Open Access Article
Open Access ArticlePreparation and antioxidant properties of tannic acid/copper ion nanozyme hybrid nanofibrous membranes†
Qiao Wu,
Jingshu Xiao,
Hu Zhuang,
Fenghai Zhao,
Ruoxi Li and
Duntie Zhang*
China Tobacco Hubei Industrial LLC, Wuhan, Hubei 430072, China
First published on 11th November 2024
Abstract
Excess free radicals can have some negative effects on human health. In this paper, a nanozyme was successfully constructed by the coordination of copper ions and tannic acid, and its structure and elemental distribution were determined by Fourier transform infrared spectroscopy, scanning electron microscopy and X-ray photoelectron spectroscopy. Free radical scavenging experiments confirmed that it possessed superoxide dismutase-like activity, catalase-like activity, and hydroxyl radical scavenging ability. The results of thermogravimetric analysis experiments demonstrated that it possessed good thermal stability. A polyacrylonitrile hybrid nanofibrous membrane loaded with Cu/TA nanozyme was successfully constructed by electrospinning technology, and the maximum scavenging rate of DPPH and ABTS radicals can reach 64.22% and 58.44%, respectively. The nanofiber membrane also exhibited the ability to protect cells from oxidative stress damage. Therefore, the hybrid nanofibrous membrane has a broad application prospect in fields such as food preservation and biomedicine.
1 Introduction
In living organisms, free radicals are usually considered as harmful substances and an important factor contributing to aging.1,2 Free radicals can be classified as endogenous and exogenous. In eukaryotic cells, endogenous free radicals are mostly produced by mitochondria.3 Electrons leaked from the mitochondrial respiratory chain react with O2 to generate superoxide radical (˙O2−), which can be further converted to hydrogen peroxide (H2O2) and hydroxyl radical (˙OH).4,5 Exogenous free radicals can be attributed to various external stimuli such as ultraviolet light, air/water pollution and toxic chemicals.6 Harmful free radicals are responsible for tissue damage and various diseases including cancer, neurodegenerative diseases, diabetes and cardiovascular diseases.7–11 Therefore, the design and fabrication of functional materials with antioxidant activity has become an important approach to scavenge free radicals and protect the host from damage.Currently, antioxidants are mainly divided into natural antioxidants and synthetic antioxidants. Antioxidant enzymes synthesized in vivo are critical parts of the antioxidant defense network.12 Superoxide dismutase (SOD), catalase and glutathione peroxidase eliminate excess free radicals in the organism to maintain homeostasis.13 Leaves, fruits and seeds of plants are the main sources of antioxidants.14 Natural antioxidants mainly contain polyphenolic compounds, vitamins and carotenoids with a wide range of biological activities and rich nutritional value. Synthetic compounds, including nitrones/nitroxides, iron ion chelators and thiols, have also been widely used for scavenging free radicals.15–17 Generally, synthetic antioxidants are more active and pure than natural antioxidants and have constant antioxidant activity, but their toxicity and safety are widely concerned.18
Nanomaterials have great potential in the field of developing novel antioxidants. Some nanomaterials, such as metal and metal oxide-based nanoparticles, exhibit free radical trapping and/or SOD-like and catalase-like activities with excellent scavenging ability of free radicals.11,19 Compared to natural enzymes, such nanomaterials exhibit tunable activity, higher stability and tolerance to harsh environments.20,21 In recent years, nanomaterials with enzyme-like activity (nanozymes) have shown promise as substitutes for natural enzymes. Many natural enzymes are metal-based enzymes and their metal catalytic activity centers are mainly copper, iron or zinc ions. Among them, copper is the active center of many proteins and enzymes and plays a crucial role in the function of several enzymes, including tyrosinase and copper zinc superoxide dismutase (SOD1).13,22,23 Therefore, it is reasonable to use copper ions to prepare nanozymes for free radical scavenging.
