Open Access Article
Open Access ArticleA low-toxicity uranyl-selective-binding linear pentapeptide sequence as a potential uranium decorporation agent†
Xiaohong Tana,
Libing Yub,
Xindan Liaob,
Chun Chenb,
Jian Chub,
Zhonghua Xiongb,
Binyuan Xiab,
Wei Tang *b,
Xijian Li
*b,
Xijian Li *b and
Yanyan Liu
*b and
Yanyan Liu *a
*a
aSchool of Safety Science and Emergency Management, Wuhan University of Technology, Wuhan, 430070, China
bInstitute of Materials, China Academy of Engineering Physics, Jiangyou, Mianyang, Sichuan 621907, China
First published on 10th December 2024
Abstract
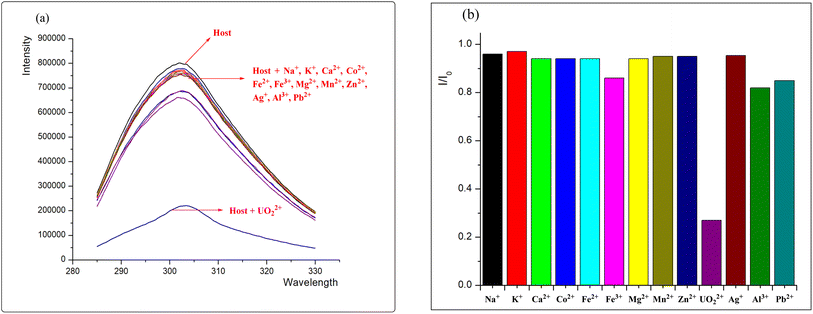
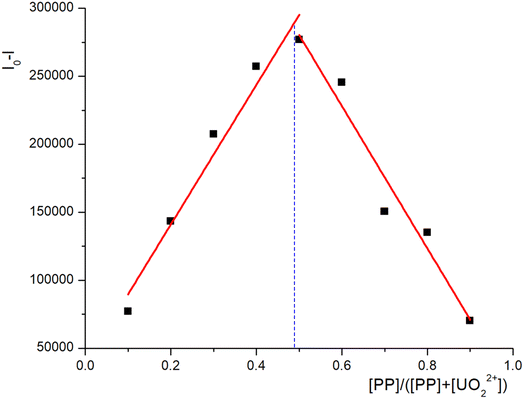
Searching for highly selective, efficient, and low-toxicity chelating agents is central to resolving uranium contamination in vivo. Peptides composed of amino acids exhibit very low toxicity for accumulation in the human body and have been proven effective in chelating actinides within the human body. Herein, we report a rationally designed short phosphorylated peptide sequence PP-B, which exhibits high affinity and selectivity for uranyl compared to other trace elements present in the body (such as Na+, K+, Ca2+, Co2+, Fe2+, Fe3+, Mg2+, Mn2+, Zn2+). The association constant for the peptide–uranyl complex is calculated to be 7.3 ×105 M−1. The result of DFT calculation shows that the phosphate group binds strongly to the UO22+ center, potentially accounting for the peptide's strong affinity towards UO22+. The results of in vivo uranyl decorporation assays reveal that PP-B has a much lower toxicity and a much higher decorporation efficiency than that of the clinically approved DTPA. These findings render PP-B a promising candidate for utilization as a novel decorporation agent.
