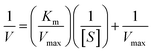Open Access Article
Open Access ArticleA simple Ag–MoS2 hybrid nanozyme-based sensor array for colorimetric identification of biothiols and cancer cells†
Yin Lia,
Yumeng Liud,
Yueqin Zhangd,
Mengmeng Donge,
Lidong Cao*bc and
Kai Jiang *b
*b
aDepartment of Dermatology, Children's Hospital, Zhejiang University School of Medicine, Hangzhou, China
bDepartment of Hepatobiliary & Pancreatic Surgery and Minimally Invasive Surgery, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, Hangzhou, China
cCollege of Mechanical Engineering, Zhejiang University, Hangzhou, China
dSchool of Public Health, Hangzhou Medical College, Hangzhou, China
eClinical Research Institute, Zhejiang Provincial People's Hospital (Affiliated People's Hospital), Hangzhou Medical College, Hangzhou, China
First published on 4th October 2024
Abstract
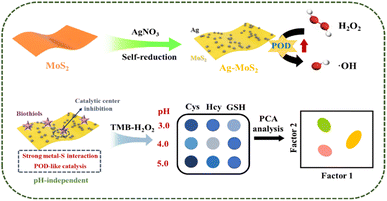
The intracellular levels of biothiols are associated with various diseases including cancer, and biothiols are regarded as tumor biomarker. Due to the similarity of the molecular structure of biothiols, the development of simple, rapid, efficient, and sensitive colorimetric sensor arrays holds great promise for clinical cancer diagnosis. Here, we developed a simple Ag–MoS2 hybrid nanozyme-based sensor array for colorimetric identification of biothiols and cancer cells. The novel Ag–MoS2 nanoprobe was synthesized in a simple and efficient way through the in situ self-reduction reaction between MoS2 and noble metal precursor. Benefiting from to the formation of heterogeneous metal structures, the peroxidase (POD)-like catalytic activity of the synthesized Ag–MoS2 hybrid nanocomposites is significantly enhanced compared to MoS2 alone. Moreover, the catalytic activity of Ag–MoS2 nanozyme was correlated with the pH of the reaction solution and the inhibitory effects of the three biothiols on the nanozyme-triggered chromogenic system differed in the specific pH environments. Therefore, each sensing unit of this electronic tongue generated differential colorimetric fingerprints of different biothiols. After principal component analysis (PCA), the developed novel colorimetric sensor array can accurately discriminate biothiols between different types, various concentrations, and different mixture proportions. Further, the sensor array was used for the colorimetric identification of real serum and cellular samples, demonstrating its great potential in tumor diagnostic applications.
1 Introduction
Biothiols are thiol-containing amino acids or oligopeptides, mainly including L-cysteine (Cys), homocysteine (Hcy) and glutathione (GSH).1,2 Biothiols are a class of small molecule compounds that play important roles in many physiological processes such as cell signaling, enzyme activities regulation, and antioxidant defences.3,4 The altered levels of biothiols have been shown to be closely associated with the development of a variety of diseases, including cardiovascular disease, neurodegenerative diseases, and even cancer.5,6 For instance, the level of biothiols in hepatocellular carcinoma cells is much higher than that in normal hepatocytes, and biothiols are considered to be a promising biomarker for liver cancer.7 Therefore, the accurate detection of biothiols is of great significance for the prevention, diagnosis and treatment of cancer.8Currently, researchers have developed various methods for the efficient detection of biothiols, such as mass spectrometry,9,10 high performance liquid chromatography,11,12 fluorescence,13,14 chemiluminescence,15,16 electrochemistry,17 and capillary electrophoresis.18 Although these quantitative detection methods for biothiols have been widely applied, they often require expensive equipment, specialized operators and additional sample pre-treatment steps, which somewhat hinder their practical application in clinical diagnosis. Compared with traditional detection methods, colorimetric sensors have the advantages of simple operation, low cost and rapid detection, which are suitable for large-scale clinical screening and on-site testing.19,20 In addition, colorimetric methods can be combined with miniaturized equipment, image capture technology and artificial intelligence algorithms, making colorimetric sensors more portable, sensitive and accurate. However, due to the high degree of similarity between various types of biothiols, traditional colorimetric sensing techniques lack the specificity to discriminate different biothiols.
Sensor array technology, also known as electronic nose/tongue, is a system that integrates multiple sensor units.21,22 These sensor units have different response characteristics to target analytes and can provide a wealth of information to identify and detect various chemical components.23,24 By designing different sensor units, each unit responds differently to the same target molecule, thus generating multidimensional information. After that, data processing is used to visually distinguish between different analytes, complex samples and mixtures. Up to now, sensor array has wide application potential for the detection of biological targets, especially in the field of disease diagnosis and health monitoring.25 For instance, Wang group used array recognition technology to achieve efficient detection of exosomes (a tumor marker) and highly sensitive and accurate screening of gastric cancer and urinary system diseases.26,27 Recently, sensor array technology has also been applied for biothiols discrimination. For example, Wang et al. exploited gold nanorods (AuNRs) with the unique localized surface plasmon resonance (LSPR) property as chromophore, and used a variety of metal ions (Hg2+, Pb2+, Cu2+, Ag+) to form a novel colorimetric sensor array for biothiols identification.28 Due to the differences in binding affinity between biothiols and different metal ions, the AuNRs exhibit unique patterns in response to various thiol molecules after etching through the Fenton reaction, forming fingerprint data for rapid, visual and accurate discrimination of biothiols. However, the above methods require multiple and complex chromogenic system designs and the use of heavy metal ions poses contamination and safety issues. Additionally, this study has not been successfully applied to intracellular biothiols detection. Further design and development of sensing probe or nanomaterial for efficient differentiation and selective detection of different intracellular biothiol molecules is crucial to advance the practical application of colorimetric sensor array technology.