Inspired by antioxidant enzymes, Cu/TA nanozyme with antioxidant activity were constructed in this study by self-assembly of copper ions and tannic acid (TA). The natural active ingredient TA, which is widely found as a water-soluble polyphenol in the barks and fruits of many plants with anti-inflammatory, antioxidant, and anti-microbial properties, was used as the reaction material.24,25 In addition, TA can chelate a variety of metal ions through coordination bonds and rapidly form stable five-element ring complexes with these ions.13,26 Although the preparation of nanoparticles using copper ions and TA has been reported in the literature,27–29 the Cu/TA nanosheet in this work were prepared by a green one-pot method. This simple synthesis process and mild reaction conditions provide an effective method for the preparation of Cu/TA nanozyme at low-cost and large-scale production. The obtained Cu/TA nanozyme combine the advantages of nanozyme and natural antioxidant, thus achieving multiple free radical scavenging abilities with the assistance of TA.30 The results showed that the Cu/TA nanozymes possessed SOD-like activity and could effectively remove ˙O2−. In addition, Cu/TA nanozymes exhibited excellent catalase-like activity and the ability to scavenge ˙OH (Fig. 1a). This synergistic antioxidant activity allows it to act as an effective antioxidant for scavenging various free radicals. In addition, the unique coordination structure endows the Cu/TA nanozyme with excellent thermal stability. The Cu/TA nanozymes were further loaded into polyacrylonitrile (PAN) nanofibrous membranes by electrospinning (Fig. 1b), and the prepared Cu/TA-PAN hybrid nanofibrous membranes (CTP membranes) exhibited good biocompatibility and antioxidant activity, which is promising for the application in the fields, such as food preservation and biomedicine.25,31,32
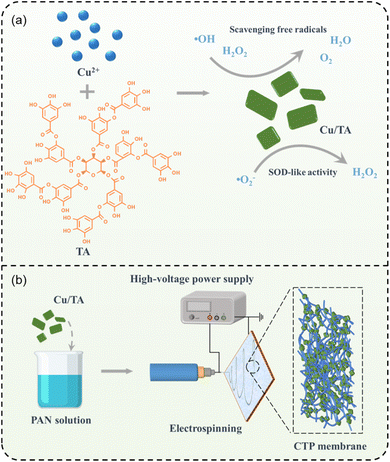 | ||
| Fig. 1 Schematic diagram of the preparation process of (a) Cu/TA nanozyme and (b) CTP nanofibrous membranes. | ||
2 Materials and methods
2.1 Materials
Tannic acid (TA) was provided by Shanghai Macklin Biochemical Technology Co., Ltd. Polyacrylonitrile (PAN, Mw = 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was purchased from DuPont. 3,3′,5,5′-tetramethyl-benzidine (TMB), 1,1-diphenyl-2-trinitrophenylhydrazine (DPPH), 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonate) (ABTS) and NaAc-HAc buffer (pH = 3.6) were purchased from Shanghai yuanye Bio-Technology Co., Ltd. Hydroxyl free radical assay kit (A018-1-1) and inhibition and produce superoxide anion assay kit (A052-1-1) were provided by Nanjing Jiancheng Bioengineering Institute. Copper(II) chloride dihydrate (CuCl2·2H2O), potassium persulfate (K2S2O8), N,N-dimethylformamide (DMF), hydrogen peroxide (H2O2, 30%) and anhydrous ethanol were purchased from Sinopharm Chemical Reagent Co., Ltd. All chemical reagents were of analytical grade and used as received without further purification.
000) was purchased from DuPont. 3,3′,5,5′-tetramethyl-benzidine (TMB), 1,1-diphenyl-2-trinitrophenylhydrazine (DPPH), 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonate) (ABTS) and NaAc-HAc buffer (pH = 3.6) were purchased from Shanghai yuanye Bio-Technology Co., Ltd. Hydroxyl free radical assay kit (A018-1-1) and inhibition and produce superoxide anion assay kit (A052-1-1) were provided by Nanjing Jiancheng Bioengineering Institute. Copper(II) chloride dihydrate (CuCl2·2H2O), potassium persulfate (K2S2O8), N,N-dimethylformamide (DMF), hydrogen peroxide (H2O2, 30%) and anhydrous ethanol were purchased from Sinopharm Chemical Reagent Co., Ltd. All chemical reagents were of analytical grade and used as received without further purification.
2.2 Preparation of Cu/TA nanozyme
According to the method reported in the literature,33 Cu/TA nanozyme was synthesized through the chelation reaction between Cu2+ and TA. Briefly, 0.025 g of TA and 0.875 g of CuCl2 were separately dissolved in 25 mL of ultrapure water. The two solutions were then mixed and adjusted to pH 7.4 using 2 M NaOH solution. The reaction mixture was stirred at room temperature for 3 h. The obtained precipitate was collected by centrifugation at 8000 rpm, washed three times with ultrapure water, and freeze-dried. The obtained Cu/TA nanozyme was stored at 4 °C.2.3 Characterization of Cu/TA nanozyme
2.4 Multienzyme-like activity of Cu/TA nanozyme