1 Introduction
Uranium is the key element for nuclear-energy production, and its utilization in many other applications has been increasing.1 Nevertheless, uranium's radioactivity and chemical toxicity pose significant risks.2–4 Uranium primarily induces health problems via poisoning kidneys (36.22%), bone (19.48%), liver (17.58%), the reproductive system (13.90%), lungs (7.24%), and the nervous system (5.58%). As an important nephrotoxicant, the site-specific accumulation of uranium in the proximal tubules may cause proximal tubular damage and renal failure. The mechanisms underlying the toxicological effects caused by uranium have been extensively studied. Recent advances have focused on oxidative stress, genetic damage, protein injury, cell apoptosis, inflammation, and metabolic disorders.5,6 The recommended clinical treatment for internal uranium contamination involves the infusion of 250 mL of sodium bicarbonate solution (1.4%). However, the intravenous injection of large quantities of sodium bicarbonate can upset the body's acid–base balance, potentially leading to hypokalemia, alkalosis, or other health issues.7–13 Diethylenetriamine pentaacetate (DTPA) is a clinically approved actinide chelator, yet its efficacy in uranium excretion is limited due to its low affinity for uranyl, poor membrane permeability, and mismatched ligand environment.14,15 Chelators containing hydroxypyridone (HOPO), terephthalamide (TAM), and catechol (CAM) functional groups have shown promise in actinide decomposition, but most CAM- and TAM-based ligands have been found to be toxic and may damage the kidney, liver and/or spleen.16–19 Based on the characteristics of porous organic, which offer tunable functional groups, high surface area, and large porosity, they are extensively applied for the extraction and purification of uranium solutions. Dr Juan Diwu's team successively synthesized carboxyl-functionalized nanoscale metal organic frameworks (nMOFs) (UiO-66-(COOH)4),20 HOPO-functionalized nMOFs (MIL-101-HOPO)21 and carboxyl-functionalized nMOFs (ZIF-71-COOH),22 as well as CON-AO obtained by attaching amidoxime (AO) groups to the channels of covalent organic sheets (CON),23 which greatly improves the selectivity and affinity for uranium. However, toxicity studies on nanomaterials are scarce, and deeper investigations into their degradation mechanisms and associated side effects on normal organs are necessary.24 Despite substantial research progress in the field of uranium prokinetics, the development of effective chelators with high selectivity and low toxicity towards uranium, possessing both high oral bioavailability and low cost for clinical use, remains a pending challenge.Uranium predominantly exists under its main oxidation state +VI with two axial oxo ligands which form the linear triatomic uranyl ion with an overall charge of +II. The ability to afford four to six equatorial ligands in octahedral, pentagonal or hexagonal bipyramidal geometries separates uranyl from most of the alkali, alkaline and transition metals.25,26 Proteins act as the primary transporters of uranyl ion in the body. Several studies have identified proteins, including transferrin, osteopontin, albumin, globulin, and fetuin, that display a strong affinity for uranyl ions.27,28 In 2014, He et al. reported that carboxylic acid side-chain ligands on proteins can form fivefold or sixfold planar coordination around the central uranyl, showing high selectivity and affinity for uranium.29 Mimicking these proteins to design small peptide ligands may be an effective strategy for rationalize uranium–protein interactions.29,30 Delangle et al. reported an engineered cyclodecapeptide with four glutamate residues to coordinate uranyl at the dioxo cation's equatorial plane, resulting in high affinity for uranyl. However, the high affinity of carboxyl groups with a variety of metal ions complicates the specific recognition of uranyl cation from numerous competing ions.31 In 2015, Wang et al. discovered that phosphorylated cyclodipeptide exhibited high selectivity and sensitivity toward uranyl ions, which was successfully employed as a fluorescent sensor for the detection of uranyl ions in river water.32 Recently, Delangle et al. employed similar strategy by using phosphate group in the cyclic peptide skeleton to achieve high affinity towards uranyl ion.33–35 Mechanism studies have showed that phosphorylated functional groups and a pre-organized structure are critical structural parameters.36,37
Cyclic peptides are structurally constrained and could result in higher thermodynamic stability in cyclic peptide–uranyl complex. Consequently, cyclic peptides generally exhibit greater affinity and selectivity for uranyl than do linear peptides.32 In contrast, linear peptides may be simpler and less expensive to utilize in practical applications. But the structure flexibility makes it a great challenge to obtain high affinity and selectivity towards uranyl. Le Clainche et al.35 synthesized a 33-amino acid peptide corresponding to the helix-loop-helix motif of the calcium binding site I of the protein calmodulin from Paramecium tetraurelia and demonstrated that mutation of two aspartic acid residues in the peptide sequence gave access to a new peptide, which was selective for the uranyl ion.
