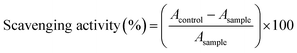Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis, characterization, biological activities, and computational studies of pyrazolyl–thiazole derivatives of thiophene†
Seema K. Bhagwata,
Tushar Janardan Pawar b,
Sayali A. Kulkarnia,
Amar A. Patila,
Rahul Ashokrao Morec,
J. Oscar C. Jimenez-Halla
b,
Sayali A. Kulkarnia,
Amar A. Patila,
Rahul Ashokrao Morec,
J. Oscar C. Jimenez-Halla d,
Juan Andres Alvarado-Salazar
d,
Juan Andres Alvarado-Salazar e,
Jose Luis Olivares-Romero
e,
Jose Luis Olivares-Romero b,
Ghazala Muteeb
b,
Ghazala Muteeb *f,
Enrique Delgado-Alvarado
*f,
Enrique Delgado-Alvarado *g and
Sachin V. Patil
*g and
Sachin V. Patil *a
*a
aDepartment of Chemistry, Research Centre HPT Arts and RYK Science College (Affiliated to S. P. Pune University), Nashik, 422005, Maharashtra, India
bRed de Estudios Moleculares Avanzados, Campus III, Instituto de Ecología, A. C., Carretera Antigua a Coatepec 351, 91073, Xalapa, Veracruz, Mexico
cDepartment of Microbiology, Dayanand Science College, Latur, 413512, Maharashtra, India
dDepartamento de Química, División de Ciencias Naturales y Exactas, Universidad de Guanajuato, Noria Alta S/N, 36050, Guanajuato, Mexico
eCarrera de Química-Farmacéutico-Biológica, Facultad de Estudios Superiores Zaragoza, UNAM, 09230 Ciudad de México, Mexico
fDepartment of Nursing, College of Applied Medical Sciences, King Faisal University, Al-Ahsa, Saudi Arabia
gMicro and Nanotechnology Research Center, Universidad Veracruzana, Blvd. Av. Ruiz Cortines No. 455 Fracc. Costa Verde, Boca del Río, Veracruz 94294, Mexico
First published on 10th December 2024
Abstract
This study reports the synthesis, characterization, and biological evaluation of a series of pyrazolyl–thiazole derivatives of thiophene. Seven compounds were synthesized and characterized using NMR spectroscopy and mass spectrometry. The antimicrobial activities of these derivatives were evaluated against various bacterial (Escherichia coli, Bacillus subtilis, Bacillus megaterium, Staphylococcus aureus) and fungal strains (Aspergillus niger, Aspergillus oryzae, Rhizopus, Candida albicans), demonstrating significant inhibition zones and low minimum inhibitory concentrations (MIC). In addition, the compounds exhibited notable antioxidant activities in DPPH and hydroxyl radical scavenging assays. Computational studies, including density functional theory (DFT) calculations and molecular docking simulations, were performed to understand the electronic properties and binding interactions of the synthesized compounds with biological targets. The molecular docking results supported the experimental findings, highlighting the potential of these pyrazolyl–thiazole derivatives as multifunctional therapeutic agents with both antimicrobial and antioxidant properties.
Introduction
The emergence of drug-resistant pathogens and the increasing prevalence of oxidative stress-related diseases have underscored the urgent need for new and effective antimicrobial and antioxidant agents. Antimicrobial resistance poses a significant global health challenge, as many conventional antibiotics are becoming less effective against resistant strains.1 This crisis has spurred the exploration of novel chemical entities capable of combating these resilient pathogens. Heterocyclic compounds have been recognized for their potential in medicinal chemistry due to their versatile chemical structures and diverse biological activities.2 Among these, thiazole, pyrazole and thiophene derivatives have garnered significant interest for their antimicrobial and antioxidant properties. Additionally, the combination of peptides with these heterocyclic compounds is an emerging area of research, offering new opportunities to develop hybrid molecules with enhanced biological activities and reduced toxicity.3Thiazole, pyrazole, and thiophene are three important five-membered heterocyclic rings widely recognized for their significant roles in medicinal chemistry. Thiazole, containing both sulfur and nitrogen atoms, is found in many bioactive molecules with antimicrobial, anti-inflammatory, and anticancer properties.4 It serves as a core structure in compounds like vitamin B1 (thiamine) and the antibiotic bacitracin, highlighting its importance in biological systems and drug design.5 Thiazole derivatives have demonstrated substantial antimicrobial activity against a wide range of pathogens, making them promising scaffolds for developing new antibiotics. Pyrazole, another five-membered ring with two adjacent nitrogen atoms, also exhibits diverse pharmacological activities, including anti-inflammatory, analgesic, antipyretic, and antitumor effects.6 The therapeutic potential of pyrazoles is underscored by their presence in well-known drugs such as celecoxib, an anti-inflammatory agent, and sildenafil, used to treat erectile dysfunction. Pyrazole rings are particularly valuable in drug design due to their ability to modulate enzyme activity, providing a basis for developing enzyme inhibitors for various diseases.7
Thiophene, a five-membered ring containing sulfur, is another versatile heterocyclic compound that has garnered attention for its biological activities. Thiophene derivatives are known for their antimicrobial, anti-inflammatory, and antitumor properties.8 They are often incorporated into drugs and biologically active molecules due to their ability to enhance the pharmacokinetic and pharmacodynamic profiles of these compounds. The presence of sulfur in thiophene contributes to its unique electronic properties, which can improve binding affinity and specificity towards biological targets. The combination of thiophene with other heterocyclic rings, such as thiazole and pyrazole, can lead to hybrid molecules with enhanced biological activities and therapeutic potential.
Together, thiazole, pyrazole, and thiophene represent a diverse group of heterocyclic compounds with broad-spectrum biological activities. Their ability to interact with various biological targets, combined with their versatile chemical structures, makes them invaluable in the development of new drugs. By exploring the potential of these rings, researchers can design hybrid molecules that leverage the unique properties of each to create more effective and targeted therapeutic agents.9
While thiophenyl–pyrazolyl–thiazole hybrids have been explored for their antimicrobial and DHFR inhibitory activities, particularly against Mycobacterium tuberculosis, our study presents a distinct approach by synthesizing a new series of these derivatives with unique structural modifications. Unlike previous studies, including the recent work by Dawood et al. (2023),10 which focused on DHFR inhibition and antimicrobial activity, our research broadens the biological scope to include antioxidant activities and explores different biological targets. Additionally, we employed advanced computational techniques to predict electronic properties and stability, providing new insights into the structure–activity relationship. These differences in chemical structure, biological evaluation, and computational analysis highlight the novelty of our work and its potential contribution to developing multifunctional agents with diverse therapeutic applications.