Recently, novel biosensing technologies based on nanomaterials have been developed and made great breakthroughs in analytical chemistry field.29,30 For instance, nanozymes are defined as a class of nanomaterials which have the natural enzyme-like catalytic functions.31 To date, various nanozymes have been successfully applied in many fields, such as biosensing, immunoassay, disease treatment and environmental monitoring.24,32,33 Since the first discovery of Fe3O4 nanoparticles with peroxidase (POD) mimetic activity by Gao et al. in 2007,34 various types of nanozymes such as noble metals, metal oxides, carbon-based nanomaterials, and metal–organic framework materials have been reported to exhibit excellent enzyme-like activity.35 As a new type of bioactive material, nanozymes have significant advantages in the field of colorimetric detection due to their low production cost, high stability, easy functionalization and adjustable catalytic activity.36,37 In recent years, several studies have reported the application of nanozyme-based colorimetric sensor array for the differentiation and detection of biothiols. For instance, Wei et al. synthesized three noble metal alloy nanozymes (AuPt, AuPd and AuPtRu) and found that the peroxidase-like activity of these gold-based alloys could be inhibited with varying degrees by different biothiols.38 Subsequently, the different nanoprobes could trigger the chromogenic system to produce various colorimetric signals for the identification and analysis of biothiols. Similarly, Bi et al. used terephthalic acid-modified graphene quantum dots (TPA@GQDs) with three transition metal ions (Fe2+, Cu2+, and Zn2+) as the sensing units for the construction of the nanozyme-based electronic tongue.39 The strong binding affinity between the different metal ions and biothiols significantly inhibited the peroxidase-like catalytic activity of TPA@GQDs, producing a cross response. The above colorimetric sensor arrays proved to have good selectivity and sensitivity, but the complex synthesis of multiple nanozymes and the cumbersome construction of the sensing units limited their practical application and clinical translation. Further simplification of the nanozymes-based colorimetric sensor array components would be of great value in the field of clinical diagnosis, and hold significant incentives for the development of functional nanozymes.
Herein, we present a simple hybrid nanozyme-based colorimetric sensor array, which can successfully differentiate and detect three common biothiols (Cys, Hcy and GSH) by easily adjusting the pH value of the nanozyme-triggered chromogenic reaction system (Scheme 1). Firstly, we prepared the novel heterogeneous Ag–MoS2 nanozyme by a simple in situ self-reduction method between MoS2 and Ag+. Benefiting from the formation of metal heterostructures, the Ag–MoS2 nanoprobe possessed excellent peroxidase-like activity, which strongly catalyzed the decomposition of H2O2, accompanied by a color change of the chromogenic substrate, 3,3′,5,5′-tetramethylbenzidine (TMB), from colorless to blue. It is noteworthy that the colorimetric reaction triggered by Ag–MoS2 nanozymes exhibits pH-dependent behavior. Specifically, the –SH of biothiols has been demonstrated to show strong interaction with metal nanostructure by the formation of metal–S bonds which can easily reduce the electron density of metal nanomaterials.40 Moreover, the affinity interaction between biothiols and metal has also been shown to be pH-dependent and further inhibits the catalytic activity.41,42 Therefore, at different pH values, the catalytic ability of Ag–MoS2 nanozymes as well as the binding effect of Ag, Mo metal center and –SH showed significant differences, resulting in different degrees of inhibition of colorimetric signals. Here, under three different pH environments, the TMB–H2O2-nanozymes chromogenic sensor forms the differential fingerprints of the three biothiols, which can be accurately distinguished from each other by principal component analysis (PCA). In addition, the constructed sensor arrays also achieved the recognition of mixtures of biothiols with different concentrations. More importantly, the fabricated sensor array of biothiols can also be applied to distinguish human serum as well as cancer cells such as hepatocellular carcinoma and normal hepatocytes, demonstrating its great potential in disease diagnosis.
2 Experimental methods
2.1. Chemicals and materials
Cysteine (Cys), Glutathione (GSH), silver nitrate trisodium (AgNO3), molybdenum disulfide (MoS2) powder, sodium acetate (NaAC), 3,3′,5,5′-tetramethylbenzidine (TMB) and polyvinylpyrrolidone (PVP) were purchased from Macklin (China). Homocysteine (Hcy), acetic acid (HAC), H2O2 (30 wt%), hydrochloric acid (HCl), and sodium hydroxide (NaOH) were purchased from Sinopharm Chemical Reagent Co., Ltd. Fetal bovine serum (FBS) was obtained from Biological Industries Corp. All the tested cells were obtained from Center for Typical Culture Collection. Dulbecco's modified Eagle's medium (DMEM) was bought from Sigma-Aldrich. Human serum was collected from healthy volunteers in Zhejiang Provincial People's Hospital according to the Ethics Committee guidelines. Ultrapure water (18.2 MΩ cm) was applied through the experiments to prepare all the solutions.2.2. Preparation of nanozymes
According to the previous reports, the MoS2 nanosheet was firstly prepared by the classical LPE method with slight modification.43 Briefly, 0.4 g of MoS2 powder was added into 40 mL 45% ethanol aqueous solution containing PVP (1 mg mL−1) and the above mixture was sonicated at 400 W for 5 h in an ice-water bath. Subsequently, the obtained MoS2 nanosheet was washed by water after centrifugation at 4000 rpm for 20 min. To prepare Ag–MoS2, 10 μL of AgNO3 (10 mM) was added into 1 mL of the MoS2 nanosheet for stirring 1 h at room temperature. After that, the Ag–MoS2 product was collected and washed three times with water by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 times to remove the excess AgNO3. Finally, the Ag–MoS2 hybrid material was obtained and stored at 4 °C for further use. For Ag NPs, 10 μL of AgNO3 (10 mM) was added into 1 mL of the PVP solution (1 mg mL−1) for stirring 4 h at room temperature. After that, the Ag–MoS2 product was collected and washed three times with water by centrifugation at 10
000 rpm for 3 times to remove the excess AgNO3. Finally, the Ag–MoS2 hybrid material was obtained and stored at 4 °C for further use. For Ag NPs, 10 μL of AgNO3 (10 mM) was added into 1 mL of the PVP solution (1 mg mL−1) for stirring 4 h at room temperature. After that, the Ag–MoS2 product was collected and washed three times with water by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 times to remove the excess PVP and AgNO3.
