Open Access Article
Open Access ArticleBlack phosphorus/silk fibroin films hamper filamentous and invasive growth of Candida albicans†
Martina Alunni Cardinali‡
a,
Debora Casagrande Pierantoni‡b,
Lucia Comez c,
Angela Contib,
Irene Chiesad,
Carmelo De Mariad,
Stefania Cortopassid,
Maria Caporali
c,
Angela Contib,
Irene Chiesad,
Carmelo De Mariad,
Stefania Cortopassid,
Maria Caporali e,
Alessandro Paciaroni
e,
Alessandro Paciaroni f,
Valeria Liberaf,
Gianluigi Cardinalib,
Paola Sassi
f,
Valeria Liberaf,
Gianluigi Cardinalib,
Paola Sassi a and
Luca Valentini
a and
Luca Valentini *gh
*gh
aDepartment of Chemistry, Biology and Biotechnology, University of Perugia, Via Elce di Sotto 8, 06123, Perugia, Italy
bDepartment of Pharmaceutical Sciences, University of Perugia, 06123 Perugia, Italy
cCNR-IOM, Istituto Officina dei Materiali, National Research Council of Italy, Via Alessandro Pascoli, 06123 Perugia, Italy
dDepartment of Ingegneria dell'Informazione and Research Center E. Piaggio, University of Pisa, Largo Lucio Lazzarino 1, 56122 Pisa, Italy
eInstitute of Chemistry of OrganoMetallic Compounds-ICCOM, National Research Council-CNR, Via Madonna del Piano 10, 50019 Sesto Fiorentino, Italy
fDepartment of Physics and Geology, University of Perugia, Via A. Pascoli, 06123, Perugia, Italy
gCivil and Environmental Engineering Department, University of Perugia, Strada di Pentima 4, 05100, Terni, Italy. E-mail: luca.valentini@unipg.it
hItalian Consortium for Science and Technology of Materials (INSTM), Via Giusti 9, 50121 Firenze, Italy
First published on 11th December 2024
Abstract
The ability of fungi and bacteria to form biofilms on surfaces poses a serious threat to health and a problem in industrial settings. In this work, we investigated how the surface stiffness of silk fibroin (SF) films is modulated by the interaction with black phosphorus (BP) flakes, quantifying the morphogenesis of C. albicans cells. Raman and infrared (IR) spectroscopies, along with scanning transmission electron microscopy, allowed us to quantify the thickness and diameter of BP flakes dispersed in the SF matrix (e.g., 5.5 nm in thickness and 20 μm in diameter), as well as an increase in beta-sheet secondary structures, resulting in the mesoscopic formation of a globular and nanofibrous surface. The formation of β-sheet crystals in the SF/BP film was correlated with a higher surface stiffness, influencing the shape of C. albicans cells and suppressing their filamentous growth. Raman spectroscopy analysis ultimately suggests an overall reduction in cell vitality and filmogenic capability of cells grown on fibroin-based films containing BP. Our results suggest that the conformational properties of SF can be suitably tuned to design optimized bioselective coatings for biomedical applications.
1. Introduction
Silk fibroin (SF) is an ancient natural biopolymer that was approved as a biomaterial by the United States Food and Drug Administration in 1993, thanks to the unique combination of properties that it offers.1 Despite this outstanding interest in SF, its inherent antibacterial activity is limited to a preclinical trial for skin wound healing,2 and it has been demonstrated only in combination with antibiotics,3 inorganic nanoparticles,4,5 antibacterial polymers,6–8 peptides, and plant extracts.9,10 The effect of SF macromolecular composition and conformational transitions on the bioactivity of microbes remains ambiguous. SF is a complex network of interconnected crystalline and non-crystalline domains. In an elegant experiment of biotechnological engineering performed on a similar material (e.g. recombinant silk spidroin), it was shown how the charge or amino acid sequence of spider silk is expected to impact crystallite size and distribution modifying the silk protein affinity for cell adhesion.11The use of regenerated Bombyx mori SF as a substrate for microbial adherence, growth, and biofilm formation represents a novel and emerging research field with potential applications in bioanalytical sciences.12 Regenerated silk can be processed into solid films with potential use as coatings for medical devices.13–15 At the nanoscale, these films self-assemble with silk I and silk II secondary conformations, which, depending on the processing parameters, result in a denser and less homogeneous packing of β-crystallites that trigger the film stiffness.16 Microbial cell adhesion and network formation are substrate stiffness dependent.17 Moreover, at the mesoscale, the organization of β-sheet crystallites influences the surface morphology with nanofibrous features.18 Consequently, the utilization of this versatile material enables the mediation of cell–cell interactions via an elastic substrate, that can drive the formation of networks sensitive to the structure of the substrate.
Along with SF, black phosphorous (BP) is a biocompatible inorganic nanomaterial.19 In our previous experiment, we demonstrated the use of SF aqueous solution as an exfoliating agent to produce BP nanosheets maintaining the stability of the flakes' dispersion into a 3D printable SF-based biomaterial for several days.20
As previously observed for SF interacting with 2D nanomaterials,21 the interaction between SF and BP can be triggered by changing the pH of the solution promoting the silk II secondary structure of SF adsorbed on BP. The secondary structure of SF chains can self-assemble on mesoscale giving rise to the formation of a nanofibrous structure.
In this study, we exploit the synergistic effect of pH and BP addition to deposit a SF coating with silk II features. We showed that exfoliated BP promotes a homogeneous β-sheet crystalline distribution in the nanofibrous surface morphology. The effect of β-sheet content and surface morphology on the proliferation of C. albicans cells was investigated and compared to growth on an uncovered substrate. The data suggest that the designed conformational transitions of SF provide new insights into bioselective coatings for biomedical applications.
