Open Access Article
Open Access ArticleModulating the pH dependent photophysical properties of green fluorescent protein†
David P. Broughton‡
 ,
Chloe G. Holod‡
,
Chloe G. Holod‡ ,
Angelica Camilo-Contreras,
Darcy R. Harris
,
Angelica Camilo-Contreras,
Darcy R. Harris ,
Scott H. Brewer
,
Scott H. Brewer * and
Christine M. Phillips-Piro
* and
Christine M. Phillips-Piro *
*
Department of Chemistry, Franklin & Marshall College, P.O. Box 3003, Lancaster, PA 17604-3003, USA. E-mail: sbrewer@fandm.edu; cpiro@fandm.edu
First published on 17th October 2024
Abstract
The photophysical properties of the β-barrel superfolder green fluorescent protein (sfGFP) arise from the chromophore that forms post-translationally in the interior of the protein. Specifically, the protonation state of the side chain of tyrosine 66 in the chromophore, in addition to the network of hydrogen bonds between the chromophore and surrounding residues, is directly related to the electronic absorbance and emission properties of the protein. The pH dependence of the photophysical properties of this protein were modulated by the genetic, site-specific incorporation of 3-nitro-L-tyrosine (mNO2Y) at site 66 in sfGFP. The altered photophysical properties of this noncanonical amino acid (ncAA) sfGFP construct were assessed by absorbance and fluorescence spectroscopies. Notably, a comparison of the pKa of the 3-nitrophenol side chain of mNO2Y incorporated in the protein relative to the phenol side chain of the tyrosine at site 66 in the native chromophore as well as the pKa of the 3-nitrophenol side chain of the free ncAA were measured and are compared. A structural analysis of the ncAA containing sfGFP construct is presented to yield molecular insight into the origin of the altered absorbance and fluorescence properties of the protein.
Introduction
The green fluorescent protein (GFP) has a β-barrel structure with an internal chromophore that is formed by cyclization of residues 65, 66, and 67 post-translationally (Fig. 1).1 This autocatalytically formed, intrinsic chromophore is responsible for photophysical properties of this protein which has been utilized for an array of various research applications.1–4 The tyrosine at site 66 is directly related to these spectroscopic attributes in addition to other key residues essential for the observed photophysical properties.1,3 Specifically, the protonation state of the phenol side chain of Y66 alters the optical properties of the protein where the protein fluoresces its characteristic green color when Y66 is in the deprotonated (phenolate) form and does not fluoresces when Y66 is in the protonated (phenol) form.1,3 Hence, numerous studies have used mutagenesis to replace Y66 with another naturally occurring amino acid to alter the wavelengths of maximum absorbance and emission in addition to the corresponding extinction coefficient and quantum yield, respectively of the protein constructs.3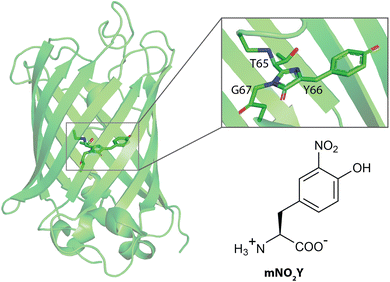 | ||
| Fig. 1 Structures of wildtype sfGFP (PDB ID 2B3P) and the noncanonical amino acid 3-nitro-L-tyrosine (mNO2Y) used in this study. The sfGFP structure has a zoom into the native chromophore formed from Threonine 65, Tyrosine 66, and Glycine 67. | ||
In addition to naturally occurring amino acids (AA), numerous noncanonical amino acids (ncAA) have been incorporated into GFP to either probe local protein environments or to modulate the photophysical properties of the protein.5–24 Site-specific spectroscopic reporters such as 4-cyano-L-phenylalanine, 4-nitro-L-phenylalanine, and 4-azidomethyl-L-phenylalanine have been successfully genetically incorporated into the superfolder green fluorescent protein (sfGFP)25 variant to probe unique local hydration states in the protein in a minimally perturbative fashion as assessed by X-ray crystallography.13–17,20 Other ncAA have been incorporated into site 66 in the internal chromophore of the GFP, replacing the native tyrosine residue, to yield an array of GFP constructs with altered photophysical properties.5,6,8,9,12,24,26,27
Here, we have utilized the known pH dependence of the photophysical properties of GFP to measure the pKa of the phenolic hydrogen of Y66 in the internal chromophore of sfGFP utilizing pH dependent absorbance and fluorescence spectroscopy.28 The subsequent modulation of the pH dependent photophysical properties of sfGFP with Y66 replaced with 3-nitro-L-tyrosine (mNO2Y, Fig. 1), which has been used to perturb the pKa in ribonucleotide reductase previously,29 was then explored with both spectroscopic techniques. This work builds upon previous literature studies exploring the replacement of Y66 with halogenated variants of tyrosine such as 3-chloro-L-tyrosine, 3-bromo-L-tyrosine, and 3-iodo-L-tyrosine in the S65T,H148D-GFP construct.9,24 Furthermore, the impact of the electron withdrawing nitro substituent was assessed by measuring the pKa of the side chain of mNO2Y both free in solution and incorporated in the internal chromophore of sfGFP. X-ray crystallography was utilized to ensure successful chromophore formation in the mNO2Y66 containing sfGFP construct (Y66mNO2Y-sfGFP), assess any protein conformation changes resulting from the ncAA incorporation, and provide a possible structural understanding for the alteration of the electronic properties of Y66mNO2Y-sfGFP relative to wt-sfGFP.
