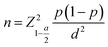Open Access Article
Open Access ArticleAntibacterial efficacy of low-dosage silver nanoparticle–sodium alginate–chitosan nanocomposite films against pure and clinical acne strains†
Bao Lam Thai Tran‡
 a,
Thanh Van Vo‡b,
Teng-Ping Chucd,
Duong Thai Bacha,
Thai Quang Nguyena,
Phuong Hong Bao Luue,
Vy Thi Thuy Tranb,
Hieu Hoang Duongb,
Nhi Hoang Nguyenf,
Gai Thi Lea,
Thu Thi Trang,
Khanh Nguyen Tuong Tranh,
Le Thi Cam Tuyenh,
Truong Ngoc Dinhij,
Ngo Ngoc Uyenj,
Tram Thi Thu Nguyenjk,
Ngoc-Van Thi Nguyenb,
Kien Trung Nguyena,
Le Thi Nhu Tran
a,
Thanh Van Vo‡b,
Teng-Ping Chucd,
Duong Thai Bacha,
Thai Quang Nguyena,
Phuong Hong Bao Luue,
Vy Thi Thuy Tranb,
Hieu Hoang Duongb,
Nhi Hoang Nguyenf,
Gai Thi Lea,
Thu Thi Trang,
Khanh Nguyen Tuong Tranh,
Le Thi Cam Tuyenh,
Truong Ngoc Dinhij,
Ngo Ngoc Uyenj,
Tram Thi Thu Nguyenjk,
Ngoc-Van Thi Nguyenb,
Kien Trung Nguyena,
Le Thi Nhu Tran *a and
Phuoc Huu Le
*a and
Phuoc Huu Le *cd
*cd
aFaculty of Medicine, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam. E-mail: ttnle@ctump.edu.vn
bFaculty of Pharmacy, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam
cCenter for Plasma and Thin Film Technologies, Ming Chi University of Technology, New Taipei City 243303, Taiwan. E-mail: phle@mail.mcut.edu.tw
dInternational PhD Program in Plasma and Thin Film Technology, Ming Chi University of Technology, New Taipei City 243303, Taiwan
eFaculty of Pharmacy, University of Medicine and Pharmacy at Ho Chi Minh City, 41-43 Dinh Tien Hoang Street, Ho Chi Minh City, Vietnam
fFaculty of Public Health, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam
gFaculty of Traditional Medicine, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam
hFaculty of Chemical Engineering, Can Tho University, 3/2 Street, Can Tho City, Vietnam
iDepartment of Physics and Biophysics, Faculty of Basic Sciences, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam
jDepartment of Biomedical Engineering, Faculty of Basic Sciences, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam
kDepartment of Chemistry, Faculty of Basic Sciences, Can Tho University of Medicine and Pharmacy, 179 Nguyen Van Cu Street, Can Tho City, Vietnam
First published on 21st October 2024
Abstract
The silver nanoparticles–sodium alginate–chitosan (AgNPs–Alg–Chi) nanocomposite film is a compelling material with demonstrated antibacterial efficacy against various pure bacterial strains. However, its potential cytotoxicity at elevated Ag doses warrants investigation. There is a notable dearth of studies assessing its antibacterial effectiveness against clinically relevant bacterial strains, notably Cutibacterium acnes. This study aims to assess the antibacterial efficacy of the low-dose AgNPs–Alg–Chi nanocomposite films on both pure bacterial strains and strains isolated from clinical samples obtained from 65 acne patients. The films were synthesized using green methods, incorporating kumquat (Citrus japonica) extract as a silver ion-reducing agent. The material characterization methods include UV-Vis and FTIR spectroscopies, SEM-EDS, XPS, cell culture, and MTT assay. We successfully fabricated the AgNPs–Alg–Chi nanocomposite films with a low-loading dose of Ag NPs (≤11 μg mL−1, and 37.8 ± 11.5 nm in size). The AgNPs–Alg–Chi nanocomposite film demonstrated comparable antibacterial efficacy to the AgNPs–Chi solution, with MIC values ranging from 3.67 to 5.50 μg mL−1 (p > 0.05) across all strains. Importantly, the AgNPs–Alg–Chi films demonstrated excellent biocompatibility with human keratinocytes (HaCaT cells), maintaining cell viability above 70%. The present AgNPs–Alg–Chi nanocomposite films synthesized by a green approach demonstrated potent antibacterial activity, making them promising for further development into suitable products for human use.
1 Introduction
According to the Institute for Health Metrics and Evaluation (IHME) in America, acne vulgaris accounted for 231 million cases globally in 2019, resulting in 4.96 million disability-adjusted life years.1 Acne significantly impacts aesthetics and social interactions, indirectly affecting patients' quality of life.2 Cutibacterium acnes (C. acnes) is a primary cause of acne vulgaris.3–5 Acne treatment options encompass antibiotics, retinoids, and adjunctive substances like benzoyl peroxide, salicylic acid, azelaic acid, and alpha-hydroxy.6 Yet, long-term use may induce dermatitis, desquamation, and hyperpigmentation.7 The rising interest in plant-based acne remedies poses questions regarding their mechanisms, adverse effects, and toxicity, warranting further investigation globally and in Vietnam.Noble metal-, carbon-based-, metal oxide-, and polymeric nanomaterials have been developed for antibacterial applications.8,9 Silver nanoparticles (AgNPs) are among the most extensively studied metal nanoparticles because of their superior conductivity, excellent light absorption, high sensitivity and resolution, strong antibacterial activity, and remarkable chemical stability.10–12
Due to the tendency of AgNPs to agglomerate, which affects dispersion stability, they should be engineered to crosslink with the polymeric matrix.13 The natural polysaccharide of chitosan and alginate enhances the material's antibacterial activity and anti-inflammatory effects.14,15 Silver nanoparticles–sodium alginate–chitosan (AgNPs–Alg–Chi) nanocomposite films are particularly appealing for biomedical applications such as drug delivery, wound dressing, tissue engineering, and antibacterial treatments.13,16–22 This is attributed to their biocompatibility, high loading capacity, favorable swelling behavior, antimicrobial activity, and cost-effectiveness.
Studies on humans have demonstrated that the utilization of AgNPs as a topical antibacterial agent was safe, and films containing AgNPs were deemed to be excellent wound dressings.14,17,18,22 Moreover, research on AgNPs' antibacterial applications has yielded promising results, particularly within the concentration range of 50 to 100 μg mL−1.23,24 Numerous investigations have demonstrated AgNPs' and AgNPs-based composite films' potent antibacterial properties against various pure bacterial strains.25–30 Meanwhile, the clinical bacterial strains have many mutations due to the natural selection process to adapt the abilities to exist harmful in the human body.31 However, the antibacterial activity of the materials on clinical bacterial strains is limited, raising the question of whether is there any difference in the antibacterial activity of a material against pure and clinical bacterial strains. In addition, these studies did not assess the minimum inhibitory concentration (MIC) value and clinical strain of bacteria for application in the clinical practice of the material. Furthermore, scant attention has been paid to microbial strains implicated in acne, notably C. acnes.
It is noteworthy that alongside its bactericidal efficacy, AgNPs may induce cytotoxic effects, with doses ≥44 μg mL−1, resulting in cell necrosis, and causing progressive membrane shrinkage after 4 to 6 hours of exposure with doses exceeding 11 μg mL−1.32 Yet, there remains a gap in understanding the bactericidal potential of AgNPs-based film at low concentrations, which circumvents cytotoxicity concerns. In addition, industrial chemicals synthesized still have limitations such as toxic chemicals, high operational costs, and energy needs.13 Therefore, it calls for a comparative study with (1) optimized design of green synthesis method for the process of AgNPs–Alg–Chi nanocomposite film (2) the antibacterial activity of a nanocomposite (i.e., AgNPs–Alg–Chi film); against pure and the isolated bacterial strains, notably C. acnes; (3) assessed the antibacterial activity and minimum inhibitory concentrations (MIC) of the materials for human safety and applications in clinical practice. Consequently, our study aims to address these gaps by assessing the antibacterial efficacy of AgNP-, Ag–Chi-solutions, and AgNPs–Alg–Chi-nanocomposite films synthesized through a green method at minimal Ag doses (≤11 μg mL−1). Specifically, we seek to evaluate the antibacterial activity of AgNPs–Alg–Chi nanocomposite films at low Ag-loading doses against pure and isolated bacterial strains derived from the clinical acne samples.