000 rpm for 3 times to remove the excess PVP and AgNO3.
2.3. Characterization instrumentation
Transmission electron microscopy (TEM) images was performed on a H-7650 (Hitachi). Energy dispersive X-ray (EDX) elemental mapping measurement was carried out by a Tecnai F30 microscopy. X-ray photoelectron spectroscopy (XPS) measurements were carried out on ESCALAB 250XI (Thermo Fisher). The size distribution was tested using a dynamic light scattering instrument (DLS, Zetasizer Nano-ZS90, Malvern). UV-vis spectra were recorded by a UV-2450 spectrometer (Shimadzu). The electron paramagnetic resonance (EPR) signals were measured using a JES-FA200 spectrometer (JEOL, Japan).2.4. Nanozyme activity test
To study the POD-like catalytic effect, 5 mM H2O2 was firstly mixed with 2.5 mM TMB in HAC-NaAc buffer (pH = 4.0, 25 °C). Next, 10 μL of nanozyme (1 mg mL−1) including Ag, MoS2 and Ag–MoS2 were added to trigger the reaction at 25 °C for 5 min and the UV-vis absorbance was recorded by UV spectrometer. To study the influence of pH values for the POD-like activity, Ag–MoS2 (10 μL, 1 mg mL−1) was added to a centrifuge tube containing HAC-NaAc buffer (pH = 4.0, 25 °C) with various pH values from 2.0 to 9.0. The catalytic kinetics was studied by monitoring the absorbance change of color-development system at 652 nm. The kinetics parameter including Michaelis–Menten constant (Km) and maximum reaction rate (Vmax) were calculated according to the double reciprocal curve of the Michaelis–Menten equation:where V is the initial reaction speed, Vmax is the maximal velocity, [S] is the H2O2 concentration, and Km is the Michaelis constant.
2.5. General procedure for colorimetric discrimination of biothiols
The general procedures for colorimetric discrimination of biothiols were carried out as follows: first, 10 μL of hybrid nanozyme (1 mg mL−1) was added to HAc-NaAc buffer at different pH values (3.0, 4.0 and 5.0), followed by the addition of different concentrations of biothiols (Cys, Hcy, and GSH). After reacted for 30 min, the mixture containing 5 mM H2O2 and 1 mM TMB was added and the absorbance change at 652 nm of the sensor systems was measured and calculated. All the experiments were repeated for 5 times. Then, the signals obtained from the sensor array were analyzed using principal component analysis (PCA) and hierarchical cluster analysis (HCA) on the original data matrix by Origin 2021 software. Further, the detection limit of sensor array for each biothiols by studying the response results of PCA analysis under low concentration according to the method of previous literature.442.6. Identification of biothiols in human serum and cell samples
In order to further investigate the practical application of the proposed sensor array for the discriminative analysis of real samples, colorimetric pattern recognition of human serum samples and cellular samples were performed separately. At first, Ag–MoS2-based sensor array was used to differentiate serum samples (200-fold dilution) spiked with three types of biothiols at the same concentration (1 μM), and followed the steps described above for colorimetric analysis and data processing. For the cell samples, a total of three common cancer cells, Hela (cervical cancer cells), HepG2 (hepatocellular carcinoma cells), MCF-7 (breast cancer cells) and normal cells, LO2 (normal hepatocytes), were selected for testing the pattern recognition ability of the colorimetric sensing array. The above cells were cultured in DMEM containing 10% FBS. After the cells had grown to the full size of the culture flasks the cells were digested with trypsin, centrifuged to discard the culture medium and washed twice with phosphate buffer solution (PBS). Next, the obtained cells were lysed by ultrasonic fragmentation and cell lysates were collected according to the literature.40 Specifically, the washed cells were suspended in 500 μL buffer, and then were lysed by ultrasonic method in ice bath (3 s each time, 2 s interval, a total of 20 times). After centrifugation, the above cell lysate was diluted 10 times with DMEM and transferred to a new test tube for further use. Then, 10 μL Ag–MoS2 (1 mg mL−1) was added to the test tube. After reacted for 30 min, the mixture containing 5 mM H2O2 and 1 mM TMB was added and the absorption change at 652 nm of the sensor systems was measured and calculated. All the experiments were repeated for 5 times. Then, the signals obtained from the sensor array were analyzed using principal component analysis (PCA) and hierarchical cluster analysis (HCA) on the original data matrix by origin 2021 software.3 Results and discussion