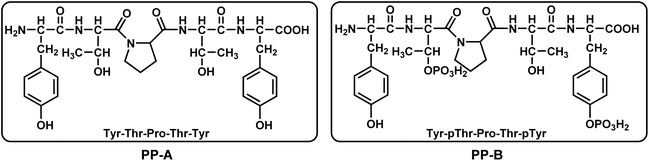
In this study, we designed the short straight-chain pentapeptide PP-B as a model peptide (Fig. 1), which is more stable compared to long linear polypeptide chains (the synthesis steps are described in ESI†). To our knowledge, the designed pentapeptide PP-B is the shortest linear peptide sequence capable of uranium decorporation. The symmetrical design of the peptide structure enhances its stability in chelation with uranyl. Tyrosine produces natural fluorescence when excited by 280 nm excitation light and can be used as a fluorescent probe. In order to improve the high selectivity and sensitivity of the peptide to uranyl ions, we phosphorylated one of the threonine and tyrosine. UO22+ tends to coordinate with 4–6 binding sites on its equatorial plane, and based on this, we plan that PP-B can provide 3–4 coordination sites (which can be provided by the phosphate, phenolic hydroxyl, and carboxyl groups on the side chain), and the other 1–2 sites can be provided by water molecules in the system. The results indicate that PP-B shows a significant improvement in affinity compared to the non-phosphorylated peptide PP-A. These findings might illuminate the importance of phosphoamino acids in uranyl binding in proteins and the relevance of considering phosphoproteins as potential uranyl targets in vivo.
2 Experimental materials and methods
2.1 Experimental materials
Uranyl nitrate was purchased from Hubei Chushengwei Chemical Ltd (Hubei, China). Diethylenetriaminepentaacetic acid (DTPA) was purchased from Macklin Biochemical Co., Ltd (Shanghai, China). 4-(2-Hydroxyethyl)1-piperazineethanesulfonic acid (HEPES, ≥99.0%) was purchased from Sangon Biotech Co., Ltd (Shanghai, China). Sodium bicarbonate and other salts solution was purchased from TCI Development Co., Ltd (Shanghai, China). HK-2 cells, fetal bovine serum (FBS) and DMEM/F-12 medium were purchased from Wuhan Procell Life Science & Technology Co., Ltd (Wuhan, China). Cell Counting Kit-8 (CCK-8) assay kit were purchased from Beyotime Biotech Inc (Shanghai, China). All reagents were analytically pure. Ultrapure water obtained using the Direct-Q5 purification system (Bausasi Technology, 18.2 MΩ cm−1 resistivity) was used in all experiments.Shimadzu LC-20AP preparative liquid chromatography (Shimadzu, Japan) was utilized in conjunction with Thermo Fisher Orbitrap Astral mass spectrometer detector (Thermo Fisher, USA) for peptide purification and structural characterization. Fluorescence spectral data for the peptides were recorded using a HORIBA Fluoromax-4 molecular fluorescence spectrometer (Horiba, France). Cell viability was measured using a luminescence detection at 450 nm on a SpectraMax i3x multifunctional enzyme marker instrument (Molecular, USA).
