Open Access Article
Open Access ArticleBioengineering of one dimensional hierarchical Cu7S4 hollow nanotubes for non-enzymatic glucose sensing applications†
Giday G. Welegergs *ab,
Abera D. Ambayec,
Mbulelo Jokazia,
Nnamdi Nwaharaa and
Tebello Nyokong
*ab,
Abera D. Ambayec,
Mbulelo Jokazia,
Nnamdi Nwaharaa and
Tebello Nyokong a
a
aInstitute for Nanotechnology Innovation, Rhodes University, Makhanda 6140, South Africa. E-mail: getgiday@gmail.com
bDebre Berhan University, Department of Chemistry, P. O. Box 445, Debre Berhan, Ethiopia
cMaterials Science and Engineering, Bio, and Emerging Technology Institute, 5954, Addis Ababa, Ethiopia
First published on 27th August 2024
Abstract
Herein, a novel and facile eco-friendly green chemistry approach has been devised at room temperature for synthesis of 1D hierarchical Cu7S4 hollow nanotubes on Cu substrate via volatile organosulfur compounds from Allium sativum L for non-enzymatic glucose detection. Field emission scanning electron microscopy (FESEM), transmission electron microscopy (TEM), X-ray diffraction (XRD), energy dispersive X-ray spectroscopy (EDX), and X-ray spectroscopy (XPS) were employed to characterize the surface morphology, structural phase, compositional, and chemical states of the obtained samples, respectively. The SEM results confirm the formation of 1D hierarchical Cu7S4 hollow nanotubes. The XRD patterns are indexed to orthogonal anilite Cu7S4 crystal planes and the EDX spectra clearly reveal the presence of Cu and S elements. XPS spectra confirms peaks of Cu 2p and S 1s core levels, which are typical characteristics of Cu(I) and S(II), respectively. The Brunauer–Emmett–Teller (BET) specific surface area for obtained Cu7S4 hollow nanotubes is 2.07 m2 g−1 with a pore size distribution of 27.90 nm. Using Cu7S4 hollow nanotubes, the detection of non-enzymatic glucose was conducted over a dynamic range of concentrations from 0.5 to 100 μmol L−1 and reveals a high sensitivity of 1058.33 μA mM−1cm−2 and a limit of detection (LOD) of 0.127 μmol L−1. The obtained results indicated that Cu7S4 hollow nanotubes are promising candidates for non-enzymatic glucose detection.
1 Introduction
Diabetes is a leading cause of death, resulting primarily from high blood glucose levels. Projections indicate that by 2045, over 783 million individuals could be affected, exceeding 10% of the global population. Therefore, rapid, responsive, and specific detection of glucose is of significant concern across various scientific and technological domains, encompassing healthcare, food, and environmental surveillance.1–7 Importantly chemical sensors are vital technologies and devices that record the physical, chemical, and/or biological changes of a material and convert them into measurable signals for various applications.8–10Most commercialized disposable glucose sensors are fabricated by use of highly optimized cocktails and these sensors are based on chronoamperometric detection of hydrogen peroxide. This has led to manufacturing of glucose test strips and commercialization of glucometer devices, that are currently available in the market. Electrochemical glucose sensors are mainly classified into glucose oxidase (GOx) enzyme-based and non-enzymatic glucose sensors. Electrochemical glucose sensors based on enzyme immobilization methods such as, sol–gel adsorption, direct adsorption, cross-linking, etc. have been widely studied and utilized in line with the cocktail methods.10–12 All types of enzyme-modified sensor electrodes necessitate complicated enzyme fixation and thus inevitably suffer from insufficient stability issues because of the nature of the enzymes due to their sensitivity to changes in humidity, pH, temperature, toxic chemicals, and interference of some electro-oxidizable species. In order to solve these problems, non-enzymatic glucose biosensors have been proposed, which have the advantages of sensitivity, high stability, fast response, affordability, portability, scalability, and ease of preparation. Electrochemical non-enzymatic biosensors involve directly depositing a sensing material onto the electrode surface, enabling unrestricted ion/electron flow and enhancing conductivities.1,10,13–23
Recently, transition metal chalcogenides (TMCs) have received great attention and exploited for a wide range of applications such as environmental screening, catalysis, photovoltaics, biomedical diagnosis and therapy because of their lower cost, natural abundance, and environmentally friendliness, higher electrical conductivity and richer redox valences.17–19 Among the transition chalcogenides, copper sulfide (CuxS, x = 1–2) is one of the most important semiconductors with a lower band gap of 1.2–2.4 eV and has an inevitable place as a promising material due to its special properties and potential applications. Interestingly, copper sulfide exists in variable stable stoichiometric phases, which have been experimentally reported and range from the copper-rich phases of chalcocite (Cu2S), digenite (Cu9S5), and roxbyite/anilite (Cu7S4) to the copper-deficient phases: covellite (CuS) and villamaninite (CuS2). All copper sulfides are essentially of p-type semiconductor nature ascribed to the existence of Cu vacancies in the crystal lattices.24–28 Copper sulfides have been explored as potential candidates for the development of non-enzymatic glucose sensors because of their interesting properties like electrical conductivity.29–31 For example, S. Radhakrishnan et al.32 successfully synthesized CuS microflower (MF) superstructure via facile solvothermal method for nonenzymatic glucose detection. This CuS MF modified electrode showed an excellent sensitivity (1007 μA mM−1 cm−2) at 0.5 V with a good anti-interference ability and long-term stability. Miaomiao Cao et al.33 also reported copper sulfides by varying the amount of Na2S precursors and Cu7S4 hollow nanospheres demonstrated a wide linear concentration response towards non-enzymatic electrochemical glucose detection from 1–2.0 mM, with a LOD of 0.023 μM and high sensitivity of (3728.7 μA mM−1 cm−2). However, the non-enzymatic glucose sensor based on hollow nanostructured CuxS has rarely been reported.
