, 33, 875–885 CrossRef CAS PubMed.
- V. R. Winrow, P. G. Winyard, C. J. Morris and D. R. Blake, Br. Med. Bull., 1993, 49, 506–522 CrossRef CAS PubMed.
- N. Liang and D. Kitts, Molecules, 2014, 19, 19180–19208 CrossRef PubMed.
- E. A. Mader, E. R. Davidson and J. M. Mayer, J. Am. Chem. Soc., 2007, 129, 5153–5166 CrossRef CAS PubMed.
- E. C. Ashby, Acc. Chem. Res., 1988, 21, 414–421 CrossRef CAS.
- E. Shalaby and S. Shanab, Afr. J. Pharm. Pharmacol., 2013, 7, 528–539 CrossRef.
- S. Ul Islam, Renewable Dyes and Pigments, Elsevier, 2024, pp. 253–269 Search PubMed.
- B. W. Meier, J. D. Gomez, O. V. Kirichenko and J. A. Thompson, Chem. Res. Toxicol., 2007, 20, 199–207 Search PubMed.
- X. Xu, A. Liu, S. Hu, I. Ares, M.-R. Martínez-Larrañaga, X. Wang, M. M.-A. Martínez, A. Anadón and M. M.-A. Martínez, Food Chem., 2021, 353, 129488 CrossRef CAS.
- J. Ham, W. Lim, S. You and G. Song, Sci. Total Environ., 2020, 702, 134775 CrossRef CAS.
- R. Liu and S. A. Mabury, Environ. Sci. Technol., 2020, 54, 11706–11719 CrossRef CAS.
- N. Kamemura, K. Oyama, K. Kanemaru, K. Yokoigawa and Y. Oyama, Toxicol. Res., 2017, 6, 922–929 CrossRef CAS PubMed.
- C. Yang, W. Lim, F. W. Bazer and G. Song, Food Chem. Toxicol., 2017, 109, 497–504 CrossRef CAS.
- A. Baran, M. Köktürk, M. Atamanalp and S. B. Ceyhun, Regul. Toxicol. Pharmacol., 2018, 94, 16–21 CrossRef CAS.
- I. P. Ejidike and P. A. Ajibade, Rev. Inorg. Chem., 2015, 35, 191–224 CAS.
- E. H. Anouar, S. Raweh, I. Bayach, M. Taha, M. S. Baharudin, F. Di Meo, M. H. Hasan, A. Adam, N. H. Ismail, J.-F. F. Weber and P. Trouillas, J. Comput.-Aided Mol. Des., 2013, 27, 951–964 CrossRef CAS PubMed.
- S. Nagar, S. Raizada and N. Tripathee, Results Chem., 2023, 6, 101153 CrossRef CAS.
- M. J. Kareem, A. A. S. Al-Hamdani, Y. G. Ko, W. Al Zoubi and S. G. Mohammed, J. Mol. Struct., 2021, 1231, 129669 CrossRef CAS.
- M. A. Said, D. L. Hughes, M. Al-Mamary, N. S. Al-Kaff and W. S. Al-Harbi, J. Mol. Struct., 2018, 1165, 305–311 CrossRef CAS.
- C. Y. Lee, A. Sharma, J. Semenya, C. Anamoah, K. N. Chapman and V. Barone, Antioxidants, 2020, 9, 189 CrossRef CAS.
- H. Keypour, M. Aidi, M. Mahmoudabadi, R. Karamian, M. Asadbegy and R. W. Gable, J. Mol. Struct., 2019, 1198, 126666 CrossRef CAS.
- J. H. Kim, H. J. Jang, W. Y. Cho, S. J. Yeon and C. H. Lee, Arabian J. Chem., 2020, 13, 1678–1684 CrossRef CAS.
- F. Sonmez, Z. Gunesli, B. Z. Kurt, I. Gazioglu, D. Avci and M. Kucukislamoglu, Mol. Diversity, 2019, 23, 829–844 CrossRef CAS.
- K. Buldurun, N. Turan, E. Bursal, A. Mantarcı, F. Turkan, P. Taslimi and İ. Gülçin, Res. Chem. Intermed., 2020, 46, 283–297 CrossRef CAS.
- M. Kumar, T. Padmini and K. Ponnuvel, J. Saudi Chem. Soc., 2017, 21, S322–S328 CrossRef CAS.
