Open Access Article
Open Access ArticleDiscrimination of mongoose hair from domestic cattle hair, human hair, and synthetic fiber using FTIR spectroscopy and chemometric analysis: a rapid, cost-effective, and field-deployable tool for wildlife forensics†
Shinta Ann Josea,
Kalaiyarasan Boopathy Thiyagarajan *a,
Chanthini Baskarb,
Rajinder Singh
*a,
Chanthini Baskarb,
Rajinder Singh c,
Dhayanithi Vasanthakumaria and
A. Udhayana
c,
Dhayanithi Vasanthakumaria and
A. Udhayana
aAdvanced Institute for Wildlife Conservation, Tamil Nadu Forest Department, Vandalur, Chennai, Tamil Nadu 600 048, India. E-mail: t.biokalai@gmail.com; aiwcrte@tn.gov.in
bSchool of Electronics Engineering, Vellore Institute of Technology, Chennai, Tamil Nadu 600 127, India
cDepartment of Forensic Science, Punjabi University, Patiala, Punjab 147 002, India
First published on 19th November 2024
Abstract
Mongoose hair is used to prepare fine brushes, which increases the demand for mongooses to be poached from the wild and brutally bludgeoned to death. Mongooses were listed as Schedule I species under the Indian Wildlife (Protection) Act 1972. Species identification of wildlife case-related samples is necessary to convict a person under this legislation. Microscopy and DNA-based techniques are commonly used to identify mongoose hair in seized brushes. However, in painting brushes, the roots, and the lower part of the hair are mostly trimmed, and only the upper part is used to make the brushes. In addition, brushes are often prepared with mixed hair from mongoose, domestic cattle, human hair, and synthetic fibre. Therefore, the identification of mongoose hair by microscopy and DNA-based techniques is restricted due to the lack of complete strands of hair and the absence of hair roots. Therefore, there is an urgent need to develop an alternative methodology for the identification of mongoose hair from seized articles. FTIR spectroscopy for forensic analysis has gained significant attention over the years because of its sensitivity, specificity, and non-destructive nature. The present study aimed to discriminate Indian grey mongoose (Herpestes edwardsii) hair from domestic cattle hair (domestic water buffalo and domestic cow), human hair, and synthetic fiber based on their chemical composition using FTIR spectroscopy and chemometric analysis. We have taken hair from four individuals for each species, namely Indian grey mongoose, domestic cattle, human hair, and synthetic fibre. The FTIR spectrum was recorded, and partial least-squares discriminant analysis (PLS-DA) was used to discriminate hair and synthetic fiber. The established PLS-DA model showed an R-square value and an RMSE (root mean square error) value of 0.9 and 0.13 respectively. Our preliminary findings have shown that FTIR spectroscopy combined with chemometrics can quickly discriminate Indian grey mongoose hair, domestic cattle hair, human hair, and synthetic fiber, providing crucial evidence for judicial proceedings.
1. Introduction
India is a megadiverse country, home to 7–8% of the world's flora and fauna. It accommodates more than 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of plants and 91
000 species of plants and 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of animals.1 Wildlife in India is protected by the Wildlife (Protection) Act 1972 (WPA), which prohibits the trade of more than 1800 species of wild animals, plants, and their derivatives.2–4 Despite strong legal protection, poaching and illegal trade have been increasing over the years, posing a significant threat to several iconic species and pushing them to the brink of extinction. Mammals are frequently poached for their skin, meat, hair, and wool, which are widely traded for different purposes, such as fur coats, leather purses, belts, shoes, carvings, etc.5,6
000 species of animals.1 Wildlife in India is protected by the Wildlife (Protection) Act 1972 (WPA), which prohibits the trade of more than 1800 species of wild animals, plants, and their derivatives.2–4 Despite strong legal protection, poaching and illegal trade have been increasing over the years, posing a significant threat to several iconic species and pushing them to the brink of extinction. Mammals are frequently poached for their skin, meat, hair, and wool, which are widely traded for different purposes, such as fur coats, leather purses, belts, shoes, carvings, etc.5,6
In recent years, mongooses (Herpestes sp.) have been poached illegally and their hair is used to produce painting and shaving brushes.7,8 The various seizures made by law enforcement agencies indicate that poaching of these animals is still regular and affects the conservation of these animals.7 For example, in 2023, the Andhra Pradesh Forest Department seized more than 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 paint brushes from several premises allegedly used by dealers in Hyderabad city, and forensic analysis confirmed that the paint brushes were prepared with Indian grey mongoose (Herpesten edwardsi);9 The Tamil Nadu Wildlife Crime Control Bureau seized 14
000 paint brushes from several premises allegedly used by dealers in Hyderabad city, and forensic analysis confirmed that the paint brushes were prepared with Indian grey mongoose (Herpesten edwardsi);9 The Tamil Nadu Wildlife Crime Control Bureau seized 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mongoose hair paint brushes from various districts of Tamil Nadu, such as Chennai, Coimbatore, and Madurai.10 The Wildlife Crime Control Bureau (WCCB), India, has conducted enforcement operations in seven Indian states from 2002 to 2021, leading to the recovery of more than 1.6 lakh mongoose hair paint brushes and 292.7 kg of raw mongoose hair.11 Additionally, the Centre for Wildlife Forensic Sciences, Advanced Institute for Wildlife Conservation, Tamil Nadu Forest Department, Vandalur, Chennai, has received several cases involving the use of mongoose hair. A study documented that there are seven mongoose species in India7 namely (1) Herpestes edwardsii, (2) Herpestes smithii, (3) Herpestes urva, (4) Herpestes palustris, (5) Herpestes javanicus, (6) Herpestes vitticolis and (7) Herpestes brachyurus. All mongoose species are protected under the Indian Wildlife (Protection) Act 1972 and classified as Schedule I species,12 and the International Union Conservation of Nature (IUCN)13 categorized mongoose species as ‘Least Concern’.
