Open Access Article
Open Access ArticleEnzyme-encapsulated metal–organic framework ZIF-8-mediated biosensor for ultrasensitive detection of urinary prostatic exosomal protein using a glucose meter†
Mutu Lina,
Qiaohong Wanga,
Yun Daia,
Jinyuan Chen *b and
Yiming Lin*c
*b and
Yiming Lin*c
aDepartment of Pharmacy, Quanzhou Maternity and Children's Hospital, Quanzhou, Fujian 362000, China
bThe Central Laboratory, Fujian Key Laboratory of Precision Medicine for Cancer, The First Affiliated Hospital, Fujian Medical University, Fuzhou, Fujian 350005, China. E-mail: chenjinyuan@fjmu.edu.cn
cDepartment of Clinical Laboratory, Quanzhou Maternity and Children's Hospital, Quanzhou, Fujian 362000, China. E-mail: linyiming0819@sina.com
First published on 1st November 2024
Abstract
A highly sensitive and efficient home-use method is impressive and promising in the self-monitoring of chronic prostatitis (CP). Herein, we developed a glucose oxidase-zeolitic imidazolate framework-8-antibody composite (GOx@ZIF-8@Ab2) to achieve highly sensitive point-of-care testing (POCT) of urinary prostatic exosomal protein (PSEP) by combining it with a personal glucometer (PGM). The developed GOx@ZIF-8@Ab2 was prepared through a simple but effective biomineralization reaction and a strong binding affinity between Zn2+ and Fc region of the antibody. This bifunctional material GOx@ZIF-8@Ab2 could not only specifically recognize the PSEP via immunoreaction but also wrap a large number of GOx molecules. Following the sandwich immunoreaction, the prepared GOx@ZIF-8@Ab2 facilitated hydrolysis of glucose in a one-to-many fashion, amplifying the detection signal and greatly improving the sensitivity of PGM detection. In comparison to those of the conventional enzyme-linked immunosorbent assay (ELISA), the detection time of the developed technique was shortened to 45 minutes and its limit of detection (0.23 pg mL−1) was reduced by 4–5 orders of magnitude. This technique was successfully utilized for detecting PSEP in human urine samples with a recovery rate of 96–116%. Owing to its rapid, ultrasensitive, specific and portable features, the as-designed platform based on GOx@ZIF-8@Ab2 and the PGM device holds great promise as a home-use POCT platform for self-monitoring of CP and other chronic diseases.
1 Introduction
Chronic prostatitis (CP) is a common disease among men, significantly impacting their overall health and quality of life.1 18.4% of men aged 20–40 years have reported symptoms of prostatitis in China, and patients with prostatitis have accounted for nearly 3–16% of cases in urological departments.2,3 Patients with CP experience urination discomfort, radiating pain, perineal swelling, erectile dysfunction, insomnia, anxiety and other symptoms, making them suffer from great mental and physical pain.4,5 Clinical diagnosis primarily relies on prostatic ultrasound examination and prostate fluid leukocyte detection, or urinary bacteria screening. These methods suffer from low resolution, high rates of false positives, reliance on specialized equipment and situations, and long processing periods.6 Consequently, there is a dearth of sensitive, efficient and home-use methods. Prostatic exosomal protein (PSEP), a subcellular structure with an average diameter of 80–150 nm, mainly exists in the Golgi-rich area of human prostate ductal epithelial cells. PSEP is discharged from the prostatic epithelial cells into the ductal lumen through exocytosis and excretion and enters the urethra to reach the urine. PSEP has strong antioxidant and antibacterial functions, which are stronger than those of neutrophils. When inflammation occurs, prostatic epithelial cell activity is abnormal, and the level of PSEP in urine is elevated.7 Therefore, PSEP has shown promise as a non-invasive tool for rapid testing of CP,8,9 with clinical multicenter studies identifying PSEP as a novel diagnostic marker strongly linked to CP.10,11With the advancement of biotechnology, immunological detection suitable for point-of-care testing (POCT) has garnered widespread attention due to its high specificity in an antigen–antibody reaction and its robustness in the complex matrix.12 Typically, POCT immunological analyses employing reasonable signal transduction mechanisms can be achieved through portable devices easily accessible for home use.13,14 The personal glucometer (PGM) stands as one of the most common portable POCT detection devices,15–17 which utilizes an immunological analysis pattern combining enzyme–antibody conjugates and has been employed for POCT analyses of various proteins.18–20 The specific principle involves the formation of an immune complex among capture antibody, target, and enzyme–antibody conjugates, followed by the addition of the enzyme's substrate after magnetic separation. The enzyme–antibody conjugate catalyzes the hydrolysis of the substrate (such as glucose and sucrose), and the PGM detects the change in glucose concentration, enabling quantitative determination of the target.21 For example, Li et al. proposed an artificial, separated, embedded nanoporous membrane cascade reaction system to improve the compatibility of nucleic acid detection for