Materials and methods
General information
Chemical reagents were purchased from Sigma-Aldrich, Hampton Research, and Thermo Fisher Scientific and used without further purification. DH10B cells were purchased from NEB. All aqueous solutions were prepared with 18 MΩ cm water.Expression and purification of wild-type and mNO2Y containing sfGFP constructs
The pBAD vector with wild-type superfolder green fluorescent protein (pBAD_wtsfGFP) containing a C-terminal six-histidine affinity tag was obtained from Dr Ryan A. Mehl (Oregon State University). Site-directed mutagenesis was used to replace the TAT codon encoding for a tyrosine residue at the 66 site, in the internal chromophore of the protein, with a TAG codon for mNO2Y incorporation via the Amber codon suppression30 resulting in the pBAD-sfGFP-66TAG construct.The pDULE vector with the engineered, orthogonal aminoacyl-tRNA synthetase for the incorporation of mNO2Y (pDULE_mNO2Y) was obtained from Dr Ryan A. Mehl (Oregon State University).31,32 The pBAD_wtsfGFP was singly transformed or the pBAD-sfGFP-66TAG and pDULE_mNO2Y vectors were co-transformed into chemically competent DH10B Escherichia coli cells. The transformed cells were used to inoculate 5 mL of non-inducing media overnight, shaking at 250 rpm at 37 °C. Aliquots (2.5 mL) of the cultured cells were used to inoculate 250 mL of autoinduction media in 500 mL baffled Erlenmeyer flasks. The media contained 100 μg mL−1 ampicillin for the singly transformed cells and 100 μg mL−1 ampicillin, 25 μg mL−1 tetracycline, and 1.1 mM mNO2Y for the co-transformed cells. The cells were harvested after 24–30 hours by centrifugation, and the expressed sfGFP protein constructs were purified through TALON cobalt ion-exchange chromatography, as previously described.33–35
The incorporation of the noncanonical amino acid mNO2Y into site 66 of sfGFP resulted in the production of the protein construct Y66mNO2Y-sfGFP while the wt-sfGFP construct contained tyrosine at site 66. The purified proteins were desalted utilizing PD10 columns into a 20 mM HEPES aqueous buffer solution (pH 7.5). The proteins were then incubated at 37 °C for 2 hours with a catalytic amount of trypsin (1 mol%) to cleave the C-terminal six-histidine affinity tag, resulting in the formation of 239-residue sfGFP protein constructs. The trypsin was deactivated with a ten-fold molar excess of phenylmethanesulfonyl fluoride (PMSF) relative to trypsin. Any uncleaved six-histidine tagged sfGFP protein was removed by TALON cobalt ion-exchange chromatography, and excess PMSF was removed as the proteins were concentrated with a 10k molecular-weight cutoff Centricon (Cytiva) in 20 mM HEPES buffer adjusted to pH 7.5 with sodium hydroxide for storage at 4 °C.
Mass spectral characterization
Prior to spectral analysis, the sfGFP constructs were desalted into water using Zeba 7k MWCO desalting columns and formic acid was added to a concentration of 0.1%. Following separation with an Acquity H series UPLC employing a Waters Protein BEH C4 300 Å 1.7 mm 1 × 50 mm column, ESI-Q-TOF mass analysis of the intact protein was performed with a Waters Xevo G2-S mass spectrometer. Raw mass spectral data from the protein charge state envelope was deconvoluted using MaxEnt1 processing as part of the Waters MassLynx V4.1 software package to obtain intact protein masses. Deconvoluted protein mass values were confirmed by manual inspection of the raw mass spectral data.Equilibrium ultraviolet-visible absorbance measurements
Room-temperature (23 °C) equilibrium UV-vis absorbance spectra in the 250–600 nm region were recorded with an Agilent Technologies Cary 100 UV-vis spectrophotometer or a Thermo Scientific Genesys 50 UV-vis spectrophotometer using a 1 cm cuvette. The spectra were recorded with one nanometer resolution and a scan speed of 600 nm min−1. mNO2Y and protein samples were dissolved in 50 mM potassium phosphate aqueous buffers with pH values ranging from 2.50–10.00 in 0.50 increments. Data analysis was performed using Igor Pro.Equilibrium fluorescence measurements
Room-temperature (23 °C) equilibrium fluorescence measurements were recorded on a HORIBA Jobin Yvon Fluoro-Max-4 spectrometer with an excitation and emission bandwidth of 1 nm. The spectra were recorded with an integration time of 0.2 seconds in a 1 cm quartz sample holder. The protein constructs were dissolved in 50 mM potassium phosphate aqueous buffers with pH values ranging from 2.50–10.00 in 0.50 increments. Data analysis was performed using Igor Pro.Side chain pKa determination