000 mongoose hair paint brushes from various districts of Tamil Nadu, such as Chennai, Coimbatore, and Madurai.10 The Wildlife Crime Control Bureau (WCCB), India, has conducted enforcement operations in seven Indian states from 2002 to 2021, leading to the recovery of more than 1.6 lakh mongoose hair paint brushes and 292.7 kg of raw mongoose hair.11 Additionally, the Centre for Wildlife Forensic Sciences, Advanced Institute for Wildlife Conservation, Tamil Nadu Forest Department, Vandalur, Chennai, has received several cases involving the use of mongoose hair. A study documented that there are seven mongoose species in India7 namely (1) Herpestes edwardsii, (2) Herpestes smithii, (3) Herpestes urva, (4) Herpestes palustris, (5) Herpestes javanicus, (6) Herpestes vitticolis and (7) Herpestes brachyurus. All mongoose species are protected under the Indian Wildlife (Protection) Act 1972 and classified as Schedule I species,12 and the International Union Conservation of Nature (IUCN)13 categorized mongoose species as ‘Least Concern’.
In general, microscopy was used to identify mongoose hair in the seized brushes. The Wildlife Institute of India (WII)14 has demonstrated the identification of Indian grey mongoose from guard hair. The Zoological Survey of India (ZSI)15 and the Wildlife Crime Control Bureau (WCCB)16 have reported microscopic structures of dorsal guard hairs for identification of mongoose species. However, the ZSI report lacks detailed descriptions of microscopic hair characters, and the WCCB report has no microscopic images. As a result, the practical application of these findings for forensic case analysis is limited. To address this issue, Sahajpal et al. (2009)7 demonstrated the identification of four mongoose species (H. edwardsii, H. smithii, H. palustris, and H. urva) by microscopic examination of the pattern of the dorsal guard hair band and discriminate functional analysis (DFA). However, all studies have exhibited the identification of mongoose species by microscopy examination of the dorsal guard hair band pattern. Still, hairs from other regions of the mongoose body (e.g., tail) are also used in brush manufacturing. Therefore, further studies must be employed and compared with the reference data to overcome this limitation. Moreover, the identification of species from hair is subject to several factors, such as the availability of complete strands of hair, the expertise of the forensic examiner, the quality and quantity of the hair, the availability of known samples (reference hair samples) and the instrumentation used for analysis.17–21 The results of microscopy analysis may sometimes not be conclusive; therefore, further study is needed to identify the species.21
Next, DNA analysis is the most common method used to detect hair at the molecular level.22 Before analysis, a forensic analyst classifies the hair based on the morphology of the hair roots to apply the appropriate analytical methods.23,24 In forensic analysis, hair samples with anagen follicular tissue are ideal for DNA analysis.25 However, up to 95% of hair collected as forensic evidence has been documented to be telogen, which is completely keratinized and contains a small amount of cellular material.26,27 To overcome this issue, Bourguignon et al. (2008)28 classified telogen hairs found at the crime scene such as type 1, without visible soft tissue; type 2, with a small amount of soft tissue attached; and type 3, with a large amount of soft tissue attached.
In most cases, the seized brushes received for forensic analysis were type 1 and DNA extraction; subsequent analysis is very challenging.7,22 However, few studies have reported the identification of species on hair shafts, though the success rate is subject to various parameters, including the content of keratin.29 In addition to this, during our forensic examination of the brushes received for analysis, we observed that the brushes prepared with mongoose hair were often mixed with hair from other domestic cattle, such as domestic water buffalo (Bubalus bubalis), domestic cow (Bos taurus), human hair, and synthetic fibers. Therefore, the identification of mongoose hair from seized brushes is a very tough task using microscopy and DNA-based techniques due to three primary reasons: (1) lack of complete hair structure (hair strands), (2) the hair samples received for analysis are telogen, and (3) the seized brushes often mixed with hair from domestic cattle, human hair, and synthetic fibres. Therefore, it is a fact that identifying mongoose hair from seized brushes and discriminating it from counterfeit (i.e., hair from domestic cattle, humans, and synthetic fibres) is a major concern, and there is an urgent need to develop robust techniques/methods to distinguish mongoose hair from counterfeit materials.