2. Experimental
2.1 Preparation of SF and SF/BP films
Silk cocoons were supplied by a local farm (Fimo srl, Milano, Italy). Black phosphorous, phosphate-buffered saline (PBS), CaCl2, FA and NaHCO3 were supplied by Sigma-Aldrich. Silk solutions were prepared as reported elsewhere.22 Briefly silk cocoons were degummed with NaHCO3 and dispersed into 5 ml FA/CaCl2 solution (CaCl2 was in weight ratio of 70/30 with respect to the silk amount (0.70 g)). SF films were produced by leaving the SF solutions to evaporate onto Petri dishes overnight with subsequent annealing at 40 °C for 2 hours. The SF film was subsequently dispersed in 100 ml of 1× PBS (pH 7.4) along with the BP using an ultrasound treatment for 1 hour (amplitude 40%, tip 13 mm, pulse 40 s/20 s), resulting in three different concentrations: 0.2, 0.1, 0.05 and 0.02 mg ml−1. The optimal concentration of BP was evaluated based on the distribution of the diameters of the flakes, analyzed using ImageJ for the equivalent diameter of the three different solutions deposited on a glass substrate (see ESI and Fig. S1†). For this analysis, a quantity of 1.85 mg of BP proved to be the most effective for exfoliation in SF and PBS. SF and SF/BP films were obtained by drop casting the PBS solutions onto a square shaped mold (1 cm × 1 cm) positioned onto fluorine tin oxide (FTO) substrate. Before casting, both solutions were sonicated for 20 minutes in an ultrasonic bath at 59 kHz and room temperature.2.2 Characterization of SF and SF/BP solutions
Contact angle measurements of the SF and SF/BP solutions in PBS were performed at room temperature using an optical tensiometer, performing a sessile drop analysis (n = 20, drop volume = 2 μl) according to the Young–Laplace law. FTO slides were tested as substrates.2.3 Film characterization
2.4 Yeast cell culture on films
A strain of the yeast species C. albicans (CMC 6549, from the CEMIN fungal collection at the University of Perugia) was used for biofilm formation. This strain was classified according to the sequencing of the ITS marker. For the preparation of the inoculum, the strain was grown in YPD medium (containing yeast extract 1%, peptone 1%, and dextrose 2% – from Difco Laboratories, Detroit, MI, USA) overnight at 37 °C under shaking conditions (120 rpm). The fungal suspension was then calibrated to an optical density of OD600 = 0.1 in the medium selected for biofilm formation. Biofilms were obtained by submerging UV-sterilized substrates in Petri dishes containing RPMI-1640 medium buffered with MOPS (3-(n-morpholino)propane sulfonic acid) to pH 7, into which the strain was inoculated. A coupon of the tested material was inoculated for each time point. After the allotted time of growth, the coupon was washed two times with sterile saline solution (0.9% NaCl) and stained with a 0.01% p/V crystal violet solution for 15 minutes at room temperature. Finally, the substrates were washed with sterile distilled water and left to dry for further analysis. Optical images of C. albicans cells on different substrates were recorded with a 3D Hirox digital microscope (HRX-01) 3000×, FOV (H)104.04 μm, resolution 0.04 μm. Changes from the circular shape of the C. albicans cells were obtained with the ImageJ program. The cells have been grouped into classes according to their circularity, i.e. the ratio between their lengths on the two axes, taking into account a binning of 0.05 between 0 and 1 (circular shape). Cell forms were counted using the ImageJ program and the percentages of the individual forms were represented graphically.2.5 Statistical analysis
GraphPad Prism 9.2.0.332 (GraphPad software, San Diego, CA, USA) was used to assess the statistical significance of all comparison studies in this work. In the statistical analysis for comparison between multiple groups, a two-way ANOVA with Tukey's post hoc analysis (multiple comparisons) was conducted with the significance threshold of ***p < 0.001.3. Results and discussion
3.1 Characterization of SF and SF/BP films
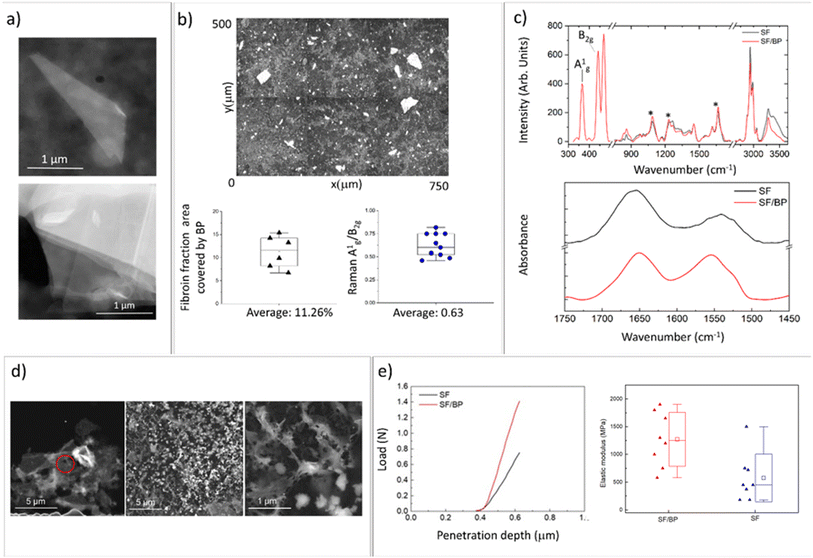