The room temperature pKa of the side chain of mNO2Y was determined by fitting the measured absorbance at the appropriate wavelength as a function of pH to eqn (1):
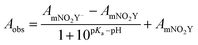 | (1) |
Crystallization, X-ray diffraction data collection, and structural refinement of Y66mNO2Y-sfGFP construct
Purified Y66mNO2Y-sfGFP was crystallized using sitting drop vapor diffusion mixing 1 μL of 35 mg mL−1 protein with 1 μL of precipitant solution (1% tryptone, 0.001 M NaN3, 0.050 M HEPES pH 7, and 20% w/v PEG 3350) allowed to equilibrate against a well containing 500 μL of precipitant solution. Crystals were looped and flash frozen in liquid nitrogen without any cryogen protectant. X-ray diffraction data were collected at APS beamline 24 ID-E at Argonne National lab on an Eiger detector to 1.51 Å resolution. Data were integrated in space group C121 and processed and scaled in XDS.36 Molecular replacement in PHASER using a search model of wt-sfGFP (PDB ID 2B3P) with the chromophore removed provided a solution with a translation-function Z-score (TFZ score) of 52.8 and a log-likelihood gain (LLG) of 3425.139.25,37The structure was refined using iterative rounds of manual refinement in COOT and automated refinement in Phenix where 6% of reflections were reserved to calculate Rfree.38,39 The unmodified chromophore was added to the structure after the first round of refinement and the nitro group was added in the third iteration using clear difference density accounting for these atoms (see ESI Fig. S2†). The cif file from ligand QCA in the Protein Databank was used to refine the chromophore in Y66mNO2Y-sfGFP. The final structure had a R and Rfree of 17.0 and 22.0% and was deposited in the Protein Databank under PDB ID 9C74. Data processing and structure refinement statistics are provided in Table 1.
| mNO2Y66-sfGFP | 9C74 |
|---|---|
| Diffraction source | 24 ID-E |
| Wavelength (Å) | 0.9792 |
| Temperature (K) | 100 |
| Crystal-to-detector distance (mm) | 180 |
| Space group | C121 |
| a, b, c (Å) | 91.81, 37.97, 68.46 |
| α, β, γ (°) | 0, 112.73, 90 |
| Mosaicity (°) | 0.115 |
| Resolution range (Å) | 61.15–1.51(1.53–1.51) |
| No. of unique reflections | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 690 (1508) 690 (1508) |
| Completeness (%) | 97.7 (90.1) |
| Multiplicity | 3.3 (3.2) |
| [I/σ(I)] | 16.4 (2.5) |
| CC1/2 | 0.999 (0.869) |
| Resolution range (Å) | 63.15–1.51 |
| Final R (Rfree) (%) | 17.0 (22.0) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| R.m.s deviations | |
| Bonds (Å) | 0.010 |
| Angles (°) | 1.165 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Ramachandran plot | |
| Most favored (%) | 98.64 |
| Allowed (%) | 1.36 |
Results and discussion
Photophysical properties of mNO2Y
The electronic absorbance spectrum of 3-nitro-L-tyrosine (mNO2Y) is sensitive to the protonation state of the phenolic hydrogen of the side chain.40,41 Hence, the electronic absorbance spectrum of mNO2Y is sensitive to pH as illustrated in Fig. 2A in the 300–550 nm region for pH values ranging from 2.50 to 10.00. Above pH 8, the ncAA exists predominantly in the phenolate (ionized) form with an absorbance band centered at 424 nm. Below pH 6, the primary absorbance band occurs at 356 nm corresponding to the neutral (phenol) form of the ncAA.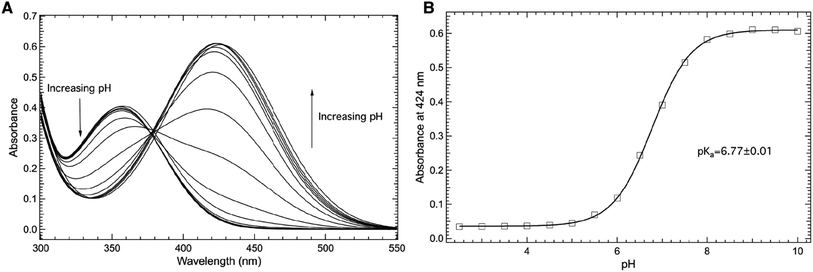 | ||
| Fig. 2 Room temperature absorbance spectra of the free noncanonical amino acid, 3-nitro-L-tyrosine (mNO2Y, (A)) as a function of pH in the 300–550 nm region. The ncAA (10 μM) was dissolved in 50 mM potassium phosphate buffers with pH values ranging from 2.50–10.00. The pH dependence of the absorbance of mNO2Y measured at 424 nm (open squares, (B)) was fit (solid curve) to eqn (1) to yield a pKa of 6.77 ± 0.01 of the 3-nitrophenolic hydrogen of mNO2Y. | ||
Fig. 2B shows the pH dependence of the absorbance measured at 424 nm (open squares) fit to eqn (1) (solid curve), which yields a pKa of 6.77 ± 0.01 for the phenolic hydrogen of mNO2Y in agreement with literature values.40,42 The determination of this pKa is the result of the correlation between the protonation state of the side chain of mNO2Y and the corresponding electronic properties of the ncAA. This correlation between protonation state and electronic properties will be used to measure the pKa of the phenolic hydrogen of either tyrosine or mNO2Y at site 66 in the internal chromophore of sfGFP (see below).