2 Materials and methods
2.1. Study design, sample size, and clinical sample collecting methods
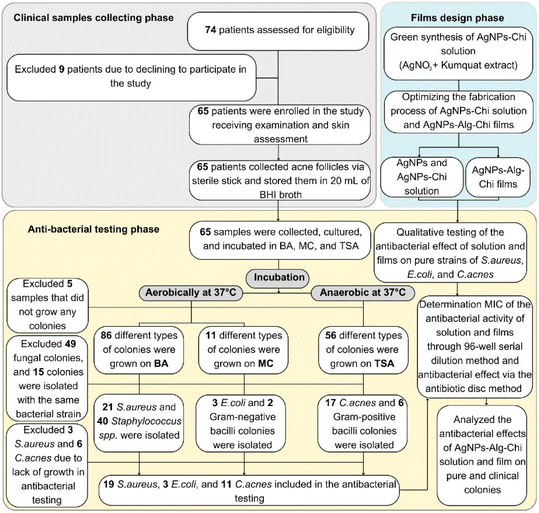
We conducted a cross-sectional descriptive study on 65 acne outpatients examined at Can Tho Dermatology Hospital from June 2022 to September 2023. Patients meeting the specified sampling and exclusion criteria were eligible for enrollment. The diagnosis of acne was made in accordance with the Vietnam Ministry of Health guidelines (2016), which are based on clinical symptoms, including the type of lesions (whiteheads, blackheads, papules, pustules, cysts) and their location, primarily in areas with high sebum secretion such as the face, back, and chest, with rare extension to the waist.33 Exclusion criteria included patients currently using systemic corticosteroids, those with diabetes, liver failure, or renal failure, individuals who had taken antibiotics within the past 2 weeks, and those who declined participation in the study.The sample size was determined using the one-proportion estimation formula.34
Referring to the 2019 Institute for Health Metrics and Evaluation (IHME) report, which indicated a global acne prevalence of 3.07%,1 we calculated a minimum required sample size of n = 46 (with p = 0.0307, α = 5%, and d = 0.05). To account for potential sample loss and mitigate biases, we enrolled 65 acne patients.
Enrolled patients underwent a clinical examination to assess signs and symptoms, and acne comedones were collected for analysis. The collection process involved several steps: (1) disinfecting the sampled acne area with 70% alcohol and allowing it to evaporate within 30 seconds, (2) using a lancet needle to puncture the acne comedo, (3) extracting the comedo and collecting it with a sterile cotton swab, (4) immediately placing the collected sample into a sterile tube containing 20 mL of Brain Heart Infusion (BHI) transport medium, (5) transporting the samples to the microbiology laboratory within 4 hours for culture.
2.2. Bacterial strains and growth conditions
We utilized pure bacterial strains for the study: S. aureus (ATCC 25923) cultured aerobically at 37 °C for 18–24 hours on Blood Agar (BA) (Merck KGaA, Germany); E. coli (ATCC 259212) cultured aerobically at 37 °C for 18–24 hours on MacConkey Agar (MC) (Merck KGaA, Germany); and C. acnes (ATCC 6919) cultured anaerobically at 37 °C for up to 72 hours on Tryptone Soya Agar (TSA) (Merck KGaA, Germany).Acne samples underwent culture on aerobic BA and MC at 37 °C for 18–24 hours and anaerobic TSA at 37 °C for 72 hours. Colonies grown on agar plates were individually subjected to gram staining and microbiological testing to identify bacterial strains (Graph S1†). The used identification agar media included (1) Mannitol Salt Agar (MSA) (Merck KGaA, Germany) for S. aureus identification, (2) Kligler Iron Agar (KIA) and (3) Sulfur Indole Motility Media (SIM) for E. coli and C. acnes identification (Graph S1†).
2.3. Green synthesis and characterization of silver nanoparticles solution, silver nanoparticles–chitosan nanocomposite solution, and silver nanoparticles–sodium alginate–chitosan nanocomposite films
To prepare the chitosan solution, dissolve chitosan in a 2% acetic acid solvent (Xilong, China) at 50 °C under vigorous magnetic stirring for 30 min. On the other hand, to obtain the Ag+ green reducing agent to Ag, a kumquat (Citrus japonica) extract was squeezed from the fresh fruit through a filter membrane and centrifuged at 6000 rpm for 10 min to separate the residue. Next, silver nitrate (0.1 mM, 520 μL, Xilong, China) and the kumquat extract were added to the as-prepared chitosan solution and heated at 90 °C for 60 minutes to synthesize silver nanoparticles–chitosan (AgNPs–Chi) solution (Fig. 1). Subsequently, an AgNPs–Alg–Chi solution was prepared by combining sodium alginate (concentration of 5% w/v, Shanghai Zhanyun Chemical, China) with the above AgNPs–Chi solution at a volume ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 under vigorous magnetic stirring for 90 min. Next, the resulting solution (20 mL) was poured into a Petri disc (diameter of 9 cm), and then the disc was annealed at 70 °C for 90 minutes in an oven to obtain an AgNPs–Alg–Chi nanocomposite film (Fig. 1). The 11 μg mL−1 AgNPs-loaded film (20 mL) was prepared from 130 μL of 0.1 mM AgNO3. For lower concentration films, such as 5.5 μg mL−1, 2.75 μg mL−1, 1.375 μg mL−1, and 0.6875 μg mL−1, the corresponding volumes were halved: 65 μL, 32.5 μL, 16.25 μL, and 8.125 μL of 0.1 mM AgNO3, respectively. Finally, we use sterile tools to precisely cut the nanocomposite film into a 6 mm-diameter circular disc, adhering to the standard size for antibiotic film in the agar diffusion method (ADM).35
1 under vigorous magnetic stirring for 90 min. Next, the resulting solution (20 mL) was poured into a Petri disc (diameter of 9 cm), and then the disc was annealed at 70 °C for 90 minutes in an oven to obtain an AgNPs–Alg–Chi nanocomposite film (Fig. 1). The 11 μg mL−1 AgNPs-loaded film (20 mL) was prepared from 130 μL of 0.1 mM AgNO3. For lower concentration films, such as 5.5 μg mL−1, 2.75 μg mL−1, 1.375 μg mL−1, and 0.6875 μg mL−1, the corresponding volumes were halved: 65 μL, 32.5 μL, 16.25 μL, and 8.125 μL of 0.1 mM AgNO3, respectively. Finally, we use sterile tools to precisely cut the nanocomposite film into a 6 mm-diameter circular disc, adhering to the standard size for antibiotic film in the agar diffusion method (ADM).35
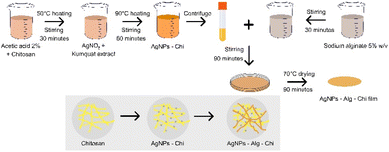 | ||
| Fig. 1 The synthesis procedure for silver nanoparticles–sodium alginate–chitosan nanocomposite film. Alg: sodium alginate; AgNPs: silver nanoparticles; Chi: chitosan. | ||
All synthetic Ag NPs-contained solutions were quality controlled using an ultraviolet-visible (UV-Vis) spectroscopy (Hitachi U-2900, Japan) by monitoring the appearance of Ag NPs plasmonic absorption peak in a range of 400–450 nm for this study, the solutions do not present the peak will be excluded from the study.
The surface morphologies of AgNP solution, Ag–Chi- and AgNPs–Alg–Chi-films were examined by scanning electron microscope (SEM, JEOL JSM-IT700HR, Japan). The size distribution of Ag nanoparticles was determined from SEM images using the freeware ImageJ. The chemical composition was analyzed by X-ray energy dispersive spectroscopy (EDS, Oxford probe) integrated with the SEM instrumentation. By combining SEM, EDS mapping, and UV-Vis spectroscopy, we conducted a comprehensive assessment of the dispersion and distribution of AgNPs within the nanocomposite films, ensuring the materials exhibited the desired properties and performance characteristics. To analyse the functional groups in AgNPs–Chi- and AgNPs–Alg–Chi-films with an AgNP dose of 11 μg mL−1, FTIR-ATR spectra of the materials were recorded using a PerkinElmer FT-IR/NIR spectrometer (Spectrum Two™, PerkinElmer Inc., Waltham, MA, USA), fitted with a Universal ATR (attenuated total reflectance) sampling accessory utilizing a diamond/ZnSe crystal. The surface chemistry of an AgNPs–Chi sample was analyzed using X-ray photoelectron spectroscopy (XPS, ThermoVG 350) with an Mg Kα X-ray source (power of 300 W, photon energy of 1253.6 eV). Curve fitting analysis was conducted using the XPSPEAK 4.1 software, assuming a Gaussian–Lorentzian peak shape, and employing Shirley background subtraction.
2.4. Antibacterial activity test
The antibacterial activity of the AgNPs–Alg–Chi nanocomposite films was assessed through three phases. First, the antibacterial activities of the synthesis solutions and the nanocomposite film were quantified by the Kirby–Bauer ADM method,35 performed on pure bacterial strains at a concentration of 100 μg mL−1 (Fig. S1†). Second, the antibacterial activity of AgNPs–Chi solution samples was quantified using the 96-well serial dilution method to determine the MIC, which is the lowest concentration of the material that inhibited bacterial growth (Fig. S2†). Third, the antibacterial activity of the AgNPs–Alg–Chi nanocomposite films with an Ag concentration adjusted to a human cell harmless concentration (≤11 μg mL−1)32 was quantified. The negative control was a dimethyl sulfoxide (DMSO) solution, while the positive control was antibiotics (Vancomycin for Gram-positive bacteria and Colistin for Gram-negative bacteria). This was done to determine the MIC of the nanocomposite film with the low Ag dosage, demonstrating the material's potential for human application. All antibacterial activity tests were conducted on Mueller Hinton Agar (MHA) (Merck KGaA, Germany).The antibacterial activity of the AgNPs–Chi solutions (concentrations of 11.0, 5.50, 2.75, 1.375, 0.688, 0.380, 0.190 μg mL−1) was evaluated by determining MIC for each bacterial strain and comparing it with the MIC of the AgNPs solution. The antibacterial effectiveness of the AgNPs–Alg–Chi nanocomposite films (concentrations of 11.0, 5.50, 2.75, 1.375, 0.688 μg mL−1) was assessed by measuring the zone of inhibition and MIC during the quantification phase. To demonstrate the antibacterial activity of the synthetic nanocomposite film, the MIC of the AgNPs–Chi solution was compared with the MIC of the nanocomposite films, analyzing their equivalence.