- D. K. Beena and D. S. Rawat, Bioorg. Med. Chem. Lett., 2013, 23, 641–645 CrossRef CAS PubMed.
- S. Baliyan, R. Mukherjee, A. Priyadarshini, A. Vibhuti, A. Gupta, R. P. Pandey and C.-M. Chang, Molecules, 2022, 27, 1326 CrossRef CAS.
- S. B. Kedare and R. P. Singh, J. Food Sci. Technol., 2011, 48, 412–422 CrossRef CAS.
- L. K. MacDonald-Wicks, L. G. Wood and M. L. Garg, J. Sci. Food Agric., 2006, 86, 2046–2056 CrossRef CAS.
- O. P. Sharma and T. K. Bhat, Food Chem., 2009, 113, 1202–1205 CrossRef CAS.
- M. Bashir, A. A. Dar and I. Yousuf, ACS Omega, 2023, 8, 3026–3042 CrossRef CAS PubMed.
- N. Patel, A. K. Prajapati, R. N. Jadeja, R. N. Patel, S. K. Patel, I. P. Tripathi, N. Dwivedi, V. K. Gupta and R. J. Butcher, Polyhedron, 2020, 180, 114434 CrossRef CAS.
- O. A. EL-Gammal, H. Alshater and H. A. El-Boraey, J. Mol. Struct., 2019, 1195, 220–230 CrossRef CAS.
- M. Yadav, S. Sharma and J. Devi, J. Chem. Sci., 2021, 133, 1–22 CrossRef.
- N. M. Abdalsahib and L. K. A. Karem, Int. J. Drug Delivery Technol., 2023, 13, 290–296 CrossRef.
- H. A. El-Boraey, O. A. El-Gammal and N. G. Abdel Sattar, J. Radioanal. Nucl. Chem., 2020, 323, 241–252 CrossRef CAS.
- M. Ikram, S. Rehman, A. Khan, R. J. Baker, T. S. Hofer, F. Subhan, M. Qayum, Faridoon and C. Schulzke, Inorg. Chim. Acta, 2015, 428, 117–126 CrossRef CAS.
- L. Liu, M. S. Alam and D. U. Lee, Bull. Korean Chem. Soc., 2012, 33, 3361–3367 CrossRef CAS.
- P. Barwa, S. Asija, Y. Deswal, N. Kumar, A. Kumar and J. Devi, Res. Chem. Intermed., 2023, 49, 3411–3440 CrossRef CAS.
- M. Aslam, I. Anis, N. Afza, M. T. Hussain, R. Mehmood, A. Hussain, S. Yousuf, L. Iqbal, S. Iqbal and I. Khan, J. Chil. Chem. Soc., 2013, 58, 1867–1871 CrossRef CAS.
- J. López-García, M. Lehocký, P. Humpolíček and P. Sáha, J. Funct. Biomater., 2014, 5, 43–57 CrossRef.
- I. V. Smolyaninov, A. I. Poddel'sky, D. A. Burmistrova, Y. K. Voronina, N. P. Pomortseva, M. A. Polovinkina, N. R. Almyasheva, M. A. Zamkova, N. T. Berberova and I. L. Eremenko, Int. J. Mol. Sci., 2023, 24, 8319 CrossRef CAS.
- J. Devi, S. Kumar, B. Kumar, S. Asija and A. Kumar, Res. Chem. Intermed., 2022, 48, 1541–1576 CrossRef CAS.
- B. Kumar, J. Devi and A. Manuja, Res. Chem. Intermed., 2023, 49, 2455–2493 CrossRef CAS.
- J. Priya and D. Madheswari, J. Biosci., 2022, 47, 29 CrossRef CAS PubMed.
- L. H. Abdel-Rahman, M. S. S. Adam, N. Al-Zaqri, M. R. Shehata, H. El-Sayed Ahmed and S. K. Mohamed, Arabian J. Chem., 2022, 15, 103737 CrossRef CAS.
- I. Rama and R. Selvameena, J. Indian Chem. Soc., 2020, 97, 2144–2154 CAS.
- H. Keypour, N. Ansari, M. Mahmoudabadi, R. Karamian, S. H. Moazzami Farida, M. Eslami Moghadam and R. William Gable, Inorg. Chim. Acta, 2020, 509, 119705 CrossRef CAS.