It is well known that electrochemical biosensing performance depends on the surface morphology of nanomaterials. Hollow nanostructured materials are the most effective in the field of diagnosis because of their unique structure and properties, which originate from their nanoscale hollow structures. Hollow nanomaterials consist of distinguishable interior cavities/voids and well-defined boundary functional shells that can effectively improve the electrocatalytic properties due to their lower density, high surface area, shell permeabilities, and sufficient space that allow for volume expansion and reduced transport paths for both mass and ion/electron transport.33–35 To the best of our knowledge, Cu7S4 hollow nanotubes using green volatile organosulfur compounds from Allium sativum L for biosensing application has not been reported yet. Allium sativum L (garlic) is a common natural product and contains highly volatile bioactive organosulfur compounds responsible both for garlic pungent odour and many of its medicinal effects.36 Herein, we explored bio-sulfurized 1D hierarchal Cu7S4 hollow nanotubes on a Cu substrate using volatile organosulfur compounds from Allium sativum L at room temperature as nano-catalysts for electrochemical non-enzymatic glucose sensing applications. The preparation, characterization, and chronoamperometric response of Cu7S4 electrode is reported in detail.
2 Experimental section
2.1 Materials and chemicals
The following reagents were employed: Copper substrates (Cu, 99.99%, Sigma-Aldrich Co., Ltd), sodium hydroxide (NaOH, 99%, Alfa-aesar Co., Ltd) D-glucose (C6H12O6, ≥96.5%, Alfa-aesar Co., Ltd), D-fructose (C6H12O6, 99%, Merck, Co., Ltd), lactose (C6H12O6, 99%, Merck, Co., Ltd), galactose (C6H12O6, 99%, Sigma-Aldrich, Co., Ltd), hydrogen peroxide (H2O2, 30–35%, Minema chemicals, Co., Ltd), uric acid (C5H4N4O3, 99%, Sigma-Aldrich, Co., Ltd), ascorbic acid (C6H8O6, 99%, Sigma-Aldrich, Co., Ltd), oxalic acid (HO2CCO2H, 99%, Sigma-Aldrich, Co., Ltd), sodium sulfide hydrate (Na2S, ≥60%, Sigma-Aldrich, Co., Ltd), sodium nitrate (NaNO3, 99%, BDH, Co., Ltd) and electrodes (Pt wire and Ag/AgCl). Allium sativum L was obtained from a local supermarket in South Africa. All reagents are analytical grade purity and were used without further purifications. Deionized water was used for all experimental works.2.2 Synthesis of Cu7S4 hollow nanotubes
Intended for pure green chemistry synthesis of Cu7S4 on Cu-substrate, bioactive volatile organosulfur compounds from Allium sativum L (garlic) were used as a source of sulfur to bio-sulfurized the surface of the Cu substrate. Prior to the experiment, copper substrates (1.0 cm × 0.12 cm × 0.5 mm) were successively sonicated in acetone, ethanol, and deionized water for 15 min each and dried by blowing of N2 gas. Typically, 60 g of Allium sativum cloves were peeled and chopped using an electrical blender being inserted it into the bottle. The sticky tape was applied on the back of the Cu substrate which was then placed on the inside of the cover of the bottle containing Allium sativum. The bottle was then tightly sealed with the cover. The distance between the chopped Allium sativum and Cu-substrate in the bottle was kept being 5 ± 1 cm and left at room temperature for 6–96 h. The obtained black Cu7S4 was dried at 60 °C for 2 h in the oven.2.3 Materials characterization
The field emission scanning electron microscopy (SEM, Zeiss Supra 40) equipped with an energy dispersive X-ray (EDX) analyzer was conducted to characterize the surface morphology and elemental composition of the biosulfurized samples. Transmission electron microscopy (TEM, Tecnai G2-F20) was carried out to record both TEM and High-resolution TEM images of the obtained samples, and selected area electron diffraction (SAED) was applied to identify the lattice structure and structural nature of the samples. X-ray diffraction (XRD, CuKα radiation (λ = 1.5405 Å, nickel filter)) and X-ray photoelectron spectroscopy (XPS, Kratos, Axis Ultra-DLD) were employed to characterize the phase structure and chemical states of the obtained samples, respectively. The specific surface area and porosity of the sample were measured at 60 °C on a micromeritics ASAP 2020 using nitrogen (N2) adsorption/desorption isotherms and Brunauer–Emmett–Teller (BET) method.2.4 Electrochemical biosensing studies
The electrochemical behaviour of biosulfurized Cu7S4 on Cu substrates as electrodes in the absence and presence of glucose was evaluated using cyclic voltammetry (CV) and chronoamperometry (CA) measurements. All electrochemical measurements were performed using an electrochemical workstation in an alkaline electrolyte (0.1 M NaOH with pH value of 13) aqueous solution using a three-electrode electrochemical cell composed of Cu7S4 on Cu-substrate as a working electrode (WE), a platinum (Pt) wire as an auxiliary/counter electrode (CE), and Ag/AgCl/(saturated KCl) as a reference electrode (RE). Cyclic voltammetry (CV) measurements were carried out in 0.1 M NaOH electrolyte under ambient conditions and recorded by varying the working electrode potential from −1.0 to 1.0 eV and scan rates of 20–280 mV s−1. The chronoamperometric (CA) studies were performed by successive addition of known amounts of glucose concentrations (0.5, 1, 5, 10, 25, 50, 75 and 100 μM) into 0.1 M NaOH at applied potential of +0.5 eV.3 Result and discussion