2.5. Cell culture and imaging
Human keratinocytes (HaCaT cells, ATCC 12191) were cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco, New York, USA) supplemented with 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. The culture medium was further enriched with 10% decomplemented fetal bovine serum (FBS; Avantor® Seradigm, Pennsylvania, USA). The cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. After 48 hours of culturing on the films, cell images were captured using a LEICA DM IL LED inverted microscope equipped with a Canon EOS 650D camera.2.6. Cell proliferation assay
The MTT assay [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich] was employed to assess the effect of AgNPs–Chi film and AgNPs–Alg–Chi films (size 5 × 5 mm2, AgNP dose of 11 μg mL−1) on cell viability.The films were placed in a 12-well plate at a cell density of 2 × 104 cells per well and incubated with HaCaT cells for 48 hours. The control samples consist of the four wells where no films were placed. Following incubation, MTT reagent was added to the cells and incubated at 37 °C for 3 hours. To dissolve the formazan crystals formed by metabolically active cells, dimethyl sulfoxide (DMSO) was added to each well. Cell proliferation was then quantified by measuring the absorbance at 570 nm, corresponding to the reduction of MTT to formazan. The results are expressed as the mean of four independent experiments.
2.7. Data bias control methods and data analysis
Data were entered and analyzed simultaneously by two researchers using SPSS 20.0 software to minimize bias. Quantitative variables with a normal distribution were described by mean ± standard deviation (SD), while those with a non-normal distribution were described using the median and interquartile range (IQR). Qualitative variables were presented as frequencies and ratios. Differences between two normally distributed quantitative variables were assessed using the independent T-test, and differences between non-normally distributed quantitative variables were evaluated using the Mann–Whitney test. Comparisons between two qualitative variables were made using the Chi-square test.2.8. Ethical approval
This study was approved by the Institutional Review Board of Can Tho University of Medicine and Pharmacy (approval number: 22.013.SV/PCT-HDDD). All participating patients were asked to fill out a consent form to participate in the study. The identities of all patients were kept confidential.3 Results and discussion
3.1. Clinical characteristics microbiological culture results of the study population
Our study involved 65 acne patients with an average age of 19.28 ± 4.05 years. Among them, 55.4% were women, and 26.15% had isolated C. acnes (Table S1†). The average duration of acne was approximately 12 months. The most common clinical symptoms were itching (58.5%) and pain (49.2%) (Table S1†). According to Karen McCoy's classification,36 the highest proportion of patients had mild acne (58.5%), and 98.5% of the patients had facial acne (Table S1†).Of the 65 acne samples collected from the clinic, 5 samples did not yield any bacterial growth, 49 exhibited fungal growth, and 15 showed cross-contamination between agar plates and were excluded from the study (Fig. 2). Among the isolated bacteria, Gram-positive cocci were the most prevalent (93.8%), S. aureus accounted for 32.3%, and C. acnes was present in 26.1% of the samples (Table 1)
| Bacteria strains | Number of isolated bacteria | Proportion per isolated bacteria number (%) | Proportion per patient (%) |
|---|---|---|---|
| a C. acnes: Cutibacterium acnes; E. coli: Escherichia coli; S. aureus: Staphylococcus aureus. | |||
| S. aureus | 21 | 23.6 | 32.3 |
| C. acnes | 17 | 19.1 | 26.1 |
| E. coli | 3 | 3.7 | 4.6 |
| Gram-positive cocci | 40 | 49.4 | 61.5 |
| Gram-positive bacillus | 6 | 6.7 | 9.2 |
| Gram-negative bacillus | 2 | 2.2 | 3.0 |
| Total | 89 | 100.0 | 136.7 |
Our study population primarily consisted of females aged 13 to 24 with mild to moderate acne severity (Table S1†). The clinical characteristics of the study group, detailed in ESI-4,† are consistent with the findings of Giavina-Bianchi et al. (2022)37 and Skroza et al. (2018).38 The prevalence of C. acnes in our study was 26.1% (Table 1), which is in line with the 27.3% prevalence reported by Miyazaki et al. (2023)39 and the 38.9% reported by Tran et al. (2019).40 Therefore, our sample population is representative of the general acne population.
3.2. Characterization of silver nanoparticles, silver nanoparticles–chitosan solution, and silver nanoparticles–sodium alginate–chitosan nanocomposite films
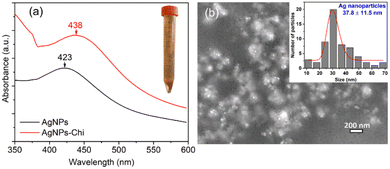
As shown in the inset of Fig. 3(a), after mixing Kumquat extract with the AgNO3–Chitosan solution for 60 minutes, the solution's color changed from colorless to yellowish-brown, indicating the formation of AgNPs. A typical UV–vis absorption spectrum of AgNPs and AgNPs–Chi solutions is presented in Fig. 3(a), exhibiting peaks at 423 nm and 438 nm, which corresponds to the surface plasmon resonance of AgNPs. Fig. 3(b) displays the AgNPs and their size distribution, ranging from 10 to 70 nm with a mean size of 37.8 ± 11.5 nm. The observed absorption peak of the AgNPs is similar to the peaks at 420 nm for AgNP sizes of 25–27 nm (ref. 41) and below 25 nm,42 and absorption peaks at 443–456 nm for AgNP sizes of 27–31 nm.10 The size of the AgNPs in this study is consistent with the reported sizes of 27.7–46.7 nm for AgNPs synthesized using tannic acid,43 and 10–60 nm for AgNPs synthesized using kumquat juice as a reducing agent.44 However, it is larger than the mean size of 14 ± 3 nm for AgNPs synthesized using AgNO3 and alginate as a natural biopolymer reducing agent at a heating temperature of 90 °C.13 The variations in Ag NP size were attributed to differences in the synthesis methods and conditions.The green synthesis of Ag nanoparticles–chitosan nanocomposite films was conducted by adding kumquat extract to aqueous solutions of silver nitrate and chitosan (Fig. 1), facilitating the reduction of Ag+ ions to Ag0. The biosynthesis of AgNPs was monitored using UV-Vis spectroscopy, revealing absorption peaks at 423 nm for AgNPs and in a range of 430–440 nm for AgNPs–Chi solution (≤11 μg mL−1). This absorption peak range is consistent with the known absorption peak of the Ag NPs,41 while chitosan solution did not show any peak in a range of 423–440 nm. This evidence collectively verifies the formation of an AgNPs-loaded Chitosan matrix.
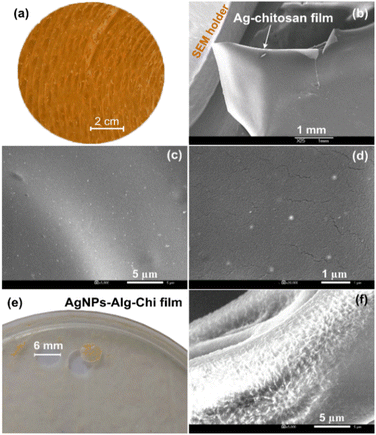
Fig. 4(a) and (b) present a photograph and a low magnification SEM image of the Ag–Chi film, revealing a yellow, rough surface and a free-standing capability. Higher magnification SEM images of the AgNPs–Chi solution in Fig. 4(c) and (d) demonstrate that low dosage of AgNPs are well-dispersed and uniformly embedded on/within the chitosan polymer. Fig. 4(e) and (f) display a photograph and an SEM image of the AgNPs–Alg–Chi film, which possesses good softness, and rougher surfaces but still retained tensile strength. In this film, the AgNPs were mostly hidden when AgNPs were primarily embedded within two polymeric alginate and chitosan materials.
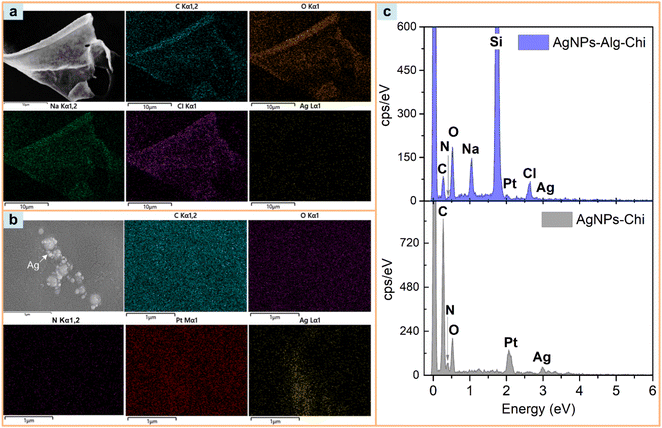
Fig. 5 presents the EDS mapping results and EDS spectra of AgNPs–Chi and AgNPs–Alg–Chi solutions prepared by dropping the solutions onto Si substrates, which were then coated with Pt for SEM imaging. In Fig. 5(a), the mapping was conducted on a representative segment of AgNPs–Alg–Chi solution. The EDS mapping spectrum confirms the presence of C, O, N, Na, Cl, Si, Pt, and Ag, with their distributions depicted in the images. As expected, the chitosan displayed elements of C, O, and N (with N originating from the amine functional group). Additionally, Ag from the AgNPs was relatively evenly distributed across the chitosan–alginate and its surrounding area (Fig. 5(a)). Moreover, the EDS mapping result for AgNPs–Chi confirms the decoration of Ag NPs on chitosan with an elemental composition containing C, O, N. These EDS results reflect the overall elemental composition of the AgNPs–Chi, while the XPS results presented in the following section will reveal the surface composition of this nanomaterial.