Successful incorporation of mNO2Y into sfGFP
The successful expression of purified wt-sfGFP and Y66mNO2Y-sfGFP was verified with ESI-Q-TOF mass spectrometry. The observed mass of fully formed wt-sfGFP was 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860.5 Daltons in agreement within experimental error of the expected mass of the protein based upon the primary sequence of the protein which is 26
860.5 Daltons in agreement within experimental error of the expected mass of the protein based upon the primary sequence of the protein which is 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860.0 Daltons. Similarly, the mass of Y66mNO2Y-sfGFP was within error of the expected mass. Specifically the observed mass was 26
860.0 Daltons. Similarly, the mass of Y66mNO2Y-sfGFP was within error of the expected mass. Specifically the observed mass was 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 905.5 Daltons while the expected mass (accounting for the Y66mNO2Y mutation) is 26
905.5 Daltons while the expected mass (accounting for the Y66mNO2Y mutation) is 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 906.0 Daltons. The formation of the mature, internal chromophore generated from the cyclization of residues 65, 66, and 67 is verified by X-ray crystallography as detailed below.
906.0 Daltons. The formation of the mature, internal chromophore generated from the cyclization of residues 65, 66, and 67 is verified by X-ray crystallography as detailed below.
Photophysical properties of wt-sfGFP and Y66mNO2Y-sfGFP
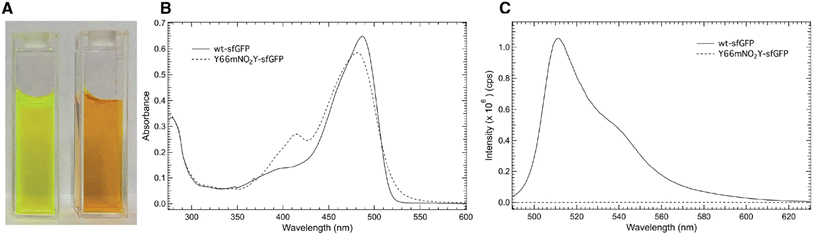
Fig. 3A shows an image of either purified wt-sfGFP (left) or Y66mNO2Y-sfGFP (right), respectively at a concentration of 10 μM at pH 7.50 in an aqueous potassium phosphate solution. The image of wt-sfGFP shows the characteristic green color25 expected of the protein while the Y66mNO2Y-sfGFP construct appears orange in color. Hence, the addition of the nitro functional group at the meta position of Y66 located in the chromophore has altered the photophysical properties of the protein. Interestingly, the incorporation of 4-nitro-L-phenylalanine into sfGFP at site 66 also showed a similar orange color.12Fig. 3B shows a comparison of the room temperature electronic absorbance spectrum of wt-sfGFP (solid curve) or Y66mNO2Y-sfGFP (dashed curve) at pH 7.50 in the 275–600 nm range. The absorbance spectrum of wt-sfGFP shows two prominent bands around 400 nm and at 487 nm. The 487 nm band is approximately 4.7 times as intense as the less prominent band centered around 400 nm. Based upon literature precedent, the band at 487 nm is due to the phenolate (ionized) form of Y66 in the internal chromophore while the 400 nm band results from the phenol (neutral) form of Y66.1,3 Similarly, the room temperature electronic absorbance spectrum of Y66mNO2Y-sfGFP reveals two prominent features. However the position of these bands have shifted relative to wt-sfGFP to 414 nm and 481 nm. The lower energy absorbance band is the more intense band similar to wt-sfGFP, however the ratio of the lower to higher absorbance band has decreased to approximately 2.2 for Y66mNO2Y-sfGFP relative to wt-sfGFP.
Fig. 3C shows the comparison of the room temperature fluorescence (emission) spectra of wt-sfGFP (solid curve) or Y66mNO2Y-sfGFP (dashed curve) at pH 7.50 in the 490–630 nm region with an excitation wavelength of 487 nm. As expected, wt-sfGFP shows its prominent emission band at 511 nm resulting in the characteristic green color of a fully mature, functioning internal chromophore of the protein seen in Fig. 3A (left).25 Interestingly, Y66mNO2Y-sfGFP does not show any measurable fluorescence under these conditions suggesting the orange color of this construct shown in Fig. 3A (right) is due to the absorbance properties rather than the emission properties of the protein. The combination of Fig. 3B and C provide quantitative verification of the effect of the inclusion of the electron withdrawing nitro substituent on the phenol ring of Y66 on the photophysical properties of the protein.