2.2 Characterization
The phosphorylated peptide was purified and characterized by high performance liquid chromatography (HPLC) and mass spectrometry (ESI-MS). High-resolution mass spectra were collected under the specific conditions (ESI+ spray voltage, 4.5 kV; nebulizer gas, 1.5 L min−1; heat block temperature, 200 °C; CDL temperature, 200 °C; detector voltage, 1.5 kV. The source settings were the same for both samples to get comparable data).2.3 Fluorescence titration experiment
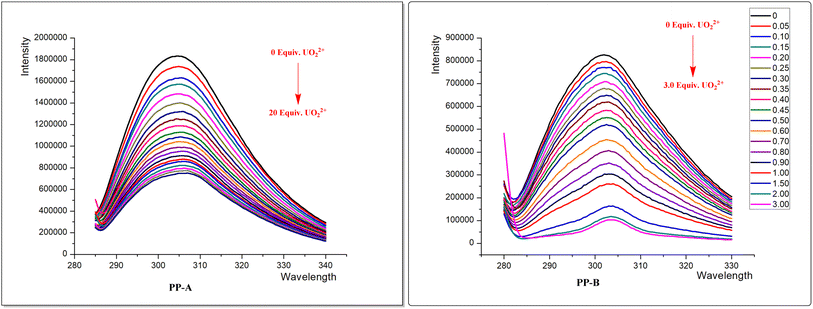
In order to verify that the introduction of the phosphate group plays an important role in enhancing the chelating ability of uranium ions, the non-phosphorylated polypeptide (PP-A) was set up as a comparison with the phosphorylated polypeptide (PP-B). The fluorescence spectra of the two polypeptides were measured in a four-side translucent quartz cuvette. A stock uranyl solution (2 mM) was prepared from uranyl nitrate in nitric acid (0.01 M). HEPES buffer (0.01 M) was prepared by dissolving solid 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid in ultrapure water. Peptide solution (20 μM) was freshly prepared by dissolving peptide solid in 0.01 M HEPES buffer, followed by pH adjustment to 7.0 with KOH. Tyrosine fluorescence quenching was followed by titration of the peptide buffered solution with a certain equivalent of uranyl. The pH was measured at the beginning and at the end of the experiment to guarantee pH stability during titration. The measurements were performed at 25 °C. Tyrosine fluorescence was excited at 275 nm (with an excitation slit of 5.0 nm) and the emission slit was adjusted (5 nm) to avoid signal saturation. The binding constants for peptide–UO22+ chelates were calculated using the BindFit v0.5 algorithm.2.4 Density functional theory (DFT) calculation
The structure was optimized by using the DFT approach as implemented in the Gaussian 16 software. The B3LYP functional38 and mixed basis set (SDD ECP basis set for U atom and the 6-31G (d, p) basis set for other atoms) were adopted for geometry optimization and frequency calculations, and the optimal geometry for each compound was determined. The DFT-D3 dispersion correction39 was applied to correct the weak interaction to improve the calculation accuracy. The SMD implicit solvation model40 was used to account for the solvation effect.2.5 In vitro cells assay
The HK-2 cell line was cultured in a medium containing a mixture of F-12 nutrient mixture (DMEM/F-12), 10% (v/v) fetal bovine serum (FBS), and 1% penicillin–streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. The cells were propagated every two days. The medium used for poisoning had the proportion of FBS adjusted from 10% to 3%. Cells for measurement were cultured in clear 96-well plates at a density of 8 × 103 HK-2 cells and incubated with 100 μL of medium per well.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 to obtain 10 mM uranyl carbonate solution. All solutions were sterilized by 0.22 μm sterilization filter. The work solutions containing chelating agent and U(VI) were prepared by diluting with culture medium to the expected concentrations for cell treatment.
8 to obtain 10 mM uranyl carbonate solution. All solutions were sterilized by 0.22 μm sterilization filter. The work solutions containing chelating agent and U(VI) were prepared by diluting with culture medium to the expected concentrations for cell treatment.3 Results and discussion
3.1 Characterization of peptides
The HPLC chromatogram of PP-A showed the peak at 9.640 minutes, with a purity of 99.64%. ESI-MS indicated that the relative molecular mass (RMM) of PP-A was 643.68. The HPLC chromatogram of PP-B showed the peak at 11.563 minutes, with a purity of 98.84%. The RMM of PP-B is 803.64. Relatively images were shown in ESI.†3.2 Mechanisms of uranyl-selective-binding linear pentapeptide
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexed with uranyl ion.