In this study, we report the synthesis, characterization, and biological evaluation of a novel series of pyrazolyl–thiazole derivatives of thiophene, which have not been previously explored in the literature. Unlike existing studies that focus primarily on individual heterocyclic rings, our work investigates hybrid molecules combining thiazole, pyrazole, and thiophene scaffolds. This unique combination is designed to exploit the complementary pharmacological properties of these rings, leading to enhanced antimicrobial and antioxidant activities. Additionally, our work incorporates comprehensive computational studies, including DFT calculations and molecular docking, to provide a deeper understanding of the electronic properties and binding interactions of these novel compounds. The insights gained from these computational studies offer valuable guidance for the rational design of more potent derivatives.
Results and discussion
Synthesis and characterization of pyrazolyl–thiazole derivatives
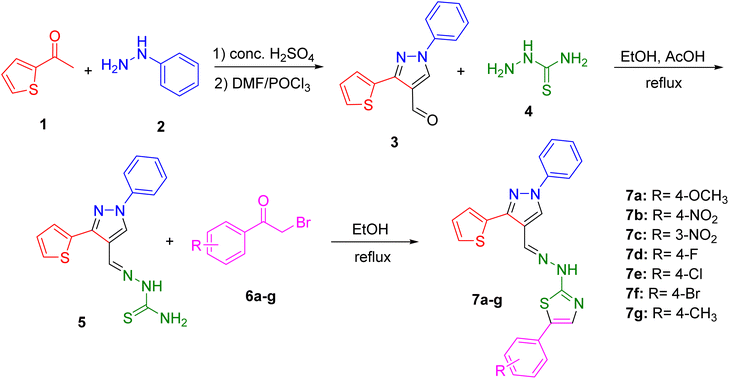
The synthesis of the pyrazolyl–thiazole derivatives of thiophene was carried out through a multi-step synthetic route, as outlined in Scheme 1. Using previously reported procedure,11 the process began with the condensation of acetyl thiophene 1 with phenyl hydrazine 2 in the presence of concentrated H2SO4. This reaction yielded the hydrazone intermediate, which was then cyclized using phosphoryl chloride (POCl3) in dimethylformamide (DMF) to form the desired pyrazole-4-carbaldehyde 3. The product was purified by recrystallization from ethanol. Next, the pyrazole-4-carbaldehyde 3 was reacted with thiosemicarbazide 4 in ethanol with acetic acid as a catalyst to form the thiosemicarbazone derivative 5. The reaction mixture was refluxed for one hour. Upon completion, the mixture was cooled to room temperature, and the solid product was filtered, washed with ethanol, and recrystallized. The final step involved the reaction of the thiosemicarbazone intermediate 5 with various substituted phenacyl bromides 6a–g in ethanol under reflux conditions. This reaction produced the final pyrazolyl–thiazole derivatives 7a–g, which were filtered, washed with ethanol, and recrystallized to obtain pure products. The synthesized derivatives included compounds with different substituents on the phenyl ring: 7a (R = 4-OCH3), 7b (R = 4-NO2), 7c (R = 3-NO2), 7d (R = 4-F), 7e (R = 4-Cl), 7f (R = 4-Br), and 7g (R = 4-CH3).The structures of the compounds 7a–g were confirmed by NMR spectroscopy and mass spectrometry. The 1H and 13C NMR spectra displayed characteristic signals corresponding to the pyrazole, thiazole, and thiophene moieties, as well as the substituents on the phenyl ring. Mass spectrometry provided molecular ion peaks that matched the expected molecular weights of the compounds 7a–g, confirming their molecular formulas.
Antimicrobial activity
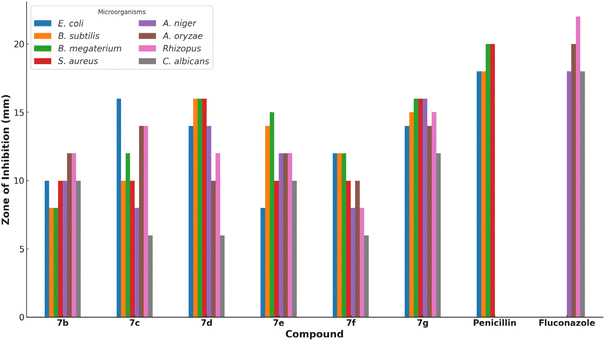
The antimicrobial activities of the synthesized pyrazolyl–thiazole derivatives 7b–g were evaluated against a panel of bacterial and fungal strains. The bacterial strains tested included Escherichia coli, Bacillus subtilis, Bacillus megaterium, and Staphylococcus aureus. The fungal strains tested included Aspergillus niger, Aspergillus oryzae, Rhizopus, and Candida albicans.The antimicrobial activity of each compound was assessed using the Kirby–Bauer disk diffusion method. In this method, disks impregnated with the synthesized compounds were placed on agar plates inoculated with the respective microbial strains. The plates were then incubated, and the zones of inhibition around each disk were measured to determine the antimicrobial effectiveness of the compounds. The results are summarized in Table 1. Compounds 7c and 7d exhibited the highest antibacterial activity, particularly against Bacillus subtilis and Bacillus megaterium, with inhibition zones of 16 mm (Fig. 1). Compound 7g also showed significant activity with inhibition zones of 15–16 mm against multiple strains.
| Compound | E. coli | B. subtilis | B. megaterium | S. aureus | A. niger | A. oryzae | Rhizopus | C. albicans |
|---|---|---|---|---|---|---|---|---|
| 7b | 10 | 8 | 8 | 10 | 10 | 12 | 12 | 10 |
| 7c | 16 | 10 | 12 | 10 | 8 | 14 | 14 | 6 |
| 7d | 14 | 16 | 16 | 16 | 14 | 10 | 12 | 6 |
| 7e | 8 | 14 | 15 | 10 | 12 | 12 | 12 | 10 |
| 7f | 12 | 12 | 12 | 10 | 8 | 10 | 8 | 6 |
| 7g | 14 | 15 | 16 | 16 | 16 | 14 | 15 | 12 |
| Penicillin | 18 | 18 | 20 | 20 | — | — | — | — |
| Fluconazole | — | — | — | — | 18 | 20 | 22 | 18 |
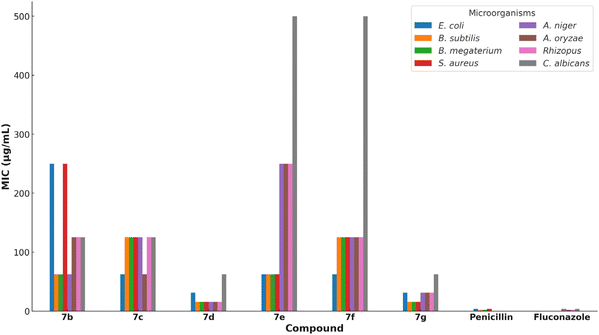
In addition to the disk diffusion assay, the minimum inhibitory concentrations (MICs) of the synthesized compounds were determined using the resazurin microtiter assay (REMA). This assay provides quantitative data on the antimicrobial potency of the compounds by measuring the lowest concentration at which visible microbial growth is inhibited. The MIC values are presented in Table 2. Compounds 7d and 7g showed the lowest MIC values, indicating their potent antimicrobial activity (Fig. 2). Notably, compound 7d exhibited MIC values of 15.63 μg mL−1 against Bacillus subtilis, Bacillus megaterium, and various fungal strains, suggesting broad-spectrum antimicrobial efficacy.