3.1 Surface morphology
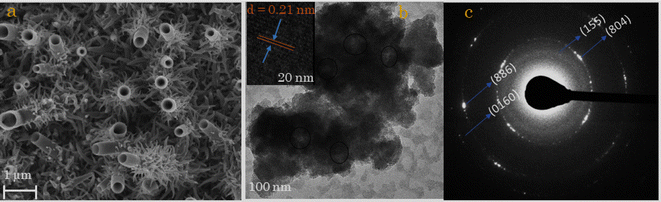
The surface morphology and microstructures of biosulfurized Cu7S4 thin films were characterized by field emission scanning electron microscopy (SEM), transmission electron microscopy (TEM) and high-resolution transmission electron microscopy (HRTEM). Fig. 1a–c shows the morphologies and microstructures of Cu7S4 thin films at 96 h reaction time. Fig. 1a reveals the biosulfurized SEM image of Cu7S4 and exhibits vertically grown one dimensional (1D) hierarchical hollow nanotubes with long depth. The growth and morphology evolution process of Cu7S4 hollow particles was obtained at different reaction intervals/stages (or from 6–96 h) as shown in Fig. S1 (ESI materials).† At 6 h reaction time, the surface of Cu substrate is covered by small nanosphere like particles. When the reaction time was prolonged to 12, 24 and then to 48 h the Cu7S4 nanoparticles are growing vertically and become bigger and longer. Further extending the reaction stage to 72 h, the vertically grown particles are created a hollow at the top. Finally, well-defined 1D hierarchal Cu7S4 hollow nanotubes with a long depth are obtained after prolonged to 96 h. The average void/hollow diameter of Cu7S4 hollow nanotubes is ≈210 nm with a shell thickness of 52 nm, and average height of the nanotubes is ≈2.09 μm. Moreover, secondary dense nano-leaves like small particles are grown and surrounded the hollow nanotubes from external at the bottom. According to the results, the growth, morphological evolution, and formation mechanism of 1D hierarchical Cu7S4 hollow nanotubes could be attributed to the Ostwald ripening mechanism for the fabrication of hollow nanostructures. This Ostwald ripening process involves the growth and recrystallization of large particles with hollow interiors by sacrificing small aggregates.37–40Further detailed structural characterizations of Cu7S4 were performed using TEM and HRTEM. Fig. 1b and c depicts the TEM images and scattered area electron diffraction (SAED) patterns of nanotubules Cu7S4 hollow nanotubules. From the TEM image in Fig. 1b, the dark edges and pale centers are observed, which provides convincing evidence of the Cu7S4 nanotubes have a hollow interior and cavities within the entire shell walls.41,42 The HRTEM image of Cu7S4 (inset of Fig. 1b) reveals a series reflection fringe with a fringe spacing is 0.21 nm, attributed to the (0160) crystal plane of orthogonal Cu7S4, which is in good agreement with a literature43 The crystallographic features of Cu7S4 were further recorded using SAED, and diffraction rings appear in the SAED patterns, as shown Fig. 1c. The uniform spots on rings confirm the perfect growth of the polycrystalline nature of Cu7S4.44,45
3.2 Structural and compositional analysis of Cu7S4 hollow nanotubes
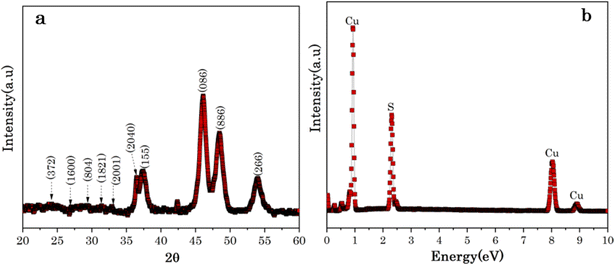
The crystal phase and structure of the obtained sample were examined using X-ray diffraction (XRD) technique. Fig. 2a shows the XRD patterns of Cu7S4 hollow nanotubes exposed to 96 h to the organosulfur compounds of Allium sativum and several well-defined diffraction peaks at 2θ values of 24.16, 26.64, 29.22, 31.18, 33.76, 35.97, 37.18, 46.10, 48.44, and 53.98° are identified corresponding to (3,7,2), (16,0,0), (8,0,4), (18,2,1), (20,0,1), (20,4,0), (1,5,5), (0,8,6), (8,8,6), and (2,6,6) crystal planes. The diffraction peaks coincide with the peaks of orthorhombic anilite Cu7S4 crystal phase (JCPDS no. 23-0958),46 and the strong diffraction peaks confirmed that the products are well-crystallized nanoparticles. The (0,8,6) peak at 2θ value of 46.10° is particularly intense, which confirms the preferential growth orientation of nanostructures with hollow-like surfaces33 (confirmed by SEM image).Fig. 2b shows the dispersive X-rays Spectroscopy (EDS) spectra of Cu7S4 hollow nanotubes and clearly reveals the presence of Cu and S elements, indicating the formation of Cu7S4. The Cu-substrates were exposed to organosulfur compounds of Allium sativum for different reaction stages from 6 to 96 h, as shown in Fig. S2 (ESI materials).† From the EDS spectra, the sulfur (S) peak located at 2.23 keV is almost non-existant for the lowest reaction stage of 6 h, while it becomes an intense peak after 12 h and increases with exposure time, even though it is dominated by the corresponding Cu peaks located at 0.91, 8.1, and 8.9 keV. Moreover, the EDS deduced relative ratio of Cu/S decrease, which confirms the S content increase with exposure time to the volatile organosulfur compounds of Allium sativum, as summarized in Table 1. Generally, the EDS analysis confirmed that the presence of continuous bio-sulfurization of Cu substrate surfaces and sulfurization saturates at Cu/S ∼1, Table 1.