1 complexed with uranyl ion.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
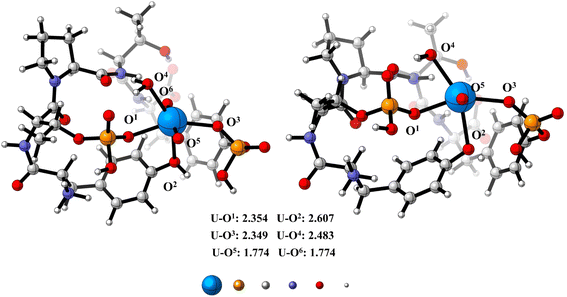
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with uranyl, we have performed DFT optimizations on the proposed UO22+–phosphate peptide complex. As is shown in Fig. 5, the U center adopts an octahedral geometry, with two oxygen atoms (UO22+) on the apical position and four oxygen atoms (two from phosphate, one from Tyr and one from water molecule) on the equatorial plane. The bond distances for the four equatorial U–O single bond range from 2.354 Å to 2.607 Å, which is in good consistency with previously reported U(VI) complexes.44 DFT calculations imply that the phosphate group binds strongly to the UO22+ center (U–O1 = 2.354 Å and U–O3 = 2.349 Å) and might explain why the phosphate peptide has strong affinity to UO22+. Relatively unstable conformation is shown in ESI.†
1 complex with uranyl, we have performed DFT optimizations on the proposed UO22+–phosphate peptide complex. As is shown in Fig. 5, the U center adopts an octahedral geometry, with two oxygen atoms (UO22+) on the apical position and four oxygen atoms (two from phosphate, one from Tyr and one from water molecule) on the equatorial plane. The bond distances for the four equatorial U–O single bond range from 2.354 Å to 2.607 Å, which is in good consistency with previously reported U(VI) complexes.44 DFT calculations imply that the phosphate group binds strongly to the UO22+ center (U–O1 = 2.354 Å and U–O3 = 2.349 Å) and might explain why the phosphate peptide has strong affinity to UO22+. Relatively unstable conformation is shown in ESI.†
3.3 In vivo uranyl decorporation
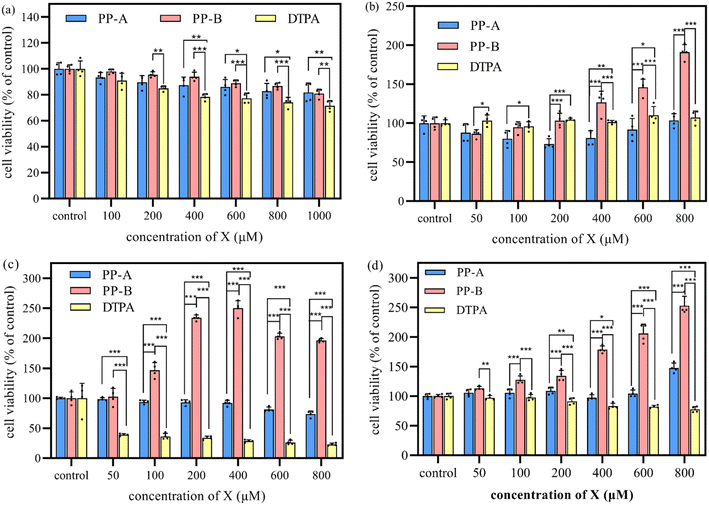
In the cytotoxicity assay, the cytotoxicity of PP-A, PP-B and DTPA exhibited a concentration-dependent increase (Fig. 6a). The survival rates of the cells remained above 80% even when the concentrations of PP-A and PP-B were increased to 1000 μM, which were significantly higher than that of DTPA, indicating that the cytotoxicity of PP-A and PP-B was significantly less than that of the clinically approved DTPA. This indicated that peptide chelators, with less toxic side effects resulting from accumulation in the human body, are bio-friendly drugs compared to DTPA.In the decorporation assay, the timely/delayed treatment group exhibited the most significant overall enhancement in cell viability (Fig. 6c and d), with a cell viability value that was 2.5 times higher when 400/800 μM PP-B solution was added compared to the control group. The addition of PP-A has little effect. However, the addition of DTPA solution not only failed to increase the cell viability of uranium, but also had a toxic effect on the cells, decreasing the cell viability, and the toxicity increased with the increase of the concentration of DTPA. Additionally, the early administration group also exhibited an enhancement in cell viability (Fig. 6b), with a cell viability value that was 1.9 times higher when 800 μM PP-B solution was added compared to the control group. The