| Scavenging efficiency (%) = (1 − Asample/Acontrol) × 100% |
| Scavenging efficiency (%) = (1 − Asample/Acontrol) × 100% |
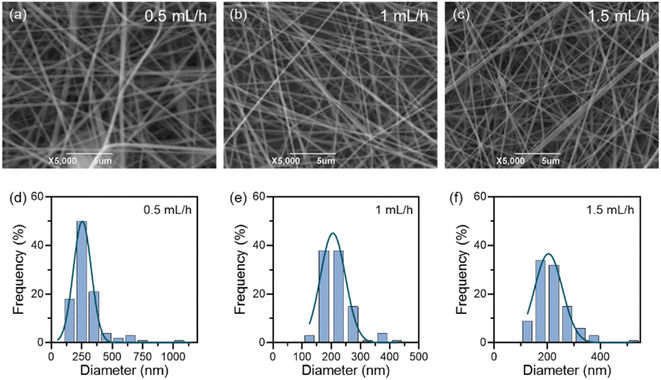
2.5 Preparation of PAN nanofibrous membranes
2.6 Preparation of CTP membranes
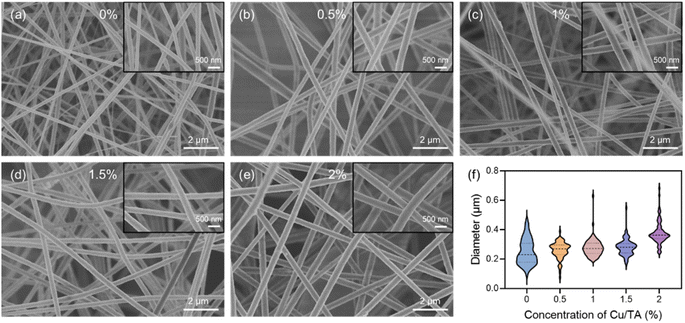
After determining the optimal concentration of the PAN solution and process parameters, Cu/TA nanozymes were added to the mixed solution at 0.5, 1, 1.5, and 2% of the PAN mass. The mixture was stirred at 60 °C until homogeneous. CTP nanofibrous membranes were then prepared using the same process parameters.2.7 Morphology of nanofibrous membranes
The samples were cut from the nanofibrous membranes, and after the samples were sprayed with gold, the surface morphology of the membranes was observed using the JSM6390LV SEM (JEOL). The fiber diameters were analyzed using Image J software.2.8 Free radical scavenging ability of nanofibrous membranes
| Scavenging efficiency (%) = (1 − A1/A0) × 100% |
| Scavenging efficiency (%) = (1 − A1/A0) × 100% |
2.9 In vitro cytocompatibility study
The in vitro cytocompatibility assay of nanofibrous membranes were conducted using the extract method. Mouse L929 fibroblast cells were cultured in HyClone™ Dulbecco's modified eagles' medium (DMEM) containing 10% fetal bovine serum and 1% penicillin-streptomycin. After sterilization with 75% ethanol solution, the nanofibrous membranes were soaked with complete medium (6 cm2 mL−1) for 24 h to make the extracts. The L929 cells were seeded at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 96-well plate and incubated for 24 h at 37 °C in a 5% CO2 atmosphere. Then, the cell culture medium was replaced with different extracts and fresh medium was used as control. After incubation for 24 h, the cell viability was detected in using the Cell Counting Kit-8 (CCK-8) method.
000 cells per well in a 96-well plate and incubated for 24 h at 37 °C in a 5% CO2 atmosphere. Then, the cell culture medium was replaced with different extracts and fresh medium was used as control. After incubation for 24 h, the cell viability was detected in using the Cell Counting Kit-8 (CCK-8) method.
2.10 In vitro oxidative stress assay
The protective effect of nanofibrous membranes on cells in H2O2 simulated oxidative stress microenvironment was investigated. The L929 cells were seeded in a 96-well plate at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and incubated for 24 h. Then, the culture medium was removed and the cells were treated with 100 μL of fresh culture medium containing 500 μM H2O2 and sterile nanofibrous membranes for 6 h. The cell viability was determined by CCK-8 assay.
000 cells per well and incubated for 24 h. Then, the culture medium was removed and the cells were treated with 100 μL of fresh culture medium containing 500 μM H2O2 and sterile nanofibrous membranes for 6 h. The cell viability was determined by CCK-8 assay.
3 Results and discussion
3.1 Synthesis and characterization of Cu/TA nanozymes
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, and the absorption peaks at 1613 cm−1 and 1537 cm−1 indicated the presence of aromatic rings with C
O groups, and the absorption peaks at 1613 cm−1 and 1537 cm−1 indicated the presence of aromatic rings with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds. In addition, the absorption peak at 1446 cm−1 was attributed to the deformation of C–C bond within the phenolic moiety, while the absorption peak at 1321 cm−1 indicated the presence of phenolic groups. The absorption peak at 1197 cm−1 was attributed to the deformation of the C–H bond, while the vibrational bands at 1100–1000 cm−1 were attributed to the deformation of the C–O and C–H bonds. Our findings are consistent with those of previous reports in the literature.35 Compared to TA, the Cu/TA nanozyme exhibited peak splitting and shifts in the FT-IR spectrum due to oxidation of TA and subsequent coordination with Cu2+. Specifically, the peak of TA at 3347 cm−1 was split into three peaks at 3516 cm−1, 3440 cm−1, and 3355 cm−1. This splitting phenomenon indicates that the coordination of TA with Cu2+ disrupted the phenolic hydroxyl HO–C vibration of TA.33 Furthermore, there was a significant shift of the absorption peaks within the fingerprint region of the FT-IR spectrum of Cu/TA nanozyme, which serves as confirmation of the coordination of TA with Cu2+.