| Reaction stages (hours) | Cu intensity | S intensity | Relative ratio Cu/S |
|---|---|---|---|
| 6 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 478.2 478.2 |
6272.7 | 4.70 |
| 12 | 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 095.0 095.0 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 431 431 |
1.91 |
| 24 | 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 938.1 938.1 |
66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 131.3 131.3 |
1.37 |
| 48 | 113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 234.8 234.8 |
96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 921.4 921.4 |
1.16 |
| 72 | 131![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 741.5 741.5 |
120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 630.2 630.2 |
1.09 |
| 96 | 149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312.2 312.2 |
139![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 419.6 419.6 |
1.06 |
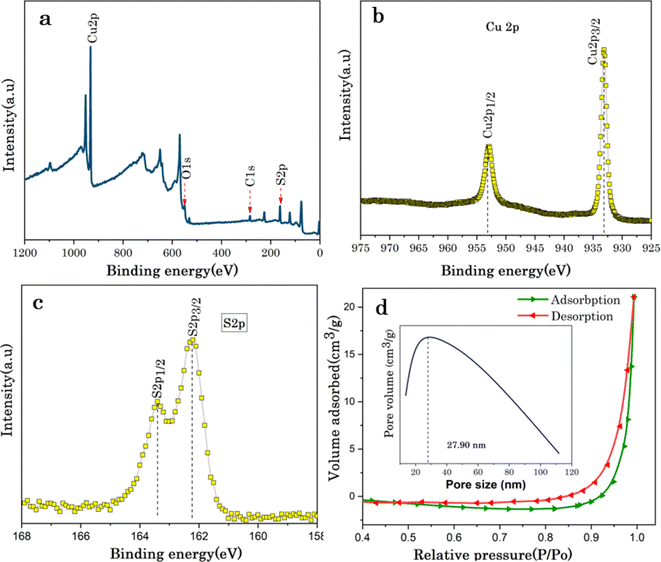
The surface chemical states and further elemental compositions of biosulfurized Cu7S4 hollow nanotubes were analysed via X-ray photoelectron spectroscopy (XPS). The Wider scan XPS survey of Cu7S4 confirms the presence of copper (Cu), sulfur (S), oxygen (O), and carbon (C) elements in Cu7S4, as displayed in Fig. 3a. Among them, O and C elements might derive from impurities and oxides on the Cu7S4 hollow nanotubules surfaces. The higher resolution XPS of Cu 2p core levels is shown in Fig. 3b, where two binding energy peaks are emerged at 953.4 and 932.2 eV, respectively are attributed to Cu 2p1/2 and Cu 2p3/2, which confirms the existence of Cu(I) in Cu7S4. From Fig. 3c, the high resolution XPS spectrum for S 2p at 163.5 and 162.2 eV binding energies which are associated to S 2p1/2 and S 2p3/2, respectively. In brief, the two XPS spectra of Cu 2p and S 2p corresponding to sulfides Cu7S4 proving the presence of Cu7S4 hollow nanotubes. Notably, the binding energies of S 2p3/2 and S 2p1/2 of Cu7S4 hollow nanotubes exhibits a slight shift to lower binding energies confirming that the higher electron configuration, resulting improved electronic conductivity of the Cu7S4 hollow nanotubes, which is highly favorable for accessing large amounts of analytes during electrochemical reactions.33
The specific surface area and pore size of Cu7S4 hollow nanotubes were analyzed from nitrogen (N2) adsorption/desorption isotherms using the Brunauer–Emmett–Teller (BET) method as shown in Fig. 3d and exhibits a type III IUPAC classification, which confirms the mesoporous structure (2–50 nm) of the sample.47 While the lower adsorption curve of the isotherm was used to measure the specific surface area, the desorption of isotherm was for pore size analysis. Based on the N2 sorption results, the calculated BET surface area of Cu7S4 is 2.07 m2 g−1, and the total pore volume is found to be 2.12 × 10−2 cm3 g−1. The pore size distribution (inset in Fig. 3d) of the sample is centered at 27.90 nm, further confirming the existence of mesopores structure for Cu7S4 hollow nanotubes. The specific surface area and pore size have significant effects on the adsorption properties and catalytic active sites of biosensor nanomaterials.48
3.3 Electrochemical non-enzymatic glucose detection
The pH impact on electrochemical glucose detection by Cu7S4 hollow nanotubes electrode was explored, and Fig. 4 shows the CV curves of Cu7S4 hollow nanotubes at pH values of 2.5, 5, 7.5, and 10 with 60 μM glucose in 0.1 M NaOH solution a scan rate of 50 mV s−1. The area of the CV current increased from pH 2.5 to 7.5; however, the redox reactions are not clearly visible, and the CV curve shape is distorted compared to the higher pH values. From the CV, the glucose oxidation reactions at pH 10 and 13 are barely detectable and the glucose oxidation peaks current increased with increasing the pH value from 10 to 13. The pH value of 13 for the 0.1 M NaOH electrolyte was determined to be the optimal experimental parameter Cu7S4 hollow nanotubes electrode for the electrochemical non-enzymatic glucose detection that will be used next.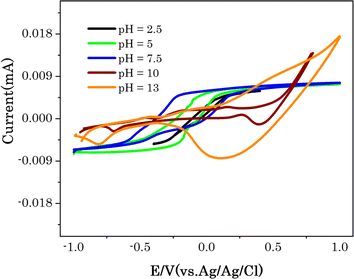 | ||
| Fig. 4 CV curves of Cu7S4 at different pH values in 0.1 M NaOH solution with 60 μM glucose at scan rate of 50 mV s−1. | ||