The application of vibrational spectroscopy for forensic purposes has gained significant attention over the years because of its sensitivity, specificity, and non-destructive nature.30 The infrared spectrum displays the vibrational characteristics of a sample based on the different absorption frequencies of the individual functional groups present in the sample (e.g., based on the chemical composition of the sample).30 In light of this, FTIR is used to analyse various materials in forensic science, including biomedical samples,31 paint,32 fingerprints,33,34 and ink.35 Furthermore, studies have documented that the identification of species from hair using Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy (ATR-FTIR).36–40 Notably, Manheim et al. (2015)41 described the identification of human, animal, and synthetic fiber by ATR-FTIR spectroscopy. Boll and co-workers (2017)42 researched the use of ATR-FTIR spectroscopy for the classification of dyed and non-dyed hairs; Bhatia et al. (2024)43 showed the identification of hairs from Royal Bengal Tiger (Panthera tigris tigris), Indian Leopard (Panthera pardus fusca) and snow Leopard (Panthera uncia) using the ATR-FTIR spectroscopic technique in combination with chemometric analysis. Hence, previous studies have well documented that species, biological, and non-biological articles can be identified by ATR-FTIR spectroscopy and chemometric analysis. Therefore, we have been motivated to employ FTIR spectroscopy combined with chemometric tools for the identification of mongoose hair from counterfeit hair (i.e., hair from domestic cattle and human).
In light of this, the current study demonstrates FTIR spectroscopy combined with chemometric tools for the identification of Indian grey mongoose hair (Herpestes edwardsii) and discriminating it from domestic cattle, such as domestic water buffalo (Bubalus bubalis), domestic cow (Bos taurus), human hair, and synthetic fiber. We selected the Indian grey mongoose for this study because it is the most common mongoose found in southern India and its proximity to human habitation.9 As a result, Indian grey mongooses are often poached and their hair is used for brush preparation. The results of this study will play an important role in the wildlife forensics in identifying Indian grey mongoose hair from seized articles.
2. Materials and methods
2.1 Collection of Indian grey mongoose hair (voucher sample)
We collected 100 hair samples (25 hair strands from each individual) from four Indian grey mongooses in our Morphometry Laboratory at Advanced Institute for Wildlife Conservation (AIWC), Chennai, Tamil Nadu, India. No sample was collected from live animals.2.2 Collection of domestic cattle hair samples
From the Kolapakkam Farm house, Chennai, India, approximately 25–30 tail hairs were collected from each of the four individuals of domestic water buffalo (Bubalus bubalis) and domestic cow (Bos taurus). Before FTIR spectroscopy analysis, the species was confirmed by molecular analysis.2.3 Human hair collection
Human hair samples were obtained for this study from four volunteers willing to donate a small amount of scalp hair. For all hair donors, only their age and biological sex were recorded; donor names and all other personally identifiable information were not collected. The donated hairs were visibly found to be natural (non-dyed) and black.2.4 Synthetic fiber
In this study, we used polyethylene terephthalate fiber. For FTIR analysis, we chopped small pieces of fiber and prepared the samples as described below.2.5 Sample preparation for FTIR spectroscopy analysis
About 25 hair strands were taken from each individual, chopped into small fragments and finely powdered using a sterile mortar and pestle. The powdered samples were then thoroughly mixed with 1.0 g of KBr powder (Merck Millipore, IR spectroscopy grade) and a 13 mm KBr pellet was prepared. The mortar and pestle were cleaned with acetone (analytical grade) and then dried before and after the preparation of the sample to avoid cross-contamination.2.6 Data analysis
3. Results and discussion
3.1 FTIR spectroscopy analysis for Indian grey mongoose hair, domestic cattle hair, human hair, and synthetic fiber
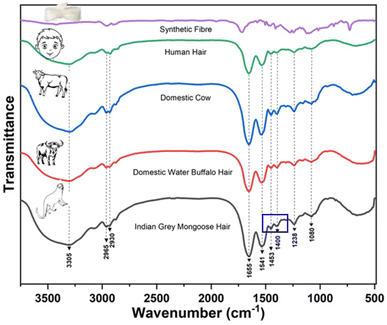
Identifying seized wild animal parts is a great challenge due to the many species involved in the illegal wildlife trade and the various forms of seized items. These articles are often made from materials derived from domestic animals and other sources (or materials); therefore, the species identification more complex using conventional methods.46 For example, CITES has documented that wild mammal skins (leather, fur) and products (hair, wool) are widely traded, often mixed with counterfeit furs, making the identification of wildlife products challenging.47 Therefore, there is an urgent need to develop alternative methods to identify intra- and interspecies variations in wild and domestic species. This will enable a better implementation of national and international laws to effectively control wildlife trade.In this