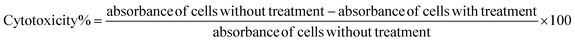Open Access Article
Open Access ArticleAntiviral activity of pyrazole derivatives bearing a hydroxyquinoline scaffold against SARS-CoV-2, HCoV-229E, MERS-CoV, and IBV propagation†
Alaa R. I. Morsy‡
a,
Sara H. Mahmoud§‡
b,
Noura M. Abou Shamab,
Walaa Arafac,
Gehad A. Yousefa,
Ahmed. A. Khalil*d and
Sayed K. Ramadan *e
*e
aCentral Laboratory for Evaluation of Veterinary Biologics (CLEVB), Agricultural Research Center, Cairo, Egypt
bCenter of Scientific Excellence for Influenza Viruses, National Research Centre (NRC), Egypt
cMicrobiology Department, Faculty of Agriculture, Cairo University, Egypt
dVeterinary Sera and Vaccines Research Institute (VSVRI), Agricultural Research Center (ARC), Cairo, Egypt. E-mail: ahme_2001@hotmail.com
eChemistry Department, Faculty of Science, Ain Shams University, Cairo, 11566, Egypt. E-mail: sayed.karam2008@sci.asu.edu.eg
First published on 2nd September 2024
Abstract
The ongoing global threat posed by coronaviruses necessitates the urgent development of effective antiviral agents. In this study, we investigated the potential of hydroxyquinoline-pyrazole candidates as antiviral agents against a range of coronaviruses, including SARS-CoV-2, MERS-CoV, and HCoV-229E. Molecular docking studies were conducted to assess the binding affinity of the synthesized compounds to key viral proteins. The compounds were prepared via condensation reactions of a pyrazolylhydrazide derivative with 2-chloro-3-formylquinoline, yielding hydrazone and pyrrolone derivatives. The cytotoxicity of compounds was evaluated using Vero E6 cells, and their antiviral activity was assessed via plaque reduction assays and viral inhibition assays using hydroxychloroquine as a positive control antiviral drug. The results revealed promising antiviral activity of the synthesized compounds against all tested coronaviruses, with selectivity indices indicating their potential as selective antiviral agents. Notably, the compounds exhibited potent inhibition of SARS-CoV-2 at lower concentrations, highlighting their promise as therapeutic candidates against this highly pathogenic virus. Likewise, the modeling pharmacokinetics approach showed its appropriate drug-likeness and bioavailability assets. These findings underscore the importance of hydroxyquinoline-pyrazole derivatives as potential antiviral agents against diverse coronaviruses, providing valuable insights for further therapeutic development.
Introduction
The Coronavirus (CoVs) family recently became well-known pathogens that can cause severe respiratory diseases with varying incidences around the globe. Among them, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) that causes COVID-19, has brought the world to the overwhelming pandemic level and caused significant socio-economic losses. While the novel coronavirus's outbreak has been somewhat curbed through vaccination and public health measures, the demand for successful antiviral medications is still relevant, especially given the further appearance of new strains and likelihood of other zoonotic spills.1,2Due to the emergence of the coronaviruses and subsequent severe acute respiratory syndrome (SARS) and Covid-19, the necessity of effective and potent antiviral agents targeting the viral proteins that are vital for the replication and pathogenesis of the virus has surged.3,4 In this respect, heterocyclic compounds have attracted much interest because of the numerous biological activities and action they exhibit as antiviral substances.5,6 Among these, the pyrazole moiety is known to be useful in drug design because it also provides several synthetic approaches and structural variations for the improvement of the pharmacological profile of the target drug.7–9 Also, the representatives of hydroxyquinoline derivatives have become the object of interest because of their activity against a wide range of viruses, tumors, and suitable pharmacokinetic properties.10,11
New research studies have been carried out regarding the antiviral activity of hydroxyquinoline and pyrazole derivatives. For example, Saadeh et al. described the recent findings on the synthesis and biological activities of 8-hydroxyquinolines as antiviral agents with a wide spectrum.12 Likewise, pyrazole derivatives are effective molecules in the fight against many viral diseases caused by viruses belonging to different families, including coronavirus. Seliem et al. synthesized pyrazolothiazole conjugates and screened them through virtual methods, and thus they showed that pyrazole class compounds have potential for hits on viral proteins of SARS-CoV-2.13
In addition, the incorporation of various heterocyclic fragments is a recent approach to the improvement of biological activity.14–16 Kandinska et al. prepared tetrahydroisoquinoline derivatives containing other heterocycles and a comparative evaluation for anti-coronavirus activity was investigated.17 This approach indicates the use of a variety of pharmacophores which is parallel to our concept of selecting both hydroxyquinoline and pyrazole core.
While several articles have been published on the pharmacological aspects of hydroxyquinoline-pyrazole derivatives, our work describes the first conceptual investigation on their antiviral profiles against coronaviruses: SARS-CoV-2 or Covid-19, MERS-CoV, and HCoV-229E. These compounds were characterized as antiviral based on the obtained SAR result from molecular modeling, synthetic and biological studies of the compounds and experiments to establish their mode of action.
Concerning the compound development and optimization of the identified leads, utilization of the computational tools based on structure–activity relationship in assist of compound optimization results in even improved antiviral efficacy and pharmacokinetic profile for further series of in vitro and in vivo studies. As a result of these findings, there are roles for a further study in vivo efficacy and toxicity in a further study in view of the modesty seen in this study. Also, it is possible to explore variations of the structure of the synthesized compounds to enhance their antiviral activity and selectivity.
Outcomes of the current research serve as the foundation for the other studies towards the investigation of the effective antiviral treatment towards coronaviruses, which is a key priority of today's healthcare systems. Thus, hydroxyquinoline-pyrazole derivatives can be regarded as a basis for the series of the new generation of multivalent antiviral agents in fighting against the new coronavirus and other similar viral infections in the future.
Materials and methods
Chemistry
Melting points were measured on a Gallenkamp electric melting point apparatus (SANYO GALLENKAMP, UK) and are uncorrected. The reactions were monitored by thin-layer chromatography (TLC) using precoated Merck Kiesel gel 60F254 aluminum backed plates obtained from Fluka, Switzerland. All reagents and solvents were purified and dried by standard techniques. The infrared spectra were recorded using potassium bromide disks on FTIR Thermo Electron Nicolet iS10 (USA) infrared spectrometer and expressed in wave number (ν, cm−1) at Faculty of Science, Ain Shams University. The 13C and 1H NMR spectra were run at 100 and 400 MHz on a Bruker NMR spectrometer (Bruker, Manufacturing & Engineering Inc., Anaheim, CA, USA) using TMS as an internal standard in DMSO at Faculty of Pharmacy, Cairo University. Chemical shifts (δ) are quoted in ppm. Mass spectrum was carried out on direct probe controller inlet part to single quadrupole mass analyzer in Thermo Scientific GCMS, MODEL ISQLT operating at 70 eV using Thermo X-Calibur software at the regional center for mycology and biotechnology (RCMB), Al-Azhar University, Cairo, Egypt. The starting pyrazolylhydrazide derivative (Hyd) was previously reported.7Condensation of the hydrazide (Hyd) with 2-chloro-3-formylquinoline
A solution of the pyrazolylhydrazide derivative (Hyd) (2.11 g, 5 mmol), 2-chloro-3-formylquinoline (0.95 g, 5 mmol) in 1,4-dioxane (20 mL) was refluxed for 30 min. The precipitated solid while heating was filtered off and recrystallized from 1,4-dioxane to produce the hydrazone derivative (HQ). When the reaction mixture was refluxed in an absolute ethyl alcohol (20 mL) including glacial acetic acid (0.1 mL) for 1 h, the precipitated solid while heating was filtered and recrystallized from ethanol/1,4-dioxane mixture (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford the pyrrolone derivative (15A).
1) to afford the pyrrolone derivative (15A).