Fig. 4 explores the pH dependence of the room temperature electronic absorbance properties of wt-sfGFP. Specifically, Fig. 4A shows the pH dependent absorbance spectra of wt-sfGFP ranging from pH 3.50 to 10.00 in the 275–600 nm region. Notably, the absorbance band at 487 nm indicative of the phenolate form of the chromophore increases with pH while the absorbance band at 393 nm (position at pH 3.50) due to the phenol form of the chromophore decreases as the pH increases. These results are not surprising since the deprotonation of the phenol side chain of Y66 is expected as the pH is increased.28
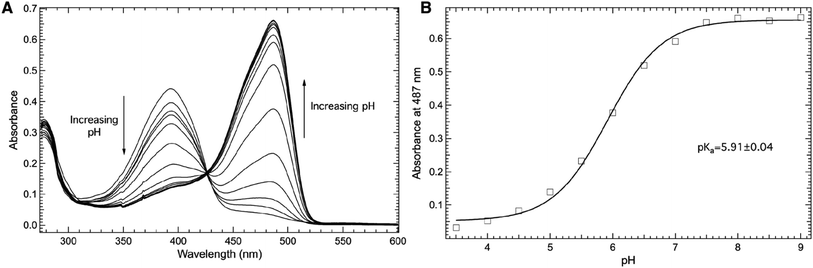 | ||
| Fig. 4 Room temperature absorbance spectra of wt-sfGFP (A) as a function of pH in the 275–600 nm region. wt-sfGFP (10 μM) was dissolved in 50 mM potassium phosphate buffers with pH values ranging from 3.50–10.00. The pH dependence of the absorbance of wt-sfGFP measured at 487 nm (open squares, (B)) was fit (solid curve) to eqn (1) to yield a pKa of 5.91 ± 0.04 of the phenolic hydrogen of tyrosine at site 66 in wt-sfGFP. | ||
The direct correlation between the phenol side chain protonation state of Y66 and the absorbance properties of the chromophore permits the pKa of the phenolic hydrogen in the chromophore of the protein to be determined.1,28 Specifically, Fig. 4B shows the pH dependence of the absorbance of the band centered at 487 nm (open squares) fit (solid curve) to eqn (1) yielding a pKa of 5.91 ± 0.04, consistent with earlier studies.27 This value is notably lower than the pKa of free tyrosine, which is the result of the involvement of Y66 in the formation of the internal chromophore with T65 and G67 and the location of the chromophore in the inside of the β-barrel structure of wf-sfGFP. It is important to note that the pKa of the phenolic hydrogen of Y66 in wt-sfGFP can also be determined using pH dependent fluorescence spectroscopy (see ESI Fig. S1†).
The impact of the introduction of the nitro group at the meta position of the phenol side chain on the pKa of the phenolic hydrogen at site 66 in the protein construct Y66mNO2Y-sfGFP is shown in Fig. 5. Specifically, Fig. 5A shows the pH dependent electronic absorbance spectra of Y66mNO2Y-sfGFP from pH 3.50 to 10.00 in the 275–600 nm region. At pH 10.00, the absorbance spectrum of Y66mNO2Y-sfGFP consists of two prominent absorbance bands centered at 414 nm and 481 nm, with the later being the more intense band.27 Notably, the absorbance band at 481 nm (pH 10.00) blue shifts to 453 nm as the pH is decreased to 3.50 while also decreasing in intensity. In contrast, the band at 414 nm (pH 10.00) red shifts to 419 nm as the pH decreases to 3.50 while growing in intensity. Similarly, the absorbance around 537 nm increases with decreasing pH. The largest absorbance change as pH varied occurred at 481 nm.
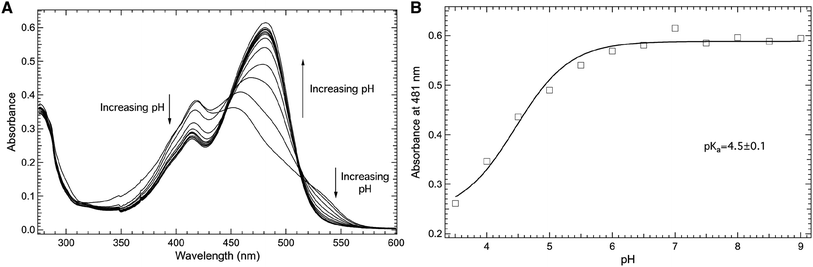 | ||
| Fig. 5 Room temperature absorbance spectra of Y66mNO2Y-sfGFP (A) as a function of pH in the 275–600 nm region. Y66mNO2Y-sfGFP (10 μM) was dissolved in 50 mM potassium phosphate buffers with pH values ranging from 3.50–10.00. The pH dependence of the absorbance of Y66mNO2Y-sfGFP measured at 481 nm (open squares, (B)) was fit (solid curve) to eqn (1) to yield a pKa of 4.5 ± 0.1 of the 3-nitrophenolic hydrogen of mNO2Y incorporated at site 66 in the protein. | ||
The pH dependence of the absorbance of Y66mNO2Y-sfGFP measured at 481 nm (open squares) as a function of pH fit to eqn (1) (solid curve) is shown in Fig. 5B. This fit reveals a pKa of 4.5 ± 0.1 of the phenolic hydrogen of mNO2Y incorporated at site 66 in the protein. It is relevant to note the larger error of this pKa relative to the pKa of free mNO2Y or Y66 in wt-sfGFP. This difference is the result of the pH dependent curve not being able to be fully sampled at low pH in Y66mNO2Y-sfGFP due to apparent protein denaturation below pH 3.5. However, the results clearly show that the pKa the 3-nitrophenolic hydrogen of mNO2Y incorporated at site 66 is lower than the corresponding pKa of tyrosine at this site. This is not a surprising result given the electron withdrawing nature of the nitro group.