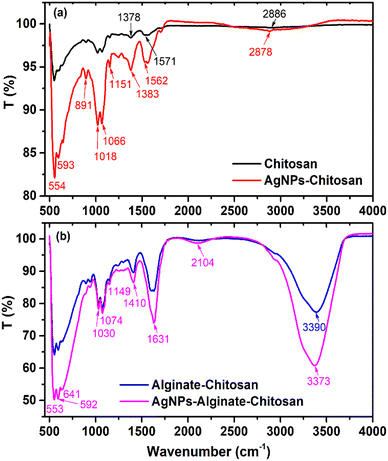
Fig. 6 presents the FTIR spectra of Chi, AgNPs–Chi, Alg–Chi, and AgNPs–Alg–Chi films. In the case of chitosan, the broad band between 2360 and 3500 cm−1 is attributed to the stretching vibrations of –OH and –NH (3200–3500 cm−1), with a central peak at 2886 cm−1 corresponding to the asymmetric stretching of –CH3 (2926 cm−1) and –CH2 (2856 cm−1) (Fig. 6(a)).45,46 Additionally, the absorption band at 1571 cm−1 is associated with N–H bending and C–N stretching of the Amide II region,47 while the peak at 1378 cm−1 can be attributed to the symmetric stretching vibration of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in carboxyl groups (Fig. 6(a)).45 The peaks observed between 1065 and 1017 cm−1 are ascribed to C–O stretching vibrations.46
O in carboxyl groups (Fig. 6(a)).45 The peaks observed between 1065 and 1017 cm−1 are ascribed to C–O stretching vibrations.46
For AgNPs–Chi films, although exhibiting stronger absorption and more intense spectra, they displayed similar absorption bands to those of chitosan. However, compared to chitosan, the FTIR absorption peaks of AgNPs–Chi showed slight shifts to lower wavenumbers, particularly in the –OH, N–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and C–N regions (Fig. 6(a)). These shifts suggest strong interactions between AgNPs and the functional groups of chitosan, providing evidence of chemical bonding and coordination within the AgNPs–Chi nanocomposite. Notably, the absorption band at 550–650 cm−1 in both chitosan and alginate–chitosan films is likely due to skeletal vibrations, particularly C–H out-of-plane bending or ring deformations within the polymer backbones of both chitosan and alginate. Although this band has been observed in some studies,46,47 it has yet to be definitively assigned. In AgNPs–Chi, this band is further intensified due to the contribution of Ag–O bonds, which fall within the same absorption range.
O, and C–N regions (Fig. 6(a)). These shifts suggest strong interactions between AgNPs and the functional groups of chitosan, providing evidence of chemical bonding and coordination within the AgNPs–Chi nanocomposite. Notably, the absorption band at 550–650 cm−1 in both chitosan and alginate–chitosan films is likely due to skeletal vibrations, particularly C–H out-of-plane bending or ring deformations within the polymer backbones of both chitosan and alginate. Although this band has been observed in some studies,46,47 it has yet to be definitively assigned. In AgNPs–Chi, this band is further intensified due to the contribution of Ag–O bonds, which fall within the same absorption range.
As shown in Fig. 6(b), the Alg–Chi and AgNPs–Alg–Chi films exhibited broad peaks at 3390 cm−1 and 3373 cm−1, respectively, corresponding to the O–H and N–H stretching vibrations,13,19 indicating the presence of hydroxyl groups in alginate and amine groups in chitosan. The peak at 1631 cm−1 is assigned to the amide I band (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), which primarily arises from the interaction between AgNPs and the carbonyl groups in both alginate and chitosan. The absorption band at 1410 cm−1 is attributed to the N–H stretching of the amide III band. Peaks at 1149 cm−1, 1074 cm−1, and 1030 cm−1 correspond to C–O–C stretching vibrations (Fig. 6(b)), originating mainly from alginate and chitosan. The peaks observed between 553 and 641 cm−1 are attributed to Ag–O bonds (Fig. 6(b)), confirming the successful incorporation of AgNPs into the Alg–Chi matrix.
O stretching), which primarily arises from the interaction between AgNPs and the carbonyl groups in both alginate and chitosan. The absorption band at 1410 cm−1 is attributed to the N–H stretching of the amide III band. Peaks at 1149 cm−1, 1074 cm−1, and 1030 cm−1 correspond to C–O–C stretching vibrations (Fig. 6(b)), originating mainly from alginate and chitosan. The peaks observed between 553 and 641 cm−1 are attributed to Ag–O bonds (Fig. 6(b)), confirming the successful incorporation of AgNPs into the Alg–Chi matrix.
The XPS analysis reveals the elemental composition and chemical states of the elements in the AgNPs–Chi nanocomposite film. The XPS survey spectrum in Fig. 7(a) shows the presence of carbon, nitrogen, oxygen, and a small peak of silver. These elemental peaks were fitted using a mixed Lorentzian–Gaussian line shape, as shown in Fig. 7(b)–(e). The C1s spectrum can be deconvoluted into C–C/C–H (284.1 eV), C–NH/C–NH2 (285.7 eV), and C–O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C–OH (287.5 eV).48 The characteristic Ag 3d peak, including Ag 3d5/2 (∼366.4 eV) and Ag 3d3/2 (∼375.3 eV),49 confirms the presence of a small amount of Ag NPs in the AgNPs–Chi nanocomposite film.
O/C–OH (287.5 eV).48 The characteristic Ag 3d peak, including Ag 3d5/2 (∼366.4 eV) and Ag 3d3/2 (∼375.3 eV),49 confirms the presence of a small amount of Ag NPs in the AgNPs–Chi nanocomposite film.
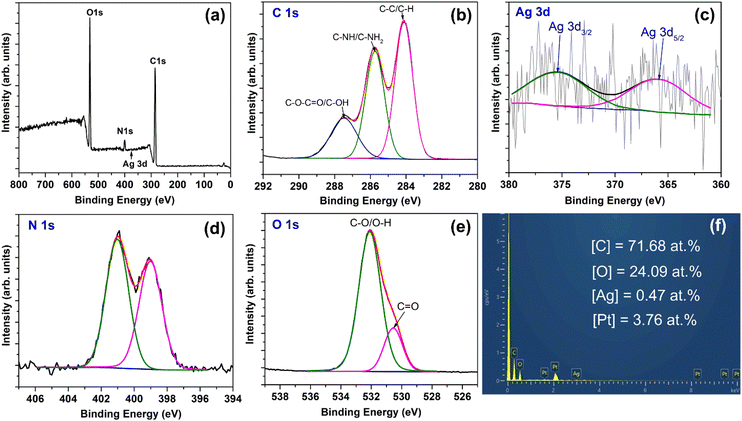 | ||
| Fig. 7 XPS spectra of AgNPs–Chi film: (a) survey spectrum, (b) C 1s, (c) Ag 3d, (d) N 1s, (e) O 1s. (f) EDS spectrum and the atomic composition of the AgNPs–Chi film. | ||
Fig. 7(f) shows the EDS spectrum, which detects C, O, Pt, and Ag elements, with the Ag atomic percentage determined to be 0.47 at%. Notably, Pt peaks appear in the EDS spectrum due to Pt coating for SEM imaging. Fig. 7(d) reveals the deconvoluted N 1s spectrum, with peaks at 399.0 eV and 401.0 eV attributed to C–NH2 and C–N bonds, respectively.50 The XPS spectrum of O1s [Fig. 7(e)] exhibits a peak at 530.5 eV corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds and a peak at 532.1 eV corresponding to C–O/O–H bonds.49 The XPS and EDS results verify the formation of the Ag–Chi composite in the nanocomposite film.
O bonds and a peak at 532.1 eV corresponding to C–O/O–H bonds.49 The XPS and EDS results verify the formation of the Ag–Chi composite in the nanocomposite film.