CRISPR-mediated, multi-enzyme reactions. They further integrated the compartmentalized cascade reaction system into a microfluidic platform and developed a simple, portable bioinspired CRISPR-MCR biosensor for HIV nucleic acid quantification by using a low-cost PGM, eliminating the need for complex instrumentation and training professionals.22 Very recently, Chen et al. reported a method for sensitive and simple detection of kanamycin using a blood PGM as the detector. Aptamers are used in sensing systems to identify kanamycin. Hairpin probe-mediated signal amplification is introduced to produce a large amount of dsDNA containing PAM sequences for CRISPR-Cas12a recognition and activation. Thus, activated Cas12a can cut ssDNA (modified with magnetic bead and invertase) to release invertase. After magnetic separation, invertase can convert sucrose into glucose, which can be quantified by a PGM. The PGM-based sensor used for kanamycin detection has a linear range of 1 pM to 100 nM with a detection limit of 1 pM.23 Liu et al. combined a smartphone app with a commercial portable PGM to build a circulating tumor cells (CTCs) instant-detection microfluidic device based on the Au@CuMOF cascade enzyme. In this microfluidic system, the Au@CuMOF signal probe and Fe3O4@SiO2 capture probe were used to dual identify and capture CTCs. Then, Au@CuMOF targets CTCs to enable the forming cellular complexes to exhibit both glucose-like oxidase and peroxidase-like activity, further catalyzing glucose reduction. Thus, a dual-mode CTC detection method using a colorimetric method and portable PGM was constructed. The linear range of the method was 4–105 cells per mL, and the detection limit was 3 cells per mL.24 Similarly, Zuo et al. developed a comprehensive, high-efficiency strategy for capturing heterogeneous CTCs from the blood of patients with non-small cell lung cancer and performing portable assays. They first employed pleated hydrogels with high biocompatibility and high specific surface area as biological interfaces to capture CTCs. Meanwhile, a mesoporous silica material containing glucose molecules and sealed by epithelial cell adhesion protein aptamers and vimentin aptamers was synthesized. When the material was bound to CTC cells on the hydrogel by the aptamers, the glucose in mesoporous silica was released. The number of CTCs captured on the gel can be reflected by the indication of PGM.25 Based on the aforementioned principle, utilizing the PGM combined with enzyme–antibody conversion technology for immuno-detection is highly desirable for achieving sensitive, accurate, and on-site detection of PSEP. However, there are still some shortcomings that need to be overcome. Firstly, the enzyme needs to be coupled with the detection antibody where a fusion protein is often prepared, which requires technical operation and a tedious pretreatment process. Additionally, the bioconjugated enzyme's signal conversion efficiency is restricted due to the limited ability of one antibody to fuse with a small number of enzyme molecules,18 making it challenging to detect urinary biomarkers at trace levels. Therefore, a facile signal amplification system containing multiple enzyme molecules is highly desired for sensitive, accurate and on-site detection of PSEP.
In this study, we developed a glucose oxidase-zeolitic imidazolate framework-8-PSEP antibody composite (GOx@ZIF-8@Ab2), to achieve highly sensitive on-site detection of PSEP when integrated with the PGM system. ZIF-8, a metal–organic framework with pores, can retain the activity of numerous enzyme molecules when encapsulated within it.26–28 The approach utilizing GOx@ZIF-8@Ab2 is distinct from prior immunological methods relying on PGM in various ways. Firstly, ZIF-8-encapsulated GOx is prepared through a facile but effective biomineralization process, rather than fussy enzyme–antibody protein conjugation or fusion, thereby rationalizing signal labeling. Secondly, ZIF-8 binds with a large number of antibody molecules through strong binding affinity between Zn2+ and the Fc region of the antibody.29–31 GOx@ZIF-8@Ab2 serves both as a proficient recognition component and an amplification unit for signals. Following the sandwich immunoreaction, the GOx@ZIF-8@Ab2 catalyst facilitates the breakdown of glucose in a one-to-many fashion, increasing the detection signal and greatly improving the sensitivity of PGM detection.28,32–35 Additionally, ZIF-8 plays a key role in this approach by acting as a versatile substance that can readily accommodate various antibodies, showcasing remarkable adaptability for diagnosing different illnesses. Compared to traditional enzyme-linked immunosorbent assay (ELISA) for detecting PSEP, this work offers significant progress: the detection time has been shortened to 45 minutes and the detection limit (0.23 pg mL−1) was reduced by 4–5 orders of magnitude.10,11 Thus, by integrating a metal–organic framework-enzyme composite system with the portable PGM immunological analysis, a straightforward and effective approach is established for the highly sensitive detection of PSEP, particularly in intimate situations or resource-limited areas related to prostatic diseases.