2-((1,3-Diphenyl-1H-pyrazol-4-yl)methylene)-N′-((2-hydroxyquinolin-3-yl)methylene)-4-oxo-4-phenylbutanehydrazide (HQ)8
Yellow crystals, mp. 330–332 °C [(ref. 8) mp. 330–332 °C], yield 77%. FTIR (ν, cm−1): 3422 (OH), 3161 (NH), 3059, 3007 (CH-aromatic), 2954, 2895 (CH-aliphatic), 1669 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1612 (C
O), 1612 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H NMR (400 MHz, DMSO-d6, δ, ppm): 12.15 (br. s, 2H, OH + NH, exchangeable), 9.14 (s, 1H, CH
N). 1H NMR (400 MHz, DMSO-d6, δ, ppm): 12.15 (br. s, 2H, OH + NH, exchangeable), 9.14 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.90 (s, 1H, C5–H pyrazole), 8.83 (s, 1H, C4–H quinoline), 8.24–8.22 (d, 1H, C5–H quinoline, J = 8 Hz), 8.00–7.98 (m, 3H, quinoline-H), 7.87–7.36 (m, 15H, aryl-H), 7.22 (s, 1H, CH
N), 8.90 (s, 1H, C5–H pyrazole), 8.83 (s, 1H, C4–H quinoline), 8.24–8.22 (d, 1H, C5–H quinoline, J = 8 Hz), 8.00–7.98 (m, 3H, quinoline-H), 7.87–7.36 (m, 15H, aryl-H), 7.22 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 3.54 (s, 2H, CH2). 13C NMR (100 MHz, DMSO, δ, ppm): 39.38 (CH2), 102.97, 104.53, 105.75, 107.48, 109.04, 110.60, 111.81, 112.51, 115.98, 118.92, 120.66, 123.26, 125.16, 126.72, 127.76, 130.19, 132.44, 133.14, 135.91, 137.99, 140.59, 142.50, 145.45, 147.70, 149.61 (C
), 3.54 (s, 2H, CH2). 13C NMR (100 MHz, DMSO, δ, ppm): 39.38 (CH2), 102.97, 104.53, 105.75, 107.48, 109.04, 110.60, 111.81, 112.51, 115.98, 118.92, 120.66, 123.26, 125.16, 126.72, 127.76, 130.19, 132.44, 133.14, 135.91, 137.99, 140.59, 142.50, 145.45, 147.70, 149.61 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 167.81 (N–C
N), 167.81 (N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 177.69 (N
O), 177.69 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH), 196.76 (C
C–OH), 196.76 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ketone). EIMS (70 eV, m/z, %): 577.04 (M+, 26.93), 541.32 (40.14), 507.46 (54.70), 483.74 (44.54), 441.67 (43.34), 411.03 (61.81), 395.83 (51.65), 376.67 (43.89), 362.02 (71.87), 348.17 (37.34), 330.01 (52.15), 308.73 (75.58), 301.27 (56.86), 217.16 (51.15), 201.28 (32.73), 177.89 (53.35), 159.40 (53.10), 140.81 (40.89), 137.26 (100.00), 130.66 (95.25), 126.75 (51.75), 113.68 (38.19), 91.72 (63.36), 74.51 (59.51). Anal. calcd for C36H27N5O3 (577.64): C, 74.86; H, 4.71; N, 12.12; found: C, 74.71; H, 4.60; N, 12.09%.
O ketone). EIMS (70 eV, m/z, %): 577.04 (M+, 26.93), 541.32 (40.14), 507.46 (54.70), 483.74 (44.54), 441.67 (43.34), 411.03 (61.81), 395.83 (51.65), 376.67 (43.89), 362.02 (71.87), 348.17 (37.34), 330.01 (52.15), 308.73 (75.58), 301.27 (56.86), 217.16 (51.15), 201.28 (32.73), 177.89 (53.35), 159.40 (53.10), 140.81 (40.89), 137.26 (100.00), 130.66 (95.25), 126.75 (51.75), 113.68 (38.19), 91.72 (63.36), 74.51 (59.51). Anal. calcd for C36H27N5O3 (577.64): C, 74.86; H, 4.71; N, 12.12; found: C, 74.71; H, 4.60; N, 12.09%.
3-((1,3-Diphenyl-1H-pyrazol-4-yl)methylene)-1-(((2-hydroxyquinolin-3-yl)methylene)amino)-5-phenyl-1,3-dihydro-2H-pyrrol-2-one (15A)8
Orange crystals, mp. 237–239 °C [(ref. 8) mp. 237–239 °C], yield 80%. FTIR (ν, cm−1): 3442 (OH), 3058 (CH-aromatic), 1699 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1617 (C
O), 1617 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H NMR (400 MHz, DMSO-d6, δ, ppm): 12.13 (s, 1H, OH, exchangeable), 9.33 (s, 1H, CH
N). 1H NMR (400 MHz, DMSO-d6, δ, ppm): 12.13 (s, 1H, OH, exchangeable), 9.33 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 9.25 (s, 1H, C5–H pyrazole), 8.89 (s, 1H, C4–H quinoline), 8.08–8.06 (d, 1H, C5–H quinoline, J = 8 Hz), 7.96–7.82 (m, 3H, quinoline-H), 7.75–7.38 (m, 15H, aryl-H), 7.23 (s, 1H, CH
N), 9.25 (s, 1H, C5–H pyrazole), 8.89 (s, 1H, C4–H quinoline), 8.08–8.06 (d, 1H, C5–H quinoline, J = 8 Hz), 7.96–7.82 (m, 3H, quinoline-H), 7.75–7.38 (m, 15H, aryl-H), 7.23 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 7.00 (s, 1H, C4–H pyrrole). 13C NMR (100 MHz, DMSO, δ, ppm): 100.78, 105.92, 108.72, 113.37, 115.38, 117.13, 119.51, 119.84, 121.92, 124.60, 126.47, 127.75, 128.74, 128.85, 128.91, 129.16, 129.31, 129.40, 130.04, 132.30, 139.43, 142.94, 145.74, 149.47, 151.46, 167.64 (C
), 7.00 (s, 1H, C4–H pyrrole). 13C NMR (100 MHz, DMSO, δ, ppm): 100.78, 105.92, 108.72, 113.37, 115.38, 117.13, 119.51, 119.84, 121.92, 124.60, 126.47, 127.75, 128.74, 128.85, 128.91, 129.16, 129.31, 129.40, 130.04, 132.30, 139.43, 142.94, 145.74, 149.47, 151.46, 167.64 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 177.69 (N
O), 177.69 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH). EIMS (70 eV, m/z, %): 559.91 (M+, 49.94), 556.79 (28.44), 548.86 (32.66), 544.26 (48.26), 541.51 (61.02), 534.86 (41.66), 512.23 (33.42), 500.07 (28.63), 486.94 (66.73), 481.86 (67.08), 474.31 (73.97), 469.29 (55.88), 459.23 (55.88), 452.99 (66.46), 447.17 (54.70), 436.17 (98.43), 416.88 (100.00), 406.74 (69.07), 391.52 (31.81), 378.58 (50.90), 367.16 (37.98), 365.82 (40.78), 353.80 (56.42), 347.06 (49.75), 330.68 (35.30), 322.21 (28.36). Anal. calcd for C36H25N5O2 (559.63): C, 77.26; H, 4.50; N, 12.51; found: C, 77.14; H, 4.38; N, 12.53%.
–OH). EIMS (70 eV, m/z, %): 559.91 (M+, 49.94), 556.79 (28.44), 548.86 (32.66), 544.26 (48.26), 541.51 (61.02), 534.86 (41.66), 512.23 (33.42), 500.07 (28.63), 486.94 (66.73), 481.86 (67.08), 474.31 (73.97), 469.29 (55.88), 459.23 (55.88), 452.99 (66.46), 447.17 (54.70), 436.17 (98.43), 416.88 (100.00), 406.74 (69.07), 391.52 (31.81), 378.58 (50.90), 367.16 (37.98), 365.82 (40.78), 353.80 (56.42), 347.06 (49.75), 330.68 (35.30), 322.21 (28.36). Anal. calcd for C36H25N5O2 (559.63): C, 77.26; H, 4.50; N, 12.51; found: C, 77.14; H, 4.38; N, 12.53%.