3.3. Antibacterial activity of silver nanoparticles–sodium alginate–chitosan nanocomposite films and silver nanoparticles–chitosan solutions
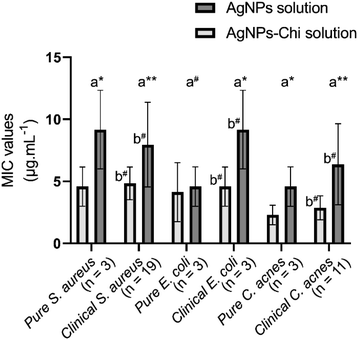
The MIC of the AgNPs–Chi solution was significantly lower than that of AgNPs for pure strains of S. aureus [4.58 ± 1.58 μg mL−1 vs. 9.17 ± 3.17 μg mL−1 (p = 0.038)], E. coli [4.13 ± 2.38 μg mL−1 vs. 4.58 ± 1.59 μg mL−1 (p = 0.096)] and C. acnes [2.29 ± 0.79 μg mL−1 vs. 4.58 ± 1.59 μg mL−1 (p = 0.038)] (Fig. 8). Additionally, there was no significant difference in the antibacterial activity of the Ag NPs and AgNPs–Chi solutions against pure and clinically isolated strains (Fig. 8). The antibacterial efficacy was assessed using the 96-well serial dilution method (performance instance in Fig. S3†) to determine the MIC of AgNPs and AgNPs–Chi solution samples prepared via the green synthesis method. Both the material solutions achieved high anti-bacterial activity, with very low MIC values (<10 μg mL−1, ranging from 2.29 to 9.17 μg mL−1) for all three types of pure and clinical bacterial strains (Fig. 8). This antibacterial performance was notably superior, with MIC values lower than those of AgNPs (MIC = 20 μg mL−1 for E. coli and MIC = 80 μg mL−1 for S. aureus) in the earlier study.51 However, the present MIC values were still higher than those reported for AgNPs–Triazole–Sulfonate copolymer (0.25 μg mL−1 for P. aeruginosa and 0.5 μg mL−1 for E. coli),52 which may be attributed to the use of a water-soluble Triazole–Sulfonate copolymer in that study. Additionally, we found that the AgNPs–Chi solution exhibited higher antibacterial performance across all bacterial strains compared to AgNPs alone, due to the synergistic effects of chitosan's intrinsic antimicrobial activity,53,54 better AgNP dispersion, and greater stability of the Ag–Chi material system (Table 3). Even at low Ag NP concentrations, both AgNPs and AgNPs–Chi demonstrated remarkably high antibacterial activities, indicating their potential for safe use in humans.The synthesized AgNPs–Alg–Chi nanocomposite films exhibited antibacterial activity with MIC values comparable to those of the AgNPs–Chi solutions for all tested strains (p > 0.05) (Table 2). For certain bacterial strains, such as clinical S. aureus and pure E. coli, the MIC values for the nanocomposite films were lower than those for the solutions. Conversely, for C. acnes and clinical E. coli, the MIC values for the nanocomposite films were higher than that for AgNPs–Chi solutions. However, these differences were not statistically significant (p > 0.05, Paired Samples T-test) (Table 2). The inhibition zones' diameters ranged from 12.27 to 13.67 mm across all experiments (Table 2).
| Bacterial strain | MIC of the AgNPs–Alg–Chi nanocomposite film (μg mL−1), mean ± SD | MIC of the AgNPs–Chi solution (μg mL−1), mean ± SD | Sterile diameter (mm), mean ± SD | pb | |
|---|---|---|---|---|---|
| a AgNPs–Alg–Chi: silver nanoparticles–sodium alginate–chitosan; AgNPs–Chi: silver nanoparticles chitosan; SD: standard deviation; C. acnes: Cutibacterium acnes; E. coli: Escherichia coli; S. aureus: Staphylococcus aureus; p: 2-tailed significance level.b Analyzing between MIC of AgNPs–Alg–Chi nanocomposite film and AgNPs–Chi solution using Paired-Samples T test.c Analyzing between pure and clinical strains of bacteria using One-Sample T test; *: p < 0.05. | |||||
| S. aureus | Pure (n = 3) | 4.58 ± 1.58 | 4.58 ± 1.58 | 13.67 ± 1.52 | — |
| Clinical (n = 19) | 4.23 ± 3.12 | 4.84 ± 1.32 | 12.52 ± 2.14 | 0.224 | |
| pc | 0.635 | 0.389 | 0.032 | ||
| E. coli | Pure (n = 3) | 3.67 ± 1.58 | 4.13 ± 2.38 | 13.67 ± 1.52 | 0.859 |
| Clinical (n = 3) | 5.50 ± 4.76 | 4.58 ± 1.58 | 13.33 ± 3.05 | 0.826 | |
| pc | 0.574 | 0.670 | 0.866 | ||
| C. acnes | Pure (n = 3) | 4.56 ± 1.62 | 2.29 ± 0.79 | 12.33 ± 2.51 | 0.205 |
| Clinical (n = 11) | 4.87 ± 3.54 | 2.87 ± 0.96 | 12.27 ± 1.73 | 0.051 | |
| pc | 0.778 | 0.072 | 0.910 | ||
The MIC of the AgNPs–Alg–Chi nanocomposite films were mostly slightly higher than those of the AgNPs–Chi solution. However, the nanocomposite films demonstrated antibacterial activity comparable to AgNPs–Chi solutions, with MIC values ranging from 3.67 to 5.50 μg mL−1, showing no significant difference (p > 0.05, Paired Samples T-test) (Table 2).
The MIC of both the Ag–Chi solution and the AgNPs–Alg–Chi nanocomposite film were significantly lower than those of other reported AgNP-based nanomaterials. For example, the MIC for Ag–organic montmorillonite nanocomposite was 313 μg mL−1 for S. aureus and 2500 μg mL−1 for E. coli (Table 3).55 Similarly, the MIC values for AgNPs–chitosan–agarose nanocomposite films were 31.25 μg mL−1 for S. aureus and E. coli, and 15.6 μg mL−1 for P. aeruginosa (Table 3).56 AgNPs synthesized from Ocimum gratissimum exhibited a MIC of 200 μg mL−1 and an inhibition zone of 14–15 mm for Acinetobacter baumannii.57 The MIC for Silver/Montmorillonite nanocomposite ranged from 15 to 75 μg mL−1 for E. coli, Salmonella spp., Pseudomonas aeruginosa, S. aureus, Listeria monocytogenes, and Bacillus cereus (Table 3).58 Table 3 summaries the AgNPs and AgNPs-based nanomaterials in this study and the relevant exciting published papers from the literature.13,20,51,52,55–61 Regardless using low AgNPs-loading concentration (≤11 μg mL−1), the AgNPs and AgNPs–Chi solutions and AgNPs–Alg–Chi nanocomposite film in this study achieved comparable or lower MIC values, but slightly smaller sterile diameter as compared to those reported in the literature (Table 3). Notably, this study reported both MIC and D values, meanwhile, it could lack one of the evaluated factors in the previous studies. It is worth noting that the antibacterial activity differences among the nanomaterials are attributed to many influent factors such as AgNP concentration, composite compositions, surface functional groups, material synthesis methods and conditions, antibacterial tests, etc.