3.1. Synthesis and characterization of Ag–MoS2
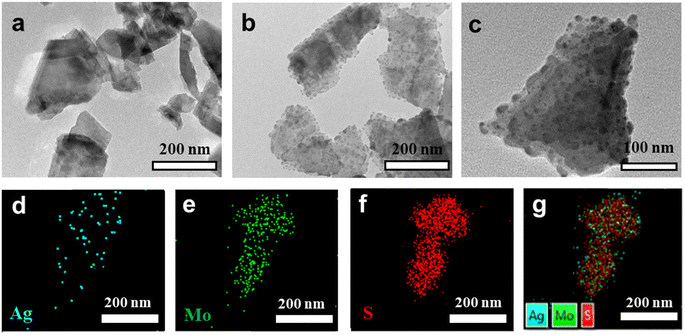
In this work, we first obtained MoS2 nanosheets with a 2D layered structure from MoS2 powders by the classical LPE method.43 Subsequently, Ag–MoS2 composites were prepared by a simple and convenient self-reduction methods. Specifically, MoS2 nanosheets reduced Ag ions in situ onto MoS2 nanosheets at room temperature to form spherical Ag nanoparticles, which led to the formation of a 0D–2D hybrid heterostructure. During the synthesis process, the defects and edges of partially unbound sulfur on the MoS2 surface were the main sites for Ag core seeding and subsequent growth. To verify the successful preparation of MoS2 and Ag–MoS2, their morphologies were observed by TEM. As revealed in Fig. 1a, the prepared MoS2 nanosheets showed irregular flakes of ∼200 nm, in agreement with other reports in the literature. Fig. 1b and c show different magnification TEM images of Ag–MoS2 nanocomposites prepared after the self-reducing method. Compared to MoS2 nanosheets, Ag–MoS2 shows the presence of obvious spherical Ag nanoparticles on MoS2 with high density, but MoS2 nanosheets remains its original morphology and size. As previously reported, the uniform and high-density noble metal particles on the 2D MoS2 support layer can significantly improve the electron transfer process and catalytic properties of metallic materials.45,46 The formation of these bimetallic hybrid structure will be beneficial to the efficient catalysis and highly sensitive sensing applications of the nanozyme. In order to resolve the composition of Ag–MoS2 in a more detailed manner, EDX elemental mapping analysis was conducted (Fig. 1d–g). The results confirmed the presence of elements such as Ag, Mo, and S in the composites, and the Ag nanoparticles were uniformly dispersed on the MoS2 nanosheets. Besides, we have studied the stability of Ag–MoS2 hybrid material both at 4 °C and room temperature (20 °C) by DLS analysis. As shown in Fig. S1,† the particle size distribution of our Ag–MoS2 nanozyme was 225 ± 9 nm, which was consistent with TEM results. In addition, it was found that the average particle size of the composite material remained stable after being stored at 4 °C and 20 °C for 14 days, respectively.Further, XPS was used to verify the elemental composition and properties of the Ag–MoS2 composites. Similar to the above mapping results, the XPS survey results confirmed the presence of Ag, Mo and S elements in Ag–MoS2 (Fig. 2a). In addition, high-resolution scans of Ag 3d, Mo 3d and S 2p were performed for the three elements, and the results are shown in Fig. 2b–d, respectively. In the high-resolution spectra of Ag 3d, two distinct peaks located at 367.28 eV and 373.28 eV were attributed to Ag 3d3/2 and Ag 3d5/2, respectively. The above results also proved that Ag exists in the composites in a monomeric state, which is a favorable valence state commonly used in nanocatalysis. The Mo and S substituents in MoS2 are generally tetravalent and divalent. In the Mo 3d spectrum, the peaks at 228.68 eV and 231.78 eV are attributed to Mo 3d5/2 and Mo 3d3/2, respectively. The binding energies of the S 2p peaks are located at 162.58 and 161.48 eV, corresponding to 2p1/2 and 2p3/2, respectively. The above results demonstrate the successful preparation of Ag–MoS2 composites.
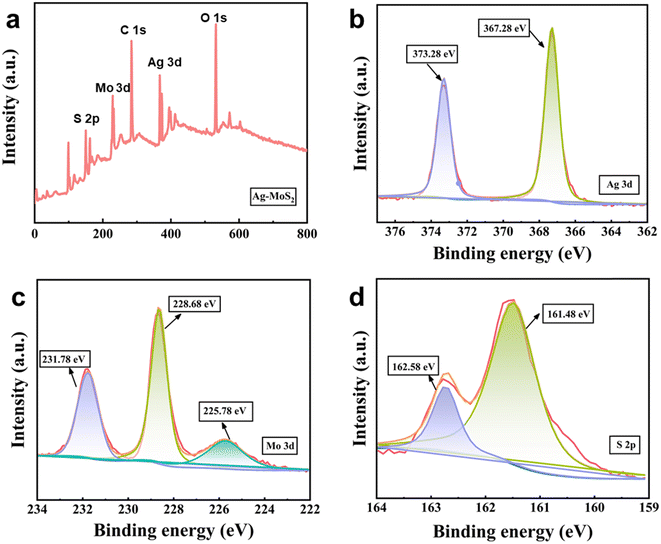 | ||
| Fig. 2 (a) The XPS survey of Ag–MoS2 hybrid material. The high-resolution XPS spectra of Ag 3d (b) Mo 3d (c) and (d) S 2p signals for Ag–MoS2. | ||
3.2. POD-like catalytic activity of Ag–MoS2
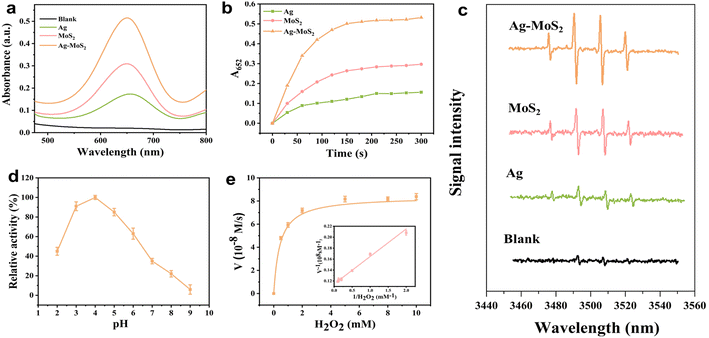
After successful preparation of novel hybrid Ag–MoS2 nanomaterials, their enzyme-like catalytic activity was carefully investigated. Here, we selected TMB as the chromogenic substrate, which can trigger the classical catalytic oxidation reaction in the presence of H2O2 to generate oxidized TMB and thus bring about a significant change both in color and absorbance. According to previous reports, both Ag and MoS2 nanomaterials were shown to have excellent catalytic properties mimicking POD. Here, for comparison, we successfully synthesized Ag NPs, which have similar morphology and size with Ag NPs in Ag–MoS2 (Fig. S2†). As indicated in Fig. 3a, after catalyzed by Ag, MoS2 or Ag–MoS2 nanozymes with the same concentration, the H2O2–TMB system showed an obvious absorption peak at 652 nm, while the same system showed almost no absorption at 652 nm in the absence of the materials. What's more, the