The pKa of the mNO2Y side chain in Y66mNO2Y-sfGFP is lower than free mNO2Y highlighting the impact of mNO2Y66 being a part of the internal chromophore of the protein in addition to the chromophore's location inside the β-barrel structure of the protein. This effect is similar to the lowering of the pKa of the phenolic hydrogen of free tyrosine compared to Y66 in wt-sfGFP for similar reasons.
Structural verification of mNO2Y incorporation, chromophore formation, and structural impact in Y66mNO2Y-sfGFP
The 1.5 Å resolution crystal structure of Y66mNO2Y-sfGFP was determined at pH 7 and provides a view into the location of the various atoms in the structure and provides for a direct comparison with the previously published wt-sfGFP structure. An alignment of the wt-sfGFP and Y66mNO2Y-sfGFP resulted in an RMSD of 0.78 Å for 224 Cα atoms and is shown in Fig. 6A, suggesting very little structural perturbation of the overall structure when the nitro group is introduced on Y66.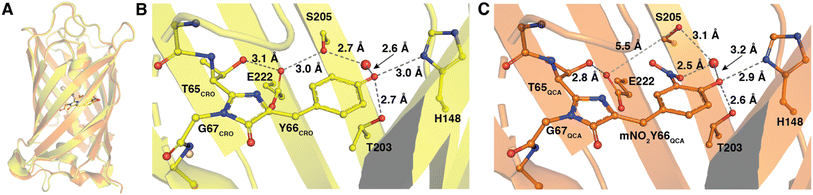 | ||
| Fig. 6 Structures of Y66mNO2Y-sfGFP and wild-type sfGFP. Alignment of Y66mNO2Y-sfGFP (orange, PDB ID 9C74) with wild-type sfGFP (yellow, PDB ID 2B3P) (A). The chromophore of wt-sfGFP (labeled CRO) with hydrogen bonds shown in dark grey dashed lines with distances (B). The chromophore in Y66mNO2Y-sfGFP (labeled QCA) and hydrogen bonds are indicated in dark grey dashed lines with distances shown. The E222–S205 distance is also shown in light grey dashed line, but is too long to be participating in a hydrogen bond (5.5 Å) (C). | ||
To prevent model bias in the area of particular interest, the chromophore was not included in the molecular replacement and was added after the first round of refinement as the native chromophore. When the native chromophore was modeled there remained unaccounted for 2Fo − Fc and +Fo − Fc electron density for the nitro group directly adjacent to the hydroxyl of the phenol, confirming the incorporation of mNO2Y at the Y66 site (see ESI Fig. S2†).
In addition to confirming mNO2Y incorporation, the electron density of the fully refined structure illustrates a fully cyclized chromophore (see ESI Fig. S2c†). Thus, mNO2Y does not inhibit the cyclization reaction, so the lack of chromophore cyclization is not the reason for the lack of fluorescence in Y66mNO2Y-sfGFP (see Fig. 3C). Notably, similar results were observed of the chromophore still forming but without fluorescing when 4-nitro-L-phenylalanine was incorporated at the Y66 site in sfGFP.12
An alignment of the wt-sfGFP chromophore and the chromophore in Y66mNO2Y-sfGFP also illustrated very little change in the location of the atoms of the chromophore (ESI Fig. S3†). However, there is a 14° rotation of the phenyl ring out of the plane with the 5 membered ring of the cyclized chromophore (see ESI Fig. S3†). This slight rotation may alter the electronics of the chromophore, which could contribute to the lack of observed fluorescence of the Y66mNO2Y-sfGFP compared to wt-sfGFP by altering the alignment of the extended π-system in the chromophore.
A closer examination of the hydrogen bonding network around the chromophores of wt-sfGFP and Y66mNO2Y-sfGFP provides some insight into the altered photophysical properties of the protein. The wt-sfGFP contains a H-bonding network from the phenolate oxygen through a structural water, S205, E222, and T65 (Fig. 6B), which stabilizes the deprotonated phenolate oxygen. In the Y66mNO2Y-sfGFP structure, the hydrogen bonding network has been disturbed by a rotation of S205 away from E222 resulting in a distance of 5.5 Å between the S205 sidechain oxygen and the closest carboxylate oxygen on the E222 side chain (Fig. 6C). The nitro group is sterically responsible for disrupting the S205–E222 hydrogen bond as it is located just adjacent to both of these residues. This H-bonding network alteration could result in the altered electronic properties of Y66mNO2Y-sfGFP relative to wt-sfGFP.
Conclusions
The pH dependent optical properties of wt-sfGFP yielded an apparent pKa of the phenolic hydrogen of the side chain of tyrosine at site 66, one of the three residues involved in the internal chromophore, in the folded protein of 5.91. The overall pH dependence of the photophysical properties and pKa is consistent with previous work.9,24,27 Here, the pH dependent electronic properties of the internal chromophore were successfully modulated by the replacement of Y66 with mNO2Y which notably adds an electronic withdrawing nitro group at the meta position of the phenol ring in the side chain of tyrosine. This mutation significantly altered the photophysical properties of the ncAA containing sfGFP construct. Notably an aqueous solution of this protein construct (Y66mNO2Y-sfGFP) appeared orange compared to the characteristic green appearance of wt-sfGFP in aqueous solution at pH 7.50. Additionally, Y66mNO2Y-sfGFP had an altered electronic absorbance spectrum with essentially no fluorescence detected compared to wt-sfGFP. Hence, the orange appearance of Y66mNO2Y-sfGFP was due to the absorptive properties of the protein rather than due to emission characteristics.The pKa of the 3-nitrophenolic hydrogen of mNO2Y in sfGFP at residue 66 was found to be 4.5, which is lower than the pKa of the Y66 side chain in wt-sfGFP. This decrease is not surprising given the electron withdrawing nature of the nitro functional group. However, this decrease is less than the decrease in the pKa value of the side chain of free mNO2Y (measured here) relative to the pKa for the side chain of free tyrosine. Hence, the incorporation of mNO2Y into the chromophore of the protein and its location in the interior of the β-barrel fold of the protein structure mitigates the impact of the nitro group on the phenolic hydrogen acidity.