- I. Bougossa, D. Aggoun, A. Ourari, R. Berenguer, S. Bouacida and E. Morallon, Chem. Pap., 2020, 74, 3825–3837 CrossRef CAS.
- A. İnan, A. B. Sünbül, M. İkiz, S. E. Tayhan, S. Bilgin, M. Elmastaş, K. Sayın, G. Ceyhan, M. Köse and E. İspir, J. Organomet. Chem., 2018, 870, 76–89 CrossRef.
- B. Iftikhar, K. Javed, M. S. U. Khan, Z. Akhter, B. Mirza and V. Mckee, J. Mol. Struct., 2018, 1155, 337–348 CrossRef CAS.
- K. Sampath and C. Jayabalakrishnan, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2015, 45, 1145–1153 CrossRef CAS.
- S. Sathiyaraj, K. Sampath, R. J. Butcher and C. Jayabalakrishnan, Transition Met. Chem., 2013, 38, 291–298 CrossRef CAS.
- J. I. Al-Hawarin, A.-A. Abu-Yamin, A. A.-A. A. Abu-Saleh, I. A. M. Saraireh, M. H. Almatarneh, M. Hasan, O. M. Atrooz and Y. Al-Douri, Materials, 2023, 16, 5160 CrossRef CAS PubMed.
- A.-A. Abu-Yamin, M. S. Abduh, S. A. M. Saghir and N. Al-Gabri, Pharmaceuticals, 2022, 15, 454 CrossRef CAS.
- R. R. Pillai, K. Karrouchi, S. Fettach, S. S. J. Armaković, S. S. J. Armaković, Y. Brik, J. Taoufik, S. Radi, M. El Abbes Faouzi and M. Ansar, J. Mol. Struct., 2019, 1177, 47–54 CrossRef CAS.
- M. Bheemarasetti, K. Palakuri, S. Raj, P. Saudagar, D. Gandamalla, N. R. Yellu and L. R. Kotha, J. Iran. Chem. Soc., 2018, 15, 1377–1389 CrossRef CAS.
- B. B. Sokmen, N. Gumrukcuoglu, S. Ugras, H. I. Ugras and R. Yanardag, J. Enzyme Inhib. Med. Chem., 2013, 28, 72–77 CrossRef CAS.
- K. Brodowska, A. Sykuła, E. Garribba, E. Łodyga-Chruścińska and M. Sójka, Transition Met. Chem., 2016, 41, 179–189 CrossRef CAS.
- D.-C. Ilies, S. Shova, V. Radulescu, E. Pahontu and T. Rosu, Polyhedron, 2015, 97, 157–166 CrossRef CAS.
- R. Manikandan, P. Viswanathamurthi, K. Velmurugan, R. Nandhakumar, T. Hashimoto and A. Endo, J. Photochem. Photobiol., B, 2014, 130, 205–216 CrossRef CAS.
- G. M. Abu El-Reash, O. A. El-Gammal and A. H. Radwan, Spectrochim. Acta, Part A, 2014, 121, 259–267 CrossRef CAS.
- S. A. Aly and S. K. Fathalla, Arabian J. Chem., 2020, 13, 3735–3750 CrossRef CAS.
- W. Jamil, S. Solangi, M. Ali, K. M. Khan, M. Taha and M. Y. Khuhawar, Arabian J. Chem., 2019, 12, 2262–2269 CrossRef CAS.
- A. G. Bharathi Dileepan, T. Daniel Prakash, A. Ganesh Kumar, P. Shameela Rajam, V. Violet Dhayabaran and R. Rajaram, J. Photochem. Photobiol., B, 2018, 183, 191–200 CrossRef CAS PubMed.
- S. Aytac, O. Gundogdu, Z. Bingol and İ. Gulcin, Pharmaceutics, 2023, 15, 779 CrossRef CAS PubMed.
- R. O. Awolope, I. P. Ejidike and H. S. Clayton, J. Appl. Pharm. Sci., 2022, 13, 132–140 Search PubMed.
- H. M. Alkahtani, A. A. Almehizia, M. A. Al-Omar, A. J. Obaidullah, A. A. Zen, A. S. Hassan and W. M. Aboulthana, Molecules, 2023, 28, 7125 CrossRef CAS PubMed.