absorbance value and catalytic rate of the Ag–MoS2 group were much higher than those of the Ag and MoS2 group (Fig. 3b). The above results suggest that the interfacial synergistic effect and electron transfer between the metals after the formation of hybrid materials will enhance the POD-like catalytic activity of MoS2 alone, achieving more efficient H2O2 decomposition and more sensitive color development. In general, the POD-like activity of metal-based nanozymes originates from the catalytically generated ˙OH radicals, which are responsible for oxidizing the colorless TMB substrate to blue oxidized TMB. To validate this, the EPR technique was utilized to investigate the formation of intermediate ˙OH radicals and DMPO was used here as a spin trapping agent. As demonstrated in Fig. 3c, compared to the blank, the three nanozymes could effectively catalyze the decomposition of H2O2 and produce ˙OH, and four peaks with the intensity ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were produced after being captured by DMPO. It is worth noting that the Ag–MoS2 composite materials exhibits the strongest EPR signal compared with the single Ag and MoS2, which may be attributed to the doping of noble metals and the synergistic effect of Ag and MoS2 in the hybrid structure. These results indicate that Ag–MoS2 has enhanced peroxidase-like catalytic activity, which can more effectively convert H2O2 to ˙OH radicals and trigger the catalytic oxidation of TMB substrates. Besides, the result is consistent with many reports that the formation of bimetallic heterostructures significantly improved the catalytic activity of nanozymes.47,48 The high-efficiency generation ability ˙OH radicals laid a good foundation for the establishment of further sensitive colorimetric sensing methods based on Ag–MoS2.
1 were produced after being captured by DMPO. It is worth noting that the Ag–MoS2 composite materials exhibits the strongest EPR signal compared with the single Ag and MoS2, which may be attributed to the doping of noble metals and the synergistic effect of Ag and MoS2 in the hybrid structure. These results indicate that Ag–MoS2 has enhanced peroxidase-like catalytic activity, which can more effectively convert H2O2 to ˙OH radicals and trigger the catalytic oxidation of TMB substrates. Besides, the result is consistent with many reports that the formation of bimetallic heterostructures significantly improved the catalytic activity of nanozymes.47,48 The high-efficiency generation ability ˙OH radicals laid a good foundation for the establishment of further sensitive colorimetric sensing methods based on Ag–MoS2.
Further, we explored the POD-like activity of the Ag–MoS2 hybrid materials at different pH values. As shown in Fig. 3d, the optimal catalytic activity of the Ag–MoS2 hybrid material was at the pH value of 4.0 and the catalytic activity is significantly inhibited under reaction conditions with pH value less than 3.0 or pH value greater than 5.0, similar to that of natural enzymes. Therefore, for a more sensitive and intuitive colorimetric analysis, pH ranges from 3.0 to 5.0 was selected as the main color development condition for the subsequent sensor array. Additionally, the long-term storage stability at both 4 °C and room temperature of the nanozyme was tested and the results showed that the composite retained over 95% catalytic activity after 14 days of storage at both temperatures (Fig. S3†). The above results show that the hybrid material has superior chemical stability, which can be effectively used as sensing probe.
To investigate the POD-like activity of the Ag–MoS2 hybrid material in more depth, the H2O2 substrate concentration was varied to study the steady-state kinetics. The results demonstrated that the catalytic process of the Ag–MoS2 nanozymes followed the standard Michaelis–Menten model well (Fig. 3e). Subsequently, the enzyme kinetic parameters for Ag–MoS2 were evaluated by Michaelis–Menten curves and Lineweave–Burk plots, and Menten constant (Km) and the maximum reaction velocity (Vmax) were calculated to be 0.4364 mM and 8.38 × 10−8 M S −1. Table 1 summarizes the detailed kinetic parameters of similar MoS2-based nanozymes. Generally, lower Km values means higher substrate affinity between H2O2 and POD-like nanozyme, while higher Vmax values mean a better catalytic ability. Notably, the Ag–MoS2 nanozyme exhibits relatively excellent Km and Vmax values compared to other materials and even natural enzymes HRP, suggesting an excellent POD-like activity for reliable and sensitive colorimetric sensor construction.