addition of PP-A and DTPA had no beneficial effect. Thus, the timely and delayed treatment with PP-B provided the optimal removal effect for uranium contamination, and early pre-treatment potentially reduced uranium burden. The use of PP-B as a promotional excretion agent significantly increased cell viability compared to PP-A and DTPA, suggesting that the phosphorylation greatly enhances the ability of PP-B to bind to uranium, whereas DTPA exhibits minimal pro-excretion activity and slightly reduced cell viability.
4 Conclusion
In conclusion, the phosphorylated peptide PP-B, bearing the well preorganized donor structure matching the uranyl coordination environment, shows a higher binding constant for UO22+ compared to the unphosphorylated peptide. This study, along with prior investigations, verifies that the incorporation of a phosphate group serves as a viable strategy to enhance the chelating ability of UO22+. In vitro cellular experiments have shown that PP-B exhibits significantly lower cytotoxicity than clinically used DTPA, with the cell survival rates consistently above 80%. PP-B demonstrated a higher uranyl-removing efficiency than PP-A and DTPA. The phosphorylated peptide is a short, straight-chain one and is facile to synthesize. Coupled with its selective binding ability, high efficiency and low cytotoxicity, the structure of this uranyl-selective-binding linear pentapeptide sequence is one of the most promising U(VI) decorporation agents and its practical application is expected to be realized in the future.Data availability
The data presented in this study are available on ESI.†Author contributions
Xiaohong Tan: methodology, experiment, software, formal analysis, investigation, writing – original draft, writing – review & editing; Libing Yu: experiment, project administration, writing – review & editing; Xindan Liao: conceptualization, methodology; Chun Chen: investigation, methodology; Jian Chu: methodology, experiment; Zhonghua Xiong: supervision, writing – review & editing; Binyuan Xia: supervision, writing – review & editing; Wei Tang: resources, writing – review & editing; Xijian Li: conceptualization, methodology, experiment, formal analysis, investigation, writing – original draft, writing – review & editing, funding acquisition, supervision, project administration.; Yanyan Liu: writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
We would like to acknowledge National Natural Science Foundation of China (22106149) for funding this work.References
- N. Hu, D. Ding, S. Li, X. Tan, G. Li, Y. Wang and F. Xu, J. Environ. Radioact., 2016, 154, 60–67 CrossRef CAS PubMed.
- É. Ansoborlo, B. Amekraz, C. Moulin, V. Moulin, F. Taran, T. Bailly, R. Burgada, M.-H. Hengé-Napoli, A. Jeanson, C. Den Auwer, L. Bonin and P. Moisy, C. R. Chim., 2008, 10, 1010–1019 Search PubMed.
- É. Ansoborlo, O. Prat, P. Moisy, C. Den Auwer, P. Guilbaud, M. Carriere, B. Gouget, J. Duffield, D. Doizi, T. Vercouter, C. Moulin and V. Moulin, Biochimie, 2006, 88, 1605–1618 Search PubMed.
- O. Carugo, J. Inorg. Biochem., 2018, 189, 1–6 CAS.