ADME analysis
The absorption, distribution, metabolism, and excretion (ADME) properties of the synthesized compounds were evaluated using the SwissADME web tool (http://www.swissadme.ch/). This free online platform allows for the prediction of physicochemical properties, lipophilicity, water solubility, pharmacokinetics, drug-likeness, and medicinal chemistry friendliness of small molecules. The following key parameters were assessed:(1) Physicochemical properties: molecular weight, number of heavy atoms, number of aromatic heavy atoms, fraction of carbons in sp3 hybridization, number of rotatable bonds, number of H-bond acceptors and donors, molar refractivity, and topological polar surface area (TPSA).
(2) Lipophilicity: consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w was calculated using five predictive models (XLOGP3, WLOGP, MLOGP, SILICOS-IT, and iLOGP).
Po/w was calculated using five predictive models (XLOGP3, WLOGP, MLOGP, SILICOS-IT, and iLOGP).
(3) Water solubility: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values were predicted using three models (ESOL, Ali, and SILICOS-IT).
S values were predicted using three models (ESOL, Ali, and SILICOS-IT).
(4) Pharmacokinetics: gastrointestinal (GI) absorption, blood–brain barrier (BBB) permeation, P-glycoprotein substrate, and interaction with cytochrome P450 isoenzymes (CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4) were evaluated.
(5) Drug-likeness: compliance with Lipinski's rule of five, Ghose, Veber, Egan, and Muegge rules was assessed.
(6) Medicinal chemistry: PAINS (Pan-Assay Interference Compounds) and Brenk alerts, along with lead-likeness were determined.
The results were interpreted using the BOILED-Egg model, which provides a graphical representation of the compounds' gastrointestinal absorption and brain penetration. Additionally, the bioavailability radar chart was used to assess the drug-likeness of the compounds based on six physicochemical properties: lipophilicity (LIPO), size, polarity (POLAR), solubility (INSOLU), flexibility (FLEX), and saturation (INSATU).
The significance of these parameters lies in their ability to predict the compounds' behavior in biological systems, their potential as drug candidates, and their likelihood of success in further development stages. This analysis provides crucial information for prioritizing compounds and guiding future optimization efforts in the drug discovery process.
Molecular modeling studies
Molecular docking study
Molecular docking studies of the compounds were performed using Molecular Operating Environment software. All minimizations were carried out until an RMSD gradient of 0.05 kcal mol−1 Å−2 with MMFF94x force field and the partial charges were automatically calculated. The structure of Proteins was downloaded from the Protein Data Bank. For docking study, the protein was prepared by Protonate 3D protocol with default options and water molecules removal. The co-crystalized ligand was used to determine the active site for docking. London dG and Triangle Matcher placement scoring function were used for docking study.In vitro methods
Viruses and cells
The SARS-CoV-2 strain (GISAID number: EPI_ISL_430819)18 from the CoV-19/Egypt/NRC-03/2020, the MERS-CoV strain NRCE-HKU270 (Genbank accession: KJ477103.2),19 and HCoV-229E were cultured and amplified in Vero E6 cells (ATCC CRL-1587) until the observation of cytopathic effects (CPE).20 The Vero E6 cells were obtained from American Type Culture Collection (ATCC). Subsequently, the viruses were quantified using TCID50 (50% tissue culture infectious dose), following established protocols.21,22 Hydroxychloroquine was used as a positive control antiviral drug.Cytotoxicity determination (CC50)
Three synthetic compounds were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich) at a concentration of 10 mg mL−1. Cytotoxic activity was tested in Vero-E6 cells using a crystal violet assay as previously described22 with minor modifications. Briefly, the cells were seeded in 96-well plates (100 μL per well at a density of 3 ×105 cells per mL) and incubated for 24 h at 37 °C in 5% CO2. After 24 h, the cells were treated with various concentrations of the compound in triplicate. At 72 h post-treatment, the supernatant was discarded, and cell monolayers were fixed with 10% formaldehyde for 1 h at room temperature (RT). The fixed monolayers were then dried thoroughly and stained with 50 μL of 0.1% crystal violet for 20 min on a bench rocker at room temperature. The monolayers were then washed, dried overnight, and the crystal violet dye in each well was dissolved in 200 μL methanol for 20 min on a bench rocker at room temperature. The absorbance of the crystal violet solutions was measured at λmax 570 nm as a reference wavelength using a multi-well plate reader. The CC50 value was calculated using nonlinear regression analysis using GraphPad Prism software (version 5.01) by plotting log concentrations of the compound versus normalized response (variable slope).The concentration that displayed 50% cytotoxicity (CC50) was calculated using a plot of percent cytotoxicity versus sample concentration.23
Inhibitory concentration determination (IC50)
The IC50 values for the compounds were determined as previously described,24 with slight modifications. 2.4 × 104 Vero-E6 cells were placed in each well of 96-well tissue culture plates and cultured overnight at 37 °C in a humidified 5% CO2 incubator. After that, the cell monolayers were rinsed once in 1× PBS. An aliquot of the SARS-CoV-2, MERS-CoV and HCoV-229E viruses25 containing 100 TCID50 were incubated with serially diluted concentrations of the tested compound and kept at 37 °C for 1 h. Another set of Vero-E6 cells were treated with virus/compound mix and co-incubated at 37 °C in a total volume of 200 μL per well. Untreated cells infected with a virus represent virus control; however, cells that have not been treated and have not been infected represent cell control. The cells were fixed with 100 μL of 10% paraformaldehyde for 20 min and stained with 0.5% crystal violet in distilled water for 15 min at room temperature after being incubated for 72 h at 37 °C in a 5% CO2 incubator. After that, 100 μL absolute methanol per well was used to dissolve the crystal violet dye and the optical density of the color was measured at 570 nm using Anthos Zenyth 200rt plate reader (Anthos Labtec Instruments, Heerhugowaard, Netherlands). The IC50 of a compound is required to reduce the virus-induced cytopathic effect (CPE) by 50%, relative to the virus control. The IC50 value was calculated using nonlinear regression analysis of GraphPad Prism software (version 5.01) by plotting log concentrations of the compounds versus normalized response (variable slope).In vitro mechanism of virus inhibition
The antiviral activity of the three synthesized compounds (hydrazide Hyd, hydrazone HQ, and pyrrolone 15A) was evaluated against SARS-CoV-2, MERS-CoV, and HCoV-229E using dose-dependent viral inhibition assays. These assays were conducted following previously established methods, with minor modifications to optimize for our specific experimental conditions.25Viral replication
Viral replication inhibition was assessed to determine the compound's ability to interfere with the coronavirus life cycle within host cells, viral dilutions of SARS-CoV-2 virus were allowed to infect Vero-E6 cells cultured at a density of 1.2 × 106 cells per well in a 6-well plate for 24 h. Each plate contains cell and virus control wells to ensure the validity of the assay and to calculate the percent of viral inhibition following treatment, respectively. The plates were incubated at 37 °C in a humidified 5% CO2 incubator for 1 h. Cell monolayers were then washed with 1× PBS. Subsequently, the different predetermined non-cytotoxic concentrations of each compound were applied, and the plates underwent a second incubation period at 37 °C in a humidified 5% CO2 incubator for 1 h. Cells were washed again and the 2% agarose overlayers were added then the plates were incubated at 37 °C in a humidified 5% CO2 incubator for 72 h. The cell monolayers were then fixed and stained using 0.1% crystal violet solution as described previously in the plaque infectivity assay.26Viral adsorption