| Compound | E. coli | B. subtilis | B. megaterium | S. aureus | A. niger | A. oryzae | Rhizopus | C. albicans |
|---|---|---|---|---|---|---|---|---|
| 7b | 250 | 62.5 | 62.5 | 250 | 62.5 | 125 | 125 | 125 |
| 7c | 62.5 | 125 | 125 | 125 | 125 | 62.5 | 125 | 125 |
| 7d | 31.25 | 15.63 | 15.63 | 15.63 | 15.63 | 15.63 | 15.63 | 62.5 |
| 7e | 62.5 | 62.5 | 62.5 | 62.5 | 250 | 250 | 250 | 500 |
| 7f | 62.5 | 125 | 125 | 125 | 125 | 125 | 125 | 500 |
| 7g | 31.25 | 15.63 | 15.63 | 15.63 | 31.25 | 31.25 | 31.25 | 62.5 |
| Penicillin | 3.9 | 1.95 | 1.95 | 3.9 | — | — | — | — |
| Fluconazole | — | — | — | — | 3.9 | 1.95 | 1.95 | 3.9 |
Antioxidant activity
The antioxidant activities of the synthesized pyrazolyl–thiazole derivatives 7b–g were evaluated using two different assays: the DPPH (2,2-diphenyl-1-picrylhydrazyl) radical scavenging assay and the hydroxyl radical scavenging assay. These assays measure the ability of the compounds to neutralize free radicals, which are implicated in various diseases due to their role in oxidative stress.| Compound | DPPH (%) | Hydroxyl (%) |
|---|---|---|
| 7b | 58.45 | 65.45 |
| 7c | 62.54 | 58.08 |
| 7d | 69.40 | 64.00 |
| 7e | 72.45 | 69.45 |
| 7f | 62.57 | 58.41 |
| 7g | 66.84 | 64.65 |
| Ascorbic acid | 81.64 | — |
| α-Tocopherol | — | 82.64 |
Computational studies
To further understand these observed biological activities, we conducted detailed computational studies. Our DFT calculations revealed variations in the HOMO–LUMO gaps among the compounds, with 7d and 7g exhibiting smaller gaps indicative of higher electronic stability and reactivity, which correlates with their enhanced antioxidant activity. Molecular docking studies identified key binding interactions between these compounds and biological targets, such as hydrogen bonding and π–π stacking, which align with the experimental antimicrobial data. Compounds 7d and 7g, which showed the most favorable binding affinities in docking studies, also demonstrated the highest antimicrobial activities in vitro. These findings underscore the utility of computational methods in predicting biological activity and guiding the design of more effective therapeutic agents.| Compound | HOMO (eV) | LUMO (eV) | ΔE (eV) |
|---|---|---|---|
| 7a | –7.10 | 0.18 | 7.28 |
| 7b | –7.66 | –0.83 | 6.83 |
| 7c | –7.60 | –0.68 | 6.92 |
| 7d | –7.34 | 0.08 | 7.42 |
| 7e | –7.38 | 0.03 | 7.41 |
| 7f | –7.39 | 0.01 | 7.40 |
| 7g | –7.22 | 0.15 | 7.37 |
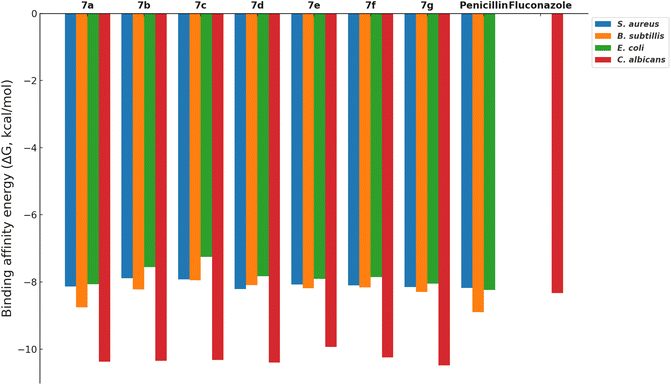
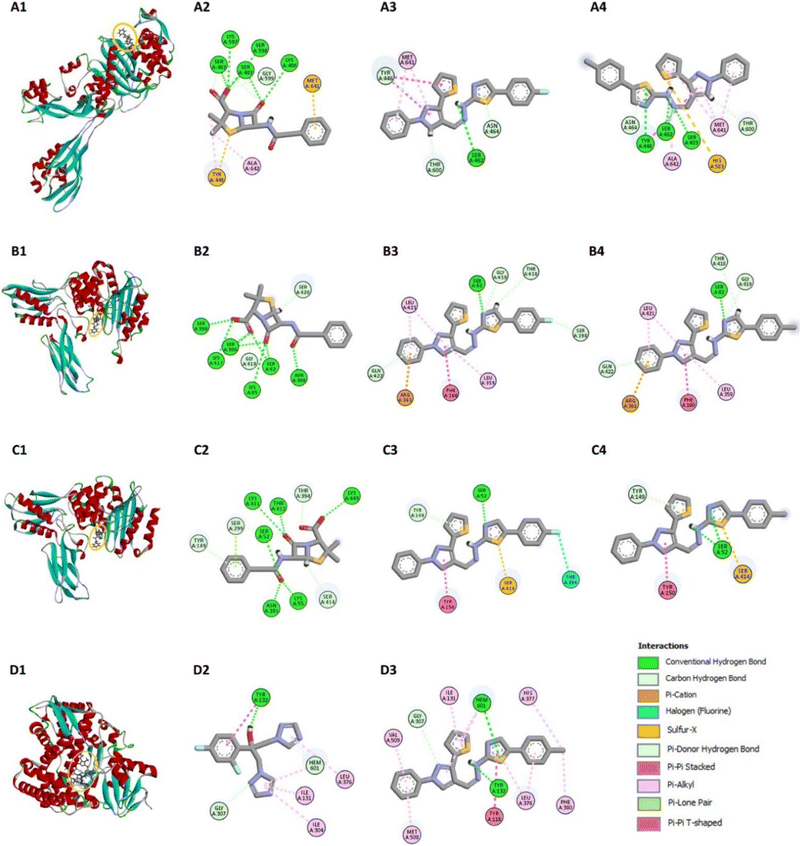
The docking simulations were performed using AutoDock Vina, where compounds 7a–g were docked into the active sites of the PBPs from Gram-positive (S. aureus and B. subtilis) and Gram-negative (E. coli) bacteria, as well as the fungal protein sterol 14α-demethylase from C. albicans. Penicillin and fluconazole were used as reference drugs in the docking studies. The binding affinities (ΔG, kcal mol−1) of the compounds were calculated, and the results, summarized in Fig. 3, reveal that the compounds generally show binding affinities comparable to or slightly better than those of the reference drugs.