3.3. Ag–MoS2 nanozyme-based sensor arrays for biothiols
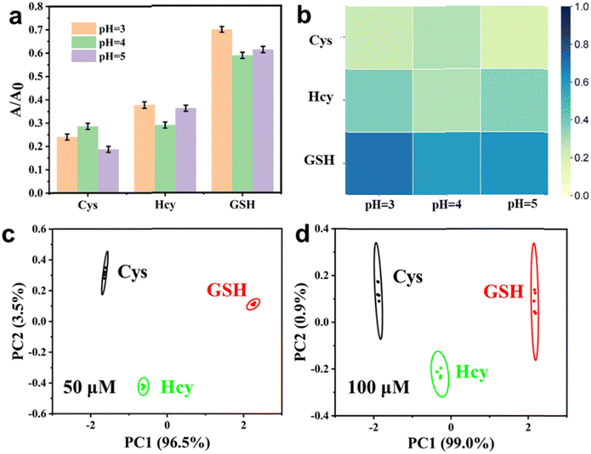
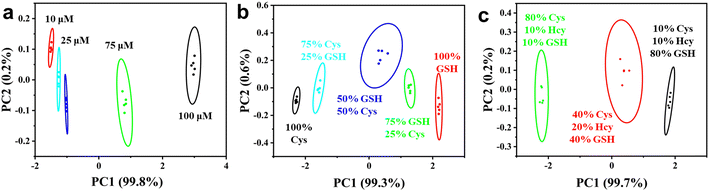
Motivated by the excellent catalytic activity of Ag–MoS2 hybrid nanomaterials, we subsequently constructed multi-channel colorimetric sensor arrays using the POD-like catalysis of H2O2–TMB chromogenic system at three different pH values (3.0, 4.0 and 5.0). Firstly, we investigated the response (A/A0) of the three-channel colorimetric sensors to three different biothiols (Cys, Hcy, and GSH) at the same concentration. As shown in Fig. 4a, unique colorimetric response signals corresponding to different biothiols (50 μM) can be observed in the reaction media at different pH, which is mainly due to the differences in the activity of Ag–MoS2 nanozymes and the different interactions between the materials and biothiols under different pH conditions. In addition, the unique response pattern of each biothiol could be more easily identified in the thermogram (Fig. 4b). After analyzed by PCA, the response patterns of three different biothiols (50 μM) were clustered into three non-overlapping regions, demonstrating the successful differentiation of three biothiols by the colorimetric sensor arrays (Fig. 4c). In addition, the fabricated sensor array could also be used to distinguish three biothiols at other concentration (100 μM), indicating that the Ag–MoS2 hybrid nanozyme-based colorimetric sensor array is a powerful tool for identifying biothiols (Fig. 4d). Compared with other sensor systems that require complex probe design, the present work achieves colorimetric differentiation of low-concentration biothiols by easily adjusting the pH value of the reaction solution with only one kind of nanozyme, which is more promising for practical applications.Then, the novel and simple colorimetric sensor array was used for the pattern recognition of the Cys at different concentrations of 10, 25, 50, 75 and 100 μM. As shown in Fig. 5a, Cys with different concentrations has been successfully identified after PCA treatment and the PC value can be used to reveal the different concentrations. Further, to confirm the quantitative analysis ability of the sensor array, the response signals induced by three biothiols with different concentrations at low concentrations were evaluated. As shown in Fig. S4,† the standard curves of Cys, Hcy, and GSH we obtained by plotting the relationship between the concentration of biothiols and score of PC1. As a result, the linear detection range were 1–25 μM for Cys, 1–10 μM for Hcy, and 5–25 μM for GSH. The detection limits calculated by conventional 3σ method were 0.72 μM for Cys, 0.89 μM for Hcy, and 3.28 μM for GSH, respectively. Subsequently, in order to further investigate the application of the novel colorimetric sensor array based on Ag–MoS2 in the discrimination of mixtures, biothiols with different molar ratios (total concentration of 100 μM) were artificially prepared and identified with the help of the developed sensor array. As shown in Fig. 5b, mixtures of two kinds of biothiols (Cys and GSH) in different proportions were successfully divided into five independent and non-overlapping groups after PCA analysis. In addition, three different proportions of biothiol mixtures were also well distinguished (Fig. 5c). The above results verify the ability of sensor array based on Ag–MoS2 nanozyme to distinguish biothiol mixtures.
3.4. Applications of the developed sensor array in real samples
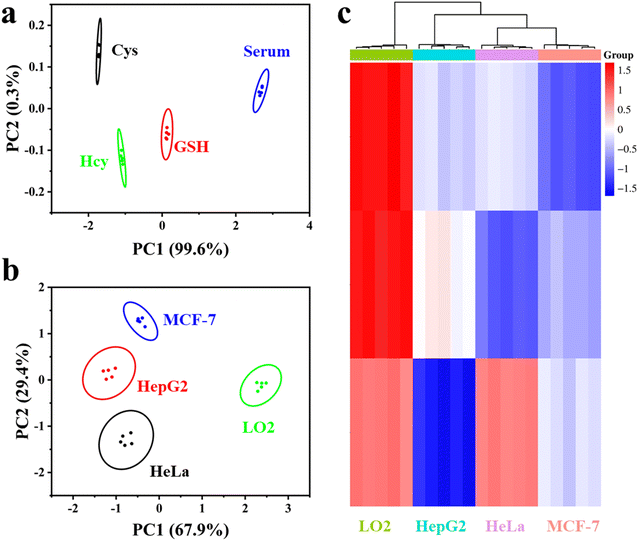
To explore the practical potential of the novel Ag–MoS2 hybrid nanozyme-based sensor array for biomedical applications, serum and cell samples were further analyzed. According to previous studies, biothiols in serum can be used as significant biomarkers.28 Here, a 200-fold dilution of serum samples mixed with the same concentration of biothiols (Cys, Hcy and GSH) was used for the study. Then, the proposed Ag–MoS2 hybrid nanozyme-based sensor array was used to distinguish the biothiols in the serum samples. As shown in Fig. 6a, other substances present in the serum did not interfere with the identification of the biothiols and blank serum sample could be well differentiated from the samples containing the three different biothiols after PCA treatment. In addition, the three kinds of biothiols showed significantly different PCA signals in serum, which demonstrated the good practical applicability of the colorimetric sensing array in complex biological matrices.The screening and identification of various types of cells is of great significance in the early and accurate diagnosis of tumors. Clinical studies have shown that there are significant differences in the content of biothiols between cancer cells and normal cells.40 Therefore, we explored the application potential of the proposed novel sensor array in the colorimetric identification of various cell samples. Herein, three common cancer cells, HepG2, Hela, MCF-7 and normal cells, LO2, were selected for testing the pattern recognition ability of the colorimetric sensing array. As shown in Fig. 6b, the four kinds of cells tested were well differentiated and aggregated into different groups without overlap after PCA by Ag–MoS2 nanozyme-based sensor array. In addition, only the PC1 value of LO2 cells was greater than 0, while the PC1 values of the other three kinds of cancer cells were negative. These results show that the sensor array can well distinguish normal cells from cancer cells. In addition, the three kinds of cancer cells can also be well distinguished by PCA analysis. Next, HCA results further confirmed that the sensor array can accurately classify cells. The above experimental results strongly support the effectiveness of the Ag–MoS2 hybrid nanozyme-based colorimetric sensor array in practical applications.