C bonds. In addition, the absorption peak at 1446 cm−1 was attributed to the deformation of C–C bond within the phenolic moiety, while the absorption peak at 1321 cm−1 indicated the presence of phenolic groups. The absorption peak at 1197 cm−1 was attributed to the deformation of the C–H bond, while the vibrational bands at 1100–1000 cm−1 were attributed to the deformation of the C–O and C–H bonds. Our findings are consistent with those of previous reports in the literature.35 Compared to TA, the Cu/TA nanozyme exhibited peak splitting and shifts in the FT-IR spectrum due to oxidation of TA and subsequent coordination with Cu2+. Specifically, the peak of TA at 3347 cm−1 was split into three peaks at 3516 cm−1, 3440 cm−1, and 3355 cm−1. This splitting phenomenon indicates that the coordination of TA with Cu2+ disrupted the phenolic hydroxyl HO–C vibration of TA.33 Furthermore, there was a significant shift of the absorption peaks within the fingerprint region of the FT-IR spectrum of Cu/TA nanozyme, which serves as confirmation of the coordination of TA with Cu2+.
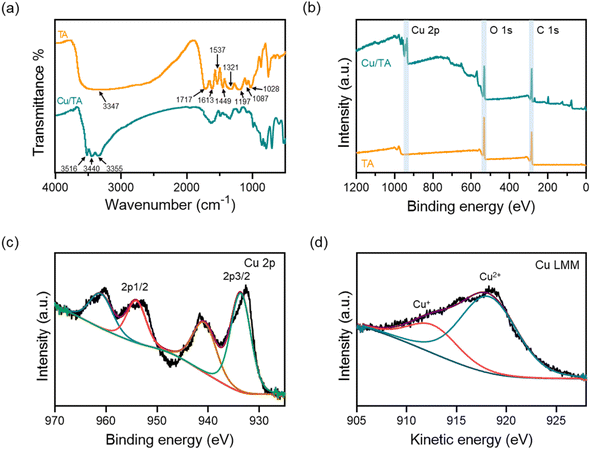 | ||
| Fig. 2 (a) FT-IR spectra of TA and Cu/TA nanozyme. (b) XPS survey spectra of TA and Cu/TA nanozyme. (c) Cu 2p high-resolution spectra and (d) Cu LMM Auger spectra of Cu/TA nanozyme. | ||
In order to understand the oxidation state of Cu in Cu/TA nanozyme, it was further characterized by XPS. As shown in Fig. 2b, the full-scan spectrum of Cu/TA nanozyme demonstrated the presence of Cu, O and C elements, indicating that the product was an inorganic-organic hybrid. The high-resolution spectrum of Cu 2p displays peaks at 933.6 eV (Cu 2p3/2) and 954.0 eV (Cu 2p1/2), accompanied by characteristic shake-up satellite peaks at 940.8 eV and 961.0 eV, respectively (Fig. 2c).36 The different valence states of Cu were distinguished using X-ray Auger spectra. As shown in Fig. 2d, the Cu LMM Auger spectrum was deconvoluted into Cu2+ and Cu+ peaks at 918.2 eV and 912.7 eV, respectively. These results indicate that Cu2+ combine with TA in a coordination form.
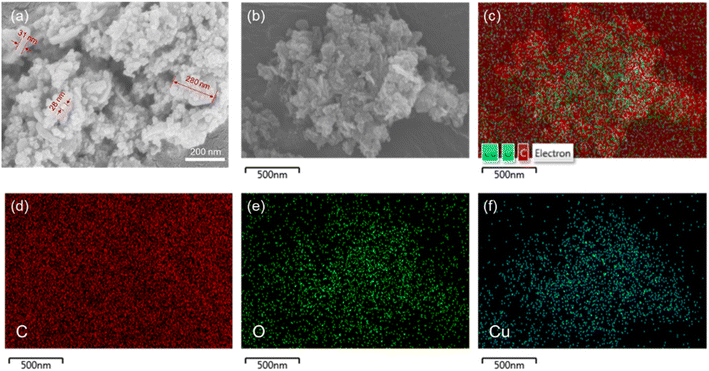
The morphology of Cu/TA nanozyme was observed using SEM (Fig. 3a). The prepared Cu/TA nanozyme exhibited two distinct morphologies. The larger structure exhibited a sheet-like morphology, with a diameter of approximately 280 nm and a thickness of approximately 31 nm. The smaller particle-like structure had a diameter of approximately 28 nm. These results are consistent with those reported in the literature.33 The EDS elemental analysis of Cu/TA nanozyme was shown in Fig. 3b–f. The presence and uniform distribution of carbon, oxygen, and copper elements on Cu/TA nanozyme also confirms the successful synthesis.