A range of non-cytotoxic concentrations were applied to the precultured Vero-E6 cells in 6 well plates (1.2 × 106 cells per well). Each plate contains cell and virus control wells to ensure the validity of the assay and to calculate the percent of viral inhibition following treatment, respectively. The plates were then incubated at 4 °C for 1 h to allow chemical adsorption onto cell receptors without active penetration. The plates were then washed with 1× PBS to remove the residual compounds. Subsequently, the countable viral dilution of SARS-CoV-2 virus was applied to allow viral adsorption/infection and another incubation period was employed at 37 °C in a humidified 5% CO2 incubator for 1 h. The cell monolayers were then washed with 1× PBS to remove the residual virus and overlayed with 2% agarose/2X DMEM overlay and incubated at 37 °C in a humidified 5% CO2 incubator for 72 h. The cell monolayers were then fixed and stained using 0.1% crystal violet solution as described previously in the plaque infectivity assay.27Virucidal effect
A simple plaque reduction assay was performed where effective concentrations of the compounds were mixed with concentrated SARS-CoV-2 virus (3–4 folds higher than the countable virus dilution). The virus/compound mixture was then incubated at room temperature for 1 h. Subsequently, ten-fold serial dilutions of the virus/compound mixture (3 or 4 times) were performed to reach a countable viral titer. The mixture dilution with countable viral titer was then applied to the Vero-E6 monolayers (1.2 × 106 cells per well) including cell and virus control wells. The plates were then incubated at 37 °C in a humidified 5% CO2 incubator for 1 h. To remove the remains of the mixture, cell monolayers were washed with 1× PBS and overlaid with 2% agarose/2X DMEM, and incubated at 37 °C in a humidified 5% CO2 incubator for 72 h. The cell monolayers were then fixed and stained, visualized using 0.1% crystal violet solution as described previously in the plaque infectivity assay.28Results
Compounds' synthesis
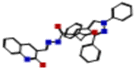
Our research was concerned with the design and construction of a wide variety of nitrogen containing heterocycles exhibiting synthetic and pharmacological importance. Noteworthy, pyrazoles have been reported as a new scaffold of good antiviral agents.8,29,30 Likewise, quinolines have attracted a tremendous attention due to their existence in some of well-known drugs.30–32 This broad spectrum of biological properties initiated our interest to examine the heterocyclic systems gathering pyrazole and quinoline scaffolds in one skeleton.Thus, the readily obtainable acid hydrazide Hyd7 was condensed with 2-chloro-3-formylquinoline under two different reaction media to produce the hydrazone HQ and pyrrolone 15A candidates encompassing each of pyrazole and quinoline scaffolds in spectacular yields (Fig. 1).8 Initially, refluxing of an equimolar mixture of pyrazolylhydrazide Hyd and the former quinoline aldehyde in 1,4-dioxane as a solvent afforded the corresponding hydrazone derivative HQ. On the other hand, when the reaction was executed in boiling ethanol containing glacial acetic acid, the pyrrolone derivative 15A was obtained as a sole product.
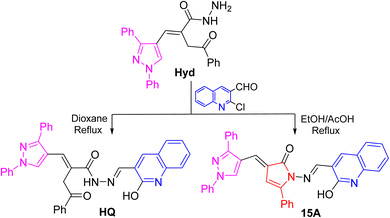 | ||
| Fig. 1 Synthesis of target antiviral agents, pyrazole-based quinoline derivatives (pyrazolylhydrazide Hyd, hydrazone HQ, and pyrrolone 15A). | ||
The chemical structures of all products were based on their analytical and spectroscopic data. The IR spectrum of quinolinylhydrazone HQ exhibited the characteristic stretching absorption bands for OH, NH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N groups at ν 3422, 3161, 1669, and 1612 cm−1, respectively. The 1H NMR spectrum of hydrazone HQ showed singlet signals for NH and CH2 protons which disappeared in that of pyrrolone 15A. Compelling evidence for the structures of compounds HQ and 15A were acquired from their 13C NMR spectra which provided the characteristic signals to these structures. Furthermore, their mass spectra exhibited the molecular ion peaks and some important abundant peaks.
N groups at ν 3422, 3161, 1669, and 1612 cm−1, respectively. The 1H NMR spectrum of hydrazone HQ showed singlet signals for NH and CH2 protons which disappeared in that of pyrrolone 15A. Compelling evidence for the structures of compounds HQ and 15A were acquired from their 13C NMR spectra which provided the characteristic signals to these structures. Furthermore, their mass spectra exhibited the molecular ion peaks and some important abundant peaks.
Formation of hydrazone HQ could be visualized to occur via elimination of water molecule followed by removal of chlorine under heating conditions. The formation of pyrrolone 15A could be explained via ring closure of hydrazone HQ. This behavior was interpreted via acid catalyzed 1,5-exo-trig cyclization. These compounds were evaluated for their antiviral activity and displayed promising potency as vide supra (100% protection against Newcastle Disease Virus).8
In silico molecular docking results
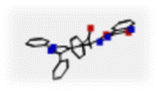
In the present study, four proteins were used for docking simulation, as follows: (i) PDB ID: 7YRZ which is the crystal structure of HCoV 229E main protease in complex with PF07321332 (Nirmatrelvir is an antiviral drug that acts as an orally active 3C-like protease inhibitor), (ii) PDB ID: 2Q6F which is the crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 (the coronavirus main protease (M(pro)), which plays a pivotal role in viral gene expression and replication through the proteolytic processing of replicase polyproteins), (iii) PDB ID: 6VGY which is the 2.05 Å resolution structure of MERS 3CL protease in complex with inhibitor 6b (hydrolase inhibitor), (iv) PDB ID: 6WUU which is the crystal structure of the SARS CoV-2 papain-like protease in complex with peptide inhibitor VIR250 (viral papain-like cysteine protease (PLpro, NSP3) is essential for SARS-CoV-2 replication and represents a promising target for the development of antiviral drugs). These proteins are used as a target to test the compounds (hydrazide Hyd, hydrazone HQ, and pyrrolone 15A).Fig. 2 illustrates the 2D diagrams validating the docking protocol by re-docking the co-crystallized ligand in the active site. The re-docking validation step successfully reproduced the experimental binding pattern of the co-crystallized ligand, confirming the suitability of the adapted docking protocol for this study. The results for each target protein are as follows:
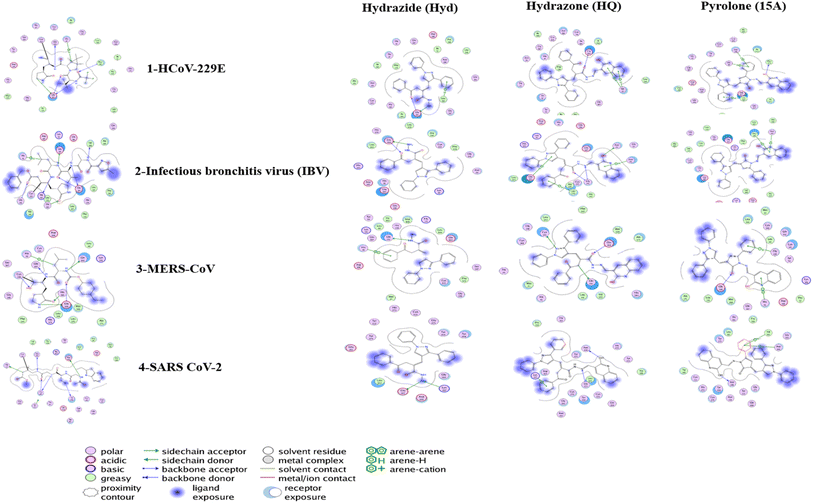 | ||
| Fig. 2 2D Diagrams showing validation of docking protocol, by docking of the co-crystallized ligand in the active site. | ||
(1) 7YRZ (HCoV 229E main protease): the docking pose showed a small RMSD of 1.139 Å compared to the co-crystallized ligand, with an energy score of −28.97 kcal mol−1. The docking pose reproduced key interactions with active site hotspots (Cys144, Gln163, Glu165, His41, and Pro188).
(2) 2Q6F (IBV main protease): the docking yielded a small RMSD of 1.09 Å and an energy score of −25.21 kcal mol−1. It accurately reproduced binding interactions with key amino acids (Val188, Glu187, His41, Gly141, His162, His161, and Glu164).
(3) 6VGY (MERS 3CL protease): the docking resulted in an RMSD of 1.929 Å and an energy score of −17.46 kcal mol−1, with binding to key hotspots (Cys148, Glu169, Gln192, Gln167, His41, and His166).