4 Conclusions
In the present study, Ag–MoS2 hybrid materials with excellent POD-like catalytic activity were obtained by in situ growth of noble metal Ag nanoparticles on MoS2 nanosheets via a simple and convenient self-reduction method. Compared with MoS2 nanozyme alone, the hybrid materials have higher POD catalytic activity due to synergy of metal co-catalysis and enhancement of electron transfer effect, and thus can be used for more sensitive colorimetric analysis. Since the competitive inhibition of POD-like activity by different biothiols on the metal-hybridized materials varies at different pH, the novel sensor array requires only one probe material for the colorimetric identification of three typical biothiols and their binary or ternary mixtures. Meanwhile, the sensor array also showed excellent discrimination ability in complex biological samples such as serum and cells. In conclusion, this work not only provides a simple and sensitive method for the differentiation of biothiols, but also broadens the application of nanozymes in colorimetric sensor arrays. Considering the simple synthesis and excellent catalytic activity of Ag–MoS2, we believe that Ag–MoS2-based colorimetric sensor arrays can combine more diversified sensing mechanisms and more ingenious designs to provide satisfactory analytical results for a wide range of diverse targets in the future, which will promote the analytical application of nanozymes in practical clinical samples.Data availability
Data will be available on request.Author contributions
Kai Jiang conceived the project and supervised the research. Kai Jiang propose the idea of methods. Yin Li and Lidong Cao designed and performed the experiments. Yumeng Liu, Yueqin Zhang and Mengmeng Dong discussed the data and check the manuscript. Yin Li and Lidong Cao wrote the paper.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by Zhejiang Province Medical and Health Technology Project (C-2018-W185 and C-2024-W1082); Zhejiang Province Traditional Chinese Medicine Technology Plan Project (C-2024-W2074); and General Projects of Zhejiang Provincial Department of Education (C-2024-G106).Notes and references
- Y. Gong, P. Wang, H. Zhai, Y. Xiao, Q. Wang, N. Ma, G. Zhang and H. Zhang, Anal. Chem., 2024, 96, 1009–1018 CrossRef CAS PubMed.
- C. Cossetti, G. Di Giovamberardino, R. Rota and A. Pastore, Nat. Commun., 2018, 9, 1588 CrossRef PubMed.
- C. K. Sen, Curr. Top. Cell. Regul., 2001, 36, 1–30 Search PubMed.
- X. Jiang, B. Du and J. Zheng, Nat. Nanotechnol., 2019, 14, 874–882 CrossRef CAS PubMed.
- J. M. An, M. Jeong, J. Jung, S. G. Yeo, S. Park and D. Kim, ACS Appl. Mater. Interfaces, 2024, 16, 4493–4504 CrossRef CAS PubMed.
- X. Cheng, H.-D. Xu, H.-H. Ran, G. Liang and F.-G. Wu, ACS Nano, 2021, 15, 8039–8068 CrossRef CAS PubMed.
- W. Ding, S. Yao, Y. Chen, Y. Wu, Y. Li, W. He and Z. Guo, Molecules, 2023, 28, 2229 CrossRef CAS PubMed.
- Y. Shen, J. Yue, W. Shi, W. Xu and S. Xu, Biosens. Bioelectron., 2020, 151, 111957 CrossRef CAS PubMed.
- M. Isokawa, T. Shimosawa, T. Funatsu and M. Tsunoda, J. Chromatogr. B, 2016, 1019, 59–65 CrossRef CAS PubMed.
- Y. Yin, B. Zhen, J. Sun, J. Ouyang and N. Na, Rapid Commun. Mass Spectrom., 2022, 36, e9291 CrossRef CAS PubMed.
- Y. Zu, J. Chromatogr. B, 2009, 877, 3358–3365 CrossRef CAS PubMed.
- J. Stachniuk, P. Kubalczyk, P. Furmaniak and R. Głowacki, Talanta, 2016, 155, 70–77 CrossRef CAS PubMed.
- J. Dai, C. Ma, P. Zhang, Y. Fu and B. Shen, Dyes Pigm., 2020, 177, 108321 CrossRef CAS.
- W. Wang, K. Wang, X.-T. Wang, R.-J. Man, C. Xu, Y.-S. Yang and H.-L. Zhu, Anal. Chim. Acta, 2021, 1177, 338786 CrossRef CAS PubMed.
- M. S. Elgawish, N. Kishikawa and N. Kuroda, Analyst, 2015, 140, 8148–8156 RSC.
- E. Akrivi, A. Vlessidis, N. Kourkoumelis, D. Giokas and G. Tsogas, Talanta, 2022, 245, 123464 CrossRef CAS PubMed.
- I. M. Mostafa, H. Liu, S. Hanif, M. R. H. S. Gilani, Y. Guan and G. Xu, Anal. Chem., 2022, 94, 6853–6859 CrossRef CAS PubMed.
- V. Dosedělová and P. Kubáň, Electrophoresis, 2024, 45, 1418–1427 CrossRef PubMed.
- V. S. Ajay Piriya, P. Joseph, S. C. G. Kiruba Daniel, S. Lakshmanan, T. Kinoshita and S. Muthusamy, Mater. Sci. Eng. C, 2017, 78, 1231–1245 CrossRef PubMed.
- Y. Zhou, X. Huang, X. Hu, W. Tong, Y. Leng and Y. Xiong, Biosens. Bioelectron., 2021, 190, 113386 CrossRef CAS PubMed.
- J. R. Askim, M. Mahmoudi and K. S. Suslick, Chem. Soc. Rev., 2013, 42, 8649–8682 RSC.
- T. Li, X. Zhu, X. Hai, S. Bi and X. Zhang, ACS Sens., 2023, 8, 994–1016 CrossRef CAS PubMed.