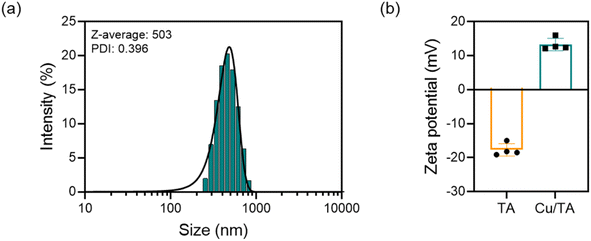
The hydrodynamic size and zeta potential of Cu/TA nanozymes were measured by DLS (Fig. 4). As shown in Fig. 4a, the hydrodynamic size of Cu/TA nanozyme was determined to be approximately 503 nm, with a PDI of 0.396. The hydrodynamic particle size incorporates the solvent layer and thus exhibits a slight discrepancy from the actual particle size. As shown in Fig. 4b, the zeta potential of Cu/TA nanozyme was 13.3 mV, which was higher than that of pure TA at −17.7 mV. This shift can be attributed to the introduction of the positively charged Cu2+, which neutralized the negatively charged phenolic hydroxyl groups of TA. The high absolute value of the zeta potential of Cu/TA nanozyme indicates that it has a stable structure and uniform dispersion in deionized water.
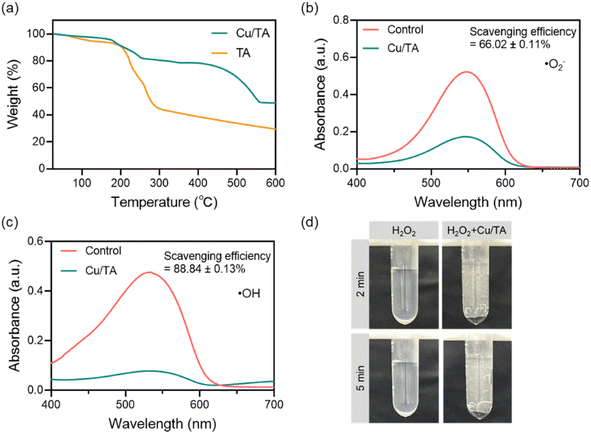
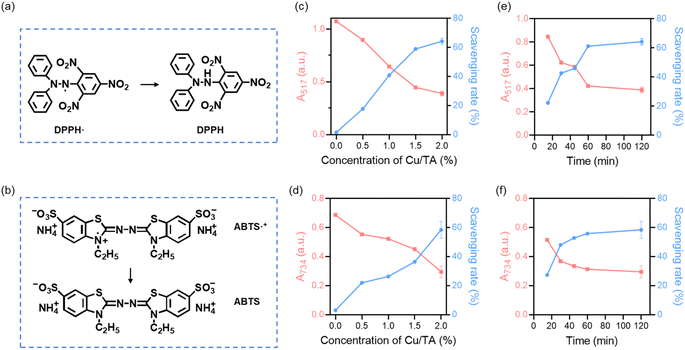
The antioxidant capacity of the obtained Cu/TA nanozyme was confirmed by a series of free radical scavenging assays (Fig. 5b–d). The ability of Cu/TA nanozyme to scavenge ˙O2− was first determined since it has a coordination active center similar to that of natural SOD. ˙O2− is one of the most destructive reactive oxygen radicals,38 and the ability of Cu/TA nanozyme to catalyze the decomposition of ˙O2− was examined using the inhibition and produce superoxide anion assay kit. During the assay, ˙O2− was generated by the reaction between xanthine and xanthine oxidase Following the addition of an electron transfer substance and Gress's reagent, the reaction system displayed a purple-red coloration and a characteristic absorption peak at 550 nm. As shown in Fig. 5b, the absorbance value of the solution at 550 nm was higher without Cu/TA nanozyme. Conversely, the presence of Cu/TA nanozyme resulted in the decomposition of ˙O2−, which led to a decrease in the absorbance value of the solution. The Cu/TA nanozyme exhibited enhanced ˙O2− scavenging ability compared to Cu2+ and pure TA (Fig. S1a†). The results indicate that the Cu/TA nanozyme exhibited SOD-like activity. pH is one of the important factors affecting the activity of nanozymes. We investigated the pH-dependent SOD-like activity of Cu/TA nanozyme. As shown in Fig. S2a in the ESI,† the optimal pH for SOD-mimicking Cu/TA nanozyme was 7.0, but it still maintained good SOD-like activity at pH 5.0 and 9.0, which could scavenge 63.7% and 61.6% of ˙O2−, respectively. As a result, Cu/TA nanozyme maintained high SOD-like activity over a wide pH range.
In addition, Cu/TA nanozyme also showed the ability to eliminate ˙OH. ˙OH is a highly oxidizing reactive oxygen radical that can cause damage to cell membranes and DNA. Therefore, the removal of ˙OH can have a positive effect on protecting cells or organisms more effectively from free radical-induced damage.38 The hydroxyl free radical assay kit was employed to investigate the scavenging efficiency of Cu2+, TA and Cu/TA nanozyme for ˙OH, which is generated by the classical Fe2+/H2O2 Fenton reaction. As shown in Fig. 5c and S1b,† Cu/TA nanozyme can eliminate more than 80% of ˙OH, which was significantly higher than that of Cu2+ and TA, showing a high ˙OH scavenging ability. In addition, Cu/TA nanozyme maintained a favorable ˙OH scavenging ability in a broad pH range (Fig. S2b†).