The SF/BP films were prepared by the liquid-assisted exfoliation of BP using SF as an exfoliating agent. Fig. 1a shows the STEM bright field image of the BP flake obtained after SF assisted exfoliation process. In Fig. 1a a STEM dark field image of a thin folded BP flake is shown together with the image of a multilayer flake. The interlayer spacing as well as the thickness of single layer are well known from literature,23 allowing for estimation of the thickness of the observed flakes. Considering 0.53 nm as interlayer spacing and that the thickness of each layer is 0.22 nm, we can roughly estimate a thickness of 5.5 nm.Fig. 1b shows the optical image of the surface of SF/BP films. Using the ImageJ software, the percentage fraction covered by BP flakes was calculated to be approximately 12% of the total surface area. In addition, the thickness of the flakes was estimated through the ratio of relative Raman intensities (Fig. 1c, top panel) of two characteristic bands, namely A1g and B2g, calculated by integrating the areas of the respective peaks at 365 and 440 cm−1. This ratio was found to be correlated by Coleman et al.24 with the thickness of the BP flakes and was used by our group in combination with AFM and STEM to verify the thickness reduction before and after the BP exfoliation process.22 In this experiment, the ratio was found to be about 0.6, consistent with STEM analysis and previous measurements of BP flakes obtained by liquid exfoliation.22
To analyze how the SF secondary structure is modified by the interaction with BP, we employed FTIR spectroscopy (Fig. 1c, right panel). Amide I (1600–1700 cm−1) and amide II (1500–1650 cm−1) FTIR bands were used to monitor the conformational changes of the SF backbone.25 Typical absorption bands of random coil are located at 1640–1654 cm−1 and 1535–1545 cm−1, whereas the bands at 1610–1630 cm−1 and 1510–1520 cm−1 are assigned to β-sheets.26–28 By focusing on the amide I region, we determined the percentages of different secondary structures (β-sheets, turns, and random coils). As shown in Fig. S2,† the addition of BP increases the fraction of the peak components in the ranges of 1620–1625 (amide II) and 1697–1703 (amide I) that are related to the antiparallel β-sheet structure.29–31 Specifically, the β-sheet content in the SF/BP film was found to be approximately 20%, which is higher than the 14% β-sheet content in the SF film.
This result was confirmed by the analysis of SF (black) and SF/BP (red) Raman spectra reported in Fig. 1c (on top). The positions of the amide I band at 1665 cm−1, amide III at 1240 cm−1 and C–C stretching at 1095 cm−1 suggest for SF film a prevalence of β-sheet conformation characteristic of silk II. Notably, in the presence of BP, the relative amount of this crystalline component of SF is enhanced, as evidenced by the higher intensity of the 1240 cm−1 peak.
The different secondary structures of SF trigger the specific interaction with the surface of both nanomaterials and adsorbing surfaces: the chain of SF derived from Bombyx mori consists of hydrophobic blocks connected by short hydrophilic spacers.32 These hydrophobic blocks contain crystalline β-sheet regions crucial for the mechanical strength (see below) and stability of silk biomaterials.32,33 So the conformational transition of SF depends on the hydrophilicity/hydrophobicity of the substrate. It has been demonstrated that hydrophobic surfaces favor random coil conformation, whereas hydrophilic surfaces favor β-sheet conformation.34,35 At pH 7.4, due to the charged state of BP,36 hydrophilic interactions between fibroin and BP dominate and enable the increase of β-sheet structures.32 To better understand this mechanism, we investigated the adsorption of fibroin on BP by scanning transmission microscopy (Fig. 1d). While the morphology of SF film prepared from pristine FA solution results in a dense and homogeneous surface (see Fig. S3†), upon redissolving in PBS we observed the formation of silk globules on BP flakes with nanofibrous morphology. SF nanofibrils and microfibrils characterized by β-sheet structures were observed to be non-toxic toward neuronal cells.37
At the mesoscale, the interaction of SF with the adsorbing surface was investigated by contact angle measurements (see Fig. S4†). Here, the electrostatic interactions of SF redissolved in PBS and adsorbed on a hydrophilic surface (i.e. FTO substrate) resulted in an increase in contact angle values, reducing the stability of random coil conformation and promoting the formation of β-sheets.