2 Experimental section
2.1 Reagents and methods
Sodium metaperiodate (NaIO4), cysteine (Cys) and glucose oxidase (GOx) were purchased from Sigma-Aldrich (Shanghai). Aladdin Biochemical Technology Co. Ltd (Shanghai) supplied the 3,3′,5,5′-tetramethylbenzidine (TMB) and 2-methylimidazole (2-MI). Sinopharm Chemical Reagent Co. Ltd provided zinc acetate hexahydrate and D-glucose. Polyvinylpyrrolidone was obtained from Macklin Biochemical Co. Ltd. Rat monoclonal antibody was obtained from Wuhan MSK Biotechnology Co., Ltd (reactivity: human). All remaining reagents were of analytical purity and utilized without further processing.2.2 Apparatus
FT-IR spectra were recorded using a Thermo Scientific Nicolet iS5 from the USA. SEM pictures were obtained using a scanning electron microscope (ultra plus, Zeiss) from Germany. Zeta potentials were measured using a Malvern Zetasizer Nano ZS device from Britain. UV-vis absorption spectra were recorded on a 2450 UV-white spectrophotometer (Shimadzu, Japan). The X-ray diffraction (XRD) patterns of ZIF-8 samples were obtained on a Rigaku diffractometer using Cu Ka (k = 1.54 Å) radiation. The N2 adsorption and desorption isotherms were obtained on an ASAP-2020 (Micromeritics, USA) sorptometer at a liquid nitrogen temperature.2.3 Synthesis of GOx@ZIF-8
GOx@ZIF-8 was synthesized with a little modification according to the previous literature.24,36 Initially, 1 mg mL−1 GOx was added in 0.5 mL 4 mg mL−1 PVP (molecular weight: 8000). After mixing for 1 minute, 0.5 milliliters of Cys (2 milligrams per milliliter) was added while stirring for an additional 30 seconds. Then, 2 milliliters of 2-MI (160 mM) and 2 milliliters of ZnC4H10O6 (40 mM) were added separately and left at 25 °C for 4 hours. Lastly, the product was purified with water and then dried to produce GOx@ZIF-8.2.4 Synthesis of GOx@ZIF-8@Ab2
Initially, 1 milligram of anti-PSEP antibody was added in 2 milliliters of deionized water, and the prepared 1 mL GOx@ZIF-8 solution (10 mg mL−1) was subsequently combined with the antibody using an ultrasonic cleaner (Shanghai Kudos, power 200 W) for 10 minutes, followed by 2 hours of mixing. After centrifuging the heterogeneous mixture at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes, it was subsequently rinsed two times with deionized water. The GOx@ZIF-8 modified with antibodies (GOx@ZIF-8@Ab2) was dispersed in deionized water and kept at 4 °C for future tests.
000 rpm for 10 minutes, it was subsequently rinsed two times with deionized water. The GOx@ZIF-8 modified with antibodies (GOx@ZIF-8@Ab2) was dispersed in deionized water and kept at 4 °C for future tests.
2.5 Synthesis of GOx-Ab2
The GOx-Ab2 conjugate was prepared according to the previous work.37 Initially, a 2 mg per mL GOx was incubated with a 100 mM sodium metaperiodate solution and left to incubate for 20 minutes at 25 °C. The remedy was subsequently filtered using a PD10 desalting column from Pharmacia that had been prepared with a 1 mM CH3COOH solution at pH 4.4. The fractions containing the protein were collected based on A280nm readings. The oxidized protein solution (1.3 mL) was combined with anti-PSEP signal antibodies (1 mg) that were previously dissolved in 125 μL of 0.1 M H2CO3 solution (pH 9.5). The above solution was mixed for 2 hours at ambient temperature, followed by the addition of 125 μL of a 4 mg mL−1 sodium borohydride solution and agitation for an additional 2 hours. The resultant GOx-Ab2 combination was cleansed using a PD10 column filled with 0.1 M PBS. The obtained protein had a concentration of 0.5 milligrams per milliliter ascertained through spectrophotometric analysis. The paired GOx-Ab2 was kept in a refrigerator at a temperature of 4 °C.2.6 Sandwich immunoassay of PSEP
First, the 1 μg mL−1 PSEP capture antibody (Ab1) was diluted through the H2CO3 solution (pH 9.5). Next, 100 microliters of the Ab1 solution were introduced into the 96-well plate and left to incubate at 4 °C overnight. Following rinsing with 10 mM PBS (pH 7.4), the plate was then treated with a PBS buffer containing 1% BSA for 10 minutes at a temperature of 25 °C. Next, 100 microliters of antigen PSEP were introduced into the microplate. After incubating for 10 minutes at 37 °C, the antigen was immobilized on the microplate surface through an immune response. After washing, these plates were incubated with 100 μL GOx@ZIF-8@Ab2 (1 μg mL−1) for 5 min at 25 °C. The immune sandwich structure of Ab1-PSEP-GOx@ZIF-8@Ab2 was achieved after eliminating the unreacted GOx@ZIF-8@Ab2.2.7 PGM detection