study, FTIR spectroscopy was used to discriminate Indian grey mongoose hair, domestic cattle hair, human hair, and synthetic fiber. Hence, we recorded the FTIR spectrum for the hair of the Indian grey mongoose, domestic cattle hairs, human hair and synthetic fibres. The findings revealed that we could visually distinguish synthetic fiber from hair (all the hair used in this study). The results suggested that through FTIR analysis, visually can be confirm seized brushes contain hair or any other materials (including other than hair origin). On another hand, a slight spectral difference, especially in the amide-II region, was observed in the hair of Indian grey mongooses compared to domestic animals (Fig. 1). Similar results were observed in the FTIR spectrum recorded for intra-species (different individuals from the same species) (Supplementary Fig. S1†). The primary biochemical component of animal fibres is keratin, a fibrous structural protein widely found in human and animal organs, including the epidermis, hoof, horn, hairs, and feathers.48 Gallagher (2009)49 detailed the protein composition of some animal fibers, while Espinoza et al. (2009),50 Manheim et al. (2016),41 Agarwal et al. (2019),51 Wang et al. (2021),48 and Xu et al. (2022)52 characterized hair proteins using spectroscopy techniques and described their chemical composition and corresponding wavenumbers (Table 1).
| S. No. | Wavelength (cm−1) | Assignment | Source | Ref. |
|---|---|---|---|---|
| 1 | 3100–3500 | Amide A (overlap of amide hydrogen N–H region and O–H region) | N–H stretching vibrations | 48–53 |
| 2 | 2850–3000 | ν(CH3) asymmetric; ν(CH3) symmetric | Saturated and unsaturated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) H, C–H region H, C–H region |
|
| 4 | 1600–1700 | Amide I (carbonyl oxygen C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O region) O region) |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations O stretching vibrations |
|
| 5 | 1500–1572 | Amide II (amide hydrogen N–H bond) | N–H formation vibrations and C–N stretching vibrations | |
| 6 | 1361–1470 | Amide III δ(CH2) (CH3) deformation | C–N region | |
| 7 | 1210–1290 | Amide III (carbonyl oxygen C–O region) | C–N and C–O stretching vibrations, N–H and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N bending vibrations C–N bending vibrations |
|
| 8 | 998–1100 | Sulphur oxygen (S–O vibration, C–H bond) | Cystine oxides |
In our study, the peak observed at 3305 cm−1 is related to the organic material amide A (primary amide) from the overlap of the N–H and O–H hydrogen amide regions (NH stretching vibrations). Based on previous studies on keratin, the peaks at 2965 cm−1 and 2930 cm−1 represent asymmetric and symmetric ν(CH3), respectively, and amide I is identified at 1655 cm−1 which corresponds to the stretching vibrations of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond (amide group). The detected peak at 1541 cm−1 is the amide II band that arises from vibrations of NH bending and C–N stretching, and the narrow peaks at 1453 cm−1 and 1400 cm−1 are due to the deformation in δ(CH2) (CH3) and δ(CH3) respectively. Similarly, a weak peak at 1238 cm−1 illustrates amide III, a combination of stretching vibrations of the C–N and C–O bonds and bending vibrations of the N–H and O
O bond (amide group). The detected peak at 1541 cm−1 is the amide II band that arises from vibrations of NH bending and C–N stretching, and the narrow peaks at 1453 cm−1 and 1400 cm−1 are due to the deformation in δ(CH2) (CH3) and δ(CH3) respectively. Similarly, a weak peak at 1238 cm−1 illustrates amide III, a combination of stretching vibrations of the C–N and C–O bonds and bending vibrations of the N–H and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N bonds. As reviewed and documented by Espinoza et al., 2009, the region between 1200 to 1000 cm−1 is associated with vibrations of the sulfur–oxygen groups of keratins, and hence the peak at 1080 cm−1 is of νs (SO) from cysteine oxide.50 Based on the FTIR spectra recorded for hair samples, we found no visual difference in the spectra (except Indian grey mongoose hair), as all hair is made of keratin proteins. Additionally, relying on visible observation of FTIR spectra can lead to observer bias. To address these issues, we used chemometric analysis to distinguish Indian grey mongoose hair from domestic cattle hair, human hair, and synthetic fiber.
C–N bonds. As reviewed and documented by Espinoza et al., 2009, the region between 1200 to 1000 cm−1 is associated with vibrations of the sulfur–oxygen groups of keratins, and hence the peak at 1080 cm−1 is of νs (SO) from cysteine oxide.50 Based on the FTIR spectra recorded for hair samples, we found no visual difference in the spectra (except Indian grey mongoose hair), as all hair is made of keratin proteins. Additionally, relying on visible observation of FTIR spectra can lead to observer bias. To address these issues, we used chemometric analysis to distinguish Indian grey mongoose hair from domestic cattle hair, human hair, and synthetic fiber.