The impact of conformational transitions on the mechanical properties of SF and BP/SF films was then investigated through AFM tests. Fig. 1e shows the force-depth penetration curves using a triangular pyramidal tip. These curves were obtained following the procedure described by Jiang et al.38 The data regarding the increase in the reduced elastic modulus after the insertion of the BP (e.g. 600 MPA for SF and 1630 MPa for SF/BP films) are consistent with both other literature data that report a similar effect with the increase of β structures39,40 and with a penetration depth of hundreds of nanometers, which is comparable to multidomain backbones associated with the hard fraction.41
3.2 SF and SF/BP films applications in microbiology
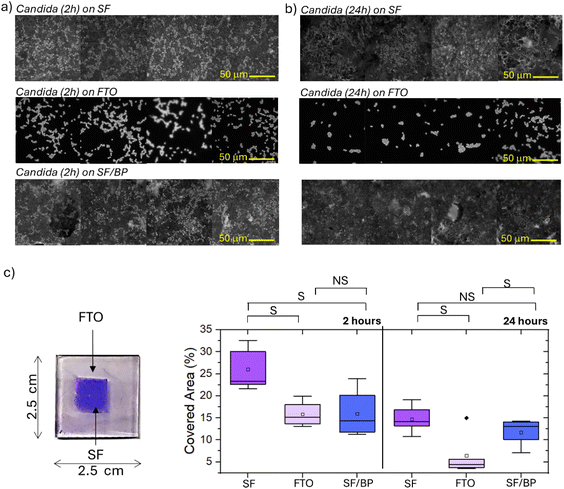
Before any microbiological assessment, it is mandatory to validate the effects of sterilization. Here, we explored whether sterilization processes (1 minute UV treatment to both sides of the FTO support using six 15 W tubes emitting light at 312 nm, with a 100 W total power of the setup and a distance of 10 mm between sample and lamps) had an impact on the secondary structures of the SF and SF/BP films on FTO. Raman spectroscopy (see Fig. S5†) revealed the stability of the protein conformation in the SF and SF/BP films and the preservation of the silk II structure after sterilization via UV irradiation. Indeed, analysis of the second derivative spectra in the amide I region (Fig. S5,† right panel), which is much more sensitive to conformational changes, shows no shift of the central minimum for fibroin, whether in the absence or presence of black phosphorus.After sterilization, the SF, SF/BP films on FTO as well as the FTO substrate were cultured with C. albicans for 2 and 24 hours, following the procedure reported in the Experimental section. Then, the different substrates cultured for 2 and 24 h were treated with crystal violet dye and observed under fluorescence microscopy, using LED light for excitation (filtered around 440 nm) and collected at about 470 nm (Fig. 2a and b). Images in Fig. 2a highlight that C. albicans cells after 2 h arrange themselves differently on the various substrates, specifically the FTO glass, the SF and the SF/BP films. For each image the fraction of covered area was quantified using ImageJ software; the results are reported in the box plots presented in Fig. 2c. In particular, the SF films promote a higher adhesion of cells to the surface after 2 h with respect to other substrates. Both FTO and BP have an inhibitory effect on cell adhesion in comparison to SF, although none of them is completely refractory. Interestingly, after 24 h (Fig. 2b and c), there was an overall decrease in the fraction of area covered by cells for all three samples. In fact, after a certain number of hours in culture, the dominant factor is likely no longer the cells' initial ability to adhere to the substrate but rather the presence of favorable conditions for growth.
The adherence of C. albicans cells to surfaces occurs almost immediately after the immersion of the substrate in the cellular suspension. It typically displays a kinetic with a rapid initial phase that lasts approximately 10 h, followed by a slower adherence rate for the next hours.36 A thick biofilm is normally produced within 24 h by active biofilm-forming strains. The ability to produce several successive layers of cells is considered one of the factors making the biofilm resistant to antifungal agents and in general one of the key features that determines the success of a strain in hospital settings.42
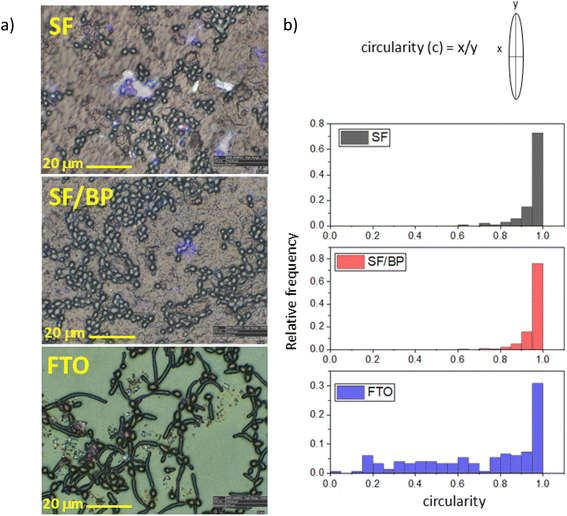
The different attitude to growth is evidenced also by the change in morphology depending on whether the cells are located on fibroin or FTO substrates. This is clearly appreciable in the optical images of SF, SF/BP films and FTO surface reported in Fig. 3a. Changes in the C. albicans morphology are reported in Fig. 3b. Round-shaped cells are observed in both SF and SF/BP samples, while in the case of FTO, they appear elongated and filamentous deviating from circular shape. Recently, it was shown that substrate properties, such as its stiffness, are known to play a crucial role in bacterial colonies.43 In particular, it was predicted that cell network formation is optimal at intermediate stiffness.43 Accordingly, we observed that C. albicans cells tend to form a branched network on an intermediate stiff SF/BP substrate (e.g. between the softer SF and the stiffest FTO). Thus, the physical properties of the underlying substrate play a crucial role in modifying the aspect of cells and function that contribute to biofilm phenotypes. The analysis of the individual forms allows the conclusion that, as the substrate changes, the number of forms changes. The percentage of the cell forms (more than 80%) had a correct shape as reported in Fig. 3. The biofilm thickness reported in Fig. S6† reflects this behavior. In particular, the profile shown in Fig. S6† is consistent with what is reported in fungal taxonomy,44 intercepting a spherical cap with a thickness of 1.4 micrometers compared to a cell thickness of 3–4 micrometers. After incubation on FTO about 30% of the cells retained their correct shape while hyphae and pseudo-hyphae accounted for 70% of the other cells.
Both the spatial arrangement of round cells and the formation of elongated structures are further highlighted by microscope image-stitching (see Fig. S7†) and by using differential interference contrast (DIC) microscopy on selected samples, whose contrast allows for clear distinction of shapes (Fig. S8–S10†). The rounded-shaped cells in SF and SF/BP have a diameter of about 3–4 micrometers, consistent with the average dimensions found in the literature for C. albicans,44 whereas the filamentous-shaped cells in FTO are about 15 micrometers long. It is worth noting that the affinity of C. albicans for fibroin was also tested after 30 minutes and after 9 hours to verify both the even faster adhesion capacity of the cultured cells and their ability to grow on the substrate. The data shown in Fig. S11† demonstrates that at very fast times adhesion is almost completely negligible and that, in line with what would be expected at intermediate times, the total number of cells on the substrate is lower than at both 2 and 24 hours.