(4) 6WUU (SARS-CoV-2 papain-like protease): the docking produced an RMSD of 1.929 Å and an energy score of −22.87 kcal mol−1, showing binding interactions with Asp164, Gly163, Gly271, Cys111, His272, and Tyr168.
These results demonstrate the reliability of the docking protocol across all four target proteins. After the validation step, three compounds were tested against the four targets and produced high scores with binding poses that are represented in Tables 1–3.
| Structure | Virus | S | rmsd_refine | E_conf. | E_place | E_score1 | E_refine | E_score2 |
|---|---|---|---|---|---|---|---|---|
 |
229E | −20.49 | 2.12 | 131.57 | −81.40 | −33.49 | −20.22 | −20.49 |
 |
IBV | −14.33 | 1.99 | 137.69 | −43.44 | −9.36 | 70.99 | −14.33 |
 |
MERS | −21.31 | 1.27 | 128.01 | −73.42 | −25.59 | 15.99 | −21.31 |
 |
SARS | −27.61 | 2.13 | 130.15 | −83.94 | −26.41 | −40.95 | −27.61 |
| Structure | Virus | S | rmsd_refine | E_conf | E_place | E_score1 | E_refine | E_score2 |
|---|---|---|---|---|---|---|---|---|
 |
229E | −20.42 | 2.83 | 118.66 | −50.81 | 10.86 | 22.03 | −21.96 |
 |
IBV | −17.23 | 2.54 | 121.39 | −71.67 | −21.39 | −13.92 | −17.23 |
 |
MERS | −22.09 | 2.91 | 120.56 | −70.54 | −24.94 | −19.34 | −22.09 |
 |
SARS | −26.53 | 2.35 | 119.87 | −76.67 | −27.98 | −37.84 | −26.53 |
| Structure | Virus | S | rmsd_refine | E_conf | E_place | E_score1 | E_refine | E_score2 |
|---|---|---|---|---|---|---|---|---|
| a S (Score): the overall docking score (lower values indicate better binding). All energy values are in kcal mol−1. rmsd_refine: root mean square deviation after refinement E_conf: conformer energy. E_place: placement energy E_score1: initial scoring function energy. E_refine: refinement energy E_score2: final scoring function energy. | ||||||||
 |
229E | −25.75 | 3.95 | 158.57 | −69.03 | −20.80 | −23.16 | −25.75 |
 |
IBV | −19.65 | 2.56 | 157.27 | −85.14 | −26.71 | −24.76 | −19.65 |
 |
MERS | −20.16 | 1.62 | 178.04 | −77.59 | −10.42 | 40.05 | −20.99 |
 |
SARS | −20.84 | 1.67 | 198.40 | −3.40 | −25.18 | 227.71 | −20.13 |
In vitro results
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg mL−1 to 1 mg mL−1. Hydroxychloroquine was used as a positive control antiviral drug. The half-maximal cytotoxic concentration (CC50) values were determined, indicating the concentration at which 50% of cell viability was compromised. For all three viruses, the compounds exhibited varying degrees of inhibitory effects, as reflected by the half-maximal inhibitory concentration (IC50) values. Selectivity indices (SI) were calculated by dividing CC50 by the corresponding IC50 for each virus. Notably, these compounds demonstrated promising selectivity against SARS-CoV-2 at lower concentrations, with increasing SI values (cf. Table 4 and Fig. 3).
000 mg mL−1 to 1 mg mL−1. Hydroxychloroquine was used as a positive control antiviral drug. The half-maximal cytotoxic concentration (CC50) values were determined, indicating the concentration at which 50% of cell viability was compromised. For all three viruses, the compounds exhibited varying degrees of inhibitory effects, as reflected by the half-maximal inhibitory concentration (IC50) values. Selectivity indices (SI) were calculated by dividing CC50 by the corresponding IC50 for each virus. Notably, these compounds demonstrated promising selectivity against SARS-CoV-2 at lower concentrations, with increasing SI values (cf. Table 4 and Fig. 3).
| Compounds | CC50 (mg mL−1) | SARS-CoV-2 | MERS-CoV | HCoV-229E | |||
|---|---|---|---|---|---|---|---|
| IC50 (mg mL−1) | SI | IC50 (mg mL−1) | SI | IC50 (mg mL−1) | SI | ||
| Hydrazide (Hyd) | 0.4913 | 0.054 | 9.09 | 0.23 | 2.1 | 0.0277 | 17.7 |
| Hydrazone (HQ) | 0.805 | 0.052 | 15.5 | 0.302 | 2.7 | 0.134 | 6.0 |
| Pyrrolone (15A) | 65.362 | 0.158 | 413 | 0.0516 | 1266 | 0.0447 | 1462 |
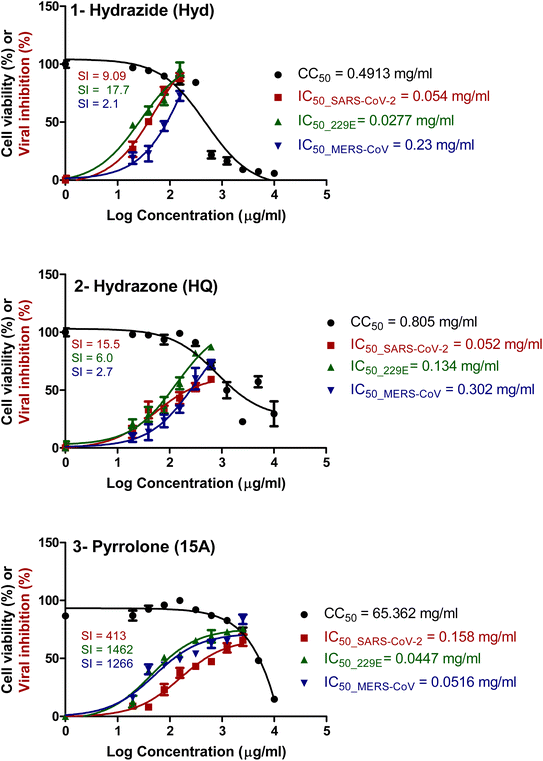 | ||
| Fig. 3 Determination of CC50 and IC50 of hydrazide (Hyd), hydrazone (HQ) and pyrrolone (15A) in Vero E6 cells against SARS-CoV-2, MERS-CoV, and HCoV-229E. Values of CC50 and IC50 were calculated using non-linear regression analysis with Graph Pad Prism software (version 5.01). | ||
Pyrazolylhydrazide (Hyd) exhibited a CC50 of 0.4913 mg mL−1, with notable inhibitory effects against SARS-CoV-2 (IC50: 0.054 mg mL−1, SI: 9.09), MERS-CoV (IC50: 0.23 mg mL−1, SI: 2.1), and HCoV-229E (IC50: 0.0277 mg mL−1, SI: 17.7). Similarly, hydrazone HQ demonstrated promising results with a CC50 of 0.805 mg mL−1 and significant inhibition against SARS-CoV-2 (IC50: 0.052 mg mL−1, SI: 15.5), MERS-CoV (IC50: 0.302 mg mL−1, SI: 2.7), and HCoV-229E (IC50: 0.134 mg mL−1, SI: 6.0). In turn, pyrrolone 15A exhibited potent antiviral activity and displayed higher cytotoxicity, necessitating further investigation. These findings underscore the potential of these derivatives as selective antiviral agents, particularly against SARS-CoV-2 and HCoV-229E, providing valuable insights for future therapeutic development.
The results indicate the percentage of inhibition observed for three different modes of action (adsorption inhibition, virucidal effect, and replication inhibition) against three coronaviruses (SARS-CoV-2, HCoV-229E, and MERS-CoV) at a concentration of 0.4 mg mL−1 of the tested compound (presumably hydrazide Hyd). These findings highlight the multifaceted antiviral mechanisms of the hydroxyquinoline-pyrazole derivatives, demonstrating their potential as broad-spectrum antiviral agents against various coronaviruses.