Compounds 7d and 7g exhibited the best binding affinities among the synthesized derivatives, correlating with their observed superior inhibitory activities as reported in the experimental assays (Table 1 and Fig. 1). Specifically, compound 7g demonstrated strong binding to the active sites of both bacterial PBPs and the fungal sterol 14α-demethylase, suggesting its potential as a broad-spectrum antimicrobial agent.
For the penicillin-binding protein PBP4 of E. coli, Kishida et al.13 have identified key residues in the active site, including Ser62, Asn154, Asp155, Met156, Thr157, Gln158, and Phe160. Compound 7d demonstrated strong hydrogen bonding with Ser62 and π–π stacking with Phe160, contributing to its inhibitory activity (Fig. 4B3). Compound 7g showed similar interactions, with additional π–alkyl interactions with Leu421 and π–cation interactions with Arg361, further strengthening its binding affinity (Fig. 4B4). These interactions align with the experimental data, where 7g exhibited significant antimicrobial activity against E. coli.
In the PBP4a of B. subtilis, the active site includes residues such as Ser52, Lys55, Asp145, Tyr150, Ser299, and Asn301.14 Compound 7d formed hydrogen bonds with Ser52 and π–π stacking with Tyr150, while also interacting with Ser414 through a sulfur-x interaction, which enhances its binding to the protein (Fig. 4C3). Compound 7g exhibited similar binding patterns, including π-donor hydrogen bonds with Tyr149 and π–π stacking with Tyr150, supporting its high binding affinity and antimicrobial efficacy (Fig. 4C4).
For the antifungal target sterol 14α-demethylase of C. albicans, azoles that inhibit CYP51 activity typically interact with iron from the heme prosthetic group.15 The active site of CYP51 includes residues such as Glu116, Leu220, Tyr243, and Ile297.15,16 Compound 7g showed a higher binding affinity (ΔG = −10.49 kcal mol−1) compared to fluconazole (ΔG = −8.33 kcal mol−1), forming multiple interactions with key residues such as Tyr118, His377, and Met508, as well as with the heme group (Fig. 4D3). Although fluconazole's interaction with the heme iron occurs at a more favorable distance, 7g's overall interaction profile suggests it could serve as a template for further modifications to enhance its antifungal potency while potentially reducing toxicity. Detailed interaction shown in ESI (pages S14–S19†).
The docking studies suggest that the synthesized pyrazolyl-thiazole derivatives, particularly 7g, have promising potential as lead compounds for further drug development. Their ability to bind effectively to both bacterial and fungal targets, coupled with favourable binding affinities, underscores their potential as dual-action antimicrobial agents.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 169 cm−1) followed by a weak electron-donor group (7g, 14
169 cm−1) followed by a weak electron-donor group (7g, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806 cm−1). This decreases progressively by using weak electron-withdrawing groups (7d (R = 4-F), 14
806 cm−1). This decreases progressively by using weak electron-withdrawing groups (7d (R = 4-F), 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 802 cm−1; 7e (R = 4-Cl), 14
802 cm−1; 7e (R = 4-Cl), 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390 cm−1; and 7f (R = 4-Br), 14
390 cm−1; and 7f (R = 4-Br), 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264 cm−1) and strong electron-withdrawing groups (7b (R = 4-NO2) 8372 cm−1; 7c (R = 3-NO2), 6386 cm−1).
264 cm−1) and strong electron-withdrawing groups (7b (R = 4-NO2) 8372 cm−1; 7c (R = 3-NO2), 6386 cm−1).
| Compound | λabs (nm) | λem (nm) |
|---|---|---|
| 7b | 320 | 430 |
| 7c | 315 | 425 |
| 7d | 318 | 428 |
| 7e | 317 | 426 |
| 7f | 319 | 429 |
| 7g | 316 | 427 |
Structure–activity relationship (SAR) analysis
The SAR analysis of the compounds 7a–g reveals how different substituents on the phenyl ring specifically influence their antimicrobial and antioxidant activities. The observed biological activities can be linked directly to the electronic effects and steric properties introduced by these substituents.The antimicrobial activity was found to be significantly influenced by the presence of electron-withdrawing groups such as nitro (NO2) and halogens (Cl, Br, F). Compounds with these substituents, particularly 7b (R = 4-NO2) and 7c (R = 3-NO2), exhibited substantial activity against B. subtilis and B. megaterium, with inhibition zones reaching up to 16 mm. The increased electrophilicity imparted by these groups likely enhances interactions with microbial targets by facilitating the formation of stronger bonds with enzyme active sites or cell membranes.
Moreover, the size and position of substituents on the phenyl ring played a crucial role in antimicrobial efficacy. Larger substituents or those at the para (4-) position, such as in 7d (R = 4-F), 7e (R = 4-Cl), and 7f (R = 4-Br), showed significant antimicrobial activity. This could be attributed to steric factors where the larger groups may enable better fitting into hydrophobic pockets of microbial enzymes, thereby enhancing binding interactions. The halogens (Cl, Br, F), being hydrophobic, further increased the overall hydrophobicity of the compounds, facilitating their penetration through microbial cell membranes. This property was particularly evident in the high activity of compounds 7d, 7e, and 7f against multiple strains.
For antioxidant activity, electron-donating groups such as methyl (CH3) significantly enhanced the radical scavenging abilities of the compounds. Compound 7g (R = 4-CH3) demonstrated notable DPPH and hydroxyl radical scavenging activities. The electron-donating nature of the methyl group stabilizes the radical form of the antioxidant by delocalizing the unpaired electron across the molecule, thus enhancing the compound's ability to neutralize free radicals. Additionally, the extended π-conjugation system present in the pyrazolyl–thiazole framework facilitates electron delocalization, further improving radical scavenging efficiency.
The antioxidant activity was also found to correlate well with the HOMO–LUMO energy gaps determined through computational studies. Compounds with larger HOMO–LUMO gaps, such as 7d and 7g, demonstrated better radical scavenging abilities, likely due to their increased electronic stability and reduced reactivity.
The dual antimicrobial and antioxidant activities of these pyrazolyl–thiazole derivatives suggest their potential as multifunctional therapeutic agents. The structural features contributing to both activities include substituents that modulate electron density and introduce steric effects, which can enhance both binding to microbial targets and radical stabilization. For example, the 4-nitro group in compound 7b and the 4-methyl group in compound 7g are associated with high activity in both antimicrobial and antioxidant assays.