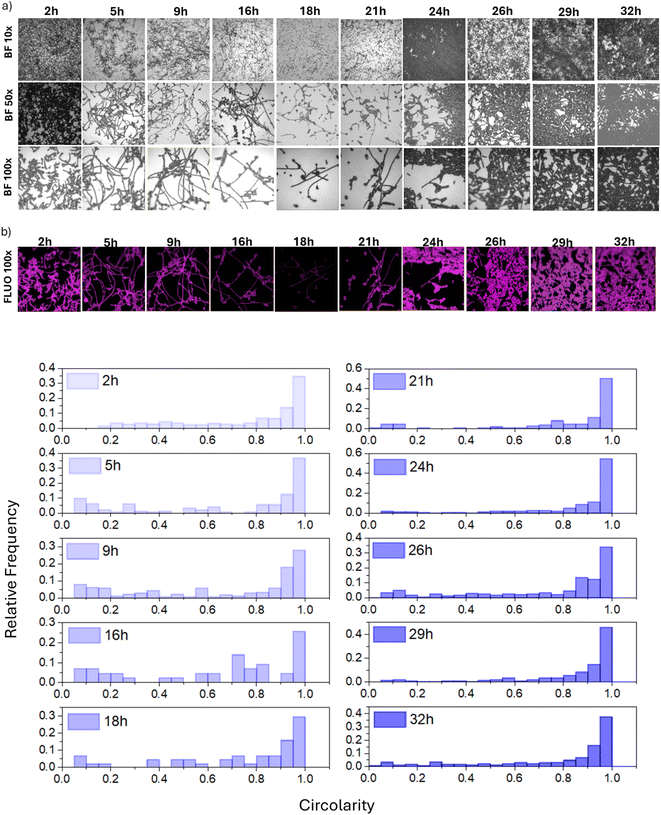
The formation of filamentous structures on FTO has been further investigated using fluorescence microscopy. Specifically, Fig. 4a shows the optical images at increasing magnifications (10×, 50× and 100×) of the FTO substrate and Fig. 4b) the fluorescence image (FLUO 100×) collected on the same area of 100× images after the crystal violet staining. Interestingly, the images demonstrate that proliferating cells primarily adopt an elongated phenotype (Fig. 4c). This phenomenon became more pronounced when monitoring the progress of the culture over time.
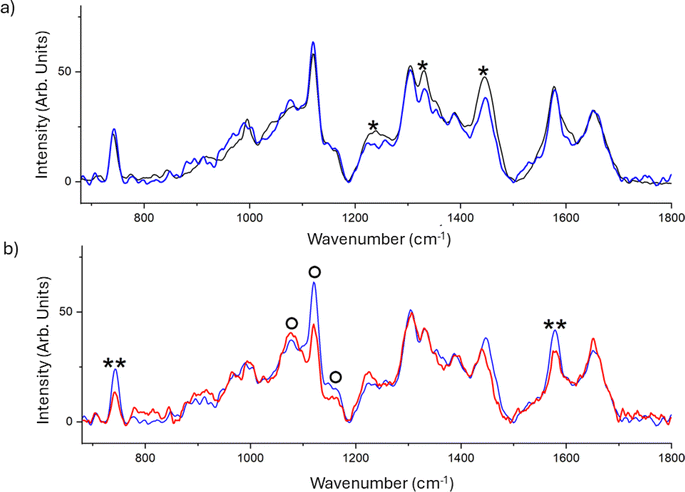
The chemical fingerprint of the different phenotypes expressed by C. albicans on the different substrates was monitored by Raman spectroscopy. This technique is widely used both for the study of different types of microorganisms and for the characterisation of biofilms.45–48 Results reported in Fig. 5a display the averaged spectra of C. albicans cells grown on FTO (black) and SF film (blue). The comparison reveals a significant decrease in the peak at 1240, 1331 and 1445 cm−1 in the spectrum of cells grown on SF. The decrease of these spectral signatures, that is attributed to the proteic constituents of the cell,49 can suggest a slowing in the metabolism that hinders the protein synthesis or a damage to the mannoproteins of the outermost layer of the cell wall.
The effect of the presence of BP dispersed in the SF film was further investigated, and the mean spectra of the C. albicans cells grown on SF (blue) and SF/BP (red) are shown in Fig. 5b. The presence of BP clearly induces significant alterations in the spectral profile of C. albicans cells, specifically: the intensity decrease of signals assigned to the reduced form of the cytochrome c at about 750 and 1585 cm−1, and an evident band-shape alteration in the spectral region between 1050 and 1150 cm−1, which contains signals from the chitin backbone in the wall of cells. Using the 532 nm excitation, we selected the resonant condition for the reduced form of mitochondrial cytochrome c, thereby increasing our sensitivity to monitor this species by enhancing the corresponding Raman signals. We observed a decrease in the bands characteristic of cytochrome c, indicating oxidative stress-induced damage, only in the presence of BP. Additionally, the altered profile in the region of C–O/C–C stretching bands of chitin, particularly the stretching of the C–O–C bond between neighboring glycosidic rings at 1070 cm−1 and the C–O–C bond within glycosidic units at 1150 cm−1, suggests a reduction in the crystallinity of the glycoprotein.49 This implies significant modifications in the cellular wall, which plays a crucial role in the cell adhesion mechanism. The underlying mechanism by which BP contributes to inducing oxidative stress in C. albicans cells may be attributed to the hydroxyl radical overproduction by the reaction between BP nanosheets and H2O2; the BP nanosheets degradation under oxidative stress conditions promotes the interaction between the BP nanosheets and cytochrome c thereby disrupting the cellular antioxidant defense system and ultimately producing toxicity.50
The chemical changes may tentatively indicate an overall reduction in cell vitality and filmogenic capability of cells when grown on the fibroin-based films, particularly in the presence of BP. In general, alterations in the region characteristic of carbohydrate signals indicate a stressed status of the planktonic cells, as also evidenced by FTIR measurements.51 It is worth noting that chemical alterations could precede morphological changes, as C. albicans on SF and SF/BP films exhibit the same cell shape on both substrates (see Fig. 3).