Adsorption inhibition, this measures the ability of the compound to prevent the virus from attaching to host cells. For SARS-CoV-2 and HCoV-229E, the inhibition percentages are 11.67%, and 12.76% indicating some level of effectiveness in preventing viral attachment. However, for MERS-CoV, the compound shows no significant adsorption inhibition.
Virucidal effect, that assesses the compound's ability to directly deactivate the virus. Notably, the compound demonstrates substantial virucidal effects against all three viruses, with percentages ranging from 20% to 73%. This suggests that the compound is highly effective at directly neutralizing the viruses. Replication inhibition, this evaluates the compound's ability to hinder viral replication within host cells. The results show moderate to high inhibition percentages across all three viruses, ranging from 10% to 55%. This indicates that the compound is effective at impeding viral replication processes.
The hydrazone HQ exhibited significant adsorption inhibition with mean values of 20.67%, 31.67%, and 9.00% for SARS-CoV-2, HCoV-229E, and MERS-CoV, respectively. Regarding the virucidal effect, mean inhibition percentages were 71.33%, 81.00%, and 26.33% for SARS-CoV-2, HCoV-229E, and MERS-CoV, respectively. Furthermore, replication inhibition showed mean values of 60.33%, 70.00%, and 16.67% for SARS-CoV-2, HCoV-229E, and MERS-CoV, respectively.
These findings suggest that hydrazone HQ with 0.8 mg mL−1 concentration effectively attenuates the coronaviruses through adsorption inhibition, virucidal effect, and replication inhibition in a dose-dependent manner. Notably, HCoV-229E exhibits the highest susceptibility to the substance across all mechanisms tested, followed by SARS-CoV-2, while MERS-CoV demonstrates comparatively lower susceptibility.
The results obtained for pyrrolone 15A at a concentration of 65 mg mL−1 demonstrate its inhibitory effects against SARS-CoV-2, HCoV-229E, and MERS-CoV across various mechanisms of action. Mean values were calculated to provide a comprehensive overview of the inhibitory activity. For adsorption inhibition, mean percentages were 19.67%, 31.33%, and 9.67% for SARS-CoV-2, HCoV-229E, and MERS-CoV, respectively. Notably, the virucidal effect was prominent, with mean inhibition percentages of 94.33%, 97.00%, and 37.67% for SARS-CoV-2, HCoV-229E, and MERS-CoV, respectively. Additionally, replication inhibition showed mean values of 70.33%, 85.33%, and 19.67% for SARS-CoV-2, HCoV-229E, and MERS-CoV, respectively. These results indicate that pyrrolone 15A exhibits remarkably potent antiviral activity against HCoV-229E and SARS-CoV-2, while demonstrating relatively lower efficacy against MERS-CoV across all tested mechanisms of action.
Discussion
This study aims at synthesizing new hydroxyquinoline-pyrazole systems and their antiviral efficacy on the recent SARS-CoV-2 and other coronaviruses such as MERS-CoV and HCoV-229E. The research topic seeks to determine the effectiveness of these compounds as antiviral agents for coronaviral diseases.A synthesis of the target compounds, which are pyrazole-based quinoline derivatives, can be easily described. Looking for the efficient construction of hydrazone and pyrrolone, the authors reacted pyrazolylhydrazide with 2-chloro-3-formylquinoline under various reaction conditions. Thus, the synthetic strategy described here correlates with the new works devoted to the elaboration of new heterocyclic derivatives with antiviral activity. For instance, Seliem et al. described the synthesis of pyrazolothiazole conjugates to act as SARS-CoV-2 inhibitors with the view of advantage of pyrazole derived molecules in seeking coronavirus.13
The molecular docking study also helps in predicting the probable interactions which the synthesized compounds may have with the important viral targets. Concerning the choice of the appropriate protein targets for docking, an emphasis on the main protease and papain-like protease in coronaviruses adds importance to the outcome. The docking scores reveal a high level of compounds' binding to the viral proteins, and the poses indicate their possible interactions, which indicates their antiviral properties. This approach is in concordance with other computational studies conducted recently by Musa et al., Where they utilized molecular docking in order to establish phenylpyrazolone-1,2,3-triazole hybrids as good inhibitors of SARS-CoV-2 main protease.33
It could also be seen that the compounds had antiviral effects of retarding the viral replication of SARS-CoV-2, MERS-CoV, and HCoV-229E and establishing the compounds as possible antiviral agents. The implication of evaluating the CC50 and IC50 enables one to determine the toxicity of a compound towards cells as well as the effectiveness of the antiviral compound. The obtained selectivity indices SI prove that the mentioned compounds enhance the selectivity toward SARS-CoV-2, mainly at relatively low concentrations. These outcomes correlate with research discussing the antiviral effect of heterocyclic compounds concerning coronaviruses. For instance, Kandinska et al. developed new tetrahydroisoquinoline hybrids containing other heterocyclic groups and compared their antiviral coronavirus potency.17
The inhibitory effects observed against several coronaviruses to support the broad-spectrum possibility of the synthesized candidates. This is extremely relevant considering that the threat of new Covid-19 variants is still present and that there is a need for universal antiviral agents. The multi-target approach that is targeting various phases of virus reproduction meets contemporary approaches to the development of drugs used to combat viruses. For example, Chang et al. have very recently published cytotoxic hexadepsipeptides and anti-coronaviral 4-hydroxy-2-pyridones from an endophytic Fusarium sp. Amalgamating the data presented in the works under analysis and the general information concerning the oligomerization of pyrroles, it is possible to assert that the search for antiviral compounds with novel chemical scaffolds will remain one of the most effective strategies for further research.34
The positive results indicated in this work call for additional future research, which should encompass in vivo evaluation of the agent's effectiveness and determination of the toxic effects of the compound. Moreover, structural modifications of the synthesized compounds could be involving an attempt to improve the antiviral activity along with selectivity. Understanding the specific mechanisms by which these compounds work against the viruses would further explain their use in the treatment of coronaviruses.
Besides, the prediction of ADME properties using SwissADME offers information on the likeliness of the compounds as drugs and information on the potential bioavailability. This approach typically assumes a greater significance in the course of the initial drug discovery phase, especially as exposed by recent studies, for example, in silico ADME/pharmacokinetics predictions were applied to estimate the property of 5-substituted 1H-tetrazole and benzoquinoline derivatives.35,36
Therefore, this work should be regarded as the investigation of the existing literature in the field of the development of potential antiviral substances against coronaviruses. The use of synthetic chemistry, molecular modeling and biologically evaluation enables for a systematic approach for screening of active molecules. The findings signify the potential use of hydroxyquinoline-pyrazole derivatives with the antiviral activity across different coronaviruses and provide the direction for further experimental studies with the view of creating means for the coronaviral infections treatment.
Modeling pharmacokinetics
The ADME (absorption, distribution, metabolism, and excretion) properties of the potent substances, which comprise their physicochemical characteristics, lipophilicity, and drug-likeness, have been predicted by SwissADME free web tool to lower the time of picking substances from an huge collection of substances in the primary phases of drug finding, biological possessions, and development for an operative drug.37 The hydrazide Hyd, hydrazone HQ, and pyrrolone 15A were found to obey with Lipinski's rule of five with a total polar surface area (TPSA) of 90.01, 109.47, and 83.61 Å2, respectively (cf. Table S1 and Fig. S1–S3 in ESI†).Also, these substances displayed appropriate physicochemical properties, which were assessed through the following six parameters: size, lipophilicity, polarity, unsaturation, insolubility, and flexibility. Regarding the absorption property, they demonstrated gastrointestinal tract (GIT) absorption owing to their being in the BOILED-EGG chart white area (Fig. S4 in ESI†). The compounds demonstrated favorable lipophilicity profiles, as indicated by their consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w values of 3.36, 5.50, and 5.83 for hydrazide (Hyd), hydrazone (HQ), and pyrrolone (15A), respectively. The bioavailability radar chart revealed promising oral bioavailability for both Hyd and HQ, with a value of 0.55. All three compounds (hydrazide Hyd, hydrazone HQ, and pyrrolone 15A) were fully encompassed within the pink area of the chart, suggesting good predicted oral bioavailability. This analysis considered key physicochemical properties including lipophilicity, polarity, molecular size, solubility, flexibility, and saturation. The results indicate that these compounds possess a balanced profile of these properties, which is favorable for oral drug candidates.