- N. Lolak, M. Boga, G. D. Sonmez, M. Tuneg, A. Dogan and S. Akocak, Pharm. Chem. J., 2022, 55, 1338–1344 CrossRef CAS.
- S. S. Hassan and P. A. Khalf-Alla, Appl. Organomet. Chem., 2020, 34, e5432 CrossRef CAS.
- H. Kizilkaya, B. Dag, T. Aral, N. Genc and R. Erenler, J. Chin. Chem. Soc., 2020, 67, 1696–1701 CrossRef CAS.
- R. Teran, R. Guevara, J. Mora, L. Dobronski, O. Barreiro-Costa, T. Beske, J. Pérez-Barrera, R. Araya-Maturana, P. Rojas-Silva, A. Poveda and J. Heredia-Moya, Molecules, 2019, 24, 2696 CrossRef CAS PubMed.
- S. Sathiyaraj, K. Sampath, R. J. Butcher, R. Pallepogu and C. Jayabalakrishnan, Eur. J. Med. Chem., 2013, 64, 81–89 CrossRef CAS.
- W. Al Zoubi, A. A. S. Al-Hamdani, S. Duraid Ahmed, H. M. Basheer, R. S. A. Al-Luhaibi, A. Dib and Y. G. Ko, J. Phys. Org. Chem., 2019, 32, e3916 CrossRef.
- I. V. Kulakov, I. V Palamarchuk, Z. T. Shulgau, T. M. Seilkhanov, Y. V Gatilov and A. S. Fisyuk, J. Mol. Struct., 2018, 1166, 262–269 CrossRef CAS.
- A. S. Alturiqi, A. M. A. Alaghaz and R. A. Ammar, J. Chin. Chem. Soc., 2017, 64, 1270–1285 CrossRef CAS.
- F. Lemilemu, M. Bitew, T. B. Demissie, R. Eswaramoorthy and M. Endale, BMC Chem., 2021, 15, 67 CrossRef CAS PubMed.
- M. M. Azaam, E.-R. Kenawy, A. S. B. El-din, A. A. Khamis and M. A. El-Magd, J. Saudi Chem. Soc., 2018, 22, 34–41 CrossRef CAS.
- I. Elaaraj, N. Moukrad, A. Bouymajane, S. Er Raouan, A. Nakkabi, O. Oulidi, F. Rhazi Filai, I. Koraichi Saad, F. Cacciola, N. El Moualij and M. Fahim, Results Chem., 2023, 5, 100787 CrossRef CAS.
- K. Subin Kumar, Results Chem., 2022, 4, 100463 CrossRef CAS.
- N. Maghraoui, D. Aggoun, B. Bouzerafa, H. Bezzi, Y. Ouennoughi, D. López, M. Fernández García, A. Ourari and M. S. Mubarak, Arabian J. Chem., 2021, 14, 103025 CrossRef CAS.
- M. Shebl, J. Coord. Chem., 2016, 69, 199–214 CrossRef CAS.
- K. Mahendra Raj, B. Vivekanand, G. Y. Nagesh and B. H. M. Mruthyunjayaswamy, J. Mol. Struct., 2014, 1059, 280–293 CrossRef CAS.
- N. El-wakiel, M. El-keiy and M. Gaber, Spectrochim. Acta, Part A, 2015, 147, 117–123 CrossRef CAS.
- N. Turan, K. Buldurun, F. Türkan, A. Aras, N. Çolak, M. Murahari, E. Bursal and A. Mantarcı, Mol. Diversity, 2022, 26, 2459–2472 CrossRef CAS.
- E. Bursal, F. Turkan, K. Buldurun, N. Turan, A. Aras, N. Çolak, M. Murahari and M. C. Yergeri, BioMetals, 2021, 34, 393–406 CrossRef CAS.
- A. Mermer, N. Demirbas, H. Uslu, A. Demirbas, S. Ceylan and Y. Sirin, J. Mol. Struct., 2019, 1181, 412–422 CrossRef CAS.
- M. N. Uddin, S. Khandaker, Moniruzzaman, Md. S. Amin, W. Shumi, Md. A. Rahman and S. M. Rahman, J. Mol. Struct., 2018, 1166, 79–90 CrossRef CAS.
- B. J. Okoli and J. S. Modise, Antioxidants, 2018, 7, 113 CrossRef.
| This journal is © The Royal Society of Chemistry 2024 |