Moreover, the variations occurring in the first two hours can only be attributed to chemical changes in the molecules already present in the cells at the time of inoculation, as no changes in gene regulation could occur within this period.
While we are not able to reasonably conclude the fungistatic rather than fungicidal mechanisms of SF/BP films, we could, at this stage, invoke a similar mechanism observed on spider silk that due to its crystalline structure makes nitrogen inaccessible52 mitigating the bacterial proliferation.53,54 Although it may be unlikely that the secondary structures of regenerated silk are solely responsible for bacterial differentiation, these structures do evidently play some role that requires further investigation.
4. Conclusion
Our results provide evidence that bacterial colonies can respond to the composition and stiffness of the growing substrate. In particular, the variation in the different components of the secondary structures of SF films offers a tunable platform for designing substrate stiffness that impacts collective colony growth. In this study, we investigated how the structural features of regenerated silk proteins affect and possibly hamper the bioactivity of microbes without additional components, such as antimicrobial agents. We showed that SF promotes the exfoliation of BP flakes; the nanofibrous feature with a higher β-sheet crystalline distribution in SF/BP film creates a bioselective surface that reduces both the proliferation of C. albicans cells and their filamentous growth. This finding paves the way for novel applications of silk fibroin, such as coatings in various biomedical applications and for the preparation of inks for bioselective 3D-printed biomaterials.Data availability
The data supporting this article have been included as part of the ESI.† Fig. S1† optical images of SF/BP in PBS. Fig. S2† curve-fitting procedure of ATR-FTIR spectra and secondary structures content of SF and SF/BP films; Fig. S3† scanning transmission electron microscopy analysis of SF film obtained from FA; Fig. S4† contact angle measurements of SF and SF/BP solutions on FTO substrates; Fig. S5† Raman spectra of SF and SF/BP before and after sterilization via UV irradiation; Fig. S6 and S7† profilometry and microscope-images of C. albicans cells; Fig. S8–S10† high resolution image of C. albicans cells on SF, SF/BP and FTO substrates; Fig. S11† fluorescence images of cells on SF and SF/BP films after the crystal violet staining.Author contribution
All authors have approved the final version of the manuscript. L. V. conceptualization: equal; funding acquisition: lead; supervision: lead; writing – original draft: equal; writing – review & editing: equal; M. A. C. data curation: lead; formal analysis: lead; investigation: lead; methodology: equal; writing – original draft: equal; writing – review & editing: equal; D. C. P. data curation: lead; formal analysis: lead; investigation: lead; methodology: equal; writing – original draft: equal; writing – review & editing: equal; L. C. investigation: equal; writing – original draft: equal; writing – review & editing: equal; A. C. investigation: equal; I. C. data curation: equal; investigation: supporting; writing – review & editing: supporting; C. D. M. funding acquisition: equal; writing – review & editing: equal; S. C. investigation: supporting; M. C. investigation: supporting; writing – review & editing: supporting; A. P. data curation: equal; methodology: equal; writing – review & editing: equal; V. L. investigation: supporting; G. C. conceptualization: equal; methodology: equal; writing – review & editing: equal; P. S. conceptualization: equal; methodology: equal; writing – review & editing: equal. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been funded by the European Union – NextGenerationEU under the Italian Ministry of University and Research (MUR) National Innovation Ecosystem grant ECS00000041 – VITALITY – CUP J97G22000170005 and CUP B43C22000470005. This study received funding from the European Union – Next-GenerationEU – National Recovery and Resilience Plan (NRRP) – Mission 4 Component 2, investment n. 1.1, Prin 2022 – Prometheus “4D printing self-deploying bio-enabled polymer scaffolds for the non-invasive treatment of bleeding intestinal ulcers”, grant: 2022BZLTTK, CUP I53D23002200006. The authors also thank SIMITECNO for the use of HIROX microscopes to acquire high-resolution optical images.References
- Y. Qi, H. Wang, K. Wei, Y. Yang, R. Y. Zheng, I. S. Kim and K. Q. Zhang, Int. J. Mol. Sci., 2017, 18, 237 CrossRef PubMed.
- W. Zhang, L. Chen, J. Chen, L. Wang, X. Gui, J. Ran, G. Xu, H. Zhao, M. Zeng, J. Ji, L. Qian, J. Zhou, H. Ouyang and X. Zou, Adv. Healthcare Mater., 2017, 6, 1700121 CrossRef.
- A. J. Choudhury, D. Gogoi, J. Chutia, R. Kandimalla, S. Kalita, J. Kotoky, Y. B. Chaudhari, M. R. Khan and K. Kalita, Surgery, 2016, 159, 539–547 CrossRef PubMed.