Po/w values of 3.36, 5.50, and 5.83 for hydrazide (Hyd), hydrazone (HQ), and pyrrolone (15A), respectively. The bioavailability radar chart revealed promising oral bioavailability for both Hyd and HQ, with a value of 0.55. All three compounds (hydrazide Hyd, hydrazone HQ, and pyrrolone 15A) were fully encompassed within the pink area of the chart, suggesting good predicted oral bioavailability. This analysis considered key physicochemical properties including lipophilicity, polarity, molecular size, solubility, flexibility, and saturation. The results indicate that these compounds possess a balanced profile of these properties, which is favorable for oral drug candidates.
Compound (Hyd) revealed a high GIT absorption while the others showed low GIT absorption. Their skin permeation (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KP) constraints were −6.18, −5.27, and −4.67 cm s−1, which given the bioactive substrates easy going to retrieve through skin. Furthermore, their cytochrome P450 isoenzymes, which accomplish a substantial role in medicines biotransformation through O-type oxidation processes, have also been anticipated. Otherwise, it is expected to access the blood–brain barrier (existed inside chart yellow area). The compounds colored red in Fig. S4† did not enter the substrate area for P-glycoprotein. A major efflux transporter is P-gp, which is capable of limiting drug oral absorption and brain penetration, so normally the nonsubstrate status is favorable for bioavailability upon oral administration and for CNS penetration.
KP) constraints were −6.18, −5.27, and −4.67 cm s−1, which given the bioactive substrates easy going to retrieve through skin. Furthermore, their cytochrome P450 isoenzymes, which accomplish a substantial role in medicines biotransformation through O-type oxidation processes, have also been anticipated. Otherwise, it is expected to access the blood–brain barrier (existed inside chart yellow area). The compounds colored red in Fig. S4† did not enter the substrate area for P-glycoprotein. A major efflux transporter is P-gp, which is capable of limiting drug oral absorption and brain penetration, so normally the nonsubstrate status is favorable for bioavailability upon oral administration and for CNS penetration.
On the other side, in terms of compliance with Lipinski's rule of five, which predicts bioavailability after oral administration, hydrazide (Hyd) did not reveal any violations, and neither did hydrazone HQ. In the case of pyrrolone (15A), it had one violation, probably because of its a bit higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of 5.83, which was a little over the recommended limit of 5. At the same time, all three compounds (Hyd, HQ, and 15A) revealed very favorable drug-like properties and were predicted to have good oral bioavailability. The log
P value of 5.83, which was a little over the recommended limit of 5. At the same time, all three compounds (Hyd, HQ, and 15A) revealed very favorable drug-like properties and were predicted to have good oral bioavailability. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values for Hyd and HQ were 3.36 and 5.50, falling within or very near to the ideal range of 1.35 to 5, likely to have good oral absorption. Although compound 15A presented higher lipophilicity, which may add advantages regarding membrane permeability, it could somewhat affect aqueous solubility. These in silico predictions taken together indicate that the physiochemical profile of the synthesized compounds is compatible with oral drug-likeness and bioavailability; the profiles of Hyd and HQ were very promising.
P values for Hyd and HQ were 3.36 and 5.50, falling within or very near to the ideal range of 1.35 to 5, likely to have good oral absorption. Although compound 15A presented higher lipophilicity, which may add advantages regarding membrane permeability, it could somewhat affect aqueous solubility. These in silico predictions taken together indicate that the physiochemical profile of the synthesized compounds is compatible with oral drug-likeness and bioavailability; the profiles of Hyd and HQ were very promising.
Conclusion
This work fills the research gap of the design, synthesis, and antiviral activity of hydroxyquinoline-pyrazole derivatives against various coronaviruses. Thus, the synergistic use of in silico molecular docking studies and in vitro antiviral assays gives a full picture of the compounds' perspective as antiviral agents.Key findings from this research include:
(1) New hydroxyquinoline-pyrazole derivatives; hydrazide (Hyd), hydrazone (HQ), and pyrrolone (15A) were synthesized and characterized.
(2) Shown to have ambivalent activity against SARS-CoV-2, as well as MERS-CoV and HCoV229E with selectivity indices, suggesting potentially selective antiviral agents.
(3) Learning of four possibilities of action, namely, adsorption inhibition, virucidal effectivity, replication hindrance, and encapsidation interference, implying that the physical chemical had a good potential of acting against a myriad of viruses.
(4) drug likeness and oral bioavailability of the synthesized compound have been found to be favorable based on the ADME prediction models.
These findings have broader implications for antiviral drug discovery and development:
(1) The present work represents a novel example of using congeneric series of hydroxyquinoline and pyrazole derived compounds to improve antiviral effect and specificity to a group of coronaviruses.
(2) The compounds discussed above have multiple target activity profiles, which is in parallel with the current trends in the antiviral drugs design, which might have certain advantages over single target ones.
(3) The conceptualization and the assessment that prioritized in this study are fast, and the layout of the study can be used as a blueprint of sorts for an accelerated antiviral drug development when new viral threats emerge.
Future research directions stemming from this work include:
(1) The extension of lead compounds aiming at improving the antiviral efficacy and selectivity.
(2) Further in vitro studies to determine the theoretical working of these compounds towards the coronavirus.
(3) Further testing of these compounds with the spectrum of viruses other to see their potentials as broad-spectrum antiviral agents.
(4) In vivo efficacy and toxicity studies to evaluate the antifungal activity and safety of the above compounds.
(5) Evaluation of the possibilities of the combined application with other antiviral drugs.
(6) The rational and knowledge-based design of new antiviral derivatives using the hydroxyquinoline-pyrazole template through the understanding of the SAR rules.
Therefore, this study advances knowledge that could be used to progress subsequent works aimed at finding the appropriate treatments for coronaviral infections. The positive results which have been achieved with these hydroxyquinoline-pyrazole derivatives, serves as aspect of curiosity about these compounds as a fresh stage of antiviral brokers. Further research and enhancement of these compounds could go a long way to improve on current strategies dealing with present and future coronavirus pandemics in the world healthcare facilities.
Ethical statement
This study was approved by the Institutional Animal Care and Use Committee at Agricultural Research Center (Approval code: ARC, CLEVB, 27, 24) and was performed in accordance with the guidelines of the National Institute of Health (NIH). All methods are reported in accordance with ARRIVE guidelines.Data availability
All data generated or analyzed during this study are included in this published article and its ESI files.†Conflicts of interest
The authors declare no competing interests.References
- C. Liu, et al., Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases, ACS Cent. Sci., 2020, 6(3), 315–331 CrossRef CAS PubMed.
- A. Dutta, A. Roy, L. Roy, S. Chattopadhyay and S. Chatterjee, Immune response and possible therapeutics in COVID-19, RSC Adv., 2021, 11, 960–977 RSC.
- Z. B. Milanovic, et al., Inhibitory activity of quercetin, its metabolite, and standard antiviral drugs towards enzymes essential for SARS-CoV-2: the role of acid–base equilibria, RSC Adv., 2021, 11, 2838–2847 RSC.
- U. Farwa and M. A. Raza, Heterocyclic compounds as a magic bullet for diabetes mellitus: a review, RSC Adv., 2022, 12, 22951–22973 RSC.
- D. A. Milenković, D. S. Dimić, E. H. Avdovićac and Z. S. Marković, Several coumarin derivatives and their Pd(ii) complexes as potential inhibitors of the main protease of SARS-CoV-2, an in silico approach, RSC Adv., 2020, 10, 35099–35108 RSC.
- J. Branković, et al., Pyrazolone-type compounds: synthesis and in silico assessment of antiviral potential against key viral proteins of SARS-CoV-2, RSC Adv., 2022, 12, 16054–16070 RSC.