The electrochemical response of Cu7S4 hollow nanotubes electrode in the absence and presence of glucose was investigated using cyclic voltammetry (CV) in 0.1 M NaOH solution as electrolyte to explore the electrocatalytic activities. All CVs were performed in the potential window between −1.0 to 1.0 eV, which covers the glucose electro-oxidation range in alkaline electrolytes. Fig. 5 shows CVs of Cu7S4 hollow nanotubes without and with 60 μM glucose in 0.1 M NaOH solution at scan rate of 20 mV s−1. In the presence of glucose, Cu7S4 hollow nanotubes electrode displayed a higher catalytic current density compared without glucose attributed to the high electro-catalytic surface area and catalytic ability of Cu7S4 hollow nanotubes. The enhanced electrocatalytic current response of Cu7S4 hollow nanotubes towards the oxidation of glucose occurs due to its hollow nanostructures, which offer a higher active surface area, good electronic conductivity, and the presence of a void-like open structure configuration (as confirmed from SEM image) in Cu7S4 hollow nanotublets allows access to large amount of glucose molecules to the inner and outer surfaces for efficient glucose oxidation.29,33
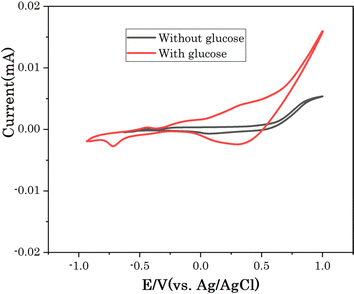 | ||
| Fig. 5 CV of Cu7S4 electrode in 0.1 M NaOH solution without and with 60 μM glucose at scan rate of 20 mV s−1. | ||
The effect of scan rate was investigated to determine the reaction kinetics of Cu7S4 hollow nanotubes in 0.1 M NaOH electrolyte containing 60 μM of glucose. Fig. 6a displays the CV responses of glucose oxidation on Cu7S4 hollow nanotubes surface for different scan rates ranging from 20 to 280 mV s−1. The oxidation (anodic) and reduction (cathodic) peak current of glucose at Cu7S4 increases with scan rates. Moreover, the reduction peak current is fully associated with the reduction of Cu(II) to Cu(I) and has shifted towards the more negative region with scan rates, suggesting a quasi-reversible electron transfer reaction of glucose.49 Both anodic peak current (Ipa) and cathodic peak current (Ipc) are plotted against the scan rates as shown in Fig. 6b and are directly proportional to the square root of scan rates. This confirms that the redox peak current on hollow Cu7S4 nanotube electrode are diffusion controlled electrochemical processes.49,50 The regression equations between the peak current and square root of the scan rates can be expressed using eqn (1) and (2) given below.51,52
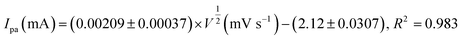 | (1) |
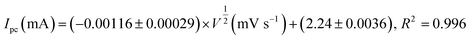 | (2) |
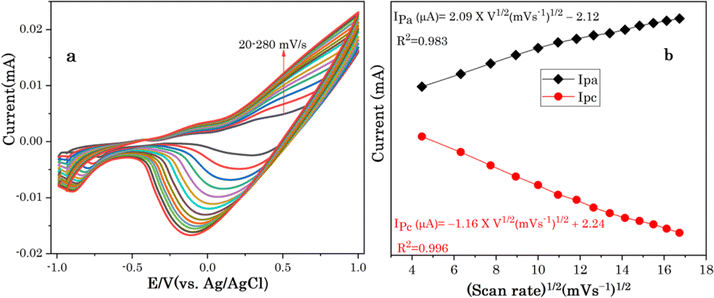 | ||
| Fig. 6 (a) CVs of Cu7S4 at different scan rates from 20 to 280 mV s−1 in a 0.1 M NaOH solution in the presence of 60 μM glucose, (b) the corresponding plots of anodic and cathodic peak currents. | ||
The non-enzymatic electrochemical sensing was investigated on Cu7S4 electrodes surface using chronoamperometry (CA) in 0.1 M NaOH solution with successive additions of known amounts of glucose concentrations at +0.5 eV. Fig. 7A and B shows the chronoamperometric responses of biosulfurized of Cu7S4 hollow nanotubes under successive injection of different glucose contents of 0.5, 1, 5, 10, 25, 50, 75 and 100 μM and the corresponding calibration curves. The electrocatalytic oxidation current of glucose exhibits a wide linear response and increases with increasing concentration of glucose in the given order from 0.5 μM to 100 μM, as shown in Fig. 7A.
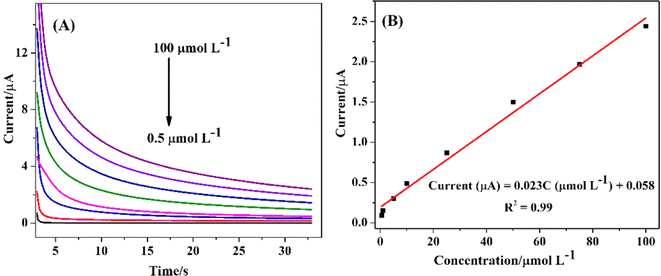 | ||
| Fig. 7 (A) Chronoamperometry response to various glucose concentrations (0.5 to 100 μmol L−1) and (B) the corresponding calibration curves for Cu7S4 hollow nanotubes. | ||
The corresponding calibration plot was obtained by plotting the amperometric current versus glucose concentrations for Cu7S4 hollow nanotubes and notably, an observable linear correlation emerged between the concentrations of glucose and the ensuing current response, as illustrated in Fig. 7B. This correlation is effectively encapsulated by a linear regression model: current (μA) = 0.058 + 0.023C (μmol L−1), indicating an acceptable coefficient of determination (R2) value of 0.99. Crucially, through eqn (3), the limit of detection (LOD) for glucose detection is robustly estimated to be 0.127 μmol L−1.