H2O2 is another important reactive oxygen species with high oxidizing capacity, which is a downstream product of ˙O2−.1 Catalase can catalyze the decomposition of H2O2 into H2O and O2.5 As shown in Fig. 5d and S1c,† no bubbles were generated in the pure H2O2 solution and the solution with incorporated TA, whereas a large number of O2 bubbles were generated after the addition of Cu/TA nanozyme. Thus, it can catalyze the decomposition of H2O2 to produce O2, confirming that Cu/TA nanozyme has catalase-like activity. These results indicate that Cu/TA nanozyme is efficient in scavenging free radicals.
The POD-like activity of Cu/TA nanozyme was evaluated by oxidizing TMB to oxTMB.39–41 As shown in Fig. S3,† in the presence of H2O2 substrate, Cu/TA nanozyme was capable of efficiently catalyzing the oxidation of TMB to produce blue oxTMB with characteristic absorption at 652 nm. In contrast, there was no significant absorbance signal in the presence of H2O2 or Cu/TA nanozyme alone. These results indicate that Cu/TA nanozyme also exhibits POD-like enzyme activity to catalyze the generation of ˙OH from H2O2.
Considering the presence of multiple Cu valence states in Cu/TA nanozyme and the inherent antioxidant properties of TA, a possible mechanism for radical scavenging by Cu/TA nanosheets was proposed. Cu(I) can be oxidized to Cu(II) by ˙O2− to produce H2O2.42 The coordination structure of Cu(II) can be converted to Cu(I) by oxidizing ˙O2− to produce O2 molecules.34 The valence state transition of Cu in Cu/TA is necessary for ˙O2− disproportionation.20,34 Cu/TA nanozyme can promote the electron transfer reaction, which inactivates ˙OH or H2O2. The pronounced CAT-like enzyme activity of Cu/TA nanozyme may be related to the Cu(I) coordination structure, which catalyzes the decomposition of H2O2 into O2 and H2O. In addition, polyphenolic ligands can facilitate the catalytic process by performing valence conversion of Cu.27,34 These results demonstrate the importance of Cu coordination with TA in determining its enzymatic activity.
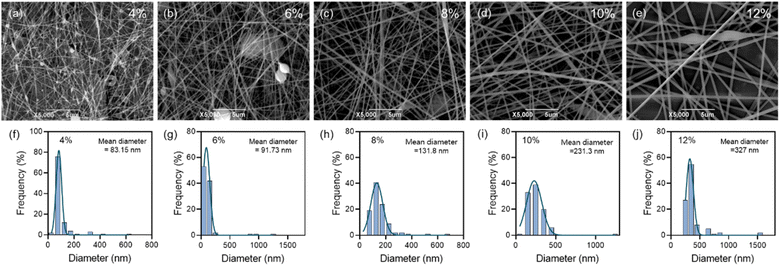
3.2 Preparation and characterization of nanofibrous membranes
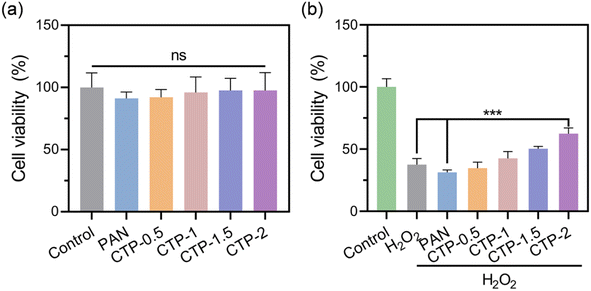
In order to simulate the in vitro ROS microenvironment, 500 μM H2O2 was used to induce oxidative stress. The L929 cells were cultured in the artificial oxidative stress environment for 6 h, and the cell viability was significantly reduced to 37% (Fig. 10b). Compared with PAN membranes, CTP membranes could significantly improve the viability of L929 cells in the H2O2-containing microenvironment, and the cell viability increased with the increase of Cu/TA nanozyme content in the nanofiber membranes. When the addition of Cu/TA nanozyme was 2%, the cell viability was reached up to 62%. The experimental results indicated that Cu/TA nanozyme in CTP membranes plays a key role in scavenging ROS and reducing cellular oxidative stress damage.