- L. Schnaider, Z. Toprakcioglu, A. Ezra, X. Liu, D. Bychenko, A. Levin, E. Gazit and T. P. Knowles, Nano Lett., 2020, 20, 1590–1597 CrossRef CAS.
- X. Fei, M. Jia, X. Du, Y. Yang, R. Zhang, Z. Shao, X. Zhao and X. Chen, Biomacromolecules, 2013, 14, 4483–4488 CrossRef CAS.
- D. Gogoi, A. J. Choudhury, J. Chutia, A. R. Pal, M. Khan, M. Choudhury, P. Pathak, G. Das and D. S. Patil, Biopolymers, 2014, 101, 355–365 CrossRef CAS.
- A. Liang, M. Zhang, H. Luo, L. Niu, Y. Feng and M. Li, Materials, 2020, 13, 285 CrossRef CAS.
- J. Lei, L. Sun, S. Huang, C. Zhu, P. Li, J. He, V. Mackey, D. H. Coy and Q. He, Am. J. Transl. Res., 2019, 11, 3919–3931 CAS.
- S. P. Miguel, D. Simões, A. F. Moreira, R. S. Sequeira and I. J. Correia, Int. J. Biol. Macromol., 2019, 121, 524–535 CrossRef CAS PubMed.
- A. Zakeri-Siavashani, M. Chamanara, E. Nassireslami, M. Shiri, M. Hoseini-Ahmadabadi and B. Paknejad, Int. J. Polym. Mater. Polym. Biomater., 2020, 71, 220–231 CrossRef.
- S. Kumari, G. Lang, E. DeSimone, C. Spengler, V. T. Trossmann, S. Lücker, M. Hudel, K. Jacobs, N. Krämer and T. Scheibel, Mater. Today, 2020, 41, 21–33 CrossRef CAS.
- S. Ghalei and H. Handa, Mater. Today Chem., 2022, 23, 100673 CrossRef CAS.
- U. Slotta, M. Tammer, F. Kremer, P. Koelsch and T. Scheibel, Supramol. Chem., 2006, 18, 465–471 CrossRef CAS.
- E. Metwalli, U. Slotta and C. Darko, Appl. Phys. A, 2007, 89, 655–661 CrossRef CAS.
- S. Wohlrab, K. Spieß and T. Scheibel, J. Mater. Chem., 2021, 22, 22050–22054 RSC.
- S. Keten, Z. Xu, B. Ihle and M. J. Buehler, Nat. Mater., 2010, 9, 359–367 CrossRef CAS PubMed.
- P. S. Noerr, J. E. Zamora Alvarado, F. Golnaraghi, K. E. McCloskey, A. Gopinathan and K. Dasbiswasas, Proc. Natl. Acad. Sci. U. S. A., 2024, 120, e2301555120 CrossRef PubMed.
- J. Li, S. Zhang, C. He and J. Ling, Int. J. Biol. Macromol., 2024, 254, 127685 CrossRef CAS PubMed.
- D. L. Childers, J. Corman, M. Edwards and J. J. Elser, BioScience, 2011, 61, 117–124 CrossRef.
- M. Alunni Cardinali, M. R. Ceccarini, I. Chiesa, S. Bittolo Bon, T. Rondini, M. Serrano-Ruiz, M. Caporali, S. Tacchi, A. Verdini, C. Petrillo, C. De Maria, T. Beccari, P. Sassi and L. Valentini, ACS Omega, 2024, 9, 17977–17988 CrossRef CAS PubMed.
- J. Burdick, C. Holland, D. Kaplan and M. Radisic, ACS Biomater. Sci. Eng., 2016, 2, 141 CrossRef CAS PubMed.
- V. Libera, R. Malaspina, S. Bittolo Bon, M. Alunni Cardinali, I. Chiesa, C. De Maria, A. Paciaroni, C. Petrillo, L. Comez, P. Sassi and L. Valentini, RSC Adv., 2024, 14, 22393 RSC.
- V. Sorkin, Y. Cai, Z. Ong, G. Zhang and Y. W. Zhang, Crit. Rev. Solid State Mater. Sci., 2016, 42, 1 CrossRef.
- D. Hanlon, C. Backes, E. Doherty, C. S. Cucinotta, N. C. Berner, C. Boland, K. Lee, A. Harvey, P. Lynch, Z. Gholamvand, S. Zhang, K. Wang, G. Moynihan, A. Pokle, Q. M. Ramasse, N. McEvoy, W. J. Blau, J. Wang, G. Abellan, F. Hauke, A. Hirsch, S. Sanvito, D. D. O'Regan, G. S. Duesberg, V. Nicolosi and J. N. Coleman, Nat. Commun., 2015, 6, 8563 CrossRef CAS.
- C. Zhang, D. Song, Q. Lu, X. Hu, D. L. Kaplan and H. Zhu, Biomacromolecules, 2012, 13, 2148–2153 CrossRef CAS.
- H. J. Jin, J. Park, V. Karageorgiou, U. J. Kim, R. Valluzzi, P. Cebe and D. L. Kaplan, Adv. Funct. Mater., 2005, 15, 1241–1247 CrossRef CAS.
- X. Hu, D. Kaplan and P. Cebe, Macromolecules, 2006, 39, 6161–6170 CrossRef CAS.
- B. P. Partlow, C. W. Hanna, J. Rnjak-Kovacina, J. E. Moreau, M. B. Applegate, K. A. Burke, B. Marelli, A. N. Mitropoulos, F. G. Omenetto and D. L. Kaplan, Adv. Funct. Mater., 2014, 24, 4615–4624 CrossRef CAS.