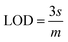 | (3) |
The analytical performances of the developed Cu7S4 hollow nanotube electrode towards non-enzymatic glucose sensors are compared with other CuxS based non-enzymatic glucose sensors previously reported using analytical parameters of linear range, limit of detection (LOD) and sensitivity as shown in Table 2. In conclusion, it is important to mention that surface morphology, high specific surface area, nanostructure porosity, and composition have crucial role in the electrocatalytic performance of nanomaterials.33,60,61
| Electrode | Linear range (μmol L−1) | LOD (μmol L−1) | Sensitivity (μA mM−1 cm−2) | Ref. |
|---|---|---|---|---|
| Cu/Cu7S4 hollow nanotubes | 0.5–100 | 0.127 | 1058.33 | This work |
| Cu2S nanoparticles | 10–3000 | 1.3 | 61.67 | 55 |
| Au-CuxS/3DCF | 1.98–976.56 | 7.62 | 59 | 56 |
| Cu@CuS | 2–8100 | 0.10 | 361.58 | 53 |
| Cu2S nanocrystal-DWCNT/GCE | 0.005–1 | 1 | 280 | 57 |
| S-rGO/CuS | 0.0001–3880 | 0.032 | 429.4 | 51 |
| CuS nanosphere-like | 1.0–1200 | 0.015 | 117.3 | 58 |
| Cu2S/MWCNT | 0.0010–1000 | 1.0 | 0.075 | 59 |
3.4 Specificity and reproducibility of Cu7S4 hollow nanotubes
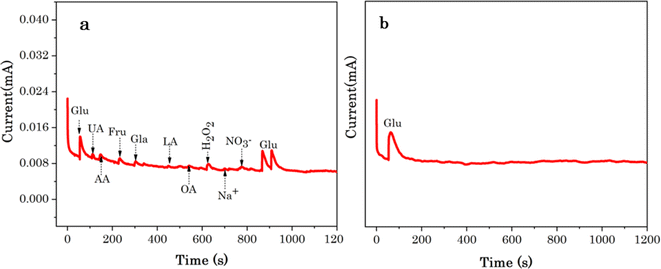
The avoidance of interfering species is still challenging for non-enzymatic glucose determination, because some organic molecules with a similar structure such as uric acid (UA), ascorbic acid (AA), oxalic acid (OA), fructose (Fru) etc., are co-existing in the biological fluids and simultaneously catalysed with glucose and generate interfering electrochemical signals.29 The selectivity of Cu7S4 hollow nanotubes was evaluated by measuring electrochemical signals in the presence of other sugars and other oxidizable interfering species. Considering that the concentration of glucose in human blood is more than 30 times of the physiological interfering compounds, thus, normal physiological concentrations of 50 μM glucose and 10 M of interferences of uric acid (UA), ascorbic acid (AA), oxalic acid (OA), Fructose (Fru), Galactose (Gla), lactose (LA), H2O2, Na+, and NO3− were orderly added into 0.1 M NaOH solution at 0.5 V as shown in Fig. 8a. The sharp response is observed for the addition of 50 μM glucose and no significant current increase is detected upon the addition of UA, AA, OA, H2O2, Na+, and NO3−, suggesting that these species do not interfere with glucose determination. Similarly, no obvious responses upon the addition of carbohydrate derivatives (i.e. Fru, Gla and La) is observed as shown Fig. 8a. The increase in current density observed again after the second injection of Glu reveals that Cu7S4 hollow nanotubes are still sensitive to Glu, and their activity is not compromised by the interferents. Thus, this study demonstrated that the hierarchical Cu7S4 hollow nanotubes reveal a high specificity or selectivity in the presence of co-interfering species toward glucose detection.The stability and reproducibility of the Cu7S4 hollow nanotubes sensor were also evaluated. Fig. 7B depicts the chronoamperometry stability of Cu7S4 hollow nanotubes which reveals a sharp response for the addition of 50 μM glucose in 0.1 M NaOH. Afterwards, a steady state current was achieved for 1200 seconds(s) confirming that the Cu7S4 hollow nanotubes glucose detecting electrode has good operational stability. Moreover, to ascertain the reproducibility of the Cu7S4 hollow nanotubes, the chronoamperometric response to the repeated addition of 50 μM D-glucose was tested by 6 repeated measurements, as shown in Fig. 9 and the proposed biosensor electrode exhibits good repeatability with a relative standard deviation (RSD) of 3.56%. The observed results confirmed that the Cu7S4 hollow nanotubes electrode has good stability and reproducibility for glucose detection.
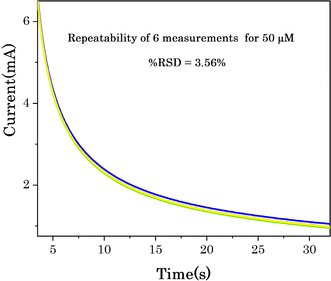 | ||
| Fig. 9 Chronoamperometric response of Cu7S4 hollow nanotubes to the repeated addition of 50 μM glucose in 0.1 M NaOH. | ||
4 Conclusion
In conclusion, low-dimensional nanomaterials with distinguishable interior hollows and shells have advantages of offering high surface areas, increased reactivities, and unique physicochemical properties for efficient biosensing applications. This work presents a novel approach for the bio-sulfurization of 1D hierarchical hollow Cu7S4 nanotubelets as electrode materials through the application of volatile organosulfur compounds. Characterization techniques confirmed the successful fabrication of hierarchical hollow nanostructures, revealing their distinct morphology and composition. The biosulfurized Cu7S4 hollow nanotube electrochemical sensor ascribed efficient performance as non-enzymatic electrochemical glucose detection. Interestingly, the detection of glucose was conducted over a dynamic range of concentrations ranges from 0.5 to 100 μmol L−1, with a limit of detection (LOD) of 0.127 μmol L−1. The Cu7S4 hollow nanotubes exhibited an excellent electro-catalytic activity towards glucose oxidation with a high sensitivity of 1058.33 μA mM−1 cm−2. Furthermore, it demonstrates also a high selectivity in the potentially co-interfering ions toward glucose determination. The inherent properties of Cu7S4 hollow nanotube surfaces, coupled with the bio-sulfurization process mediated by volatile organosulfur compounds of Allium sativum L contribute to enhanced sensitivity and selectivity towards glucose detection. This innovative green bio-approach holds significant promise for the development of advanced glucose sensing technologies with potential applications in biological samples.Data availability
The data supporting this article are available within the paper and the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Department of Science and Innovation (DSI) and National Research Foundation (NRF), South Africa through DSTI/NRF South African Research Chairs Initiative for Professor of Medicinal Chemistry and Nanotechnology (UID 62620), DSI/Mintek Nannotechnology Innovation Centre, and Rhodes University (for materials and postdoctoral research fund for G. G. W.), South Africa. The authors give credit to ithemba labs MRD, Cape town, South Africa for the characterizations.References
- Y. Huo, R. Li, S. Xiu, Y. Wang, L. Zhang, A. Jin and B. Quan, Diamond Relat. Mater., 2023, 140, 110455–110463 CrossRef.