After the immune complex was formed, 100 μL of 25 mM glucose in CH3COOH–CH3COONa solution (pH 5.0) was added to each well. Following a 30 minutes incubation at 37 °C, the glucose levels were assessed using PGM analysis. The glucose consumption (ΔGlu) was calculated by using the initial glucose concentration and subtracting the glucose concentration after the catalytic reaction.2.8 Real urine sample tests
Urine samples were collected from healthy volunteers. Next, the urine sample was diluted by a factor of 100 using 10 mM PBS (pH 7.4). In real samples, PSEP detection was performed similar to that described in the previous section. In short, 10, 20, and 30 pg mL−1 of standard PSEP were added to a 1% diluted urine sample.3 Results and discussion
3.1 Principle of the proposed GOx@ZIF-8@Ab2-based PGM immunoassay
Theoretically, GOx@ZIF-8@Ab2 serves as a bifunctional material with both catalytic activity and recognition of PSEP protein. This immunoassay principle is shown in Fig. 1. By sequentially adding a PESP-capturing antibody, PSEP protein, and PESP signal antibody modified GOx@ZIF-8 (GOx@ZIF-8@Ab2) onto the microplate, we can easily construct a sandwich-like immunosensor. With the addition of the glucose substrate, GOx catalyzes the hydrolysis into gluconic acid and hydrogen peroxide. Based on the amount of glucose consumed as tested by a glucose meter, we thus can achieve quantitative detection of PSEP protein.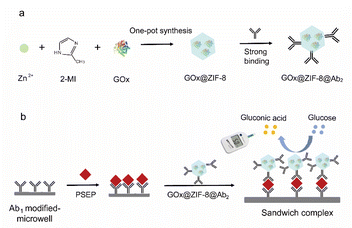 | ||
| Fig. 1 Illustration of the GOx@ZIF-8@Ab2 synthesis process (a) and the concept behind PGM immunoassay (b). | ||
3.2 Preparation and characterization of ZIF-8, GOx@ZIF-8@Ab2 and GOx@ZIF-8
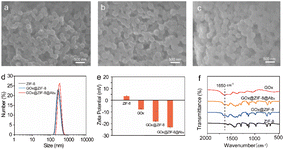
GOx@ZIF-8 was synthesized using a self-assembly method similar to metallothionein. Initially, GOx, PVP and Cys were assembled together to create Cys/PVP/GOx.28 Cys hastened the creation of a prenucleation center containing Zn2+ and 2-MI near the glucose oxidase, facilitating the in situ mineralization of glucose oxidase by ZIF-8. The antibody was tailored to ZIF-8 through the high-affinity binding of its histidine-rich Fc region to zinc atoms of ZIF-8. To verify the preparation of ZIF-8, we investigated the XRD patterns for simulated ZIF-8 (JCPDS 00-062-1030) and the as-synthesized ZIF-8 (Fig. S1†). The obvious crystal peaks at 2θ = 10.23°, 12.55°, 14.56°,16.28°, 17.88° and 19.36° well-matched with those from the standard card of ZIF-8 (JCPDS No. 00-062-1030), which corresponded to the reflections planes of ZIF-8 (002), (112), (022), (013), (222) and (123), respectively. This result indicated the successful synthesis of ZIF-8. Fig. 2a and S2a† display the consistent polyhedral shape of ZIF-8 nanoparticles with an average diameter of ∼260 nm. Following the incorporation of GOx into ZIF-8, the morphology and dimensions of GOx@ZIF-8 (Fig. 2b, S2b and S3†) and GOx@ZIF-8@Ab2 (Fig. 2c) remained unchanged in comparison to ZIF-8 nanoparticles, suggesting that the presence of GOx did not disrupt the integrity of ZIF-8. DLS analysis showed that ZIF-8 and GOx@ZIF-8 had similar hydrodynamic sizes, while GOx@ZIF-8@Ab2 were approximately 468 nm in size with a PDI of 0.336 in aqueous solution (Fig. 2d). Furthermore, the zeta potential findings for ZIF-8, GOx, GOx@ZIF-8, and GOx@ZIF-8@Ab2 were +3.6 mV, −7.9 mV, −18.2 mV, −23.2 mV, respectively, indicating the in situ mineralization of GOx in ZIF-8 and the binding of Ab2 to the ZIF-8 surface (Fig. 2e). The FT-IR findings additionally confirmed the in situ mineralization of GOx within the ZIF-8 nanoparticles (Fig. 2f). The distinctive peaks of GOx between 1640 cm−1 and 1660 cm−1 were attributed to the amide I, representing the stretching mode of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. GOx@ZIF-8@Ab2 and GOx@ZIF-8 exhibited amide I absorption bands, which clearly showed that GOx was trapped within ZIF-8. To further verify that GOx is wrapped in ZIF-8, we performed a calculation of the GOx loading rate. In the GOx@ZIF-8 synthesis process, precipitation (GOx@ZIF-8) and supernatant were obtained by centrifugation. The supernatant contains unreacted GOx. Thus, we calculated the amount of glucose consumed by precipitation of GOx@ZIF-8 and the supernatant Gox. The loading rate of GOx was calculated as 42.7% according to eqn (1) (Table S1).† Besides, the incorporation of GOx and Ab2 did not obviously alter the pore-size distribution of the ZIF-8 matrix (Fig. S4†). All of the above results indicated the successful preparation of GOx@ZIF-8-Ab2.