3.2 Chemometric modelling for discrimination of Indian grey mongoose hair, domestic cattle hair, human hair, and synthetic fiber
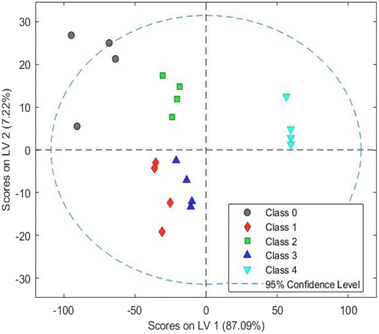
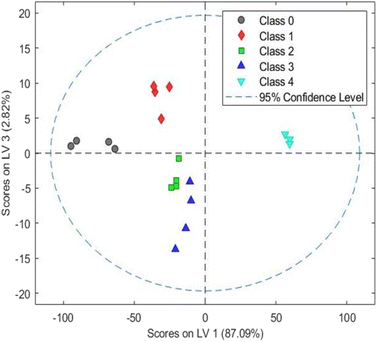
The PLS-discriminant analysis (PLS-DA) method is commonly used for predictive, descriptive modelling, and discriminative variable selection.54 PLS-DA has various applications in various fields, such as forensic science, banking, medical diagnosis, food analysis, metabolomics, and soil science.54–58 Keeping in view, this study used PLS-DA analysis to differentiate between Indian grey mongoose hair, domestic cattle hair, human hair, and synthetic fiber based on their intensity of IR radiation absorption by biochemical composition of each hair sample.The PLS-DA model was calculated as three latent variables (LV), namely LV1 (87.09%), LV2 (7.22%), and LV3 (2.82%); consequently, it contributed to the separation of the five groups. Score plots between LVs are shown in Fig. 2 and 3. The plot clearly showed species wise cluster and differentiation; the latent variables effectively highlighted the differences in the FTIR spectra recorded for each hair sample. The developed model has shown an R-square value of 0.9 and an RMSE value (root mean square error) of 0.13. Hence, the PLS-DA analysis has successfully discriminated the Indian grey mongoose hair, domestic cattle hair, human hair, and synthetic fiber based on the IR transmittance variations of the chemical compositions present in the hair samples (especially keratin protein).
Proteins are made of hundreds of amino acids folded into a well-defined structure that is stabilized by thousands of interactions. Infrared (IR) spectroscopy can reveal the structure of proteins via vibrational resonances from the polypeptide backbone and/or side chains. Such resonances depend on the protein structure and local interactions (such as hydrogen bonding between amino acids).59 In light of this, studies documented that FTIR spectroscopy analysis of polypeptides and proteins reveals nine characteristic IR absorption bands: amide A, B, and I–VII. The amide I and II bands are the most prominent vibrational bands of the protein backbone. The amide I band (1700–1600 cm−1) is the most sensitive spectral region to the protein's secondary structural components and is primarily due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch vibrations of the peptide linkages. On the other hand, the amide II band is mainly derived from in-plane NH bending and the CN stretching vibration.48–53
O stretch vibrations of the peptide linkages. On the other hand, the amide II band is mainly derived from in-plane NH bending and the CN stretching vibration.48–53
In this regard, the current study aimed to discriminate the Indian grey mongoose hair, domestic cattle hairs, human hair, and synthetic fibres based on their biochemical composition using FTIR analysis. The FTIR analysis of hair showed the characteristic peaks at 1655 cm−1 (amide I), 1541 cm−1 (amide II), and peaks at 1453 cm−1 and 1400 cm−1 are due to deformation in δ(CH2) (CH3) and δ(CH3) respectively.60 These spectral peaks were attributed to hair keratin protein (abundant in the outer layer of skin, hair, and nails) and similar peaks were observed in all the hair samples, excluding slight differences in Indian grey mongoose hair. Studies reported that in the protein analysis through IR spectroscopy, the amide I band is sensitive to the secondary structure and is not strongly influenced by side chains of proteins. In contrast, the IR absorption properties vary in the α-helices, β-sheets, random coils, and loops (i.e., protein secondary structure elements) due to hydrogen bonding of different strength from one peptide –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group to H–N– group of a neighboring amino acid. The strength of the hydrogen bond plays a vital role in the absorption frequency of the C
O group to H–N– group of a neighboring amino acid. The strength of the hydrogen bond plays a vital role in the absorption frequency of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibration in the amide I, corresponding to different secondary structure segments within the protein.61–63
O vibration in the amide I, corresponding to different secondary structure segments within the protein.61–63
Considering that keratin protein is present in all the hair used in this study, the amino acid compositions of hairs vary from species to species. Subsequently, the hydrogen bonding patterns change within keratin proteins (keratin in different hair types). As a result, it impacted in IR absorption properties of keratin proteins present in each hair type. The small variations in amide I and amide II regions of hair cannot be identified by the naked eye. However, PLS-DA analysis identified the difference in IR absorption properties in the amide I, amide II, and amide III regions of hair and discriminated.63 At this movement, the current study demonstrated discrimination of mongoose hair, domestic cattle, human hair, and synthetic fiber using FTIR spectroscopy combined with chemometric analysis.