| Nanomaterial | Route | Synthesis method | Ag NP size (nm) and shape | Antibacterial activity | Study | ||
|---|---|---|---|---|---|---|---|
| MIC of solution (μg mL−1) | MIC of film (μg mL−1) | Sterile diameter (D) (mm) | |||||
| a AgNPs: silver nanoparticles, AgNO3: silver nitrat, Alg: sodium alginate, AG: agarose, Cur: curcumin, Chi: chitosan, NaBH4: sodium borohydride, Na-VSA: vinylsulfonic acid sodium, VT: 1-vinyl-1,2,4-triazole, TSC: trisodium citrate, TPP: polyanionic sodiumtripolyphosphate, OMMT: organic montmorillonite, MMT: montmorillonite, A. baumannii: Acinetobacter baumannii, B. cereus: Bacillus cereus, B. subtilis: Bacillus subtilis, C. albicans: Candida albicans, C. acnes: Cutibacterium acnes; E. coli: Escherichia coli, E. aerogene: Enterobacter aerogenes, E. faecalis: Enterococcus faecalis, P. aeruginosa: Pseudomonas aeruginosa, S. aureus: Staphylococcus aureus, S. enterica: Salmonella enterica, L. monocytogenes: Listeria monocytogenes. | |||||||
| AgNPs | Green | AgNO3/Citrus japonica | 37.8 ± 11.5 spherical | S. aureus: 9.17 ± 3.17; E. coli: 4.58 ± 1.59; C. acnes: 4.58 ± 1.59 | — | — | Our study |
| AgNPs–Chi | Green | AgNO3/Citrus japonica/Chi | 37.8 ± 11.5 spherical | S. aureus: 4.58 ± 1.58; E. coli: 4.13 ± 2.38; C. acnes: 2.29 ± 0.79 | — | — | |
| AgNPs–Alg–Chi | Green | AgNO3/Citrus japonica/Alg/Chi | 37.8 ± 11.5 spherical | — | S. aureus: 4.58 ± 1.58; E. coli: 3.67 ± 1.58; C. acnes: 4.56 ± 1.62 | S. aureus: 13.67 ± 1.52, E. coli: 13.67 ± 1.52, C. acnes: 12.33 ± 2.51 | |
| AgNPs | Chemical | AgNO3/NaBH4/TSC | 5–30 spherical | S. aureus: 80; E. coli: 20 | — | E. coli: 12.4 | Agnihotri et al. (2014)51 |
| AgNPs | Green | AgNO3/Ocimum gratissimum | 73.24–87.89 rod | A. baumannii: 32 | A. baumannii: 200 | 14–15 | Gautam et al. (2023)57 |
| AgNPs–Chi | Green | AgNO3/Chi | 10.0 ± 5.4 spherical | S. aureus: 4.96; E. coli: 2.48 | — | — | Biao et al. (2017)20 |
| AgNPs | Biological | AgNO3/Penicillium polonicum | 10–30 spherical | S. enterica: 7.81 | — | — | Neethu et al. (2018)59 |
| AgNPs | Chemical | AgNO3/VT/Na-VSA | 3–12 spherical | P. aeruginosa: 0.25; E. coli: 0.5; E. faecalis: 4 | — | — | Pozdnyakov et al. (2022)52 |
| CurNPs–Chi | Green | Cur/Chi/TTP | 134.37 ± 1.99 spherical | S. aureus: 400; C. albicans: 400 | — | — | Ma et al. (2020)60 |
| AgNPs–OMMT | Chemical | AgNO3/OMMT | 10–20 spherical | — | S. aureus: 313; E. coli: 2500; C. albicans: 62 | — | Zhang et al. (2018)55 |
| AgNPs–Chi–AG | Green | AgNO3/Chitosan/AG | 2–7 spherical | – | S. aureus: 31.25; E. coli: 15.1; P. aeruginosa: 15.1 | — | Ghasemzadeh et al. (2021)56 |
| AgNPs–MMT | Chemical | Ag NO3/OMMT/NaBH4 | 12–30 spherical | E. coli: 30 ± 0.25; P. aeruginosa: 15 ± 0.47; Salmonella spp.: 30 ± 0.31; S. aureus: 45 ± 0.29; L. monocytogenes: 75 ± 0.4; B. cereus: 60 ± 0.53 | – | E. coli: 15; P. aeruginosa: 17; Salmonella typhi: 15; S. aureus: 13; L. monocytogenes: 11; B. cereus: 12 | El-Sherbiny et al. (2023)58 |
| AgNPs–Alg–Chi | Green | AgNO3/Alg/Chi | 9 ± 2 spherical | – | – | E. coli: 17; B. subtilis: 16 | Oe et al. (2023)61 |
| AgNPs–Alg–Chi | Green | AgNO3/Alg/Chi | 14 ± 3 spherical | – | – | E. coli: 15 ± 3; E. aerogene: 14 ± 2; P. aeruginosa: 28 ± 5; B. cereus: 59 ± 4; E. faecalis: 47 ± 4 | Sharma et al. (2012)14 |
The 6 mm-diameter AgNPs–Alg–Chi film were 13.67 ± 1.52 mm for S. aureus, 13.67 ± 1.52 mm for E. coli, and 12.33 ± 2.51 mm for C. acnes following the incubation period, indicating substantial antimicrobial activity relative to the film's original size (Fig. S4†). This result was comparable to or slightly smaller than the sterile ring diameters in previous studies. For instance, Oe et al. (2023)61 synthesized AgNPs–Alg–Chi nanocomposite films, achieving a sterile ring diameter of 17 ± 1 mm for the film with a 10 mM AgNO3 concentration. This sterile ring diameter is larger than that of our materials, which can be attributed to the larger film-cutting size, higher silver concentration, and smaller silver nanoparticle size (9 ± 2 nm vs. 37.8 ± 11.5 nm). Sharma et al. (2012)13 also synthesized AgNPs–Alg–Chi nanocomposite films and observed an E. coli sterile diameter of 15 ± 0.3 mm for 10 mm-diameter films (Table 3). These findings suggest that the AgNPs–Alg–Chi nanocomposite films developed in this study hold significant promise for biomedical applications such as antibacterial treatments,62–64 wound healing,14,17,65 anti-inflammatory therapies,15,66 and facial masks for acne treatment in dermatology.
Numerous studies have demonstrated that AgNP-based materials with antimicrobial properties offer promising alternatives for combating microbial resistance in clinical settings12 by utilizing multiple mechanisms of action (Fig. 9):
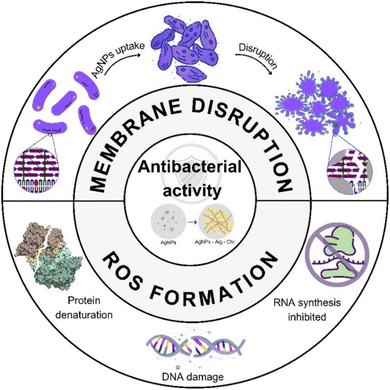 | ||
| Fig. 9 Schematic illustration of the proposed antibacterial mechanisms for AgNPs, Ag–Chi and Ag–Alg–Chi nanomaterials. | ||
– Surface binding: AgNPs attach to the bacterial cell membrane, compromising its integrity.67 This results in increased permeability, cell wall penetration, intracellular infiltration, and cytoplasmic leakage, which are the primary mechanisms responsible for disrupting Gram-positive bacteria.67,68
– Reactive oxygen species (ROS) formation: AgNPs catalyze the formation of reactive oxygen species, including hydrogen peroxide, superoxide anions, and hydroxyl radicals, which serve as the primary mechanism for disrupting Gram-negative bacteria.69
– Protein and enzyme denaturation: these materials cause the denaturation of microbial proteins and enzymes and alter DNA replication, thereby hindering cell division and reproduction.70
– Respiratory chain and signal transduction alteration: AgNPs disrupt the cellular respiratory chain and/or signal-transduction pathways.71
– Synergistic effect with chitosan and alginate: literature reports indicate a synergistic effect between AgNPs and chitosan. Chitosan can control the release of Ag+ ions, prolonging the antimicrobial effect over time.72
For the primary acne-causing pathogen, Gram-positive cocci, the antibacterial activity of AgNPs against Gram-positive bacteria is influenced by several factors. The thicker peptidoglycan layer in Gram-positive bacteria provides some protection, but AgNPs can still penetrate and disrupt the cell wall's integrity. Additionally, Gram-positive bacteria may be more susceptible to ROS-mediated damage due to their lower antioxidant defenses. These combined effects significantly enhance the antibacterial efficacy of AgNPs, making AgNPs–Alg–Chi composites highly effective against a broad spectrum of bacteria. This suggests they could be a valuable nonspecific initial treatment for infected lesions or for preventing infections.
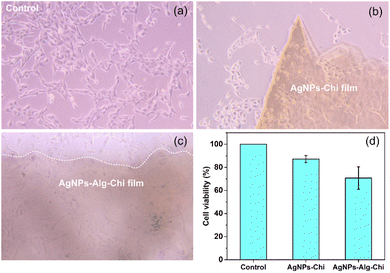
Optical microscope images confirm that human keratinocytes (HaCaT cells) can survive in proximity to and beneath the AgNPs–Chi and AgNPs–Alg–Chi films without being noticeably affected or killed (Fig. 10a–c). However, no cell growth was observed directly on the films, likely due to several factors, including the surface hydrophobicity of the polymers, the lack of bioactive cues, and the dissimilar mechanical properties between the polymer-based films and the cells, all of which are necessary for proper cell attachment and growth. Fig. 10(d) shows the MTT assay results, where the AgNPs–Chi film exhibited high cell viability of 87.1 ± 3.1%, while the AgNPs–Alg–Chi film demonstrated a lower cell viability of 70.8 ± 9.7%. The exact reason for the reduced HaCaT cell viability on the AgNPs–Alg–Chi film remains unclear. Since Alg–Chi nanoparticles have only shown mild toxicity to HaCaT cells, even at high concentrations,15 the lower cell viability may be attributed to the slightly higher release rate of AgNPs in the Alg–Chi matrix compared to the Chi-only matrix. A faster or greater release of AgNPs could increase the local concentration of silver ions, potentially enhancing cytotoxic effects and thus lowering cell viability. Overall, since materials with cell viability ≥70% are considered non-toxic and compatible with cell growth,73 both the AgNPs–Chi and AgNPs–Alg–Chi films exhibit good biocompatibility and are promising candidates for dermatological applications.
3.4. Strengths, novelty, and limitations of the study
Our study was meticulously designed, with the sample size calculated using a formula to ensure the representativeness of the population. The film fabrication process and antibacterial activity testing were clearly described, making them easily reproducible. The results demonstrate the potential of AgNPs–Alg–Chi nanocomposite films for clinical applications, particularly in antibacterial treatments and wound healing at non-cytotoxic concentrations. Potential uses include facial masks or medical gauze for healing after trauma, insect bites, and post-operative care. Long-term use of antibiotics can lead to side effects such as dermatitis, desquamation, and hyperpigmentation. In contrast, our material, with the gentle properties of chitosan and alginate, offers a promising foundation for developing further treatments, particularly for acne. However, to support broader clinical applications, larger studies with randomized controlled trial designs are necessary to validate these findings. Additionally, our results showed no significant difference in antibacterial activity between clinical bacterial strains and pure strains. Therefore, this study offers valuable insights into the antibacterial efficacy of AgNPs-based materials against both pure and clinical strains, providing a useful reference for future research while potentially simplifying research protocols.Our study has some limitations, including a small sample size collected from a single hospital in one area. A study with a larger sample size across multiple regions would provide a more comprehensive characterization of the bacteriological and epidemiological profiles of acne patients. Additionally, we did not evaluate other properties of the nanocomposite film, such as hydration capacity. Moreover, the antibacterial effect of the nanocomposite films may decrease over time, which was not assessed in our study. Future research should investigate the time-dependent reduction in antimicrobial activity and establish proper storage methods for optimal application.