O. GOx@ZIF-8@Ab2 and GOx@ZIF-8 exhibited amide I absorption bands, which clearly showed that GOx was trapped within ZIF-8. To further verify that GOx is wrapped in ZIF-8, we performed a calculation of the GOx loading rate. In the GOx@ZIF-8 synthesis process, precipitation (GOx@ZIF-8) and supernatant were obtained by centrifugation. The supernatant contains unreacted GOx. Thus, we calculated the amount of glucose consumed by precipitation of GOx@ZIF-8 and the supernatant Gox. The loading rate of GOx was calculated as 42.7% according to eqn (1) (Table S1).† Besides, the incorporation of GOx and Ab2 did not obviously alter the pore-size distribution of the ZIF-8 matrix (Fig. S4†). All of the above results indicated the successful preparation of GOx@ZIF-8-Ab2.
3.3 Immunoassay based on GOx@ZIF-8@Ab2 and PGM
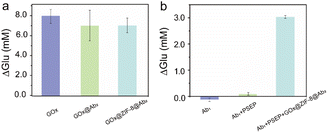
GOx exhibits specific catalysis in decomposing glucose molecules into C6H10O7 and H2O2 molecules. To ascertain the catalytic activity of GOx@ZIF-8@Ab2, we synthesized a GOx-Ab2 composite using the NaIO4 oxidation method, and compared the glucose consumption by GOx@ZIF-8@Ab2, pure glucose oxidase, and GOx-Ab2 at the same mass (40 μg of GOx). As illustrated in Fig. 3a, the glucose consumption by GOx-Ab2 and GOx@ZIF-8@Ab2 were 6.94 mM and 6.88 mM, respectively, sufficiently demonstrating that the presence of ZIF-8@GOx enzyme coupled with PSEP antibody minimally affects the catalytic activity, maintaining over 99% activity. Compared to pure GOx enzyme, the glucose consumption of GOx-Ab2 and GOx@ZIF-8@Ab2 exhibited reductions of 11.5% and 12.7%, respectively, possibly due to steric hindrance by antibodies impeding glucose molecules accessing the active center. Thus, our bio-mineralization method of loading antibodies onto ZIF-8 effectively maintains its activity, offering a simpler and more efficient approach compared to chemical synthesis methods, with great application prospects. The preserved activity of GOx may be attributed to the porous structure of ZIF-8, which does not obstruct sufficient reaction between glucose molecules and the enzyme's active site.To demonstrate the successful recognition of PSEP protein by the synthesized GOx@ZIF-8@Ab2, we compared its glucose consumption with that of a separately modified PSEP-capturing antibody and the PSEP-capturing antibody combined with PSEP. As shown in Fig. 3b, there was no significant glucose consumption in the former two cases, whereas the sandwich-like structure based on GOx@ZIF-8@Ab2 exhibited significant glucose consumption, indicating efficient recognition of PSEP protein. Therefore, we have successfully developed a novel sensing system for PSEP detection using a portable glucose meter.