However, the number of samples was limited to only four in the current study; therefore, we were unable to validate the results. The study showed that FTIR spectroscopy combined with chemometric analysis could discriminate Indian grey mongoose hair, domestic cattle (domestic water buffalo and domestic cow), synthetic fiber, and human hair. The results indicate that this method can be used to identify Indian grey mongoose hair from seized articles and can discriminate it from domestic cattle hair, human hair, and synthetic fiber.
Furthermore, it is believed that FTIR spectroscopy combined with chemometric analysis will facilitate the rapid identification of Indian grey mongoose from seized articles and will be cost-effective compared to molecular techniques. Furthermore, creating an FTIR spectral library for Indian grey mongoose hair could be beneficial, as it could be utilized in the field, and border force and law enforcement officials can use it to identify mongoose hair in seized articles. This study showed positive signs for the identification of mongoose hair in seized articles, including mixed mongoose hair from domestic cattle, human hair, and synthetic fiber. However, to enhance the forensic applications of this methodology, additional studies should be performed to validate it with a large number of hair samples, which will facilitate the construction of a comprehensive classification and discriminant model.
Our study recorded the FTIR spectra of samples made with potassium bromide (KBr) pellets. As a result, we used a fine powder of 25 hair samples to record the spectra of one sample (in triplicate). Due to the non-availability of hair samples, we are unable to use a high volume of samples for this study. Consequently, the data generated are inadequate to validate this methodology. Furthermore, India is home to seven species of mongoose; our study specifically focused on identifying Indian grey mongoose hair. Therefore, future studies should aim to identify and differentiate the other mongoose species. Despite these limitations, our proof-of-concept study marks a significant advance in species discrimination based on hair analysis. The results obtained from this study will have practical applications in wildlife forensic investigations.
4. Conclusions
In wildlife forensics, discriminating or identifying individual species from an unknown sample is necessary to convict a person under the law. The current study attempted to identify Indian grey mongoose hair and distinguish it from domestic cattle hair, such as domestic water buffalo, domestic cow, human hair, and synthetic fiber, using FTIR spectroscopy combined with chemometric analysis. The preliminary results of this study documented that FTIR spectroscopy, combined with chemometric analysis, is a powerful tool for discriminating Indian grey mongoose hair from other animal hair and has great potential for forensic applications. However, the current study is carried out with a minimum sample size; as a result, we were unable to validate the results. Therefore, to apply these methods to forensic practice, we will improve the experimental setup with a high volume of samples in the following research to resolve the problem stated above.Ethical approval
The Principal Chief Conservator of Forests & Chief Wildlife Warden, Tamil Nadu Forest Department, Government of Tamil Nadu, has permitted to use wild animal hair samples for this study. Human hair samples were collected from volunteers after the project scientist explained the purpose of the hair collection and the importance of the project. Domestic cattle hair was collected from the farm in the presence of the Institute's veterinary doctor, and the project scientist explained the purpose of the hair collection to the farm owner.Data availability
All of the data generated in this study is presented in this manuscript.Author contributions
SAJ: conducted all the experiments, analyzed the FTIR data, and prepared the graphs. CB: chemometric analysis. RS: assisted in discussions and edited the manuscript. KBT: developed the idea and research methodology, analyzed the data, drafted the original manuscript and supervised. DV: logistic support. AU: funding arrangements. All authors have read and approved the manuscript for publication.Conflicts of interest
The authors declare that they have no conflicts of interest. The authors are solely responsible for the content and writing of this manuscript.Acknowledgements
The authors would like to express their gratitude to the Government of Tamil Nadu for their financial support under the ‘Modernization of Forest Force’ scheme. The authors would also like to acknowledge Ms M. Prema, Laboratory Assistant at AIWC, for her assistance in sample preparations. Furthermore, the authors thank all human volunteers who provided hair samples for this study.References
- Biodiversity and Conservation, https://ebooks.inflibnet.ac.in/esp03/chapter/current-status-of-biodiversity-in-india/, accessed 20 May 2024 Search PubMed.
- L. Wilson-Wilde, Forensic Sci. Med. Pathol., 2010, 6, 221–222 CrossRef PubMed.
- M. Anagnostou and B. Doberstein, Ambio, 2022, 51, 1615–1631 CrossRef PubMed.
- M. GoI, Facc Minist. Environ., 1972, 1–218 Search PubMed.