Furthermore, the nearly coplanar arrangement of the pyrazolyl and thiazole rings, as revealed by the geometry optimization studies, facilitates π–π interactions, which are crucial for both antimicrobial binding and radical stabilization. The molecular docking results also support this observation, as compounds 7d and 7g formed strong π–π stacking interactions with key residues in the active sites of target proteins, correlating with their high experimental activities.
Conclusions
In conclusion, the synthesized pyrazolyl–thiazole derivatives of thiophene demonstrated significant antimicrobial and antioxidant activities, making them promising candidates for further development as multifunctional therapeutic agents. The SAR analysis revealed that specific substituents on the phenyl ring significantly influence the biological activities of these compounds, with electron-withdrawing groups enhancing antimicrobial efficacy and electron-donating groups improving antioxidant properties. Computational studies provided valuable insights into the electronic properties and binding interactions of the compounds, further supporting the experimental results. The dual antimicrobial and antioxidant activities of these derivatives underscore their potential in combating drug-resistant pathogens and oxidative stress-related diseases. Future studies could incorporate solvent-phase calculations to explore the influence of solvent effects on the electronic and structural properties of the compounds.Experimental section
Materials and methods
All reagents and solvents were purchased from commercial suppliers and used without further purification unless otherwise noted. Analytical thin-layer chromatography (TLC) was performed on silica gel plates, and the compounds were visualized using UV light. Column chromatography was carried out using silica gel (60–120 mesh). Melting points were determined using a capillary melting point apparatus and are uncorrected.Synthesis of pyrazolyl–thiazole derivatives
The synthetic route for the pyrazolyl–thiazole derivatives of thiophene 7b–g is outlined in Scheme 1.Recrystallization. The crude product was dissolved in hot ethanol (approximately 20 mL per gram of product) and allowed to cool to room temperature. The solution was further chilled in an ice bath to promote crystallization. The resulting crystals were filtered, washed with cold ethanol, and dried under vacuum to obtain the purified product.
Recrystallization. The crude product was dissolved in a minimum amount of hot ethanol, and petroleum ether was added slowly to the solution. The mixture was then kept at 0 °C to promote crystallization. The resulting crystals were filtered, washed with cold petroleum ether, and dried under vacuum to obtain the purified product.
Characterization of synthesized compounds
The structures of the synthesized compounds were confirmed by NMR spectroscopy and mass spectrometry. 1H and 13C NMR spectra were recorded on a 400 MHz NMR spectrometer using CDCl3 or DMSO-d6 as solvents. Chemical shifts (δ) are reported in parts per million (ppm) relative to the internal standard tetramethylsilane (TMS). The peak assignment for NMR spectra was done and mentioned in the characterization for each compound. Mass spectra were recorded on an ESI-MS spectrometer.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N),7.2–7.6 (m, 5H, phenyl ring), 7.94 (d, 2H, Ar-H), 7.74 (d, 2H, Ar-H), 6.95–7.15 (m, 3H, thiophene), 3.76 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6): δ 55, 102, 114, 116, 119, 127, 128, 129, 130, 134, 139, 145, 159, 168; MS (EI, 70 eV): m/z (%): 458 (M + H, 100).
N),7.2–7.6 (m, 5H, phenyl ring), 7.94 (d, 2H, Ar-H), 7.74 (d, 2H, Ar-H), 6.95–7.15 (m, 3H, thiophene), 3.76 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6): δ 55, 102, 114, 116, 119, 127, 128, 129, 130, 134, 139, 145, 159, 168; MS (EI, 70 eV): m/z (%): 458 (M + H, 100).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.5–7.9 (m, 5H, phenyl ring), 8.29 (d, 2H, Ar-H), 7.96 (d, 2H, Ar-H), 7.24–7.40 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 108, 116, 119, 124, 126, 127, 128, 129, 130, 134, 139, 145, 146, 168; MS (EI, 70 eV): m/z (%): 473 (M + H, 100).
N), 7.5–7.9 (m, 5H, phenyl ring), 8.29 (d, 2H, Ar-H), 7.96 (d, 2H, Ar-H), 7.24–7.40 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 108, 116, 119, 124, 126, 127, 128, 129, 130, 134, 139, 145, 146, 168; MS (EI, 70 eV): m/z (%): 473 (M + H, 100).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.3–7.9 (m, 4H, Ar-H), 7.58–7.85 (m, 5H, phenyl ring), 7.25–7.56 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 104, 115, 116, 119, 127, 128, 129, 130, 134, 139, 145, 168; MS (EI, 70 eV): m/z (%): 473 (M + H, 100).
N), 8.3–7.9 (m, 4H, Ar-H), 7.58–7.85 (m, 5H, phenyl ring), 7.25–7.56 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 104, 115, 116, 119, 127, 128, 129, 130, 134, 139, 145, 168; MS (EI, 70 eV): m/z (%): 473 (M + H, 100).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.4–7.8 (m, 5H, phenyl ring), 8.39 (d, 2H, Ar-H), 7.94 (d, 2H, Ar-H), 7.23–7.40 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 106, 116, 117, 118, 119, 120, 122, 125, 126, 127, 128, 129, 130, 132, 134, 135, 136, 139, 148, 168; MS (EI, 70 eV): m/z (%): 446 (M + H, 100).
N), 7.4–7.8 (m, 5H, phenyl ring), 8.39 (d, 2H, Ar-H), 7.94 (d, 2H, Ar-H), 7.23–7.40 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 106, 116, 117, 118, 119, 120, 122, 125, 126, 127, 128, 129, 130, 132, 134, 135, 136, 139, 148, 168; MS (EI, 70 eV): m/z (%): 446 (M + H, 100).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.4–7.6 (m, 5H, phenyl ring), 7.97 (d, 2H, Ar-H), 7.88 (d, 2H, Ar-H), 7.24–7.41 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 104, 116, 119, 127, 128, 129, 130, 134, 135, 145, 168; MS (EI, 70 eV): m/z (%): 462 (M + H, 100).
N), 7.4–7.6 (m, 5H, phenyl ring), 7.97 (d, 2H, Ar-H), 7.88 (d, 2H, Ar-H), 7.24–7.41 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 104, 116, 119, 127, 128, 129, 130, 134, 135, 145, 168; MS (EI, 70 eV): m/z (%): 462 (M + H, 100).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.5–7.7 (m, 5H, phenyl ring), 7.90 (d, 2H, Ar-H), 7.72 (d, 2H, Ar-H), 7.09–7.48 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 116, 118, 121, 126, 127, 129, 130, 131, 138, 145, 167; MS (EI, 70 eV): m/z (%): 506 (M + H, 100).