3.4 Optimization of experimental conditions
Optimizing the reaction conditions is essential for maximizing the immunosensor's detection performance of PSEP. As shown in Fig. 4a, when the incubation time between GOx@ZIF-8@Ab2 and PSEP antigen increased from 5 min to 10 min, the glucose consumption increased and maximized at 10 min. Therefore, we chose 10 min for further incubation. Additionally, there was a notable rise in glucose utilization as the concentration of GOx@ZIF-8@Ab2 elevated, peaking at 5 mg mL−1 (see Fig. 4b). No further increase in glucose consumption can be observed, so we attributed this to the saturation of GOx@ZIF-8@Ab2 relative to the PSEP antigen. Therefore, a concentration of 5 mg mL−1 GOx@ZIF-8@Ab2 was determined to be the most effective for the subsequent immune tests. In addition, the catalytic time was closely related to the glucose consumption. Fig. 4c demonstrated that glucose consumption increased with catalytic time and leveled off after 30 minutes. Therefore, a duration of 30 minutes was chosen for the following experiment.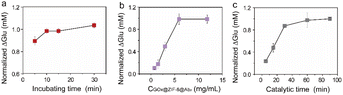 | ||
| Fig. 4 Effects of incubation time (a), GOx@ZIF-8@Ab2 concentration (b) and catalytic time (c) on the PGM-based immunoassay. | ||
3.5 Evaluation of the PGM immunosensor based on GOx@ZIF-8@Ab2
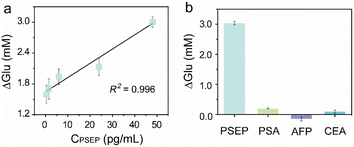
The GOx@ZIF-8@Ab2-based PGM immunosensor's performance was evaluated under ideal conditions by monitoring glucose consumption at varying PSEP concentrations. Fig. 5a demonstrates that glucose consumption rose as the PSEP concentration increased, indicating a strong linear correlation between PSEP concentration ranging from 0.375 pg mL−1 to 48 pg mL−1 (R2 = 0.996). The limit of detection (LOD) was determined to be 0.23 pg mL−1 (S/N = 3.3, where S stands for the abbreviation of signal, and N stands for the abbreviation of noise). A comparison between the present immunosensor and other previous PSEP detection methods is summarized in Table S2.† The findings showed that the proposed immunsensor outperformed other methods in detecting PSEP, with a significantly lower LOD value compared to previous approaches, highlighting the exceptional sensitivity of the study.Examination of the effects of various common proteins was performed to assess the specificity of the immunosensor (Fig. 5b). Glucose consumption did not show significant changes when detecting prostate-specific antigen (PSA), alpha-fetoprotein (AFP), and carcinoembryonic antigen (CEA). However, a noticeable increase in glucose consumption was observed for 48 pg per mL PSEP antigen, confirming the high specificity of the immunosensor. Furthermore, the reliability of this immunosensor was analysed by conducting five parallel tests to measure the glucose consumption values of PSEP at concentrations of 6 pg mL−1 and 48 pg mL−1 (Table S3†), and the RSDs were 14% and 5%, respectively. Although the storage temperature influenced the performance of the immunosensor (Fig. S5†), after three weeks of storage at 4 °C the glucose consumption value for 48 pg per mL PSEP remained at 107.8% (Fig. S6†). The findings confirmed the excellent consistency and durability of the immunosensor.
3.6 Real-sample analysis
The actual sample was obtained from the hospital. After obtaining the sample, we performed a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm centrifugation for 15 min to remove the urine residue so as not to interfere with the detection results. The supernatant after centrifugation was divided and placed in the −20 °C refrigerator for use. When the urine sample was redissolved, it was diluted to the desired concentration using 10 mM PBS (pH 7.4). The reliability of the immunoassay for detecting PSEP in actual samples was tested by spiking various concentrations of PSEP into diluted urine samples for recovery experiments. With the addition of different concentrations of the PSEP standard into the human urine sample, the recoveries were calculated. Table 1 displays that the experiments yielded satisfactory results, with a recovery rate between 96% and 116% and an RSD below 10.0%, demonstrating the immunosensor's reliability for analyzing real samples. Thus, this work provides a promising solution to urinary PSEP detection, which holds a high potential in home-use CP monitoring by using the portable PGM.
000 rpm centrifugation for 15 min to remove the urine residue so as not to interfere with the detection results. The supernatant after centrifugation was divided and placed in the −20 °C refrigerator for use. When the urine sample was redissolved, it was diluted to the desired concentration using 10 mM PBS (pH 7.4). The reliability of the immunoassay for detecting PSEP in actual samples was tested by spiking various concentrations of PSEP into diluted urine samples for recovery experiments. With the addition of different concentrations of the PSEP standard into the human urine sample, the recoveries were calculated. Table 1 displays that the experiments yielded satisfactory results, with a recovery rate between 96% and 116% and an RSD below 10.0%, demonstrating the immunosensor's reliability for analyzing real samples. Thus, this work provides a promising solution to urinary PSEP detection, which holds a high potential in home-use CP monitoring by using the portable PGM.
| Sample | Spiked (pg mL−1) | Found (pg mL−1) | Recovery (%, n = 3) | RSD (%, n = 3) |
|---|---|---|---|---|
| 1 | 0 | 8.0 | — | 6.1 |
| 2 | 10 | 17.6 | 96 | 9.7 |
| 3 | 20 | 27.2 | 96 | 7.2 |
| 4 | 30 | 42.9 | 116 | 4.0 |
4 Conclusions
In summary, this work demonstrated an immune-sensing platform constructed by GOx@ZIF-8@Ab2 and PGM for sensitive, specific and rapid detection of PSEP. Furthermore, the proposed platform was developed for the accurate detection of PSEP in urine samples. We demonstrated that GOx@ZIF-8 was rapidly synthesized in a simple biomineralizing manner with no significant reduction in enzymatic activity after the loading of the capture antibody. Because ZIF-8 can wrap a large number of GOx molecules to catalyze more glucose hydrolysis in a one-to-many manner, the detection sensitivity of this method could be greatly improved. For the detection of PSEP protein in solution, the glucose consumption (ΔGlu) followed a linear relationship with the PSEP concentration, and the LOD was evaluated as low as 0.23 pg mL−1 for PSEP. The detection time was only 45 minutes and the LOD was the lowest value reported so far. In addition, the platform was used for the recovery test of PSEP in actual urine solution, and the results were satisfactory. This immunosensor has substantial advantages including high sensitivity and specificity, fast response, ease of universality and portability, offering an affordable and accessible solution for the detection of PSEP. By changing the antibody molecules, this platform holds great promise for the detection of other reproductive system disease-related markers in the future and could be potentially translated into a home-use platform for the self-monitoring of chronic disease.Ethical statement
This study conformed to the ethical guidelines of the Declaration of Helsinki and was approved by the Ethical Committee of Quanzhou Maternity and Children's Hospital. Human urine samples were collected from Quanzhou Maternity and Children's Hospital and did not have any identifying information about any of the participants, and all participants provided written informed consent.Data availability
The authors confirm that the data supporting this study's conclusions are available within the article and its ESI.† Further information and raw data are available upon request.Author contributions
Mutu Lin: investigation, data curation, writing the original draft. Qiaohong Wang: formal analysis, data curation, writing refinement. Yun Dai: formal analysis. Jinyuan Chen: supervision, validation, project administration, writing – reviewing and editing. Yiming Lin: project administration, data curation, writing – reviewing and editing. All authors contributed to the data analysis, revising and approving the final manuscript to be published.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by grants from Fujian Provincial Society of Laboratory Medicine and National (Fujian) Genetic Testing Technology Application Demonstration Center (2023LHYC040) and Quanzhou City Science and Technology Program of China (Grant No. 2021C052R).Notes and references
- A. Yebes, C. Toribio-Vazquez, S. Martinez-Perez, J. M. Quesada-Olarte, A. Rodriguez-Serrano, M. Álvarez-Maestro and L. Martinez-Piñeiro, Curr. Neurol. Rep., 2023, 24, 241–251 CAS.
- Z. Zhang, Z. Li, Q. Yu, C. Wu, Z. Lu, F. Zhu, H. Zhang, M. Liao, T. Li, W. Chen, X. Xian, A. Tan and Z. Mo, Andrology, 2015, 3, 1119–1124 CrossRef CAS PubMed.
- F. U. Khan, A. U. Ihsan, H. U. Khan, R. Jana, J. Wazir, P. Khongorzul, M. Waqar and X. Zhou, Biomed. Pharmacother., 2017, 94, 1064–1076 CrossRef PubMed.
- Z.-Y. Hao, H.-J. Li, Z.-P. Wang, J.-P. Xing, W.-L. Hu, T.-F. Zhang, X.-S. Zhang, J. Zhou, S. Tai and C.-Z. Liang, J. Androl., 2011, 32, 496–501 CrossRef PubMed.
- S.-D. Chung, C.-C. Huang and H.-C. Lin, J. Affective Disord., 2011, 134, 404–409 CrossRef PubMed.
- F. U. Khan, A. U. Ihsan, H. U. Khan, R. Jana, J. Wazir, P. Khongorzul, M. Waqar and X. Zhou, Biomed. Pharmacother., 2017, 94, 1064–1076 CrossRef PubMed.
- X. Feng, M. Zhang, L. Zhang, H. Hu, L. Zhang, X. Zhang, S. Fan and C. Liang, Int. Urol. Nephrol., 2020, 52, 225–232 CrossRef CAS PubMed.
- L. Yin, Y. Tang, A. Pan, L. Yang, X. Zhu and Y. Liu, Medicine, 2019, 98, e16848 CrossRef CAS PubMed.
- Q. Ge and J. Lou, Lab. Med., 2023, 54, 212–214 CrossRef PubMed.
- X. Li, T. Jiang, F. Liu, X. Shao, Y. Xu, W. Sheng and W. Sun, Urol. Int., 2017, 100, 112–118 CrossRef CAS PubMed.
- W. Liang, Z. Wu, G. Zhang, W. Chen, X. Hu, J. Yang, J. Meng, Y. Zeng, H. Li and X. Shang, Transl. Androl. Urol., 2020, 9, 2218–2226 CrossRef PubMed.
- S. Perveen, A. Negi, V. Gopalakrishnan, S. Panda, V. Sharma and R. Sharma, Clin. Chim. Acta, 2023, 538, 139–156 CrossRef CAS PubMed.
- F. Ranjbari, A. Nosrat, F. Fathi and A. Mohammadzadeh, Clin. Chim. Acta, 2024, 558, 118670 CrossRef CAS PubMed.
- R. Gupta, P. Gupta, S. Wang, A. Melnykov, Q. Jiang, A. Seth, Z. Wang, J. J. Morrissey, I. George, S. Gandra, P. Sinha, G. A. Storch, B. A. Parikh, G. M. Genin and S. Singamaneni, Nat. Biomed. Eng., 2023, 7, 1556–1570 CrossRef CAS PubMed.