- A. K. Rana and N. Kumar, Biodivers. Conserv., 2023, 32, 1473–1491 CrossRef PubMed.
- A. Mozer and S. Prost, Forensic Sci. Int. Anim. Environ., 2023, 3, 100064 CrossRef.
- V. Sahajpal, S. P. Goyal, R. Raza and R. Jayapal, Sci. Justice, 2009, 49, 205–209 CrossRef PubMed.
- Wildlife Trust of India, https://www.wti.org.in/news/trade-in-mongoose-hair-illegal-from-today, accessed 20 May 2024 Search PubMed.
- Wildlife Trust of India, https://www.wti.org.in/news/huge-seizures-of-mongoose-hair-brushes-in-hyderabad, accessed 20 May 2024 Search PubMed.
- The Hindu, https://www.thehindu.com/news/national/tamil-nadu/tamil-nadu-wildlife-crime-control-bureau-seizes-14000-paint-brushes-made-of-mongoose-hair/article67238995.ece, accessed 20 May 2024 Search PubMed.
- Wildlife Trust of India, https://www.wti.org.in/news/massacre-of-the-mongoose-operation-to-dismantle-trade-in-mongoose-hair-brushes, accessed 20 May 2024 Search PubMed.
- India Code, https://www.indiacode.nic.in/bitstream/123456789/1726/1/a1972-53.pdf, accessed 20 May 2024 Search PubMed.
- IUCN Red List of Threatened Species, https://www.iucnredlist.org/search?query=mongoose%26searchType=species, accessed 20 May 2024 Search PubMed.
- A. Bahuguna, V. Sahajpal, S. P. Goyal, S. K. Mukherjee and V. Thakur, Species Identification from Guard Hair of Selected Indian Mammals, Wildlife Institute of India, 2010 Search PubMed.
- M. Kamalakannan, C. Venkatraman and K. Chandra, Tricho-taxonomy of indian mammals: carnivores, artiodactyls, primates, giant squirrels, lagomorphs and pangolins, Zool. Survey India, 2019, 1–227 Search PubMed.
- Wildlife Crime Control Bureau, A Manual on Wildlife Species in Trade, 2011, http://wccb.gov.in/WriteReadData/userfiles/file/AManualonWildlifeSpeciesintrade.pdf, accessed 21 May 2024 Search PubMed.
- J. K. De, S. Chakraborty and R. Chakraborty, Mammalia, 1998, 62, 285–296 CrossRef.
- A. Linacre and S. S. Tobe, Investig. Genet., 2011, 2, 1–9 CrossRef PubMed.
- M. S. Dahiya and S. K. Yadav, J. Forensic Res., 2013, 4, 178 Search PubMed.
- L. A. Hausman, Sci. Mon., 1930, 30, 258–277 Search PubMed.
- Z. Zafarina and S. Panneerchelvam, Malays. J. Med. Sci. MJMS, 2009, 16, 35 Search PubMed.
- V. Sahajpal and S. P. Goyal, DNA Fingerprinting: Advancements and Future Endeavors, 2018, 89–127 Search PubMed.
- Z. Liu, H. Simayijiang, Q. Wang, J. Yang, H. Sun, R. Wu and J. Yan, Int. J. Legal Med., 2023, 137, 613–633 CrossRef PubMed.
- S. L. Koch, S. R. Tridico, B. A. Bernard, M. D. Shriver and N. G. Jablonski, Am. J. Hum. Biol., 2020, 32, e23316 CrossRef PubMed.
- E. M. Brooks, M. Cullen, T. Sztydna and S. J. Walsh, Aust. J. Forensic Sci., 2010, 42, 115–122 CrossRef.
- J. Edson, E. M. Brooks, C. McLaren, J. Robertson, D. McNevin, A. Cooper and J. J. Austin, Forensic Sci. Int. Genet., 2013, 7, 180–188 CrossRef CAS PubMed.
- C. A. Linch, J. Forensic Sci., 2009, 54, 346–349 CrossRef CAS PubMed.
- L. Bourguignon, B. Hoste, T. Boonen, K. Vits and F. Hubrecht, Forensic Sci. Int. Genet., 2008, 3, 27–31 CrossRef CAS PubMed.
- K. S. Grisedale, G. M. Murphy, H. Brown, M. R. Wilson and S. K. Sinha, Int. J. Legal Med., 2018, 132, 107–115 CrossRef PubMed.
- J. Manheim, Honors thesis, University at Albany, 2015.
- S. G. Kazarian and K. L. A. Chan, Biochim. Biophys. Acta, Biomembranes, 2006, 1758, 858–867 CrossRef CAS.
- C. Muehlethaler, G. Massonnet and P. Esseiva, Forensic Sci. Int., 2011, 209, 173–182 CrossRef CAS PubMed.