- B. Tao, H. Zheng, J. Li, F. Miao and P. Zhang, Mater. Sci. Eng., B, 2017, 298, 116895 CrossRef.
- D. Das, A. Das, M. Reghunath and K. K. Nanda, Green Chem., 2017, 19, 1327–1335 RSC.
- S. Kumar, M. B. Gawande, I. Medřík, M. Petr, O. Tomanec, V. Kupka, R. S. Varma and R. Zbořil, Green Chem., 2020, 22, 5619–5627 RSC.
- S. Yu, R. D. Webster, Y. Zhou and X. Yan, Catal. Sci. Technol., 2017, 7, 2050–2056 RSC.
- P. Bhavani, D. P. Kumar, H. S. Shim, P. Rangappa, M. Gopannagari, D. A. Reddy, J. K. Song and T. K. Kim, Catal. Sci. Technol., 2020, 10, 3542–3551 RSC.
- W. Lu, R. Zhang, X. Zhang, Y. Shi, Y. Wang and H. Shi, Analyst, 2023, 148, 5469–5475 RSC.
- A. D. Ambaye, T. G. Kebede, B. Ntsendwana and E. N. Nxumalo, Synth. Met., 2023, 299, 117452 CrossRef.
- N. Njoko, M. Louzada, J. Britton, S. Khene, T. Nyokong and P. Mashazi, Colloids Surf., B, 2020, 190, 110981 CrossRef PubMed.
- M. Wei, Y. Qiao, H. Zhao, J. Liang, T. Li, Y. Luo, S. Lu, X. Shi, W. Lu and X. Sun, Chem. Commun., 2020, 56, 14553–14569 RSC.
- P. Wu, J. Fan, Y. Tai, X. He, D. Zheng, Y. Yao, S. Sun, B. Ying, Y. Luo, W. Hu, X. Sun and Y. Li, Food Chem., 2024, 447, 139018 CrossRef CAS PubMed.
- D.-W. Hwang, S. Lee, M. Seo and T. D. Chung, Anal. Chim. Acta, 2018, 1033, 1–34 CrossRef CAS PubMed.
- S. Wang, L. Zhao, R. Xu, Y. Ma and L. Ma, Electroanal. Chem., 2019, 853, 113527 CrossRef CAS.
- J. Lv, C. Kong, Y. Xu, Z. Yang, X. Zhang, S. Yang, G. Meng, J. Bi, J. Li and S. Yang, Sens. Actuators, B, 2017, 248, 630–638 CrossRef.
- L. Goodnight, D. Butler, T. Xia and A. Ebrahimi, Biosensors, 2021, 11, 409 CrossRef PubMed.
- H. Siampour, S. Abbasian, A. Moshaii and A. R. Amirsoleimani, Sci. Rep., 2022, 12, 18945 CrossRef.
- C. Wei, X. Zou, Q. Liu, S. Li, C. Kang and W. Xiang, Electrochim. Acta, 2020, 334, 135630 CrossRef.
- O. Adeniyi, N. Nwahara, D. Mwanza, T. Nyokong and P. Mashazi, Sens. Actuators, B, 2021, 348, 130723 CrossRef CAS.
- M. Bisht, S. K. Thayallath, P. Bharadwaj, G. Franklin and D. Mondal, Green Chem., 2023, 25, 4591–4624 RSC.
- F. Zhou, C. You, Q. Wang, Y. Chen, Z. Wang, Z. Zeng, X. Sun, K. Huang and X. Xiong, Electroanal. Chem., 2020, 876, 114477 CrossRef CAS.
- G. G. Welegergs, G. G. Welegergs, H. G. Gebretnisae, M. G. Tsegay, Z. Y. Nuru, S. Dube and M. Maaza, Infrared Phys. Technol., 2011, 113, 103619 CrossRef.
- G. G. Welegergs, Z. M. Mehabaw, H. G. Gebretinsae, M. G. Tsegay, L. Kotsedi, Z. Khumalo, N. Matinisie, Z. T. Aytuna, S. Mathur, Z. Y. Nuru, S. Dube and M. Maaza, Infrared Phys. Technol., 2023, 104820 CrossRef CAS.
- X. Li, J. Cao, L. Yang, M. Wei, X. Liu, Q. Liu, Y. Hong, Y. Zhou and J. Yang, Dalton Trans., 2019, 7, 2442–2454 RSC.
- M. P. Ravele, O. A. Oyewo, S. Ramaila, L. Mavuru and D. C. Onwudiwe, Catalysts, 2021, 11, 1238 CrossRef CAS.
- B. Janani, R. Balakrishnaraja, A. M. Elgorban, A. H. Bahkali, R. S. Varma, A. Syed and S. S. Khan, J. Environ. Manage., 2023, 326, 116615–116626 CrossRef PubMed.
- S. Farhadi and F. Siadatnasab, Mater. Res. Bull., 2016, 83, 345–353 CrossRef.
- S. Saeed, N. Rashid, R. Hussain, M. A. Malik, P. O'Brien and W.-T. Wong, J. Chem., 2023, 37, 3214–3221 Search PubMed.
- G. G. Welegergs, N. Numan, S. Dube, Z. Nuru, N. Botha, K. Cloete, S. Azizi, I. Madiba, M. Akbari, R. Morad, M. G. Tsegay, H. G. Gebretinsae, C. Mtshali, Z. Khumalo, F. Ezema, A. Krief, A. Gibaud, M. Henini, M. P. Seopela, M. Chaker and M. Maaza, Nanoscale Horiz., 2023, 2, 1–27 Search PubMed.