A structural analysis of Y66mNO2Y-sfGFP confirmed the formation of a mature chromophore including mNO2Y, a slight rotation of the phenyl ring, and a modulation of the S205–E222 hydrogen bond length relative to wt-sfGFP, likely due to sterics relating to the introduction of a nitro group in the internal chromophore. This ring alignment and hydrogen bond length modulation coupled with the electron withdrawing nature of the nitro group helps to provide a molecular basis for the modulated pH dependent electronic properties of Y66mNO2Y-sfGFP relative to wt-sfGFP.
The current work highlights the ability to modulate the pH dependent optical properties of sfGFP through the replacement of a key tyrosine residue with mNO2Y. This work has potential future applications in other proteins by using this substitution to alter enzymatic activity, protein gas binding affinity, and/or optical properties.
Data availability
The protein crystal structure of Y66mNO2Y-sfGFP has been deposited to the Protein Databank and assigned PDB ID 9C74. The PDB DOI for this deposition is DOI: https://doi.org/10.2210/pdb9c74/pdb.Author contributions
D. P. Broughton: investigation, C. G. Holod: investigation, A. Camilo-Contreras: investigation, D. R. Harris: investigation, S. H. Brewer: conceptualization, formal analysis, funding acquisition, supervision, visualization, writing, C. M. Phillips-Piro: conceptualization, formal analysis, funding acquisition, supervision, visualization, writing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Science Foundation (CAREER-1847937) and Henry Dreyfus Teacher-Scholar Awards (TH-15-009 and TH-21-007). The Hackman Fund (F&M) and Eyler Fund (F&M) provided additional student support. Data for the crystal structure were collected at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank Kenneth Hess (F&M) for the collection and analysis of mass spectral data; and Lisa Mertzman (F&M) and Julie Gemmel (F&M) for administrative support.References
- R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509–544 CrossRef CAS.
- J. Lippincott-Schwartz, E. Snapp and A. Kenworthy, Nat. Rev. Mol. Cell Biol., 2001, 2, 444–456 CrossRef CAS.
- M. Zimmer, Chem. Rev., 2002, 102, 759–781 CrossRef CAS.
- D. M. Chudakov, M. V. Matz, S. Lukyanov and K. A. Lukyanov, Physiol. Rev., 2010, 90, 1103–1163 CrossRef CAS.
- L. Wang, J. Xie, A. A. Deniz and P. G. Schultz, J. Org. Chem., 2003, 68, 174–176 CrossRef CAS.
- D. D. Young, S. Jockush, N. J. Turro and P. G. Schultz, Bioorg. Med. Chem. Lett., 2011, 21, 7502–7504 CrossRef CAS.
- S. C. Reddington, S. Driezis, A. M. Hartley, P. D. Watson, P. J. Rizkallah and D. D. Jones, RSC Adv., 2015, 5, 77734–77738 RSC.
- S. C. Reddington, P. J. Rizkallah, P. D. Watson, R. Pearson, E. M. Tippmann and D. D. Jones, Angew. Chem., Int. Ed., 2013, 52, 5974–5977 CrossRef CAS.
- L. M. Oltrogge and S. G. Boxer, ACS Cent. Sci., 2015, 1, 148–156 CrossRef CAS.
- M. G. Romei, C.-Y. Lin, I. I. Mathews and S. G. Boxer, Science, 2020, 367, 76–79 CrossRef CAS.
- J. K. Villa, H. A. Tran, M. Vipani, S. Gianturco, K. Bhasin, B. L. Russell, E. J. Harbron and D. D. Young, Molecules, 2017, 22, 1194 CrossRef.
- G. M. Olenginski, J. Piacentini, D. Harris, N. Runko, B. M. Papoutsis, J. Alter, K. Hess, S. H. Brewer and C. M. Phillips-Piro, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2021, 77, 1010–1018 CrossRef CAS.
- N. Maurici, N. Savidge, B. U. Lee, S. H. Brewer and C. M. Phillips-Piro, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2018, 74, 650–655 CrossRef CAS.
- A. B. Dippel, G. M. Olenginski, N. Maurici, M. T. Liskov, S. H. Brewer and C. M. Phillips-Piro, Acta Crystallogr., Sect. D: Struct. Biol., 2016, 72, 121–130 CrossRef CAS.
- C. G. Bazewicz, M. T. Liskov, K. J. Hines and S. H. Brewer, J. Phys. Chem. B, 2013, 117, 8987–8993 CrossRef CAS.