- M. Szafarska, M. Woźniakiewicz, M. Pilch, J. Zięba-Palus and P. Kościelniak, J. Mol. Struct., 2009, 924, 504–513 CrossRef.
- N. J. Crane, E. G. Bartick, R. S. Perlman and S. Huffman, J. Forensic Sci., 2007, 52, 48–53 CrossRef CAS PubMed.
- W. Dirwono, J. S. Park, M. R. Agustin-Camacho, J. Kim, H.-M. Park, Y. Lee and K.-B. Lee, Forensic Sci. Int., 2010, 199, 6–8 CrossRef CAS.
- K. Virkler and I. K. Lednev, Anal. Chem., 2009, 81, 7773–7777 CrossRef CAS.
- H. Panayiotou, PhD thesis, Queensland University of Technology, 2004.
- S. Brandes, PhD thesis, Queensland University of Technology, 2009.
- P. M. J. Barton, PhD thesis, Queensland University of Technology, 2011.
- D. Kurouski and R. P. Van Duyne, Anal. Chem., 2015, 87, 2901–2906 CrossRef CAS PubMed.
- J. Manheim, K. C. Doty, G. McLaughlin and I. K. Lednev, Appl. Spectrosc., 2016, 70, 1109–1117 CrossRef CAS PubMed.
- M. S. Boll, K. C. Doty, R. Wickenheiser and I. K. Lednev, Forensic Chem., 2017, 6, 1–9 CrossRef CAS.
- D. Bhatia, C. P. Sharma, S. Sharma and R. Singh, Sci. Justice, 2024, 64, 314–321 CrossRef.
- L. Mariey, J. P. Signolle, C. Amiel and J. Travert, Vib. Spectrosc., 2001, 26, 151–159 CrossRef CAS.
- D. Ballabio and V. Consonni, Anal. Methods, 2013, 5, 3790–3798 RSC.
- V. Sharma, C. P. Sharma, V. P. Kumar and S. P. Goyal, Forensic Sci. Int., 2016, 266, 226–233 CrossRef.
- Using the CITES Appendices – UNIA, https://cites.unia.es/cites/file.php/1/trainers/E-GCBriefcase-05.ppt, accessed 20 May 2024 Search PubMed.
- X. Wang, Z. Shi, Q. Zhao and Y. Yun, Materials, 2021, 14, 1–15 CAS.
- W. Gallagher, W. Xu, J. Xia, S. Min and Y. Xiong, Spectrochim. Acta, Part A, 2022, 274, 121034 CrossRef.
- E. O. Espinoza, B. W. Baker, T. D. Moores and D. Voin, Endanger. Species Res., 2009, 9, 239–246 CrossRef.
- V. Agarwal, A. G. Panicker, S. Indrakumar and K. Chatterjee, Int. J. Biol. Macromol., 2019, 133, 382–390 CrossRef CAS.
- W. Xu, J. Xia, S. Min and Y. Xiong, Spectrochim. Acta, Part A, 2022, 274, 121034 CrossRef CAS.
- B. A. McGregor, X. Liu and X. G. Wang, J. Text. Inst., 2018, 109, 813–822 CrossRef CAS.
- L. C. Lee, C.-Y. Liong and A. A. Jemain, Analyst, 2018, 143, 3526–3539 RSC.
- R. G. Brereton, Chemometrics for Pattern Recognition, John Wiley & Sons Ltd., Chichester, England, 2009 Search PubMed.
- L. C. Soares, J. O. Alves, L. A. Linhares, F. B. E. Filho and M. P. F. Fontes, Microchem. J., 2017, 133, 258–264 CrossRef CAS.
- J. Trevisan, P. P. Angelov, P. L. Carmichael, A. D. Scott and F. L. Martin, Analyst, 2012, 137, 2302–2312 RSC.
- L. Wu, B. Du, Y. V. Heyden, L. Chen, L. Zhao, M. Wang and X. Xue, Trac, Trends Anal. Chem., 2017, 86, 25–38 CrossRef CAS.
- Y. Shai, Biochim. Biophys. Acta, 2013, 1828, 2306–2313 CrossRef CAS.
- S. Saravanan, D. K. Sameera, A. Moorthi and N. Selvamurugan, Int. J. Biol. Macromol., 2013, 62, 481–486 CrossRef CAS PubMed.
- B. C. Powell and G. E. Rogers, Hair Keratin: Composition, Structure and Biogenesis, Springer, Berlin, Heidelberg, 1986, pp. 695–721 Search PubMed.
- Z. Ganim, H. S. Chung, A. W Smith, L. P Deflores, K. C. Jones and A. Tokmakoff, Acc. Chem. Res., 2008, 41, 432–441 CrossRef CAS.
- Á. I. López-Lorente and B. Mizaikoff, Anal. Bioanal. Chem., 2016, 408, 2875–2889 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06981a |
| This journal is © The Royal Society of Chemistry 2024 |