4 Conclusion
In summary, Cu/TA nanozymes constructed by self-assembly of Cu2+ and TA exhibited inherent SOD-like activity, catalase-like activity, and ˙OH scavenging ability. These unique properties enable the Cu/TA nanozyme to effectively scavenge various free radicals. The Cu/TA nanozyme hybrid nanofibrous membranes were further made by electrospinning technique, and the results showed that the diameter of the composite fibers increased significantly after the introduction of Cu/TA nanozyme into the PAN fibers. In addition, the CTP composite membranes possessed excellent antioxidant capacity and were able to protect cells from oxidative stress damage. These results demonstrate the broad prospects of Cu2+/TA self-assembled metal nanozymes in artificial enzyme development and open new avenues for the development of novel antioxidant films.Data availability
All data generated or analyzed during this study are included in this published article. The data supporting this article have been included in the main article and the ESI.†Author contributions
Qiao Wu: conceptualization, methodology, data curation, investigation, formal analysis, writing – original draft. Jingshu Xiao: methodology, investigation, software. Hu Zhuang: investigation, software. Fenghai Zhao & Ruoxi Li: methodology. Duntie Zhang: writing – review & editing, supervision, resources.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The authors would like to thank the School of Life and Health Sciences of Hubei University of Technology for providing instruments and services.References
- K. B. Beckman and B. N. Ames, Physiol. Rev., 1998, 78, 547–581 CrossRef CAS PubMed.
- X.-Q. Wang, W. Wang, M. Peng and X.-Z. Zhang, Biomaterials, 2021, 266, 120474 CrossRef.
- E. Cadenas, Mol. Aspects Med., 2004, 25, 17–26 CrossRef.
- M. L. Circu and T. Y. Aw, Free Radicals Biol. Med., 2010, 48, 749–762 CrossRef PubMed.
- J. Zhao, F. Guo, L. Hou, Y. Zhao and P. Sun, J. Controlled Release, 2023, 355, 273–291 CrossRef PubMed.
- Y. F. Mustafa, Indian J. Clin. Biochem., 2024, 39, 154–167 CrossRef PubMed.
- S. Prasad, S. C. Gupta and A. K. Tyagi, Cancer Lett., 2017, 387, 95–105 CrossRef PubMed.
- U. S. Srinivas, B. W. Q. Tan, B. A. Vellayappan and A. D. Jeyasekharan, Redox Biol., 2019, 25, 101084 CrossRef.
- E. R. Stadtman and B. S. Berlett, Drug Metab. Rev., 1998, 30, 225–243 CrossRef.
- C. Zhang, X. Wang, J. Du, Z. Gu and Y. Zhao, Advanced Science, 2021, 8, 2002797 CrossRef CAS PubMed.
- S. Liang, X. Tian and C. Wang, J. Inflammation Res., 2022, 15, 6307–6328 CrossRef CAS PubMed.
- B. Halliwell, Nat. Rev. Mol. Cell Biol., 2024, 25, 13–33 CrossRef CAS PubMed.
- J. Liu, X. Han, T. Zhang, K. Tian, Z. Li and F. Luo, J. Hematol. Oncol., 2023, 16, 116 CrossRef CAS PubMed.
- A. Augustyniak, G. Bartosz, A. Čipak, G. Duburs, L. U. Horáková, W. Łuczaj, M. Majekova, A. D. Odysseos, L. Rackova, E. Skrzydlewska, M. Stefek, M. Štrosová, G. Tirzitis, P. R. Venskutonis, J. Viskupicova, P. S. Vraka and N. Žarković, Free Radical Res., 2010, 44, 1216–1262 CrossRef CAS.
- H. J. Forman and H. Zhang, Nat. Rev. Drug Discovery, 2021, 20, 689–709 CrossRef CAS.
- D. Genovese, A. Baschieri, D. Vona, R. E. Baboi, F. Mollica, L. Prodi, R. Amorati and N. Zaccheroni, ACS Appl. Mater. Interfaces, 2021, 13, 31996–32004 CrossRef CAS.
- P. Ghezzi, Redox Biol., 2021, 44, 102001 CrossRef CAS PubMed.
- M. Stoia and S. Oancea, Antioxidants, 2022, 11, 638 CrossRef CAS.
- S. Zhou, H. Cai, X. He, Z. Tang and S. Lu, Coord. Chem. Rev., 2024, 500, 215536 CrossRef CAS.
- S. Lin, Y. Cheng, H. Zhang, X. Wang, Y. Zhang, Y. Zhang, L. Miao, X. Zhao and H. Wei, Small, 2020, 16, 1902123 CrossRef CAS.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef PubMed.
- X. Yu, Y. Wang, J. Zhang, J. Liu, A. Wang and L. Ding, Adv. Healthcare Mater., 2024, 13, 2302023 CrossRef PubMed.
- Y. Tang, Y. Han, J. Zhao, Y. Lv, C. Fan, L. Zheng, Z. Zhang, Z. Liu, C. Li and Y. Lin, Nano-Micro Lett., 2023, 15, 112 CrossRef.
- H. Jafari, P. Ghaffari-Bohlouli, S. V. Niknezhad, A. Abedi, Z. Izadifar, R. Mohammadinejad, R. S. Varma and A. Shavandi, J. Mater. Chem. B, 2022, 10, 5873–5912 RSC.
- W. Zhang, S. Roy, P. Ezati, D.-P. Yang and J.-W. Rhim, Trends Food Sci. Technol., 2023, 136, 11–23 CrossRef.