N), 7.5–7.7 (m, 5H, phenyl ring), 7.90 (d, 2H, Ar-H), 7.72 (d, 2H, Ar-H), 7.09–7.48 (m, 3H, thiophene); 13C NMR (100 MHz, DMSO-d6): δ 116, 118, 121, 126, 127, 129, 130, 131, 138, 145, 167; MS (EI, 70 eV): m/z (%): 506 (M + H, 100).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.38–7.67 (m, 5H, phenyl ring), 7.97 (d, 2H, Ar-H), 7.75 (d, 2H, Ar-H), 7.21–7.25 (m, 3H, thiophene), 2.27 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ 103, 116, 119, 126, 127, 128, 129, 130, 134, 137, 138, 139, 145, 147, 168; MS (EI, 70 eV): m/z (%): 442 (M + H, 100).
N), 7.38–7.67 (m, 5H, phenyl ring), 7.97 (d, 2H, Ar-H), 7.75 (d, 2H, Ar-H), 7.21–7.25 (m, 3H, thiophene), 2.27 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ 103, 116, 119, 126, 127, 128, 129, 130, 134, 137, 138, 139, 145, 147, 168; MS (EI, 70 eV): m/z (%): 442 (M + H, 100).Antimicrobial activity
The antimicrobial activities of the synthesized pyrazolyl–thiazole derivatives 7b–g were evaluated against a panel of bacterial and fungal strains. The bacterial strains tested included E. coli, B. subtilis, B. megaterium, and S. aureus. The fungal strains tested included A. niger, A. oryzae, Rhizopus, and C. albicans.Antioxidant activity
The antioxidant activities of the synthesized pyrazolyl–thiazole derivatives 7b–g were evaluated using the DPPH (2,2-diphenyl-1-picrylhydrazyl) radical scavenging assay and the hydroxyl radical scavenging assay.19Computational studies
Gaussian 09 software was used to optimize the geometries and compute the electronic properties of the synthesized compounds 7b–g.20 Geometry optimizations at gas-phase with no constraints were done using the long-range corrected ωB97X-D hybrid functional along with Ahlrichs' def2-TZVPP basis set. All geometry optimizations were performed in the gas phase to focus on intrinsic molecular properties and reduce computational complexity. This approach is commonly employed in similar studies and provides reliable insights into electronic and structural properties.21 The absence of imaginary frequencies in the vibrational analysis confirmed that the optimized structures were true minima on the potential energy surface. Key computational results, including HOMO–LUMO energy values, thermodynamic properties, and absorption/emission spectra, are detailed in the results section and ESI.†The molecular docking studies were carried out in the Molegro Virtual Docker software. The validation of the molecular docking method was performed by re-docking 12 methods based on the combination of 4 scoring functions and 3 search algorithms (data show in ESI†). In each method, 10 runs, a maximum of 1500 iterations and a population of 50 poses, a search sphere of 10 Å and a grid of 0.20 Å were performed. The method was chose using an RMSD value <2.0 Å as a criterion.22 The binding energy was obtained in the PROtein binDIng enerGY prediction (PRODIGY) server and Discovery Studio BIOVIA was used to visualize the ligand–receptor interactions.
Data availability
All data supporting the findings of this study are available within the article and its ESI† files. Additional datasets generated and analyzed during the current study, including raw NMR spectra, mass spectrometry data, and detailed computational outputs, are available from the corresponding author upon reasonable request.Author contributions
Conceptualization, S. V. P.; methodology, S. K. B., S. A. K., A. A. P., R. A. M., G. M., and S. V. P.; software, T. J. P., J. O. C. J. H, J. A. A. S., and J. L. O. R.; validation, S. K. B., T. J. P., R. A. M., J. L. O. R., and S. V. P.; formal analysis, S. K. B, S. V. P.; investigation, S. K. B., T. J. P., S. A. K., A. A. P., R. A. M. E. D. A., and S. V. P.; resources, S. V. P., G. M. and E. D. A.; data curation, T. J. P. and S. V. P.; writing—original draft preparation, T. J. P., R. A. M., J. O. C. J. H., J. A. A. S.; writing—review and editing, T. J. P., J. O. C. J. H., J. L. O. R. and S. V. P.; visualization, E. D. A. and S. V. P.; supervision, R. A. M. and S. V. P.; project administration, S. V. P.; funding acquisition, G. M., E. D. A. and S. V. P. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia [Grant No. KFU242665]. T. J. P. thanks CONAHCYT for the postdoctoral fellowship [MOD.ORD.10/2023-I1200/331/2023].Notes and references
- For selected reviews and articles on antimicrobial resistance, see: (a) P. Butaye, E. V. Duijkeren, J. Prescott and S. Schwarz, Antimicrobial resistance in bacteria from animals and the environment, Vet. Microbiol., 2014, 171, 269–272 CrossRef PubMed; (b) R. Urban-Chmiel, A. Marek, D. Stępień-Pyśniak, K. Wieczorek, M. Dec, A. Nowaczek and J. Osek, Antibiotic Resistance in Bacteria-A Review, Antibiotics, 2022, 11, 1079 CrossRef CAS; (c) M. Chatzopoulou and L. Reynolds, The Role of Antimicrobial Restrictions in Bacterial Resistance Control: A Systematic Literature Review, J. Hosp. Infect., 2020, 104, 125–136 CrossRef CAS.
- (a) P. K. Chandole, T. J. Pawar, J. L. Olivares-Romero, S. R. Tivari, B. G. Lara, H. Patel, I. Ahmad, E. Delgado-Alvarado, S. V. Kokate and Y. Jadeja, Exploration of novel cationic amino acid-enriched short peptides: design, SPPS, biological evaluation and in silico study, RSC Adv., 2024, 14, 17710–17723, 10.1039/d3ra08313f; (b) S. R. Tivari, S. V. Kokate, J. L. Belmonte-Vázquez, T. J. Pawar, H. Patel, I. Ahmad, M. S. Gayke, R. S. Bhosale, V. D. Jain, G. Muteeb, E. Delgado-Alvarado and Y. Jadeja, Synthesis and Evaluation of Biological Activities for a Novel 1,2,3,4-Tetrahydroisoquinoline Conjugate with Dipeptide Derivatives: Insights from Molecular Docking and Molecular Dynamics Simulations, ACS Omega, 2023, 8, 48843–48854, DOI:10.1021/acsomega.3c05961.
- (a) S. Singh, S. Tehlan and P. Kumar Verma, Pyrazole scaffolds having promising pharmacological potential: a review, Mini-Rev. Med. Chem., 2023, 23, 2142–2165, DOI:10.2174/1389557523666230320164203; (b) M. Asif, M. Almehmadi, A. A. Alsaiari and M. Allahyani, Diverse pharmacological potential of different substituted pyrazole derivatives, Curr. Org. Synth., 2024, 21, 858–888, DOI:10.2174/0115701794260444230925095804.