- X. Yan, Y. Gu, C. Li, B. Zheng, Y. Li, T. Zhang, Z. Zhang and M. Yang, Anal. Methods, 2018, 10, 381–388 RSC.
- H. T. Zhang, G. Wu and X. H. Chen, Mater. Chem. Phys., 2006, 98, 298–303 CrossRef CAS.
- L. Qian, J. Mao, X. Tian, H. Yuan and D. Xiao, Sens. Actuators, B, 2013, 176, 952–959 CrossRef CAS.
- S. Radhakrishnan, H.-Y. Kim and B.-S. Kim, Sens. Actuators, B, 2016, 233, 93–99 CrossRef CAS.
- M. Cao, H. Wang, P. Kannan, S. Ji, X. Wang, Q. Zhao, V. Linkov and R. Wang, Appl. Surf. Sci., 2019, 492, 407–416 CrossRef.
- X. Li, N. Li, Y. Gao and L. Ge, Chin. J. Catal., 2022, 43, 679–707 CrossRef.
- L. Cong, H. Xie and J. Li, Adv. Energy Mater., 2017, 7, 1601906 CrossRef.
- Z. Xu, Z. Qiu, Q. Liu, Y. Huang, D. Li, X. Shen, K. Fan, J. Xi, Y. Gu, Y. Tang, J. Jiang, J. Xu, J. He, X. Gao, Y. Liu, H. Koo, X. Yan and L. Gao, Nat. Commun., 2018, 9, 3713 CrossRef PubMed.
- B. Zhao, G. Shao, B. Fan, W. Zhao, Y. Xie and R. Zhang, J. Mater. Chem. A, 2015, 3, 10345–10352 RSC.
- M. Cao, H. Lian and C. Hu, Nanoscale, 2010, 2, 2619–2623 RSC.
- W.-S. Wang, L. Zhen, C.-Y. Xu, J.-Z. Chen and W.-Z. Shao, ACS Appl. Mater. Interfaces, 2019, 1, 780–788 CrossRef.
- J.-X. Cui, W.-S. Wang, L. Zhen, W.-Z. Shao and Z.-L. Chen, CrystEngComm, 2012, 14(20), 7025–7030 RSC.
- G. Cheng and A. R. Hight Walker, Anal. Bioanal. Chem., 2010, 396, 1057–1069 CrossRef CAS PubMed.
- M. Basu, A. K. Sinha, M. Pradhan, S. Sarkar and T. Pal, J. Phys. Chem. C, 2011, 115, 12275–12282 CrossRef CAS.
- Y. Jiang, S. Zhang, Q. Ji, J. Zhang, Z. Zhang and Z. Wang, J. Mater. Chem. A, 2014, 2, 4574–4579 RSC.
- W. Xu, S. Zhu, Y. Liang, Z. Li, Z. Cui, X. Yang and A. Inoue, Sci. Rep., 2015, 18125 CrossRef CAS.
- J. Liu and D. Xue, J. Mater. Chem., 2011, 21, 223–228 RSC.
- J. Hu, J. Yang, S. Yi, X. Zhou and S. Chen, J. Energy Storage, 2024, 77, 109901 CrossRef.
- C. Nethravathi, R. Ragesh Nath, J. T. Rajamathi and M. Rajamathi, ACS Omega, 2019, 4, 4825–4831 CrossRef PubMed.
- Z. Huang, L. Wang, H. Wu, H. Hu, H. Lin, L. Qin and Q. Li, J. Alloys Compd., 2022, 896, 163045 CrossRef.
- L. Jayasingha, C. Jayathilak, R. Kumar, K. Ohar, M. Kaumal, S. Gunewardene, D. Dissanayake and S. Jayanetti, Electrochim. Acta, 2020, 329, 135177 CrossRef CAS.
- W. Lu, Y. Sun, H. Dai, P. Ni, S. Jiang, Y. Wang, Z. Li and Z. Li, Sens. Actuators, B, 2016, 231, 860–866 CrossRef CAS.
- N. Karikalan, R. Karthik, S.-M. Chen, C. Karuppiah and A. Elangovan, Sci. Rep., 2017, 7, 2494 CrossRef PubMed.
- A. D. Ambaye, K. K. Kefeni, T. G. Kebede, B. Ntsendwana, S. B. Mishra and E. N. Nxumalo, J. Electroanal. Chem., 2022, 919, 116542 CrossRef CAS.
- X. Zhang, L. Wang, R. Ji, L. Yu and G. Wang, Electrochem. Commun., 2012, 24, 53–56 CrossRef.
- Q. Chen, D. Chu, L. Yan, H. Lai, X. Q. Chu, D. Ge and X. Chen, New J. Chem., 2021, 45, 10031–10039 RSC.
- S. K. Maji, A. K. Dutta, G. R. Bhadu, P. Paul, A. Mondal and B. Adhikary, J. Mater. Chem. A, 2013, 1, 4127–4134 CAS.
- L. N. T. Mai, S. Radhakrishnan and J. Mathiyarasu, Electrochim. Acta, 2017, 246, 544–552 CrossRef.
- Y. Myung, D. M. Jang, Y. J. Cho, H. S. Kim, J. Park, J.-U. Kim, Y. Choi and C. J. Lee, J. Phys. Chem. C, 2009, 113, 1251–1259 CrossRef CAS.
- W. Wang, L. Zhang, S. Tong, X. Li and W. Song, Biosens. Bioelectron., 2009, 25, 708–714 CrossRef PubMed.
- H. Lee, S. W. Yoon, E. J. Kim and J. Park, Nano Lett., 2007, 7, 778–784 CrossRef.
- J. Ding, S. Ji, H. Wang, J. Key, D. J. Brett and R. Wang, J. Power Sources, 2018, 374, 48–54 CrossRef.
- J. Zhang, Y. Xu and B. Zhang, Chem. Commun., 2014, 50, 13451–13453 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05199h |
| This journal is © The Royal Society of Chemistry 2024 |