- D. M. Byler and H. Susi, Biopolymers, 1986, 25, 469–487 CrossRef CAS PubMed.
- J. L. R. Arrondo, A. Muga, J. Castresena and F. M. Goni, Prog. Biophys. Mol. Biol., 1993, 59, 23–56 CrossRef CAS PubMed.
- A. Dong, P. Huang and W. Caughey, Biochemistry, 1990, 29, 3303–3308 CrossRef CAS PubMed.
- S. Tadepalli, H. Hamper, S. Hyun Park, S. Cao, R. R. Naik and S. Singamaneni, ACS Biomater. Sci. Eng., 2016, 2, 1084–1092 CrossRef CAS PubMed.
- Y. J. Lin, W. Z. Cao and T. O. Yang, Sustainable Environ. Res., 2019, 29, 22 CrossRef CAS.
- G. Ramage, K. Vandewalle, B. L. Wickes and J. L. López-Ribot, Rev. Iberoam. De. Micol., 2001, 18, 163–170 CAS.
- D. Wilson, R. Valluzzi and D. Kaplan, Biophys. J., 2000, 78, 2690–2701 CrossRef CAS PubMed.
- C. Holland, K. Numata, J. Rnjak-Kovacina and F. P. Seib, Adv. Healthcare Mater., 2019, 8, 1800465 CrossRef.
- C.-Z. Zhou, F. Confalonieri, M. Jacquet, R. Perasso, Z.-G. Li and J. Janin, Proteins:Struct., Funct., Genet., 2001, 44, 119–122 CrossRef CAS.
- C. Jiang, X. Wang, R. Gunawidjaja, Y.-H. Lin, M. K. Gupta, D. L. Kaplan, R. R. Naik and V. V. Tsukruk, Adv. Funct. Mater., 2007, 17, 2229 CrossRef CAS.
- D. M. Ebenstein and K. J. Wahl, J. Mater. Res., 2006, 21, 2035 CrossRef CAS.
- D. M. Ebenstein, J. Park, D. L. Kaplan and K. J. Wahl, MRS Online Proc. Libr., 2005, 841, 57 CAS.
- H. Shulha, C. Wong, D. L. Kaplan and V. V. Tsukruk, Polymer, 2006, 47, 5821 Search PubMed.
- L. Corte, L. Roscini, C. Colabella, C. Tascini, A. Leonildi, E. Sozio, F. Menichetti, M. Merelli, C. Scarparo, W. Meyer, G. Cardinali and M. Bassetti, Sci. Rep., 2016, 6, 26860 CAS.
- M. E. Asp, M.-T. Ho Thanha, D. A. Germanna, R. J. Carrolla, A. Franceski, R. D. Welch, A. Gopinathd and A. E. Patteson, PNAS Nexus, 2022, 1, 1–13 CAS.
- M. Staniszewska, M. Bondaryk, E. Swoboda-Kopec, K. Siennicka, G. Sygitowicz and W. Kurzatkowski, Braz. J. Microbiol., 2013, 44, 813 CAS.
- M. Alunni Cardinali, D. Casagrande Pierantoni, S. Caponi, L. Corte, D. Fioretto and G. Cardinali, Biophys. Chem., 2019, 254, 106249 CrossRef CAS PubMed.
- S. Mattana, M. Alunni Cardinali, S. Caponi, D. Casagrande Pierantoni, L. Corte, L. Roscini, G. Cardinali and D. Fioretto, Biophys. Chem., 2017, 229, 123–129 CrossRef CAS.
- L. Roscini, A. Vassiliou, L. Corte, D. Casagrande Pierantoni, V. Robert, C. Tascini, S. Mattana, M. Alunni Cardinali, S. E. Orfanos, D. Fioretto and G. Cardinali, Infect. Dis. Ther., 2018, 7, 27–34 CrossRef PubMed.
- S. Caponi, L. Liguori, A. Giugliarelli, M. Mattarelli, A. Morresi, P. Sassi, L. Urbanelli and C. Musio, Biophys. Chem., 2013, 182, 58–63 CrossRef CAS PubMed.
- G. Pezzotti, M. Kobara, T. Nakaya, H. Imamura, N. Miyamoto, T. Adachi, T. Yamamoto, N. Kanamura, E. Ohgitani and E. Marin, Int. J. Mol. Sci., 2022, 23, 5359 CrossRef CAS PubMed.
- Z. Fan, S. Wu, Z. An, Y. Wang, B. Xu, X. Wang, Y. Xu, H. Li, G. Duan, S. Zhang and X. Tian, Colloid Interface Sci. Commun., 2024, 62, 2215 Search PubMed.
- L. Corte, M. Tiecco, L. Roscini, S. De Vincenzi, C. Colabella, R. Germani, C. Tascini and G. Cardinali, PLoS One, 2015, 10, e0115275 CrossRef.
- Y. Termonia, Macromolecules, 1994, 27, 7378–7381 CrossRef CAS.
- N. Dinjaski, D. Ebrahimi, Z. Qin, J. E. M. Giordano, S. Ling, M. Buehler and D. L. Kaplan, J. Tissue Eng. Regener. Med., 2018, 12, e97–e105 CrossRef CAS.
- Q. Lu, B. Zhang, M. Li, B. Zuo, D. L. Kaplan, Y. Huang and H. Zhu, Biomacromolecules, 2011, 12, 1080–1086 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05126b |
| ‡ Shared first authorship. |
| This journal is © The Royal Society of Chemistry 2024 |