Large-scale production may introduce variations in properties due to differences in equipment and process parameters, which our study did not assess under industrial mass production conditions. However, the next step includes verifying the effectiveness, safety, and consistency of large-scale production, focusing on critical parameters and dosage uniformity. Despite these limitations, our primary goal of evaluating the antibacterial activity and clinical application of the film at non-cytotoxic concentrations was met. Our findings show that low doses (<11 μg mL−1) of the films exhibit relatively strong antibacterial activity.
3.5. Future outlook
The future of AgNPs–Alg–Chi nanocomposite films in medical applications is promising, particularly for antimicrobial gauze and facial masks. As demand for antimicrobial products grows, scaling up production of these nanocomposites will be essential, requiring solutions to manufacturing challenges, ensuring product consistency, and meeting regulatory standards. Collaboration between researchers, manufacturers, and regulatory bodies will be crucial for efficient commercialization. These materials hold significant potential for wound dressings by providing enhanced protection against infections, promoting faster healing, and reducing the need for frequent changes. Future research will aim to optimize the balance between antimicrobial effectiveness and biocompatibility, ensuring safety for prolonged skin contact.Environmental considerations, especially regarding the disposal of AgNP-containing products, will also be critical. Research into eco-friendly production methods and recycling protocols will be necessary to minimize environmental impact. Overall, AgNPs–Alginate–Chitosan nanocomposites have the potential to revolutionize infection control in healthcare, but ongoing research and innovation will be key to achieving this while ensuring safety, scalability, and sustainability.
4 Conclusions
We successfully synthesized AgNPs and AgNPs–Chi solutions, and AgNPs–Alg–Chi nanocomposite films with an average Ag nanoparticle size of 37.8 ± 11.5 nm via a facile green approach process. Among bacteria isolated from acne patients, Gram-positive cocci were the most prevalent (93.8%), with S. aureus accounting for 32.3% and C. acnes for 26.1%. The MIC values of AgNPs solution for S. aureus, E. coli, and C. acnes were 9.17 ± 3.17, 4.58 ± 1.59, and 4.58 ± 1.59 μg mL−1, respectively. MIC values for AgNPs–Chi solution (AgNPs–Alg–Chi film) were 4.58 ± 1.58 μg mL−1 (4.58 ± 1.58 μg mL−1) for S. aureus, 4.13 ± 2.38 μg mL−1 (3.67 ± 1.58 μg mL−1) for E. coli, and 2.29 ± 0.79 μg mL−1 (4.56 ± 1.62 μg mL−1) for C. acnes. Additionally, the AgNPs–Alg–Chi film exhibited substantial sterile zones with diameters ranging from 12.33 to 13.67 mm for S. aureus, E. coli, and C. acnes, significantly exceeding the original film's 6 mm diameter. The study also compared bacterial strains isolated from acne samples with pure bacterial strains and found no significant differences. This result provides a valuable reference for future research, indicating that pure bacterial strains can effectively represent clinical strains in studies. The synthesized low-dosage AgNPs, AgNPs–Chi solution, and AgNPs–Alg–Chi nanocomposite film demonstrated strong antibacterial activity against both pure and clinical strains. Importantly, the AgNPs–Chi and AgNPs–Alg–Chi films exhibited good biocompatibility with human keratinocytes (HaCaT cells, viability >70%). Looking ahead, these materials hold promise for biomedical applications such as antibacterial treatments, wound healing, anti-inflammatory therapies, and facial masks for acne treatment in dermatology.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
The manuscript was done through the contributions of all authors. All authors have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the National Science and Technology Council of Taiwan under grant number NSTC 113-2222-E-131-003 (P. H. Le). The authors acknowledge the support of Can Tho University of Medicine and Pharmacy, Vietnam, and Ming Chi University of Technology, Taiwan, in facilitating this study.References
- Global Health Metrics, Acne Vulgaris – Level 3 Cause. Health Metrics and Evaluation (IHME), The University of Washington, Seattle WA, USA, 2019, pp. 3–4 Search PubMed.
- E. Mallon, J. N. Newton, A. Klassen, S. L. Stewart-Brown, T. J. Ryan and A. Y. Finlay, Br. J. Dermatol., 1999, 140, 672–676 CrossRef.
- Y.-G. Kim, J.-H. Lee, S. Park and J. Lee, Microbiol. Spectrum., 2022, 10(1), e020566 Search PubMed.
- N. Weber, K. Biehler, K. Schwabe, B. Haarhaus, K.-W. Quirin, U. Frank, C. M. Schempp and U. Wölfle, Molecules, 2019, 24(2), 223 CrossRef.
- C. Jantarat, P. Sirathanarun, T. Chuchue, A. Konpian, G. Sukkua and P. Wongprasert, Sci. Pharm., 2018, 86(1), 8 CrossRef.
- J. J. Leyden, K. J. McGinleya and B. Vowels, Dermatology, 1998, 196(1), 55–58 CrossRef CAS.
- S. H. Kennedy, Y. Manevich and J. Biaglow, Biochem. Biophys. Res. Commun., 1995, 212(1), 118–125 CrossRef CAS.
- J.-W. Xu, K. Yao and Z.-K. Xu, Nanoscale, 2019, 11(18), 8680–8691 RSC.
- S. Yougbaré, C. Mutalik, G. Okoro, I.-H. Lin, D. I. Krisnawati, A. Jazidie, M. Nuh, C.-C. Chang and T.-R. Kuo, Int. J. Nanomed., 2021, 26(16), 5831–5867 CrossRef.
- K. V. Alex, P. T. Pavai, R. Rugmini, M. S. Prasad, K. Kamakshi and K. C. Sekhar, ACS Omega, 2020, 5(22), 13123–13129 CrossRef PubMed.
- S. Chen, Y. Quan, Y.-L. Yu and J.-H. Wang, ACS Biomater. Sci. Eng., 2017, 3(3), 313–321 CrossRef CAS PubMed.
- M. A. Polinarski, A. L. B. Beal, F. E. B. Silva, J. Bernardi-Wenzel, G. R. M. Burin, G. I. B. de Muniz and H. J. Alves, Part. Part. Syst. Charact., 2021, 38(4), 2100009 CrossRef CAS.
- S. Sharma, P. Sanpui, A. Chattopadhyay and S. S. Ghosh, RSC Adv., 2012, 2(13), 5837–5843 RSC.
- K. Chen, F. Wang, S. Liu, X. Wu, L. Xu and D. Zhang, Int. J. Biol. Macromol., 2020, 148, 501–509 CrossRef CAS PubMed.
- A. J. Friedman, J. Phan, D. O. Schairer, J. Champer, M. Qin, A. Pirouz, K. Blecher-Paz, A. Oren, P. T. Liu, R. L Modlin and J. Kim, J. Invest. Dermatol., 2013, 133(5), 1231–1239 CrossRef CAS.
- G. C. Porter, D. R. Schwass, G. R. Tompkins, S. K. R. Bobbala, N. J. Medlicott and C. J. Meledandri, Carbohydr. Polym., 2021, 1(251), 117017 CrossRef.
- S. Alven and B. A. Aderibigbe, Pharmaceutics, 2024, 16(3), 327 CrossRef CAS PubMed.
- K. Varaprasad, T. Jayaramudu, V. Kanikireddy, C. Toro and E. R. Sadiku, Carbohydr. Polym., 2020, 236, 116025 CrossRef CAS.
- S. Li, W. Lu, Q. Tang, Q. Xiao, Y. Kang, L. Hu, Y. Huang, C. Peng and H. Yang, Chem. Eng. Sci., 2024, 285, 119547 CrossRef CAS.
- L. Biao, S. Tan, Y. Wang, X. Guo, Y. Fu, F. Xu, Y. Zu and Z. Liu, Mater. Sci. Eng., C, 2017, 76, 73–80 CrossRef CAS.
- A.-G. Niculescu and A. M. Grumezescu, Nanomaterials (Basel), 2022, 12(2), 186 CrossRef CAS.
- T. T. T. Nguyen, N. T. K. Tran, T. Q. Le, T. T. A. Nguyen, L. T. M. Nguyen and T. V. Tran, Alexandria Eng. J., 2023, 69, 419–430 CrossRef.
- Y. Li, P. Leung, L. Yao, Q. W. Song and E. Newton, J. Hosp. Infect., 2006, 62(1), 58–63 CrossRef CAS.
- C. B. Hiragond, A. S. Kshirsagar, V. V. Dhapte, T. Khanna, P. Joshi and P. V. More, Vacuum, 2018, 156, 475–482 CrossRef CAS.
- T. T. Ha, T. B. Q. Tran, H. V. T. Luong, N. H. Vo, T. X. C. Nguyen and T. T. Nguyen, Can Tho Univ. J. Sci., 2018, 54, 96–104 Search PubMed.
- H. Ghasemzadeh, S. Afraz, M. Moradi and S. Hassanpour, Int. J. Biol. Macromol., 2021, 179, 532–541 CrossRef CAS.
- N. Aziz, M. Faraz, R. Pandey, M. Shakir, T. Fatma, A. Varma, I. Barman and R. Prasad, Langmuir, 2015, 31(42), 11605–11612 CrossRef CAS.