- P. C. Jurs, G. Bakken and H. McClelland, Chem. Rev., 2000, 100, 2649–2678 CrossRef CAS PubMed.
- H. Lv, X. Ma, G. Zhang, H. Wang, X. Hai and S. Bi, Biosens. Bioelectron., 2024, 258, 116370 CrossRef CAS PubMed.
- Z. Li, J. R. Askim and K. S. Suslick, Chem. Rev., 2018, 119, 231–292 CrossRef PubMed.
- N. Wu, X.-Y. Zhang, J. Xia, X. Li, T. Yang and J.-H. Wang, ACS Nano, 2021, 15, 19522–19534 CrossRef CAS PubMed.
- X.-W. Zhang, M.-X. Liu, M.-Q. He, S. Chen, Y.-L. Yu and J.-H. Wang, Anal. Chem., 2021, 93, 6437–6445 CrossRef CAS PubMed.
- D. Yuan, J. J. Liu, H. H. Yan, C. M. Li, C. Z. Huang and J. Wang, Talanta, 2019, 203, 220–226 CrossRef CAS PubMed.
- Y. Jia, M. Zhu, X. Zhang, D. Jia, T. Tian, B. Shi, Z. Ru, H. Ma, Y. Wan and Q. Wei, Anal. Chem., 2024, 96(25), 10116–10120 CrossRef CAS PubMed.
- Y. Jia, X. Zhang, X. Kuang, D. Fan, X. Sun, X. Ren, H. Ma, D. Wu and Q. Wei, Anal. Chem., 2024, 96(31), 12593–12597 CrossRef CAS PubMed.
- H. Wang, K. Wan and X. Shi, Adv. Mater., 2019, 31, 1805368 CrossRef CAS PubMed.
- L. Huang, D. W. Sun, H. Pu and Q. Wei, Compr. Rev. Food Sci. Food Saf., 2019, 18, 1496–1513 CrossRef PubMed.
- A. Hasnat, S. Hasan, S. Bano, S. Sultana, A. O. Ibhadon and M. Z. Khan, J. Mater. Chem. B, 2023, 11, 6762–6781 RSC.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang and S. Perrett, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- Y. Ai, Z. N. Hu, X. Liang, H. b. Sun, H. Xin and Q. Liang, Adv. Funct. Mater., 2022, 32, 2110432 CrossRef CAS.
- B. Unnikrishnan, C.-W. Lien, H.-W. Chu and C.-C. Huang, J. Hazard. Mater., 2021, 401, 123397 CAS.
- X. Ren, D. Chen, Y. Wang, H. Li, Y. Zhang, H. Chen, X. Li and M. Huo, J. Nanobiotechnol., 2022, 20, 92 CrossRef CAS PubMed.
- J. Lin, Q. Wang, X. Wang, Y. Zhu, X. Zhou and H. Wei, Analyst, 2020, 145, 3916–3921 RSC.
- X. Zhu, T. Li, X. Hai and S. Bi, Biosens. Bioelectron., 2022, 213, 114438 CrossRef CAS PubMed.
- W. Duan, Z. Qiu, S. Cao, Q. Guo, J. Huang, J. Xing, X. Lu and J. Zeng, Biosens. Bioelectron., 2022, 196, 113724 CrossRef CAS PubMed.
- M. H. Conklin and M. R. Hoffmann, Environ. Sci. Technol., 1988, 22, 899–907 CrossRef CAS PubMed.
- Y. Zhang, Z. Lu, S. Zhang, H. Chen and H. Ma, Soft Matter, 2011, 7, 11496–11500 RSC.
- X. Liu, Z. Chen, T. Wang, X. Jiang, X. Qu, W. Duan, F. Xi, Z. He and J. Wu, ACS Nano, 2022, 16, 6916–6928 CAS.
- X. Wei, Y. Wang, Y. Zhao and Z. Chen, Biosens. Bioelectron., 2017, 97, 332–337 CAS.
- S. Su, C. Zhang, L. Yuwen, J. Chao, X. Zuo, X. Liu, C. Song, C. Fan and L. Wang, ACS Appl. Mater. Interfaces, 2014, 6, 18735–18741 CAS.
- X. Yu, Y. Sun, J. Hu, J. Wang, X. Zhuang, S. Zhang, H. Ren, H. Qiu, Y. Zhang and Y. Hu, ACS Appl. Nano Mater., 2022, 6, 685–694 CrossRef.
- Y. Cheng, Y. D. Xia, Y. Q. Sun, Y. Wang and X. B. Yin, Adv. Mater., 2024, 36, 2308033 CrossRef CAS PubMed.
- X. Liu, X. Liang, J. Yu, K. Xu, J.-W. Shen, W. Duan and J. Zeng, Trac. Trends Anal. Chem., 2023, 117386 CrossRef CAS.
- W. Yin, J. Yu, F. Lv, L. Yan, L. R. Zheng, Z. Gu and Y. Zhao, ACS Nano, 2016, 10, 11000–11011 CrossRef CAS PubMed.
- L. Feng, L. Zhang, S. Zhang, X. Chen, P. Li, Y. Gao, S. Xie, A. Zhang and H. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 17547–17556 CrossRef CAS PubMed.
- S. R. Ali and M. De, ACS Appl. Mater. Interfaces, 2022, 14, 42940–42949 CrossRef CAS PubMed.
- F. Wei, X. Cui, Z. Wang, C. Dong, J. Li and X. Han, Chem. Eng. J., 2021, 408, 127240 CrossRef CAS PubMed.
- Z. Y. Liao, Y. M. Xia, J. M. Zuo, T. Wang, D. T. Hu, M. Z. Li, N. N. Shao, D. Chen, K. X. Song and X. Yu, Adv. Healthcare Mater., 2022, 11, 2101698 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05409a |
| This journal is © The Royal Society of Chemistry 2024 |