- E. E. Smith, B. Y. Linderman, A. C. Luskin and S. H. Brewer, J. Phys. Chem. B, 2011, 115, 2380–2385 CrossRef CAS.
- E. M. Tookmanian, E. E. Fenlon and S. H. Brewer, RSC Adv., 2015, 5, 1274–1281 RSC.
- E. M. Tookmanian, C. M. Phillips-Piro, E. E. Fenlon and S. H. Brewer, Chem.–Eur. J., 2015, 21, 19096–19103 CrossRef CAS.
- C. G. Bazewicz, J. S. Lipkin, E. E. Smith, M. T. Liskov and S. H. Brewer, J. Phys. Chem. B, 2012, 116, 10824–10831 CrossRef CAS.
- B. Lee, B. M. Papoutsis, N. Y. Wong, J. Piacentini, C. Kearney, N. A. Huggins, N. Cruz, T. T. Ng, K. H. Hao, J. S. Kramer, E. E. Fenlon, P. S. Nerenberg, C. M. Phillips-Piro and S. H. Brewer, J. Phys. Chem. B, 2022, 126, 8957–8969 CrossRef CAS.
- J. T. First, J. D. Slocum and L. J. Webb, J. Phys. Chem. B, 2018, 122, 6733–6743 CrossRef CAS.
- J. D. Slocum, J. T. First and L. J. Webb, J. Phys. Chem. B, 2017, 121, 6799–6812 CrossRef CAS.
- J. D. Slocum and L. J. Webb, J. Am. Chem. Soc., 2016, 138, 6561–6570 CrossRef CAS.
- C.-Y. Lin and S. G. Boxer, J. Phys. Chem. B, 2020, 124, 9513–9525 CrossRef CAS.
- J.-D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2006, 24, 79–88 CrossRef.
- J. Chang, M. G. Romei and S. G. Boxer, J. Am. Chem. Soc., 2019, 141, 15504–15508 CrossRef CAS.
- B. G. Oscar, L. Zhu, H. Wolfendeen, N. D. Rozanov, A. Chang, K. T. Stout, J. W. Sandwisch, J. J. Porter, R. A. Mehl and C. Fang, Front. Mol. Biosci., 2020, 7, 131 CrossRef CAS.
- M. Kneen, J. Farinas, Y. Li and A. S. Verkman, Biophys. J., 1998, 74, 1591–1599 CrossRef CAS.
- K. Yokoyama, U. Uhlin and J. Stubbe, J. Am. Chem. Soc., 2010, 132, 8385–8397 CrossRef CAS.
- J. Xie and P. G. Schultz, Nat. Rev. Mol. Cell Biol., 2006, 7, 775–782 CrossRef CAS.
- R. B. Cooley, J. L. Feldman, C. M. Driggers, T. A. Bundy, A. L. Stokes, P. A. Karplus and R. A. Mehl, Biochemistry, 2014, 53, 1916–1924 CrossRef CAS.
- H. Neumann, J. L. Hazen, J. Weinstein, R. A. Mehl and J. W. Chin, J. Am. Chem. Soc., 2008, 130, 4028–4033 CrossRef CAS.
- S. J. Miyake-Stoner, C. A. Refakis, J. T. Hammill, H. Lusic, J. L. Hazen, A. Deiters and R. A. Mehl, Biochemistry, 2010, 49, 1667–1677 CrossRef CAS.
- R. Nathani, P. Moody, M. E. B. Smith, R. J. Fitzmaurice and S. Caddick, ChemBioChem, 2012, 13, 1283–1285 CrossRef CAS.
- J. L. Seitchik, J. C. Peeler, M. T. Taylor, M. L. Blackman, T. W. Rhoads, R. B. Cooley, C. Refakis, J. M. Fox and R. A. Mehl, J. Am. Chem. Soc., 2012, 134, 2898–2901 CrossRef CAS.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS.
- D. Liebschner, P. V. Afonine, M. L. Baker, G. Bunkóczi, V. B. Chen, T. I. Croll, B. Hintze, L. W. Hung, S. Jain, A. J. McCoy, N. W. Moriarty, R. D. Oeffner, B. K. Poon, M. G. Prisant, R. J. Read, J. S. Richardson, D. C. Richardson, M. D. Sammito, O. V. Sobolev, D. H. Stockwell and P. D. Adams, Acta Crystallogr., Sect. D: Struct. Biol., 2019, 75, 861–877 CrossRef CAS.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS.
- V. De Filippis, R. Frasson and A. Fontana, Protein Sci., 2006, 15, 976–986 CrossRef CAS.
- J. F. Riordan, M. Sokolovsky and B. L. Vallee, Biochemistry, 1967, 6, 358–361 CrossRef CAS.
- T. Pálla, E. Fogarasi, B. Noszál and G. Tóth, Chem. Biodiversity, 2019, 16, e1900358 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Room temperature pH dependent fluorescence spectra of wt-sfGFP, refinement of mNO2Y incorporated chromophore in sfGFP, and the alignment of the cyclized chromophores of Y66mNO2Y-sfGFP and wt-sfGFP. See DOI: https://doi.org/10.1039/d4ra05058d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |