Open Access Article
Open Access ArticleSingle GC base pair recognition by a heterocyclic diamidine: structures, affinities, and dynamics†
Edwin N. Ogbonnaa,
J Ross Terrella,
Ananya Paul a,
Abdelbasset A. Farahatabc,
Gregory M. K. Poon
a,
Abdelbasset A. Farahatabc,
Gregory M. K. Poon a,
David W. Boykina and
W. David Wilson
a,
David W. Boykina and
W. David Wilson *a
*a
aDepartment of Chemistry and Centre of Diagnostics and Therapeutics, Georgia State University, Atlanta, GA 30303-3083, USA. E-mail: wdw@gsu.edu; Fax: +1 404-413-5505; Tel: +1 404-413-5503
bDepartment of Pharmaceutical Organic Chemistry, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt
cMaster of Pharmaceutical Science Program, California North State University, Elk Grove, CA 95757, USA
First published on 18th September 2024
Abstract
The recognition of specific genomic arrangements by rationally designed small molecules is fundamental for the expansion of targeted gene expression. Here, we report the first X-ray crystal structures that demonstrate single G (guanine) recognition by a highly selective diamidine (DB2447) in a mixed DNA sequence. The study presents detailed structural information on the mechanism of single G recognition by D2447 and its various interactions in the DNA minor groove. Molecular dynamics and binding studies were used to evaluate the details of our reported structures. The study provides structural insight and resources necessary for understanding single G selection in genomic sequences.
Introduction
The discovery of heterocyclic cationic compounds that possess specific recognition principles for DNA has precipitated the emergence of a library of natural and synthetic small molecules that can bind nucleic acids and affect gene expression. The classical AT bp binders, netropsin, and distamycin, were prominent in providing insight into DNA-small molecule interactions in the minor groove through the elucidation of the molecular structure of their DNA complexes.1–6 As a consequence, the rational design of minor groove binders with specificity for AT base pairs (bps) has become one of the major strategies to target nucleic acid, leading to the development of an extensive array of efficacious compounds for both biotechnology and therapeutic applications.7–9 Aromatic diamidines are cationic compounds, of which many are strong AT binders, with a broad significance in biomedical research and disease therapeutics. Synthetic AT binders, DAPI and Hoechst 33258, are used as agents for biological staining.10–12 Pentamidine and furamidine have potent antiparasitic properties against a range of infective agents and showed promising results against trypanosome disease (sleeping sickness) in human clinical trials.13–16 The recognition principle of AT binders involves the donation of hydrogen bonds, by the compound, to an acceptor oxygen (O) of thymine or nitrogen (N) of adenine on the floor of the DNA minor groove. Therefore, with a greater abundance of structural information on AT-specific diamidine binders and their interactions with AT-rich DNA sequences, improved design modules have yielded compounds with better selectivity for AT bp, and hence, greater drug efficacy.17–20 For example, prodrug forms of furamidine have shown increased oral absorptivity and stronger potency against animal trypanosomes and malaria in humans.21 Other analogs of furamidine and diamidines have shown greater effectiveness against a host of parasitic infections.22 The successes of AT binders are demonstrated. However, to a considerable extent, these successes were also predicated upon a successful structural characterization of their interactions with DNA at the molecular level.23Despite the advances in the research of AT binding diamidine molecules, there remained a limitation in the potential for GC base pairs in the genome of a target organism. To overcome this limitation and enhance selective DNA recognition, hydrogen bond acceptor groups that bond with the NH2 of guanine on the floor of the DNA minor groove were designed.18,24 These developments led to new diamidine modules, and a series of novel and potent GC-specific binders have been synthesized.24,25 The biological importance of having distinct classes of compounds that can recognize different arrangements of AT and GC bps cannot be overemphasized. The human genome with mixed bp DNA sequences can be selectively targeted with these compounds. Nonetheless, a major problem that persists in the study of GC-specific diamidine binders is the lack of available structural information.26,27 Such information is critical to understanding how different GC-specific binders interact with DNA and how their binding affects the overall B-DNA structure. With little to no structural resources for GC-specific diamidine compounds, the design of improved GC-specific compounds, as has been reported for their AT binding counterparts, remains elusive. No crystal structures depicting GC-specific DNA recognition by diamidines have been reported.28 To overcome this challenge, we screened an extensive collection of diamidine binders (DB2447,29 DB2277,30,31 DB2457![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31) specific to GC under our crystallization conditions. We obtained a high-resolution crystal structure for the pyridyl-linked diamidine, DB2447 (Scheme 1) which we report here.
31) specific to GC under our crystallization conditions. We obtained a high-resolution crystal structure for the pyridyl-linked diamidine, DB2447 (Scheme 1) which we report here.
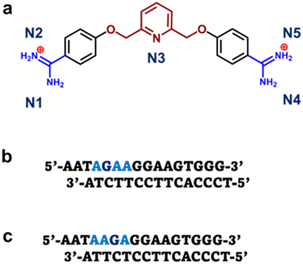 | ||
| Scheme 1 (a) Structure of pyridyl bis-methoxybenzamidine (DB2447). (b) The 5′-AGAA-3′ single G sequence with overhanging ends. (c) The 5′-AAGA-3′ single G sequence with overhanging ends. | ||
Small molecules like netropsin, distamycin, and diamidines have all been reported to successfully bind DNA–protein complexes.32–35 We reported pertinent information of diamidine binding to the DNA complex of an ETS transcription factor.36 These reports show the molecular target for the action of a minor groove binder need not be DNA itself, but a DNA–protein complex. Also, the main objective of designing customized small molecules is to enhance their drug-like interactions with DNA complexes in infectious organisms or cancer cells. Therefore, this report was an excellent opportunity to use a selective GC binder, DB2447, to capture a single GC bp DNA recognition in a DNA–protein context. X-ray crystallography provided the ideal methodology to explore this opportunity. Studies show the promoter region of some transcription factors as ideal binding sites for numerous minor groove binders.36 The reported AT-specific diamidines bind strongly to the AT-rich flanking sequences of the ETS transcription factor PU.1, an essential protein for haematopoiesis and cell fate decisions in human cells.36,37 PU.1 protein has been reported to enhance DNA crystallization and provided insight into DNA structure–function studies.38 Therefore, for this study, the PU.1 protein was selected as the ideal cofactor to facilitate both DNA crystallization and the study of single GC bp DNA recognition with DB2447.
Here, we report the first high-resolution crystal structure that captures a single GC bp recognition by diamidines. Two single G DNA sequences were used in this study: 5′-AATAGAAGGAAGTGGG-3′ and 5′-AATAAGAGGAAGTGGG-3′. The difference between the two DNA sequences is the position of their single G bp (5′-A1ATAG5AAGG-3′ and 5′-A1ATAAG6AGG-3′). This single G shift was to evaluate the preciseness and selectivity of DB2447. The results from both single G DNA complex structures delineate the specific recognition principles of DB2447 and how the compound interacts with the shape of the minor groove. The results of the molecular dynamics and binding studies of DB2447 provide support for the crystal structures and validate the unique properties of DB2447.
Results
X-ray structure of the DB2447-bound PU.1-AGAA complex
We report the high-resolution X-ray crystal structures of single G containing DNA complexes: bound and unbound with DB2447. The PU.1-5′-AATAGAAGGAAGTGGG-3′ complex with DB2447 (PDB ID: 8VDH) is shown in Fig. S1.† The PU.1 protein binds to the major groove of the DNA and stabilizes the structure, while DB2447 selectively binds to the minor groove. The PU.1-AGAA-DB2447 complex is the first high-resolution X-ray crystal structure demonstrating a single G recognition by diamidines. The PU.1-AGAA-DB2447 structure diffracted to a resolution of 1.64 Å with an Rfree value of 20.8%. DB2447 is bound in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 manner with the 5′-AGAA-3′ DNA. The structure shows the pyridyl N3 of DB2447 targets explicitly a single G in the floor of the minor groove at the 5′ side of the PU.1 binding site, forming a strong direct hydrogen bond with the NH2 of G5 at 2.8 Å (Fig. 1a). This unique targeting of a single G in the minor groove by the hydrogen bond accepting N3 of DB2447 validates our current and previously reported biophysical and MD experimental results.29
1 manner with the 5′-AGAA-3′ DNA. The structure shows the pyridyl N3 of DB2447 targets explicitly a single G in the floor of the minor groove at the 5′ side of the PU.1 binding site, forming a strong direct hydrogen bond with the NH2 of G5 at 2.8 Å (Fig. 1a). This unique targeting of a single G in the minor groove by the hydrogen bond accepting N3 of DB2447 validates our current and previously reported biophysical and MD experimental results.29
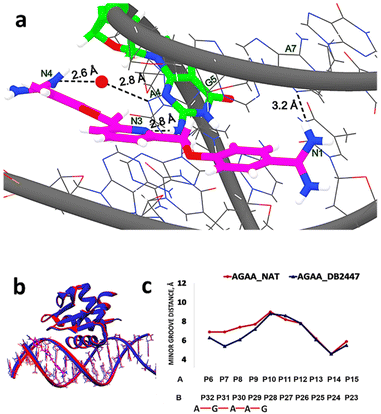 | ||
| Fig. 1 (a) Significant bonding distances between 5′-AATAGAAGGAAGTGGG-3′ (wire) and DB2447 (stick and wire, carbon in magenta, nitrogen in blue) (PDB ID: 8VDH). The single G recognition, a strong direct H-bond between pyridyl N3 of DB2447 with N–H of G5 residue (stick and wire, carbon in green, nitrogen in blue) is 2.8 Å. H-bond between the amidine N1 and N3 of A7 is 3.2 Å. Amidine N4 forms an interfacial water-mediated H-bond with N3 of A4, N4–O–H2 is 2.6 Å and O–H2–N3 of A4 is 2.8 Å. (b) Overlay of the structure of DB2447 bound 5′-AATAGAAGGAAGTGGG-3′ complex (dark blue) with native (red) (PDB ID: 8V9N). PU.1 protein is bound to the major groove in both DNA complexes while DB2447 is bound in the minor groove. (c) Minor groove width difference between AGAA-Native (red) and AGAA-DB2447 (blue). | ||
For the purpose of simplicity, PU.1-AGAA and PU.1-AAGA complexes will also be referred to as “AGAA” and “AAGA”, respectively. A significant recognition strategy of diamidines is the direct hydrogen bonding between the amidine NH2 (N1, N2, N4, N5 from Scheme 1) with O2 of thymine (T) or N3 of adenine (A) in the floor of the minor groove. The PU.1-AGAA-DB2447 structure reveals a strong direct H-bond between amidine N1 and N3 of A7 on one end of the DB2447 (Fig. 1a). However, on the other end of DB2447, the compound (N4) uses an interfacial water molecule for contact with DNA. Although most classical diamidines form strong direct H-bonds with DNA, some have been reported to form indirect H-bonds with DNA using an interfacial water molecule.23,39 The amidine N4 utilizes interfacial water molecule to form an H-bond with N3 of A4 (–NH⋯O–H⋯N). Our results show that DB2447 is one of the special compounds that can make direct and indirect hydrogen bond contacts with DNA. Furthermore, our results suggest that DB2447 binding affects the structure of 5′-AGAA-3′. An overlay of the bound and unbound (PDB ID: 8V9N) structures of PU.1-AGAA (Fig. 1b) shows compression of the DB2447-bound minor groove (blue in Fig. 1b).
This compression of the minor groove width is elucidated in Fig. 1c. DB2447 binding compresses the minor groove. The DB2447-bound minor groove is compressed by at least 1.5 Å between phosphates P7 and P31 (blue in Fig. 1c). Beyond the binding region of DB2447, the changes in the minor groove distances are negligible.
X-ray structure of the DB2447-bound PU.1-AAGA complex
The PU.1-5′-AATAAGAGGAAGTGGG-3′ complex with DB2447 (PDB ID: 8VDI) is shown in Fig. S2.† The single G was shifted from the 5th position in 5′-AGAA-3′ DNA to the 6th position in 5′-AAGA-3′ DNA. The PU.1-AAGA-DB2447 structure diffracted to a resolution of 1.93 Å with an Rfree value of 20.1%. DB2447 was bound in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 manner to the PU.1-5′-AAGA-3′ complex. Fig. 2a shows the unique recognition principle of DB2447 for a single G residue. The pyridyl N3 of DB2447 forms a strong direct hydrogen bond with the NH2 of G6 (2.9 Å). The amidine N1 forms a direct hydrogen bond with the O2 of T27 at a distance of 3.3 Å. On the other end of DB2447, multiple interactions are observed. There is a direct hydrogen bond interaction between amidine N4 and O2 of T30 (3.5 Å), and an indirect hydrogen bond interaction between N4 and N3 of A31. The compound utilizes interfacial water to form a hydrogen bond contact with the N3 of A31. The amidine N4 forms an H-bond with an interfacial water molecule (–NH⋯O–H) at a distance of 3.6 Å, while the water molecule contacts the DNA (O–H⋯O
1 manner to the PU.1-5′-AAGA-3′ complex. Fig. 2a shows the unique recognition principle of DB2447 for a single G residue. The pyridyl N3 of DB2447 forms a strong direct hydrogen bond with the NH2 of G6 (2.9 Å). The amidine N1 forms a direct hydrogen bond with the O2 of T27 at a distance of 3.3 Å. On the other end of DB2447, multiple interactions are observed. There is a direct hydrogen bond interaction between amidine N4 and O2 of T30 (3.5 Å), and an indirect hydrogen bond interaction between N4 and N3 of A31. The compound utilizes interfacial water to form a hydrogen bond contact with the N3 of A31. The amidine N4 forms an H-bond with an interfacial water molecule (–NH⋯O–H) at a distance of 3.6 Å, while the water molecule contacts the DNA (O–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) T) at 2.7 Å.
T) at 2.7 Å.
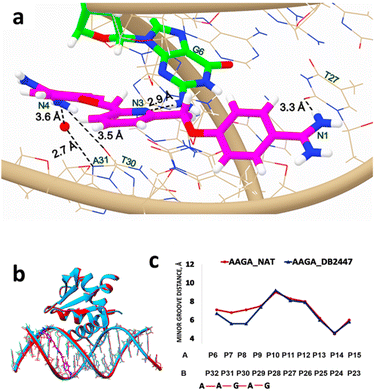 | ||
| Fig. 2 (a) Significant bonding distances between 5′-AATAAGAGGAAGTGGG-3′ (wire) and DB2447 (PDB ID: 8VDI) (stick and wire, carbon in magenta, nitrogen in blue). The single G recognition, a strong direct H-bond between pyridine N3 of DB2447 with N–H of G6 residue (stick and wire, carbon in green, nitrogen in blue) is 2.9 Å. H-bond between the amidine N1 and O2 (in red) of T27 is 3.3 Å. The amidine N4 forms a direct H-bond interaction with O2 of T30, a distance of 3.5 Å. N4 also forms an interfacial water-mediated bond with N of A31, N4–O–H2 IS 3.6 Å and O–H2–N is 2.7 Å. (b) Overlay of the structure of DB2447 bound to the 5′-AATAAGAGGAAGTGGG-3′ complex (sky blue) with native (red) (PDB ID: 8E4H). PU.1 protein is bound to major groove in both DNA complexes. DB2447 bound in the minor groove of AAGA-DB2447. (c) Minor groove width difference between AAGA-Native (red) and AAGA-DB2447 (blue). | ||
DB2447 recognizes a single G residue similar to PU.1-AGAA. However, one major difference observed in AGAA is that both amidines (N1 and N4) of DB2447 contact the 5′-AGAA-3′ or forward strand (Fig. 1a), while both amidines (N1 and N4) in 5′-AAGA-3′ each contact the complementary or reverse strand (3′ → 5′). In AGAA, the N1 and N4 amidine contacts the N3 of A4 and A7, respectively, of the 5′-AGAA-3′ strand. In contrast, the N1 amidine in AAGA contacts the O2 of T27, and the N4 amidine contacts both the O2 of T30 and the N3 of A31, respectively, which is the complementary strand. It is noteworthy to mention that previous reports of AT bp binding diamidines often show one amidine end (NH2) contacting the 5′ → 3′ strand while the other end contacts the complementary strand.19,20,23,37 However, DB2447 appears to be an exception as both amidine groups (N1, N4) bind to the same strand whether in AGAA or AAGA. We propose that this exceptional feature has been observed due to the high degree of flexibility of DB2447. Planar flexibility is a favorable property for a minor groove binder for interactions with the groove shape. Crystal structure and MD results from previous reports have shown that diamidines can systematically orient themselves to make the most optimum contact in the minor groove structures.23 The crystal structures of AGAA and AAGA demonstrate that DB2447 is flexible enough to make favorable additional contacts in its recognition of a single G residue. DB2447 binding to the minor groove of 5′-AAGA-3′ compresses the groove width in comparison to the unbound complex (PDB ID: 8E4H) (Fig. 2b). The minor groove is compressed by 1.5 Å at P8 (Fig. 2c). Like 5′-AGAA-3′, the changes in the minor groove distance become negligible away from the region of DB2447 binding.
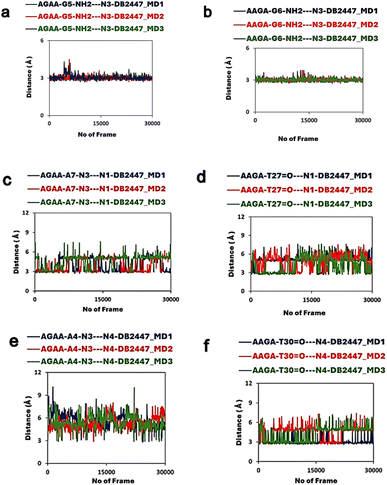
Molecular dynamics (MD) simulation studies of the DB2447 binding in the minor groove of the PU.1-AGAA and PU.1-AAGA complexes
To expand on the observations of our ligand-DNA complex crystal structures, we conducted a 600 ns MD simulation study to understand the dynamic behaviours of DB2447 binding in the minor groove of PU.1-AGAA and PU.1-AAGA (Fig. 3). The simulation study provides further comprehension of the properties of DB2447 single G recognition (Fig. 3). A significant observation of the MD simulation experiment is that N3 of DB2447 in the minor groove of AGAA shows and maintains the principle single G recognition contact with NH2 of G5 while the N1 and N4 amidine contact the N3 of A7 and A4 respectively (Fig. 3a–d). The strong direct H-bond of N3 with NH2 of G5 (2.6 Å–3.4 Å) remains constant for the entirety of the MD simulation (600 ns) (Fig. 3b). Again, this further underlines the excellent selectivity of DB2447 for a single G residue. In contrast, N1 and N4 of DB2447 do not maintain a consistent bonding distance with the DNA. Fig. 3c shows that the average distance of N4 throughout the simulation experiment is 4.95 Å. This is because, at various periods of the MD simulation, N4 does not have the optimum distance to maintain a strong direct contact with N3 of A4, and thus, will require interfacial water for mediated DNA contact (3.4 Å–5.8 Å). The rotation of the DB2447 bonds allows the amidine to have alternating NH groups (N4 and N5) contacting the DNA throughout the entire simulation. The amidine bond rotation causes the NH groups (N4 and N5) at one end of DB2447 to maintain an overall strong interaction with DNA. Fig. 3d shows that N1 can make strong direct contact with N3 of A7 (2.6 Å–3.4 Å) and maintains that contact for much longer periods during the entire MD simulation experiment as the average distance of N1 to N3 of A7 was 3.2 Å. The simulation results show that N1 requires interfacial water (3.4 Å–5.8 Å) but at a less consistent period than N4. Of course, like the former amidine end, bond rotation means that N2 can alternate with N1 in making direct contact with the DNA (Fig. 3d). Other interactions of DB2447 include those of the phenyl C–H's (C12 and C15) with DNA (Figure S3†).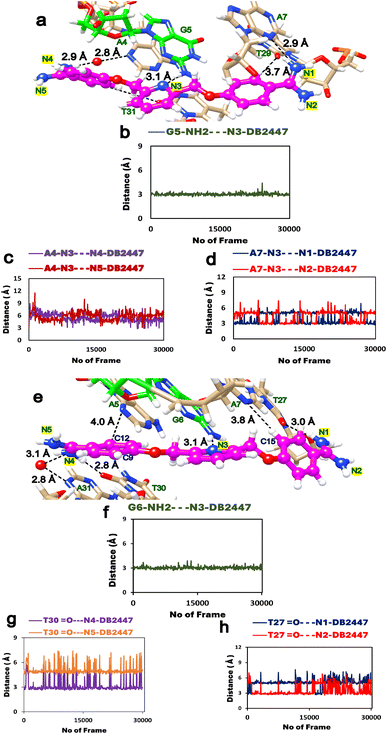 | ||
| Fig. 3 (a) Snapshot of MD simulations of the PU.1-AGAA-DB2447 complex; DB2447 (magenta, ball, and stick) bound to the single G recognition site with diamidine groups (N1/N2 and N4/N5) contacting the DNA. (b) N3 maintains a strong H-bond (black, dashed lines) with NH2 of G5. (c) N4 and N5 making alternating contacts with A4. N4 makes contact using an interfacial water (3.4 Å–5.8 Å). Favorable C–H interactions are observed between C12⋯A5 and C15⋯A7 (Figure S3†). (d) N1 and N2 making alternating contacts with A7. (e) Snapshot of MD simulations of the PU.1-AAGA-DB2447 complex. Color scheme same as Fig. 3a. (f) N3 maintains a strong H-bond (black, dashed lines) with NH2 of G6 (g) N4 amidine makes contact with O2 of T30 throughout the simulation experiment but no amidine bond rotation was observed. (h) N1 contact with O2 of T27 show only a transient amidine rotation for the entire period of simulation. N4 and N1 make strong direct contacts (2.6 Å–3.4 Å) and also utilize interfacial water (3.4 Å–5.8 Å) in their interactions with DNA throughout the MD simulation. | ||
The MD experiment for AAGA (Fig. 3e) shows the N3 of DB2447 making the principle recognition contact with the single G (Fig. 3f). This strong H-bond contact (2.6 Å–3.4 Å) remains stabilized throughout the simulation. More so, this shows DB2447 to be an excellent selection molecule for a single G in an AAGA sequence. N4 maintains an average distance of 3 Å for the simulation. This suggests a predominant strong direct H-bond contact (2.6 Å–3.4 Å) between N4 and O2 of T30 throughout the simulation, as observed in Fig. 3g. Although Fig. 3g does show N4 amidine vibrating away from O2 of T30 at certain periods of the experiment, no bond rotation was observed. The average distance of N1 for the entire simulation was 4 Å. Fig. 3h suggests that N1 required interfacial water (3.4 Å–5.8 Å) for contact with O2 of T27 about 90% of the simulation time. This means the N2 amidine, for long periods of the simulation, did not alternate with N1 to make DNA contact. Fig. 3h shows a rotation of the amidine bond for a transitory period during the simulation (frames 1–300 and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357–18
357–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 079). Also observed from Fig. 3e is a strong direct H-bond interaction between N4 and N3 of A31. Although a strong direct DNA contact for N4 with A31, Figure S4† suggests the average distance of N4 from A31 also allows N4 to use interfacial water for indirect contact (3.4 Å–5.8 Å) with the DNA. This is corroborated by the crystal structure of AAGA (Fig. 2a). Overall, the results of the entire MD simulation experiment show the three major H-bond interactions that facilitate the binding of DB2447 at the AGAA and AAGA minor grooves at N1, N3, and N4 respectively. For the entire simulation with both sequences, N3 always maintains a strong and persistent H-bond with the single G (Table S2†). At the same time, N1 and N4 both have periods of strong direct H-bond with DNA and periods of interfacial water-assisted DNA contact.
079). Also observed from Fig. 3e is a strong direct H-bond interaction between N4 and N3 of A31. Although a strong direct DNA contact for N4 with A31, Figure S4† suggests the average distance of N4 from A31 also allows N4 to use interfacial water for indirect contact (3.4 Å–5.8 Å) with the DNA. This is corroborated by the crystal structure of AAGA (Fig. 2a). Overall, the results of the entire MD simulation experiment show the three major H-bond interactions that facilitate the binding of DB2447 at the AGAA and AAGA minor grooves at N1, N3, and N4 respectively. For the entire simulation with both sequences, N3 always maintains a strong and persistent H-bond with the single G (Table S2†). At the same time, N1 and N4 both have periods of strong direct H-bond with DNA and periods of interfacial water-assisted DNA contact.
In addition to the former MD experiment (MD1), two more MD simulation experiments (MD2 and MD3) were conducted for both AGAA and AAGA, respectively. These subsequent experiments used the same MD parameters as MD1. Fig. 4a and b shows the remarkable consistency of the principle recognition property of DB2447 for single G in both AGAA and AAGA. In both sequences, N3 maintains a strong direct H-bond contact (between 2.9 Å to 3.1 Å) with the single G throughout the MD2 and MD3 experiments. In Fig. 4c, the MD2 and MD3 experiments for the N1 in AGAA and AAGA show that N1 makes a strong direct H-bond with N3 of A7 and can also use interfacial water assistance for DNA contact. The average distances of N1 in AGAA (N1⋯N3 of A7) and AAGA (N1⋯O2 of T27) for MD2 and MD3 are 4.5 Å, 4.2 Å (MD2) and 4.2 Å, 2.8 Å (MD3). Fig. 4c and d show that N1 vibrates away at varying distances from the DNA in the multiple simulations, allowing N2 to contact DNA.
The MD simulation results for N4, Fig. 4e and f, show an interesting observation. Fig. 4e shows the N4 of DB2447 for AGAA, which uses interfacial water for DNA contact for long periods of the simulation. The average distances were 4.4 Å and 3.9 Å for MD1 and MD2, respectively. N4 of AAGA also uses interfacial water significantly throughout the simulation, with average distances of 3.9 Å for MD1 and 3.9 Å for MD2. Also, the increasing distance of N4 from DNA shows a bond rotation that allows N5 to alternately make direct H-bond contact with O2 of T30. Again, despite the longer bonding distances of N4 in AGAA and AAGA, N4 always retain a strong direct H-bond contact with DNA at certain periods of the simulation. Both N1 and N4 of AGAA and AAGA make strong direct contact with DNA or use interfacial water for stability in all the simulations.
Both structural and MD results highlight the unique properties of DB2447. The stability of the principle H-bond formation and the dynamic amidine group interactions provide potency and stability for the compound as it interacts with the shape of the minor groove. As a result, DB2447 does not show any sliding along the shape of the minor groove throughout the entire simulation. Additional polar interactions are observed between the DB2447 phenyl rings and DNA. For example, C12⋯A5 and C15⋯A7 in AGAA (Fig. 3a and Figure S3†) show a consistent polar molecular interaction (3.5 Å to 4.0 Å) for the entire simulation studies. The favorable C12 and C15 interactions with the N3 of A5 and A7 of the DNA, respectively, further stabilize the overall B-DNA structure. These interactions are similarly observed in AAGA.
Biosensor surface plasmon resonance (SPR) determination of binding affinities and kinetics
As DB2447 has been reported to be selective for a single G residue, the binding equilibrium constant for DB2447 was evaluated to characterize their interaction with -AGAA- and -AAGA- sites. This was done using SPR methods with chip-immobilized streptavidin capture of biotin-labelled hairpin DNA on a sensor chip surface. The DB2447 compound was injected over the chip surface in a buffer solution at 100 mM NaCl. Given the single G shift between both binding sites, DB2447 has measurably different binding affinities (Figure S5†). Table S3† shows the binding of DB2447 in a single binding site of 5′-TAGAA-3′ has a KA = 5.4 × 106 M−1 (KD = 185 nM) at 0.1 M NaCl. In comparison, the DB2447 binding in 5′-TAAGA-3′ has a binding affinity KA = 3.5 × 106 M−1 (KD = 286 nM), which is over 1.5 times weaker than the 5′-TAGAA-3′ binding site. The results show the difference in binding affinities could be related to structure. The structure of AAGA has a flanking G residue that causes a greater expansion of the minor groove from P7 to P8 (Fig. 2c) compared to P7 to P8 of AGAA
that causes a greater expansion of the minor groove from P7 to P8 (Fig. 2c) compared to P7 to P8 of AGAA  (Fig. 1c).
(Fig. 1c).
Discussion
DB2447 binds in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mode to PU.1-AGAA with the 2Fo-Fc map showing a singular binding orientation (Fig. S6a†). The symmetrical nature of DB2447 makes the pyridine group the center of symmetry. The nitrogen of the pyridine accepts the NH2 from the single G to delineate the recognition principle of DB2447. The binding of a single G in AGAA by DB2447 ranges across five residues (5′-TAGAA-3′) in the minor groove. The central N3 of DB2447 recognizes the single G in 5′-TA
1 mode to PU.1-AGAA with the 2Fo-Fc map showing a singular binding orientation (Fig. S6a†). The symmetrical nature of DB2447 makes the pyridine group the center of symmetry. The nitrogen of the pyridine accepts the NH2 from the single G to delineate the recognition principle of DB2447. The binding of a single G in AGAA by DB2447 ranges across five residues (5′-TAGAA-3′) in the minor groove. The central N3 of DB2447 recognizes the single G in 5′-TA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AA-3′, while the linking methoxybenzamidine groups span across the four flanking residues. The DB2447 conformation in the minor groove is such that there is a strong direct H-bond at one diamidine end, while the other amidine utilizes interfacial water molecules for DNA contact (Fig. 1a). Similar binding conformation in the minor groove as DB2447 has been reported for some specially designed diamidines.23 The consequence of DB2447 binding is that one amidine end in the minor groove protrudes closer to the minor groove floor, while the other amidine end slightly extends outwards.
AA-3′, while the linking methoxybenzamidine groups span across the four flanking residues. The DB2447 conformation in the minor groove is such that there is a strong direct H-bond at one diamidine end, while the other amidine utilizes interfacial water molecules for DNA contact (Fig. 1a). Similar binding conformation in the minor groove as DB2447 has been reported for some specially designed diamidines.23 The consequence of DB2447 binding is that one amidine end in the minor groove protrudes closer to the minor groove floor, while the other amidine end slightly extends outwards.
DB2447 binds similarly in the AAGA complex. The optimum length of the N3 H-bond in both structures emphasizes the high selectivity of DB2447 for a single G (Fig. 1a and 2a). The strong direct H-bond formation by N3 of DB2447 appears to lock in the single G recognition in both AGAA and AAGA structures. In the bound -AAGA- structure, N3 forms a direct H-bond with the 5′-AAGA-3′ strand of the DNA, while N1 and N4 both contact the complementary strand, respectively. In contrast, the bound AGAA structure shows that the N1, N3, and N4 do not contact the complementary strand (Fig. 1a).
The amidine groups (N1 and N4) in AGAA and AAGA can make strong direct H-bonds with the DNA and use interfacial water for assisted interactions with DNA (Fig. 1a and 2). The behaviour of these amidine groups of DB2447, as observed in the structural results, has been substantially corroborated by the MD simulation experiment results (Fig. 3 and 4). Therefore, the MD simulation results help to explain the determined hydrogen bond distances of the crystal structures. For example, the N4 amidine in the AAGA complex can make direct contact with the DNA (Fig. 2a), though the distance (3.5 Å) is slightly greater than the acceptable limit. A longer H-bond is also observed for the interfacial water contact by N4 of DB2447 (3.6 Å) (Fig. 2a). The MD simulation results show the interacting amidine groups N1 and N4 of DB2447 and surrounding water molecules to be highly dynamic (Fig. 3a and b). The combination of the amidine bond rotations and dynamic water account for varying bonding distances with the DNA, including but not limited to the slightly longer H-bond distances at the amidine N4 end. Furthermore, the MD simulation results show that at different states of DB2447 binding, the N4 amidine can alternate between making direct contact with the DNA (N4⋯T30) or using interfacial water to make DNA contact (N4⋯A31). One suggestion for the dynamics observed at the N4 amidine is the subsequent widening of the groove width from a narrower A7 to a wider G8. This widening of the minor groove decreases the proximity for a favorable interaction between amidine and DNA, possibly leading to less stable amidine–water interactions. However, the MD simulation results suggest DB2447 has no fixed conformations for DNA contacts at the amidine end. The DB2447 amidine groups behaved differently at different periods of the simulation to maximize contact with DNA. This means that amidine group dynamics play a major role in the stability of DB2447 in the minor grooves of these sequences. Except for the principle recognition at N3, the nature and context of the interactions of DB2447 with the DNA can change with time. The dynamic interactions at play at the amidine ends of DB2447 are necessary for DB2447 binding and stability at the minor groove. As the principal single G recognition contact at N3 is stabilized, the N3 acts like the pivot of the molecule, as the planar components of DB2447 confer flexibility across the compound, especially at its terminal ends. This unique quality of DB2447 allows for an overall stabilized interaction that fits with the shape of the minor groove. A combination of MD and crystal structure results suggests that interfacial water is necessary for DB2447 binding in the minor groove. The necessity of an interfacial water molecule for assisted DB2447 binding is supported by the fact that the DB2447-bound structures do not show other surrounding water molecules at the terminal ends of the diamidine. This suggests the presence of an interfacial water molecule at the location of the DNA-DB2447 interface could be considered critical for DB2447 binding and stability.
The binding studies show that DB2447 has a stronger binding affinity for AGAA than AAGA. In contrast to crystal structure and MD studies, SPR can quantitatively measure the binding affinity of DB2447. As it appears, AGAA seems to offer a favorable binding motif for DB2447 (KD = 185 nM) than AAGA (KD = 286 nM). These results suggest there maybe structural implications associated with DB2447 binding because of the single G shift. As discussed earlier, the subsequent widening of the AAGA minor groove at the A7-G8 junction (about 2 Å) would appear to diminish H-bond contact, not enhance it. This might help explain the greater binding affinity of DB2447 for AGAA. Although the structure and MD results show an overall favorable DB2447 affinity for the DNA, our SPR results provide a more quantitative evaluation that captures the relative affinities of DB2447 in both single G DNA sequences.
Further structural considerations for DB2447 binding are observed within the minor groove. The single G in the AGAA DNA is located just along the first DNA turn (Figure S1†), and as a result, the G5 residue in AGAA is deeper into the minor groove than G6 in AAGA (Fig. 1a and 2a). As a consequence, DB2447 orients itself to make the most optimum contact with the 5′-AGAA-3′ strand because of the position of NH2 of G5. This is not the case with AAGA, as the single G is located slightly away from the DNA turn. These results emphasize single G recognition as the guiding factor for DB2447 binding in the minor groove. Although the N3 selectivity for single G remained consistent in both structures, the manner of DB2447-binding was affected by the position of the single G. The increased widening of the groove width in AAGA and the position of DNA turn in AGAA seem to favor a stronger DB2447 binding in AGAA. Hence, a stronger binding affinity is reported for the AGAA structure. The helical parameters of both DNA complexes were evaluated to determine the effect of DB2447 binding on the local base pair geometry of AGAA and AAGA. The resulting analysis showed no significant changes in the local base geometry of the AGAA and AAGA complexes compared to their native structures (Figure S7 and S8†).
Conclusions
The results of this study demonstrate that DB2447 is highly selective for a single G residue in a mixed DNA context. The detailed structural, MD, and SPR data provide rational support for our understanding of single G recognition. Notwithstanding the relative binding affinities of DB2447 for both AGAA and AAGA DNAs, the selectivity of DB2447 for a single G in both structures was unaffected. The discussions elaborated in this report provided further comprehension of the properties and dynamics of GC-specific binders. Indeed, these findings are relevant and significant to current advances in biotechnology and therapeutic applications.Data availability
We declare that subsequent experimental data and pertinent information source regarding this manuscript are available in the ESI section.† Additionally, raw data from MD simulation and SPR binding experiments are available at https://doi.org/10.7910/DVN/EWTNNX.Author contributions
The authors confirm contributions to the paper as follows: E. N. O., J. R. T., A. P. and W. D. W. designed the study and experiments. A. A. F. and D. W. B. synthesized the compound for this study. G. M. K. provided the protein for the crystallization experiments. E. N. O., J. R. T., A. P. and W. D. W. wrote the manuscript. All authors have read and agreed to the work presented on this manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank the National Institutes of Health Grant GM111749 (W. D. W and D. W. B) for their financial support. This research used resources from beamline 17-ID-2 of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704, beamline 22-BM of the Advanced Photon Source, a DOE Science User Facility under Contract No. DE-AC02-06CH11357, and Beamline 8.2.2 of the Advanced Light Source, a DOE Science User Facility under contract No. DE-AC02-05CH11231, is supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01.References
- R. E. Dickerson and M. L. Kopka, J. Biomol. Struct. Dyn., 1985, 2, 423–431 CrossRef PubMed.
- M. L. Kopka, C. Yoon, D. Goodsell, P. Pjura and R. E. Dickerson, J. Mol. Biol., 1985, 183, 553–563 CrossRef CAS PubMed.
- Antibiotics III: Mechanism of Action of Antimicrobial and Antitumor Agents, ed. J. W. Corcoran and F. E. Hahn, Springer, New York, 1975, pp. 79–100 Search PubMed.
- C. Zimmer and U. Wahnert, Prog. Biophys. Mol. Biol., 1986, 47, 31–112 CrossRef CAS PubMed.
- J. G. Pelton and D. E. Wemmer, Biochemistry, 1988, 27, 8088–8096 CrossRef CAS PubMed.
- G. V. Gurskaya, S. L. Grokhovsky, A. L. Zhuze and B. P. Gottikh, Biochim. Biophys. Acta, 1979, 563, 336–342 CrossRef CAS PubMed.
- W. D. Wilson, F. A. Tanious and A. Mathis, Biochimie, 2008, 90, 999–1014 CrossRef CAS PubMed.
- E. A. Dickie, F. Giordani, M. K. Gould, P. Maser, C. Burn, J. C. Mottram, S. P. S. Rao and M. P. Barrett, Trop. Med. Infect. Dis., 2020, 5, 29 CrossRef PubMed.
- T. Wenzler, S. Yang, O. Braissant, D. W. Boykin, R. Brun and M. Z. Wang, Antimicrob. Agents Chemother., 2013, 57, 5330–5343 CrossRef CAS PubMed.
- L. F. De Castro and M. Zacharias, J. Mol. Recognit., 2002, 15, 209–220 CrossRef PubMed.
- W. D. Wilson, F. A. Tanious, H. J. Barton, R. L. Jones, L. Strekowski and D. W. Boykin, J. Am. Chem. Soc., 1989, 111, 5008–5010 CrossRef CAS.
- M. Battaglia, D. Pozzi, S. Grimaldi and T. Parasassi, Biotech. Histochem., 1994, 69, 152–156 CrossRef CAS PubMed.
- M. Sands, M. A. Kron and R. B. Brown, Rev. Infect. Dis., 1985, 7(5), 625–6344 CrossRef CAS PubMed.
- F. Doua, T. W. Miezan, J. R. S. Singaro, F. B. Yapo and T. Baltz, Am. J. Trop. Med. Hyg., 1996, 55, 586–588 CAS.
- E. M. De Souza, R. Menna-Barreto, T. C. Araújo-Jorge, A. Kumar, Q. Hu, D. W. Boykin and M. N. Soeiro, Parasitology, 2006, 133, 75–79 CrossRef CAS PubMed.
- A. A. Farahat, A. Kumar, T. Wenzler, R. Brun, A. Paul, P. Guo, W. D. Wilson and D. W. Boykin, Eur. J. Med. Chem., 2023, 252, 115287 CrossRef CAS PubMed.
- R. Nanjunda and W. D. Wilson, Protoc Nucleic Acid Chem, Wiley, New York, chapter 8: unit 8.8, 2012 Search PubMed.
- Y. Chai, A. Paul, M. Rettig, W. D. Wilson and D. W. Boykin, J. Org. Chem., 2014, 79, 852–866 CrossRef CAS PubMed.
- S. Mallena, M. Lee, C. Bailly, S. Neidle, A. Kumar, D. W. Boykin and W. D. Wilson, J. Am. Chem. Soc., 2004, 126, 13659–13669 CrossRef CAS PubMed.
- E. N. Ogbonna, A. Paul, J. R. Terrell, Z. Fang, C. Chen, G. M. Poon, D. W. Boykin and D. W. Wilson, Bioorg. Med. Chem., 2022, 68, 116861 CrossRef CAS PubMed.
- M. Z. Wang, J. Y. Saulter, E. Usuki, Y. L. Cheung, M. Hall, A. S. Bridges, G. Loewen, O. T. Parkinson, C. E. Stephens, J. L. Allen, D. C. Zeldin, D. W. Boykin, R. R. Tidwell, A. Parkinson, M. F. Paine and J. E. Hall, Drug Metab. Dispos., 2006, 34, 1985–1994 CrossRef CAS PubMed.
- M. P. Barnett, C. G. Gemmel and C. J. Suckling, Pharmacol. Ther., 2013, 139, 12–23 CrossRef PubMed.
- E. N. Ogbonna, A. Paul, A. J. Farahat, J. R. Terrell, E. Mineva, V. Ogbonna, D. W. Boykin and W. D. Wilson, ACS Bio Med Chem Au, 2023, 3, 335–348 CrossRef CAS PubMed.
- A. Paul, Y. Chai, D. W. Boykin and W. D. Wilson, Biochemistry, 2015, 54, 577–587 CrossRef CAS PubMed.
- A. Paul, P. Guo, D. W. Boykin and W. D. Wilson, Molecules, 2019, 24, 946 CrossRef PubMed.
- A. Paul, A. Kumar, R. Najunda, A. A. Farahat, D. W. Boykin and W. D. Wilson, Org. Biomol. Chem., 2015, 15, 827–835 RSC.
- P. Guo, A. Paul, A. Kumar, N. K. Harika, S. Wang, A. A. Farahat, D. W. Boykin and W. D. Wilson, Chem. Commun., 2017, 53, 10406–10409 RSC.
- N. Harika, M. Germann and W. D. Wilson, Chem.–Eur. J., 2017, 23, 17612–17620 CrossRef CAS PubMed.
- A. Paul, A. A. Farahat, D. W. Boykin and W. D. Wilson, Life, 2022, 12, 681 CrossRef CAS PubMed.
- N. K. Harika and W. D. Wilson, Biochemistry, 2018, 57, 5050–5057 CrossRef CAS PubMed.
- A. Paul, P. Guo, D. W. Boykin and W. D. Wilson, Molecules, 2019, 24, 1946 CrossRef PubMed.
- M. Kotecha, J. Kluza, G. Wells, C. C. O'Hare, C. Foni, R. Mantovani, P. W. Howard, P. Morris, D. E. Thurston, J. A. Hartley and D. Hochhauser, Mol. Cancer Ther., 2008, 7, 1319–1328 CrossRef CAS PubMed.
- P. Peixoto, Y. Liu, S. Depauw, M. P. Hilderbrand, D. W. Boykin, C. Bailly, W. D. Wilson and M. H. David-Cordonnier, Nucleic Acid Res., 2008, 36, 2241–2253 CrossRef PubMed.
- M. A. Grant, R. M. Baron, A. A. Macais, M. D. Layne, M. A. Perrella and A. C. Rigby, Biochem. J., 2009, 418, 103–112 CrossRef CAS PubMed.
- R. M. Baron, S. Lopez-Guzman, D. F. Riascos, A. A. Macais, M. D. Layne, G. Cheng, C. Harris, S. W. Chung, R. Reeves, U. H. vonAndrian and M. A. Parrella, PLoS One, 2010, 5, e10656 CrossRef PubMed.
- M. Munde, S. Wang, A. Kumar, C. E. Stephens, A. A. Farahat, D. W. Boykin, W. D. Wilson and G. M. Poon, Nucleic Acid Res., 2014, 42, 1379–1390 CrossRef CAS PubMed.
- I. Antony-Debre, A. Paul, J. Leite, k. Mitchell, H. M. Kim, L. A. Carvajal, T. I. Todorova, K. Huang, A. Kumar, A. A. Farahat, B. Bartholdy, S. R. Narayanagari, J. Chen, A. Ambesi-Impiombato, A. A. Ferrando, I. Mantzaris, E. Gavathiotis, A. Verma, B. Wall, D. W. Boykin, W. D. Wilson, G. M. K. Poon and U. Steidl, J. Clin. Invest., 2005, 127, 4297–4313 CrossRef PubMed.
- J. R. Terrell, S. J. Taylor, A. L. Schneider, Y. Lu, T. N. Vernon, S. Xhani, R. H. Gumpper, M. Luo, W. D. Wilson, U. Steidl and G. M. K. Poon, Cell Rep., 2023, 42, 112671 CrossRef CAS PubMed.
- D. Wei, W. D. Wilson and S. Neidle, J. Am. Chem. Soc., 2013, 135, 1369–1377 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC PDB: 8VDH, 8VDI, 8V9N, 8E4H. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ra05957c |
| This journal is © The Royal Society of Chemistry 2024 |