- Z. Guo, W. Xie, J. Lu, X. Guo, J. Xu, W. Xu, Y. Chi, N. Takuya, H. Wu and L. Zhao, J. Mater. Chem. B, 2021, 9, 4098–4110 RSC.
- Y. Chen, X. Yang, K. Li, J. Feng, X. Liu, Y. Li, K. Yang, J. Li and S. Ge, ACS Nano, 2024, 18, 7024–7036 CrossRef PubMed.
- D. Li, J. Li, S. Wang, Q. Wang and W. Teng, Adv. Healthcare Mater., 2023, 12, 2203063 CrossRef PubMed.
- Y. Huang, Y. Chen, G. Cheng, W. Li, H. Zhang, C. Yu, J. Fang, J. Zuo, Y. Li, L. Xu and D. Sun, Int. J. Nanomed., 2024, 19, 231–245 CrossRef CAS PubMed.
- Y. Xu, Y. Luo, Z. Weng, H. Xu, W. Zhang, Q. Li, H. Liu, L. Liu, Y. Wang, X. Liu, L. Liao and X. Wang, ACS Nano, 2023, 17, 18732–18746 CrossRef CAS PubMed.
- V. Venezia, C. Prieto, M. Verrillo, M. Grumi, B. Silvestri, G. Vitiello, G. Luciani and J. M. Lagaron, Int. J. Biol. Macromol., 2024, 263, 130210 CrossRef CAS PubMed.
- S. Tie, Q. Zhang, Y. Zhao, Y. Wu, D. Liu, L. Zhao and S. Gu, RSC Adv., 2024, 14, 7572–7581 RSC.
- Y. Wang, X. Wang, J. Wang, P. Zeng, P. Yang and A. Zhao, Mater. Today Commun., 2024, 38, 108130 CrossRef CAS.
- M. Wen, T. Wang, N. Li, Y. Wu, L. Zhang, Y. Xue and L. Shang, Adv. Funct. Mater., 2024, 2403634 CrossRef CAS.
- B. Muhoza, S. Xia and X. Zhang, Food Hydrocolloids, 2019, 97, 105174 CrossRef CAS.
- Y. Huang, H. Zhong, C. Jiang, J. Yang, J. Zhang, F. Zhao and C. Liu, Particuology, 2024, 84, 126–135 CrossRef CAS.
- Z. Guo, R. Shen, Y. Chen, Y. Huo, J. Li, L. Zhou and J. Hu, Adv. Funct. Mater., 2024, 2402633 CrossRef CAS.
- F. Xu, Y. Tang, H. Wang, H. Deng, Y. Huang, C. Fan, J. Zhao, C. Lin and Y. Lin, Small, 2022, 18, 2201205 CrossRef CAS.
- F. Zhu, Y. Yu, Z. Yu, H. Qiu, G.-P. Lu, Z. Chen, J. Hu and Y. Lin, Small, 2024, 20, 2311848 CrossRef CAS.
- Q. Fu, N. Wang, C. Zhou and X. Su, Talanta, 2024, 266, 124991 CrossRef CAS PubMed.
- F. Mo, S. Zhong, T. You, J. Lu and D. Sun, ACS Appl. Mater. Interfaces, 2023, 15, 52114–52127 CAS.
- T. Liu, B. Xiao, F. Xiang, J. Tan, Z. Chen, X. Zhang, C. Wu, Z. Mao, G. Luo, X. Chen and J. Deng, Nat. Commun., 2020, 11, 2788 CrossRef CAS PubMed.
- S. Luo, A. Saadi, K. Fu, M. Taxipalati and L. Deng, J. Sci. Food Agric., 2021, 101, 6355–6367 CrossRef CAS PubMed.
- J. Xi, S. Shahab and R. Mirzaeifar, RSC Adv., 2022, 12, 29162–29169 RSC.
- H. Salimi-Kenari, M. Barari, S. R. Nabavi, A. Mousavi Anjeh and S. R. Hosseini, Polym.-Plast. Technol. Mater., 2023, 62, 294–305 CAS.
- H. Gade, S. Nikam, G. G. Chase and D. H. Reneker, Polymer, 2021, 228, 123902 CrossRef CAS.
- Z. Sun, M. Li, Z. Jin, Y. Gong, Q. An, X. Tuo and J. Guo, Int. J. Biol. Macromol., 2018, 120, 2552–2559 CrossRef CAS PubMed.
- A. Refate, Y. Mohamed, M. Mohamed, M. Sobhy, K. Samhy, O. Khaled, K. Eidaroos, H. Batikh, E. El-Kashif, S. El-Khatib and S. Mehanny, Heliyon, 2023, 9, e17051 CrossRef CAS PubMed.
- W. Zuo, M. Zhu, W. Yang, H. Yu, Y. Chen and Y. Zhang, Polym. Eng. Sci., 2005, 45, 704–709 CrossRef.
- İ. Gulcin, Arch. Toxicol., 2020, 94, 651–715 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05314a |
| This journal is © The Royal Society of Chemistry 2024 |