- Q. Wang, Y. He, S. He, S. Yu, Y. Jiang and F. Wang, Chem. Commun., 2023, 59, 1345–1348 RSC.
- H. Dai, J. Zhang, Y. Wu, J. Zhao, C. Liu and Y. Cheng, Anal. Chem., 2024, 96, 1789–1794 CrossRef CAS PubMed.
- E. Amalfitano, M. Karlikow, M. Norouzi, K. Jaenes, S. Cicek, F. Masum, P. Sadat Mousavi, Y. Guo, L. Tang, A. Sydor, D. Ma, J. D. Pearson, D. Trcka, M. Pinette, A. Ambagala, S. Babiuk, B. Pickering, J. Wrana, R. Bremner, T. Mazzulli, D. Sinton, J. H. Brumell, A. A. Green and K. Pardee, Nat. Commun., 2021, 12, 724 CrossRef CAS PubMed.
- B. Lin, D. Liu, J. Yan, Z. Qiao, Y. Zhong, J. Yan, Z. Zhu, T. Ji and C. J. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 6890–6897 CrossRef CAS PubMed.
- E. K. Leonard, M. Aller Pellitero, B. Juelg, J. B. Spangler and N. Arroyo-Currás, J. Am. Chem. Soc., 2022, 144, 11226–11237 CrossRef CAS PubMed.
- D. Huang, Z. Shi, J. Qian, K. Bi, M. Fang and Z. Xu, Biotechnol. Bioeng., 2021, 118, 1568–1577 CrossRef PubMed.
- Y. Xiang and Y. Lu, Anal. Chem., 2012, 84, 4174–4178 CrossRef CAS PubMed.
- Z. Li, N. Uno, X. Ding, L. Avery, D. Banach and C. Liu, ACS Nano, 2023, 17, 3966–3975 CrossRef CAS PubMed.
- J. Chen, G. Shi and C. Yan, Sci. Total Environ., 2023, 872, 162279 CrossRef CAS PubMed.
- X. Liu, Y. Fang, X. Chen, W. Shi, X. Wang, Z. He, F. Wang and C. Li, Biosens. Bioelectron., 2024, 251, 116105 CrossRef CAS PubMed.
- Y. Zuo, W. Lu, Y. Xia, J. Meng, Y. Zhou, Y. Xiao, L. Zhu, D. Liu, S. Yang, Y. Sun, C. Li and Y. Yu, ACS Sens., 2023, 8, 187–196 CrossRef CAS PubMed.
- G. Chen, X. Kou, S. Huang, L. Tong, Y. Shen, W. Zhu, F. Zhu and G. Ouyang, Angew. Chem., Int. Ed., 2020, 59, 2867–2874 CrossRef CAS PubMed.
- S. Huang, G. Chen and G. Ouyang, Chem. Soc. Rev., 2022, 51, 6824–6863 RSC.
- F. Shao, Y. Wu, Z. Tian and S. Liu, Biomaterials, 2021, 274, 120869 CrossRef CAS PubMed.
- Q. Zhang, J. Liang, A. Bongers, J. J. Richardson, K. Liang and Z. Gu, Adv. Sci., 2023, 10, 2206546 CrossRef CAS PubMed.
- J. Huang, L. Jiao, W. Xu, Q. Fang, H. Wang, X. Cai, H. Yan, W. Gu and C. Zhu, ACS Appl. Mater. Interfaces, 2021, 13, 33383–33391 CrossRef CAS PubMed.
- D. Wei, D. Xiong, N. Zhu, Y. Wang, X. Hu, B. Zhao, J. Zhou, D. Yin and Z. Zhang, Anal. Chem., 2022, 94, 12981–12989 CrossRef CAS PubMed.
- Y. Cao, F. Mo, Y. Liu, Y. Liu, G. Li, W. Yu and X. Liu, Biosens. Bioelectron., 2022, 198, 113819 CrossRef CAS PubMed.
- A. Rodriguez-Abetxuko, M. C. Morant-Miñana, M. Knez and A. Beloqui, ACS Omega, 2019, 4, 5172–5179 CrossRef CAS.
- X. Zhang, F. Zhang, Z. Lu, Q. Xu, C. Hou and Z. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 25565–25571 CrossRef CAS PubMed.
- T.-O. Knedel, E. Ricklefs, C. Schlüsener, V. B. Urlacher and C. Janiak, ChemistryOpen, 2019, 8, 1337–1344 CrossRef CAS PubMed.
- G. Chen, S. Huang, X. Kou, S. Wei, S. Huang, S. Jiang, J. Shen, F. Zhu and G. Ouyang, Angew. Chem., Int. Ed., 2019, 58, 1463–1467 CrossRef CAS PubMed.
- D. B. Papkovsky, T. C. O'Riordan and G. G. Guilbault, Anal. Chem., 1999, 71, 1568–1573 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05460a |
| This journal is © The Royal Society of Chemistry 2024 |