- M. Alavi and M. Rai, Appl. Microbiol. Biotechnol., 2019, 103(21–22), 8669–8676 CrossRef CAS PubMed.
- A. Hernández-Rangel, P. Silva-Bermudez, B. L. España-Sánchez, E. Luna-Hernández, A. Almaguer-Flores, C. Ibarra, V. I. Garcia-Perez, C. Velasquillo and G. Luna-Barcenas, Mater. Sci. Eng., C, 2019, 94, 750–765 CrossRef.
- S. Hajji, S. B. Khedir, I. Hamza-Mnif, M. Hamdi, I. Jedidi, R. Kallel, S. Boufi and M. Nasri, Biochim. Biophys. Acta, Gen. Subj., 2019, 1863(1), 241–254 CrossRef CAS PubMed.
- S. E. Luria and M. Delbrück, Genetics, 1943, 28(6), 491–511 CrossRef CAS.
- P. Gopinath, S. K. Gogoi, A. Chattopadhyay and S. S. Ghosh, Nanotechnology, 2008, 19(7), 075104 CrossRef CAS.
- Vietnam Ministry of Health, Guide to Diagnosing and Treating Dermatological Diseases, Medical Publishing House, Hanoi, Vietnam, 2016 Search PubMed.
- M. A. Pourhoseingholi, M. Vahedi and M. Rahimzadeh, Gastroenterol. Hepatol. Bed Bench., 2013, 6(1), 14–17 Search PubMed.
- J. J. Biemer, Ann. Clin. Lab. Sci., 1971, 3(2), 135–140 Search PubMed.
- Vietnam Ministry of Health, Guidelines for diagnosis and treatment of dermatological diseases: Issued together with Decision No. 75/QD-BYT dated January 13, 2015, https://kcb.vn/upload/2005611/20210723/Huong-dan-chan-doan-dieu-tri-Da-lieu.pdf.
- M. Giavina-Bianchi, M. F. D. Azevedo and E. Cordioli, J. Prim. Care Commun. Health, 2022, 13, 1–8 Search PubMed.
- N. Skroza, E. Tolino, A. Mambrin, S. Zuber, V. Balduzzi, A. Marchesiello, N. Bernardini, I. Proietti and C. Potenza, J. Clin. Aesthet. Dermatol., 2018, 11(1), 21–25 Search PubMed.
- A. N. Miyazaki, M. J. C. Salles, G. V. Gonçalves, L. H. G. Conte, T. G. de Oliveira, A. B. N. Santili, M. N. L. Kurihara, I. N. M. Santos and L. A. da Silva, Rev. Bras. Ortop. (Sao Paulo), 2023, 58(2), 257–264 CrossRef PubMed.
- H. G. Tran, H. D. Tran, C. B. Huynh, T. T. T. Nguyen and B. V. Huynh, J. Med. Assoc. Thailand, 2019, 102, 1–5 Search PubMed.
- E. Hermosilla, M. Díaz, J. Vera, M. J. Contreras, K. Leal, R. Salazar, L. Barrientos, G. Tortella and O. Rubilar, Int. J. Mol. Sci., 2023, 24(3), 2318 CrossRef CAS PubMed.
- A. Regiel, S. Irusta, A. Kyzioł, M. Arruebo and J. Santamaria, Nanotechnology, 2013, 24(1), 015101 CrossRef PubMed.
- T. Y. Kim, S.-H. Cha, S. Cho and Y. Park, Arch. Pharmacal Res., 2016, 39(4), 465–473 CrossRef CAS PubMed.
- T. Tsuji, T. Kakita and M. Tsuji, Appl. Surf. Sci., 2003, 206(1–4), 314–320 CrossRef CAS.
- Q. Li, R. Ai, J. Fan, X. Fu, L. Zhu, Q. Zhou, L. Chen, W. Ma, Y. Li and L. Liu, Mater. Today Commun., 2024, 38, 107927 CrossRef CAS.
- M. Buşilă, V. Muşat, T. Textor and B. Mahltig, RSC Adv., 2015, 5, 21562–21571 RSC.
- Y. Xie, X. Liao, J. Zhang, F. Yang and Z. Fan, Int. J. Biol. Macromol., 2018, 119, 402–412 CrossRef CAS.
- R. Nadra and L. Aljerf, Int. J. Nanomanuf., 2019, 15(3), 269–289 CrossRef.
- N. Shaikh, N. N. Som, P. K. Jha and P. Pamidimukkala, Int. J. Biol. Macromol., 2023, 253(Pt 7), 127444 CrossRef CAS PubMed.
- K. Kasinathan, K. Marimuthu, B. Murugesan, S. Samayanan, S. J. Panchu, H. C. Swart and S. R. I. Savariroyan, Int. J. Biol. Macromol., 2021, 178, 270–282 CrossRef CAS PubMed.
- S. Agnihotria, S. Mukherjiabc and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- A. Pozdnyakov, A. Emel'yanov, A. Ivanova, N. Kuznetsova, T. Semenova, Y. Bolgova, S. Korzhova, O. Trofimova, T. Fadeeva and G. Prozorova, Pharmaceutics, 2022, 14(1), 206 CrossRef CAS.
- J. Li and S. Zhuang, Eur. Polym. J., 2020, 138, 109984 CrossRef CAS.
- A. Guarnieri, M. Triunfo, C. Scieuzo, D. Ianniciello, E. Tafi, T. Hahn, S. Zibek, R. Salvia, A. D. Bonis and P. Falabella, Sci. Rep., 2022, 12(1), 8084 CrossRef CAS.
- L. Zhang, J. Chen, W. Yu, Q. Zhao and J. Liu, J. Nanomater., 2018, 2018(1), 6190251 Search PubMed.
- H. Ghasemzadeh, S. Afraz, M. Moradi and S. Hassanpour, Int. J. Biol. Macromol., 2021, 179, 532–541 CrossRef CAS.
- D. Gautam, K. G. Dolma, B. Khandelwal, M. Gupta, M. Singh, T. Mahboob, A. Teotia, P. Thota, J. Bhattacharya, R. Goyal, S. M. R. Oliveira, M. L. de Pereira, C. Wiart, P. Wilairatana, K. Eawsakul, M. Rahmatullah, S. S. Saravanabhavan and V. Nissapatorn, PeerJ, 2023, 11, e15590 CrossRef.
- M. M. El-Sherbiny, R. P. Devassy, M. E. El-Hefnawy, S. T. Al-Goul, M. I. Orif and M. H. El-Newehy, Molecules, 2023, 28(9), 3699 CrossRef CAS.
- S. Neethu, S. J. Midhun, M. A. Sunil, S. Soumya, E. K. Radhakrishnan and M. Jyothis, J. Photochem. Photobiol., B, 2018, 180, 175–185 CrossRef CAS PubMed.
- S. Ma, D. Moser, F. Han, M. Leonhard, B. Schneider-Stickler and Y. Tan, Carbohydr. Polym., 2020, 241, 116254 CrossRef CAS PubMed.
- T. Oe, D. Dechojarassri, S. Kakinoki, H. Kawasaki, T. Furuike and H. Tamura, J. Funct. Biomater., 2023, 14(4), 199 CrossRef CAS.
- H. Y. Atay, Functional Chitosan, 2020, vol. 6, pp. 457–489 Search PubMed.
- E. O. Ogunsona, R. Muthuraj, E. Ojogbo, O. Valerio and T. H. Mekonnen, Appl. Mater. Today, 2020, 18, 100473 CrossRef.
- H. Gong, K. Zhang, C. Dicko, L. Bülow and L. Ye, ACS Appl. Nano Mater., 2019, 2(3), 1655–1663 CrossRef CAS.
- S. Wichai, P. Chuysinuan, S. Chaiarwut, P. Ekabutr and P. Supaphol, J. Drug Delivery Sci. Technol., 2019, 51, 662–671 CrossRef CAS.
- S. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7(13), e1701503 CrossRef.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, Coord. Chem. Rev., 2018, 357, 1–17 CrossRef CAS.
- R. Zhao, M. Lv, Y. Li, M. Sun, W. Kong, L. Wang, S. Song, C. Fan, L. Jia, S. Qiu, Y. Sun, H. Song and R. Hao, ACS Appl. Mater. Interfaces, 2017, 9(18), 15328–15341 CrossRef CAS.
- J. Chen, F. Wang, Q. Liu and J. Du, Chem. Commun., 2014, 50(93), 14482–14493 RSC.
- J. Xiong, Y. Cao, H. Zhao, J. Chen, X. Cai, X. Li, Y. Liu, H. Xiao and J. Ge, ACS Nano, 2022, 16(11), 19013–19024 CrossRef CAS.
- Y. Qing, L. Cheng, R. Li, G. Liu, Y. Zhang, X. Tang, J. Wang, H. Liu and Y. Qin, Int. J. Nanomed., 2018, 13, 3311–3327 CrossRef CAS PubMed.
- B. Zhao, Q. Zhou, C. Lou, X. Jin and W. Li, Int. J. Biol. Macromol., 2021, 287–292 CrossRef CAS PubMed.
- S. C. Boca, M. Potara, A. M. Gabudean, A. Juhem, P. L. Baldeck and S. Astilean, Cancer Lett., 2011, 311, 131–140 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05180g |
| ‡ These two authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2024 |