- M. Ma, R. Wang, L. Xu, M. Xu and S. Liu, Environ. Int., 2020, 145, 106107 CrossRef CAS PubMed.
- J. C. Liandong Zhang, B. Xia, Z. Xiong, S. Zhang and W. Tang, Toxics, 2022, 10, 575 CrossRef.
- J. W. Howland, At. Energy Biophys. Biol. Med., 1948, 1, 174 CAS.
- Y. Ohmachi, T. Imamura, M. Ikeda, E. Shishikura, E. Kim, O. Kurihara and K. Sakai, J. Toxicol. Pathol., 2015, 28, 65–71 CrossRef CAS PubMed.
- M. H. Bhattacharyya, B. D. Breitenstein, H. Metivier, B. A. Muggenburg, G. N. Stradling and V. Volf, Radiat. Prot. Dosim., 1992, 41, 1–42 CAS.
- T. Fujii, A. Udy, E. Licari, L. Romero and R. Bellomo, J. Crit. Care, 2019, 51, 184–191 CrossRef CAS PubMed.
- T. Y. Yang, H. M. Lin, H. Y. Wang, M. H. Chuang, C. C. Hsieh, K. T. Tsai and J. Y. Chen, Clin. J. Am. Soc. Nephrol., 2024, 19, 959–969 CrossRef.
- D. P. Hickman, Radiat. Prot. Dosim., 2010, 141, 215–216 CrossRef.
- National Health Commission of the People's Republic of China, Medical Management Standard for Internal Contamination of Radionuclides, WS/T 583—2017, Standards Press of China, Beijing, 2017 Search PubMed.
- E. Fattal, N. Tsapis and G. Phan, Adv. Drug Delivery Rev., 2015, 90, 40–54 CrossRef CAS.
- A. E. V. Gorden, J. Xu, K. N. Raymond and P. Durbin, Chem. Rev., 2003, 103, 4207–4282 CrossRef CAS.
- Y. Bao, D. Wang, Z. Li, Y. Hu, A. Xu, Q. Wang, C. Shao and H. Chen, Toxicol. Appl. Pharmacol., 2013, 269, 17–24 CAS.
- P. Durbin, B. Kullgren, J. Xu and K. Raymond, Health Phys., 1997, 72, 865–879 CrossRef CAS.
- Q. Zhang, B. Jin, T. Zheng, X. Tang, Z. Guo and R. Peng, Inorg. Chem., 2019, 58, 14626–14634 CAS.
- B. Chen, S. Hong, X. Dai, X. Li, Q. Huang, T. Sun, D. Cao, H. Zhang, Z. Chai, J. Diwu and S. Wang, J. Am. Chem. Soc., 2022, 144, 11054–11058 CrossRef CAS PubMed.
- D. X. Wang, L. Chen, Z. Bai, D. D. Zhang, J. Guan, Y. Zhang, C. Shi and P. D. J. Diwu, Angew. Chem., 2021, 133, 1670–1674 CrossRef.
- M. Chen, L. Lang, L. Chen, X. Wang, C. Shi, Q. Sun, Y. Xu, J. Diwu and S. Wang, Chin. J. Chem., 2022, 40, 2054–2060 CrossRef CAS.
- L. Chen, X. Wang, M. Chen, Q. Sun, Y. Chen, X. Zhang, R. Hong, Y. Xu, J. Guan, S. Hong, D. Cao, T. Sun, X. Li, L. Chen and J. Diwu, Adv. Healthcare Mater., 2023, 12, 2300510 CrossRef CAS.
- L. Chen, R. Bai, X. Wang, Y. Zhang, L. He, M. Zhang, Y. Chong, Z. Chai, X.-F. Wang and J. Diwu, ACS Appl. Bio Mater., 2020, 3, 8731–8738 CrossRef CAS PubMed.