- S. K. Ramadan, S. S. Shaban and A. I. Hashem, Facile and expedient synthesis and anti-proliferative activity of diversely pyrrolones bearing 1,3-diphenylpyrazole moiety, Synth. Commun., 2020, 50(2), 185–196 CrossRef CAS.
- A. R. Morsy, S. K. Ramadan and M. M. Elsafty, Synthesis and antiviral activity of some pyrrolonyl substituted heterocycles as additives to enhance inactivated Newcastle disease vaccine, Med. Chem. Res., 2020, 29, 979–988 CrossRef CAS.
- S. K. Rampadan, et al., Some pyrimidohexahydroquinoline candidates: synthesis, DFT, cytotoxic activity evaluation, molecular docking, and in silico studies, RSC Adv., 2024, 14(23), 16584–16599 RSC.
- B. Y. Li, et al., Quinoline, with the active site of 8-hydroxyl, efficiently inhibits Micropterus salmoides rhabdovirus (MSRV) infection in vitro and in vivo, J. Fish Dis., 2022, 45(6), 895–905 CrossRef CAS PubMed.
- S. K. Ramadan, H. S. Abd-Rabboh, N. M. Gad, W. S. I. Abou-Elmagd and D. Haneen, Synthesis and characterization of some chitosan-quinoline nanocomposites as potential insecticidal agents, Polycyclic Aromatic Compounds, 2023, 43(8), 7013–7026 CrossRef CAS.
- H. A. Saadeh, K. A. Sweidan and M. S. Mubarak, Recent advances in the synthesis and biological activity of 8-hydroxyquinolines, Molecules, 2020, 25(18), 4321 CrossRef CAS.
- A. F. Seliem, Synthesis and Virtual Screening of Pyrazolothiazole Conjugates as Promising SARS-CoV-2 Inhibitors, ChemistrySelect, 2023, 8(19), e202300605 CrossRef CAS.
- A. S. Elgubbi, E. A. E. El-Helw, A. Y. Alzahrani and S. K. Ramadan, Synthesis, computational chemical study, antiproliferative activity screening, and molecular docking of some thiophene-based oxadiazole, triazole, and thiazolidinone derivatives, RSC Adv., 2024, 14(9), 5926–5940 RSC.
- A. M. Abdelrahman, A. A. Fahmi, S. A. Rizk and E. A. E. El-Helw, Synthesis, DFT and Antitumor Activity Screening of Some New Heterocycles Derived from 2,2'-(2-(1,3-Diphenyl-1H-pyrazol-4-yl)ethene-1,1-diyl)bis(4H-benzo[d][1,3]oxazin-4-one), Polycyclic Aromatic Compounds, 2023, 43(1), 721–739 CrossRef CAS.
- M. M. Kaddah, A. A. Fahmi, M. M. Kamel, S. A. Rizk and S. K. Ramadan, Rodenticidal Activity of Some Quinoline-Based Heterocycles Derived from Hydrazide-Hydrazone Derivative, Polycyclic Aromatic Compounds, 2023, 43(5), 4231–4241 CrossRef CAS.
- M. I. Kandinska, et al., Synthesis of Novel 1-Oxo-2,3,4-trisubstituted Tetrahydroisoquinoline Derivatives, Bearing Other Heterocyclic Moieties and Comparative Preliminary Study of Anti-Coronavirus Activity of Selected Compounds, Molecules, 2023, 28(3), 1495 CrossRef CAS PubMed.
- A. Kandeil, et al., Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Africa and Middle East, Viruses, 2019, 11(8), 717 CrossRef CAS PubMed.
- O. Kutkat, et al., Robust antiviral activity of commonly prescribed antidepressants against emerging coronaviruses: in vitro and in silico drug repurposing studies, Sci. Rep., 2022, 12(1), 12920 CrossRef CAS PubMed.
- M. R. Gomaa, et al., Incidence, household transmission, and neutralizing antibody seroprevalence of Coronavirus Disease 2019 in Egypt: Results of a community-based cohort, PLoS Pathog., 2021, 17(3), e1009413 CrossRef CAS PubMed.
- L. J. Reed and H. Muench, A simple method of estimating fifty per cent endpoints, Am. J. Epidemiol., 1938, 27(3), 493–497 CrossRef.
- M. Feoktistova, P. Geserick and M. Leverkus, Crystal violet assay for determining viability of cultured cells, Cold Spring Harb. Protoc., 2016, 2016(4), 343–346 Search PubMed.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65(1–2), 55–63 CrossRef CAS PubMed.
- A. Mostafa, et al., FDA-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2, Pharmaceuticals, 2020, 13(12), 443 CrossRef CAS PubMed.
- A. Kandeil, et al., Coding-complete genome sequences of two SARS-CoV-2 isolates from Egypt, Microbiol. Resour. Announce., 2020, 9(22), 004899 Search PubMed.
- F. G. Hayden, K. Cote and R. G. Douglas-Jr, Plaque inhibition assay for drug susceptibility testing of influenza viruses, Antimicrob. Agents Chemother., 1980, 17(5), 865–870 CrossRef CAS PubMed.
- M. Kuo, M. H. Browning and M. L. Penner, Do lessons in nature boost subsequent classroom engagement? Refueling students in flight, Front. Psychol., 2018, 8, 2253 CrossRef PubMed.
- M. Zhang, T. Tanaka and M. Ikura, Calcium-induced conformational transition revealed by the solution structure of apo calmodulin, Nat. Struct. Biol., 1995, 2(9), 758–767 CrossRef CAS PubMed.
- W. S. I. Abou-Elmagd, et al., Synthesis and anti H5N1 activities of some pyrazolyl substituted N-heterocycles, Nat. Sci., 2016, 14(9), 115 Search PubMed.
- E. A. E. El-Helw, A. R. Morsy and A. I. Hashem, Evaluation of some new heterocycles bearing 2-oxoquinolyl moiety as immunomodulator against highly pathogenic avian influenza virus (H5N8), J. Heterocycl. Chem., 2021, 58(4), 1003–1014 CrossRef CAS.
- S. K. Ramadan, et al., Synthesis, SAR studies, and insecticidal activities of certain N-heterocycles derived from 3-((2-chloroquinolin-3-yl)methylene)-5-phenylfuran-2(3H)-one against Culex pipiens L. larvae, RSC Adv., 2022, 12(22), 13628–13638 RSC.
- M. M. Kaddah, et al., Synthesis and biological activity on IBD virus of diverse heterocyclic systems derived from 2-cyano-N'-((2-oxo-1,2-dihydroquinolin-3-yl) methylene)acetohydrazide, Synth. Commun., 2021, 51(22), 3366–3378 CrossRef CAS.
- A. Musa, et al., Phenylpyrazolone-1,2,3-triazole hybrids as potent antiviral agents with promising SARS-CoV-2 main protease inhibition potential, Pharmaceuticals, 2023, 16(3), 463 CrossRef CAS PubMed.
- S. Chang, et al., Nocaviogua A and B: two lipolanthines from root-nodule-associated Nocardia sp, Front. Chem., 2023, 11, 1233938 CrossRef CAS PubMed.
- A. El-Sewedy, et al., One-pot synthesis, computational chemical study, molecular docking, biological study, and in silico prediction ADME/pharmacokinetics properties of 5-substituted 1 H-tetrazole derivatives, Sci. Rep., 2023, 13(1), 17869 CrossRef CAS PubMed.
- E. A. E. El-Helw, M. Asran, M. E. Azab, M. H. Helal, A. Y. Alzahrani and S. K. Ramadan, Synthesis and in silico studies of certain benzo[f]quinoline-based heterocycles as antitumor agents, Sci. Rep., 2024, 14, 15522 CrossRef CAS PubMed.
- C. A. Lipinski, Lead-and drug-like compounds: the rule-of-five revolution, Drug Discovery Today: Technol., 2004, 1(4), 337–341 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04728a |
| ‡ Equal contributing first authors. |
| § Current affiliation: Texas Biomedical Research Institute, Texas, USA. |
| This journal is © The Royal Society of Chemistry 2024 |