- For application of thiazoles in pharmaceutical, see: (a) C. Tratrat, M. Haroun, I. Xenikakis, K. Liaras, E. Tsolaki, P. Eleftheriou, A. Petrou, B. Aldhubiab, M. Attimarad, K. N. Venugopala, S. Harsha, H. S. Elsewedy, A. Geronikaki and M. Soković, Design, Synthesis, Evaluation of Antimicrobial Activity and Docking Studies of New Thiazole-based Chalcones, Curr. Top. Med. Chem., 2019, 19, 356–375, DOI:10.2174/1568026619666190129121933; (b) I. Apostolidis, K. Liaras, A. Geronikaki, D. Hadjipavlou-Litina, A. Gavalas, M. Soković, J. Glamočlija and A. Ćirić, Synthesis and biological evaluation of some 5-arylidene-2-(1,3-thiazol-2-ylimino)-1,3-thiazolidin-4-ones as dual anti-inflammatory/antimicrobial agents, Bioorg. Med. Chem., 2013, 21, 532–539, DOI:10.1016/j.bmc.2012.10.046; (c) S. A. F. Rostom, I. M. El-Ashmawy, H. A. Abd El Razik, M. H. Badr and H. M. A. Ashour, Design and synthesis of some thiazolyl and thiadiazolyl derivatives of antipyrine as potential non-acidic anti-inflammatory, analgesic and antimicrobial agents, Bioorg. Med. Chem., 2009, 17, 882–895, DOI:10.1016/j.bmc.2008.11.035; (d) N. Y. Guerrero-Pepinosa, M. C. Cardona-Trujillo, S. C. Garzón-Castaño, L. A. Veloza and J. C. Sepúlveda-Arias, Antiproliferative activity of thiazole and oxazole derivatives: a systematic review of in vitro and in vivo studies, Biomed. Pharmacother., 2021, 138, 111495, DOI:10.1016/j.biopha.2021.111495; (e) S. Sinha, S. L. Manju and M. Doble, Chalcone-Thiazole Hybrids: Rational Design, Synthesis, and Lead Identification against 5-Lipoxygenase, ACS Med. Chem. Lett., 2019, 10, 1415–1422, DOI:10.1021/acsmedchemlett.9b00193; (f) G. A. Hampannavar, R. Karpoormath, M. B. Palkar, M. S. Shaikh and B. Chandrasekaran, Dehydrozingerone Inspired Styryl Hydrazine Thiazole Hybrids as Promising Class of Antimycobacterial Agents, ACS Med. Chem. Lett., 2016, 7, 686–691, DOI:10.1021/acsmedchemlett.6b00088.
- M. T. Chhabria, S. Patel, P. Modi and P. S. Brahmkshatriya, Thiazole: A Review on Chemistry, Synthesis and Therapeutic Importance of its Derivatives, Curr. Top. Med. Chem., 2016, 16, 2841–2862, DOI:10.2174/1568026616666160506130731.
- For application of pyrazoles in pharmaceutical, see: (a) A. Ansari, A. Ali, M. Asif and Shamsuzzaman, Review: biologically active pyrazole derivatives, New J. Chem., 2017, 41, 16–41, 10.1039/c6nj03181a; (b) H. A. M. Soliman, M. Hamed, J.-S. Lee and A. E.-D. H. Sayed, Protective effects of a novel pyrazolecarboxamide derivative against lead nitrate induced oxidative stress and DNA damage in Clarias gariepinus, Environ. Pollut., 2019, 247, 678–684, DOI:10.1016/j.envpol.2019.01.074; (c) H. A. Mohamed, Y. A. Ammar, G. A. M. Elhagali, H. A. Eyada, D. S. Aboul-Magd and A. Ragab, In Vitro Antimicrobial Evaluation, Single-Point Resistance Study, and Radiosterilization of Novel Pyrazole Incorporating Thiazol-4-one/Thiophene Derivatives as Dual DNA Gyrase and DHFR Inhibitors against MDR Pathogens, ACS Omega, 2022, 7, 4970–4990, DOI:10.1021/acsomega.1c05801; (d) O. Ebenezer, M. Shapi and J. A. Tuszynski, A Review of the Recent Development in the Synthesis and Biological Evaluations of Pyrazole Derivatives, Biomedicines, 2022, 10, 1124, DOI:10.3390/biomedicines10051124; (e) D. Becerra, R. Abonia and J.-C. Castillo, Recent Applications of the Multicomponent Synthesis for Bioactive Pyrazole Derivatives, Molecules, 2022, 27, 4723, DOI:10.3390/molecules27154723; (f) G. Ouyang, X.-J. Cai, Z. Chen, B.-A. Song, P. S. Bhadury, S. Yang, L.-H. Jin, W. Xue, D.-Y. Hu and S. Zeng, Synthesis and Antiviral Activities of Pyrazole Derivatives Containing an Oxime Moiety, J. Agric. Food Chem., 2008, 56, 10160–10167, DOI:10.1021/jf802489e.
- (a) F. Azimi, H. Azizian, M. Najafi, F. Hassanzadeh, H. Sadeghi-aliabadi, J. B. Ghasemi, M. A. Faramarzi, S. Mojtabavi, B. Larijani, L. Saghaei and M. Mahdavi, Design and synthesis of novel quinazolinone-pyrazole derivatives as potential α-glucosidase inhibitors: structure–activity relationship, molecular modeling and kinetic study, Bioorg. Chem., 2021, 114, 105127, DOI:10.1016/j.bioorg.2021.105127; (b) Y. Y. Tu, J. Sonnenberg, K. F. Lewis and C. S. Yang, Pyrazole-induced cytochrome P-450 in rat liver microsomes: an isozyme with high affinity for dimethylnitrosamine, Biochem. Biophys. Res. Commun., 1981, 103, 905–912, DOI:10.1016/0006-291X(81)90896-2.
- (a) R. Shah and P. K. Verma, Therapeutic importance of synthetic thiophene, Chem. Cent. J., 2018, 12, 137, DOI:10.1186/s13065-018-0511-5; (b) O. V. Khoroshilova, K. E. Borovkova, L. R. Nikiforova, J. V. Salmova, A. O. Taraskin, D. V. Spiridonova and A. V. Vasilyev, TfOH-promoted multichannel transformations of trifluoromethyl side chain substituted thiophene and furan families to access antimicrobial agents, New J. Chem., 2023, 47, 18492–18516, 10.1039/d3nj02990e; (c) M. Özgür, M. Yılmaz, H. Nishino, E. Ç. Avar, H. Dal, A. T. Pekel and T. Hökelek, Efficient syntheses and antimicrobial activities of new thiophene containing pyranone and quinolinone derivatives using manganese(III) acetate: the effect of thiophene on ring closure–opening reactions, New J. Chem., 2019, 43, 5737–5751, 10.1039/c9nj00054b; (d) K. K. Mak, Z. Shiming, O. Epemolu, A. T. Dinkova-Kostova, G. Wells, I. G. Gazaryan, R. Sakirolla, Z. Mohd and M. R. Pichika, Synthesis and Anti-Inflammatory Activity of 2-Amino-4,5,6,7-tetrahydrobenzo[b]thiophene-Derived NRF2 Activators, ChemistryOpen, 2022, 11, e202200181, DOI:10.1002/open.202200181.