- Y. Sun, L. Zheng, Y. Yang, X. Qian, T. Fu, X. Li, Z. Yang, H. Yan, C. Cui and W. Tan, Nano-Micro Lett., 2020, 12, 103 CrossRef CAS.
- J. Vanhorn and H. Huang, Coord. Chem. Rev., 2006, 250, 765–775 CrossRef CAS.
- A. Garai and P. Delangle, J. Inorg. Biochem., 2020, 203, 110936 CrossRef CAS PubMed.
- S. Safi, G. Creff, A. Jeanson, L. Qi, C. Basset, J. Roques, P. L. Solari, E. Simoni, C. Vidaud and C. Den Auwer, Chemistry, 2013, 19, 11261–11269 CrossRef CAS.
- M. Starck, N. Sisommay, F. A. Laporte, S. Oros, C. Lebrun and P. Delangle, Inorg. Chem., 2015, 54, 11557–11562 CrossRef CAS PubMed.
- L. Zhou, M. Bosscher, C. Zhang, S. Özçubukçu, L. Zhang, W. Zhang, C. J. Li, J. Liu, M. P. Jensen, L. Lai and C. He, Nat. Chem., 2014, 6, 236–241 CrossRef CAS PubMed.
- M. R. Beccia, S. Sauge-Merle, N. Brémond, D. Lemaire, P. Henri, C. Battesti, P. Guilbaud, S. Crouzy and C. Berthomieu, Biomolecules, 2022, 12, 1703 CrossRef CAS.
- C. Lebrun, M. Starck, V. Gathu, Y. Chenavier and P. Delangle, Chem.–Eur. J., 2014, 20, 16566–16573 CrossRef CAS.
- C.-T. Yang, J. Han, M. Gu, J. Liu, Y. Li, Z. Huang, H.-Z. Yu, S. Hu and X. Wang, Chem. Commun., 2015, 51, 11769–11772 RSC.
- M. Starck, F. A. Laporte, S. Oros, N. Sisommay, V. Gathu, P. L. Solari, G. Creff, J. Roques, C. Den Auwer, C. Lebrun and P. Delangle, Chem.–Eur. J., 2017, 23, 5281–5290 CrossRef CAS PubMed.
- C. L. Fanny, A. Laporte, C. Vidaud and P. Delangle, Chem.–Eur. J., 2019, 25, 8570–8578 CrossRef.
- L. L. Clainche and C. Vita, Environ. Chem. Lett., 2006, 4, 45–49 CrossRef.
- L. Abou-Zeid, A. Pell, M. Garcia Cortes, H. Isnard, P. Delangle and C. Bresson, Anal. Chim. Acta, 2023, 1242, 340773 CrossRef CAS.
- R. Pardoux, S. Sauge-Merle, N. Bremond, M. R. Beccia, D. Lemaire, C. Battesti, P. Delangle, P. L. Solari, P. Guilbaud and C. Berthomieu, Inorg. Chem., 2022, 61, 20480–20492 CrossRef CAS.
- D. A. Wappett and L. Goerigk, J. Chem. Theory Comput., 2023, 19, 8365–8383 CrossRef CAS.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104 CrossRef.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS.
- P. W. Durbin, Health Phys., 2008, 95, 465–492 CrossRef CAS.
- M. Starck, N. Sisommay, F. A. Laporte, S. Oros, C. Lebrun and P. Delangle, Inorg. Chem., 2015, 54, 11557–11562 CrossRef CAS.
- F. Laporte, Y. Chenavier, A. Botz, C. Gateau, C. Lebrun, S. Hostachy, C. Vidaud and P. Delangle, Angew. Chem., Int. Ed., 2022, 61, e202203198 CrossRef CAS PubMed.
- A. Bavelloni, M. Piazzi, M. Raffini, I. Faenza and W. L. Blalock, IUBMB Life, 2015, 67, 239–254 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06173j |
| This journal is © The Royal Society of Chemistry 2024 |