- (a) B. Yu, S. Zhou, L. Cao, Z. Hao, D. Yang, X. Guo, N. Zhang, V. A. Bakulev and Z. Fan, Design, Synthesis, and Evaluation of the Antifungal Activity of Novel Pyrazole–Thiazole Carboxamides as Succinate Dehydrogenase Inhibitors, J. Agric. Food Chem., 2020, 68, 7093–7102, DOI:10.1021/acs.jafc.0c00062; (b) N. Harikrishna, A. M. Isloor, K. Ananda, A. Obaid and H.-K. Fun, 1,3,4-Trisubstituted pyrazole bearing a 4-(chromen-2-one) thiazole: synthesis, characterization and its biological studies, RSC Adv., 2015, 5, 43648–43659, 10.1039/C5RA04995D; (c) S. Bondock, W. Fadaly and M. A. Metwally, Synthesis and antimicrobial activity of some new thiazole, thiophene and pyrazole derivatives containing benzothiazole moiety, Eur. J. Med. Chem., 2010, 45, 3692–3701, DOI:10.1016/j.ejmech.2010.05.018.
- D. H. Dawood, M. M. Sayed, S. T. K. Tohamy and E. S. Nossier, New Thiophenyl-pyrazolyl-thiazole Hybrids as DHFR Inhibitors: Design, Synthesis, Antimicrobial Evaluation, Molecular Modeling, and Biodistribution Studies, ACS Omega, 2023, 8, 39250–39268, DOI:10.1021/acsomega.3c04736.
- R. Vanjari, T. Guntreddi, S. Kumar and K. N. Singh, Sulphur promoted C(sp3)–C(sp2) cross dehydrogenative cyclisation of acetophenone hydrazones with aldehydes: efficient synthesis of 3,4,5-trisubstituted 1H-pyrazoles, Chem. Commun., 2015, 51, 366–369, 10.1039/c4cc08210a.
- D. Lim and N. C. J. Strynadka, Structural basis for the β lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus, Nat. Struct. Mol. Biol., 2002, 9, 870–876, DOI:10.1038/nsb858.
- H. Kishida, S. Unzai, D. I. Roper, A. Lloyd, S. Y. Park and J. R. H. Tame, Crystal Structure of Penicillin Binding Protein 4 (dacB) from Escherichia coli, both in the Native Form and Covalently Linked to Various Antibiotics, Biochemistry, 2006, 45, 783–792, DOI:10.1021/bi051533t.
- E. Sauvage, C. Duez, R. Herman, F. Kerff, S. Petrella, J. W. Anderson, S. A. Adediran, R. F. Pratt, J. Frère and P. Charlier, Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide, J. Mol. Biol., 2007, 371, 528–539, DOI:10.1016/j.jmb.2007.05.071.
- T. Y. Hargrove, L. Friggeri, Z. Wawrzak, A. Qi, W. J. Hoekstra, R. J. Schotzinger, J. D. York, F. P. Guengerich and G. I. Lepesheva, Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis, J. Biol. Chem., 2017, 292, 6728–6743, DOI:10.1074/jbc.M117.778308.
- M. V. Keniya, M. Sabherwal, R. K. Wilson, M. A. Woods, A. A. Sagatova, J. D. A. Tyndall and B. C. Monk, Crystal Structures of Full-Length Lanosterol 14α-Demethylases of Prominent Fungal Pathogens Candida albicans and Candida glabrata Provide Tools for Antifungal Discovery, Antimicrob. Agents Chemother., 2018, 62, e01134, DOI:10.1128/AAC.01134-18.
- G. G. Mandawad, R. D. Kamble, S. V. Hese, R. A. More, R. N. Gacche, K. M. Kodam and B. S. Dawane, An efficient synthesis of isoxazoline libraries of thiophene analogs and its antimycobacterial investigation, Med. Chem. Res., 2014, 23, 4455, DOI:10.1007/s00044-014-1016-y.
- (a) S. S. Mahurkar, S. S. Makone and R. A. More, An efficient and recyclable catalyst for synthesis of 1,3-diphenyl-3-(phenyl thio)propan-1-one derivatives and their antibacterial evaluation, Chem.-Biol. Interact., 2019, 9, 277–284 CAS; (b) R. More, G. Sanap, A. Siddiqui, S. Dhutekar, S. Patil, V. Diwan, M. Lokare, A. Ingle and P. Maske, Antibacterial, antioxidant, phytochemicals evaluation of five Indian medicinal plants, World J. Pharm. Res., 2018, 7, 892–898 Search PubMed.
- R. A. More, G. B. Sanap, M. A. Karale, Y. P. Sarnikar and R. N. Gacche, Antioxidant and cytotoxicity profile of the selected alcoholic beverages widely consumed in the Maharashtra State of India, Indian J. Public Health Res. Dev., 2020, 11, 607–612 Search PubMed.
- (a) A. D. Becke, Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functionals, J. Chem. Phys., 1997, 107, 8554–8560, DOI:10.1063/1.475007; (b) Q. Wu and W. Yang, Empirical correction to density functional theory for van der Waals interactions, J. Chem. Phys., 2002, 116, 515–524, DOI:10.1063/1.1424928; (c) S. Grimme, Semiempirical GGA-type density functional constructed with a long-range dispersion correction, J. Comput. Chem., 2006, 27, 1787–1799, DOI:10.1002/jcc.20495; (d) J.-D. Chai and M. Head-Gordon, Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620, 10.1039/b810189b.
- (a) L. K. Zaitri and S. M. Mekelleche, DFT and TD-DFT study on quadratic NLO response and optoelectronic activity in novel Y-shaped imidazole-based push-pull chromophores, J. Mol. Model., 2021, 27, 136, DOI:10.1007/s00894-021-04764-7; (b) A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648, DOI:10.1063/1.464913; (c) C. Lee, W. Yang and R. G. Parr, Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785, DOI:10.1103/PhysRevB.37.785.
- B. A. Hernández Alba, E. Galicia Galicia, R. Retana Ugalde, J. E. Mendieta Wejebe and J. A. Alvarado Salazar, In silico screening based on chemoinformatics and molecular docking of trypanothione reductase inhibitors from Trypanosoma cruzi, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-2cl8l.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06228k |
| This journal is © The Royal Society of Chemistry 2024 |