Open Access Article
Open Access ArticleRational design and synthesis of novel phenyltriazole derivatives targeting MRSA cell wall biosynthesis†
Mohamed M. Elsebaei *a,
Hany G. Ezzata,
Ahmed M. Helala,
Mohamed H. El-Shershabya,
Mohammed S. Abdulrahmanb,
Moaz Alsedawy.b,
Ahmed K. B. Aljohani
*a,
Hany G. Ezzata,
Ahmed M. Helala,
Mohamed H. El-Shershabya,
Mohammed S. Abdulrahmanb,
Moaz Alsedawy.b,
Ahmed K. B. Aljohani c,
Mohammed Almaghrabic,
Marwa Alsulaimanyc,
Basmah Almohaywid,
Read Alghamdic,
Samar F. Miskie,
Arafa Musaf and
Hany E. A. Ahmed
c,
Mohammed Almaghrabic,
Marwa Alsulaimanyc,
Basmah Almohaywid,
Read Alghamdic,
Samar F. Miskie,
Arafa Musaf and
Hany E. A. Ahmed *a
*a
aPharmaceutical Chemistry Department, Faculty of Pharmacy, Al-Azhar University, Nasr City 11884, Cairo, Egypt. E-mail: m.elsebaei@azhar.edu.eg; heahmad@azhar.edu.eg
bMicrobiology and Immunology Department, Faculty of Pharmacy, Al-Azhar University, Nasr City 11884, Cairo, Egypt
cPharmacognosy and Pharmaceutical Chemistry Department, Pharmacy College, Taibah University, Al-Madinah Al-Munawarah 41477, Saudi Arabia. E-mail: Akjohani@taibahu.edu.sa
dDepartment of Pharmaceutical Chemistry, College of Pharmacy, King Khalid University, Abha 61421, Saudi Arabia
eDepartment of Pharmacology and Toxicology, College of Pharmacy, Taibah University, Medina 42353, Saudi Arabia
fDepartment of Pharmacognosy, College of Pharmacy, Jouf University, Sakaka, Aljouf 72341, Saudi Arabia
First published on 20th December 2024
Abstract
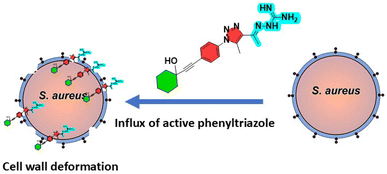
Antimicrobial resistance in methicillin-resistant Staphylococcus aureus (MRSA) is a major global health challenge. This study reports the design and synthesis of novel phenyltriazole derivatives as potential anti-MRSA agents. The new scaffold replaces the thiazole core with a 1,2,3-triazole ring, enhancing antimicrobial efficacy and physicochemical properties. A series of derivatives were synthesized and evaluated, with four compounds (20, 23, 29 and 30) showing significant activity against MRSA (MIC ≤ 4 μg mL−1). Compound 29 emerged as the most promising candidate, showing rapid bactericidal activity and superior performance over vancomycin in time-kill assays. It exhibited selective toxicity against bacterial cells, minimal cytotoxicity in human cell lines and low hemolytic activity. Mechanistic studies showed that compound 29 targets the bacterial cell wall by binding to penicillin-binding protein 2a (PBP2a), disrupting cell wall integrity. Additionally, it showed strong anti-biofilm activity and reduced MRSA biofilms by up to 40%. Preliminary pharmacokinetic profiles suggested a favorable profile, including a prolonged plasma half-life and good oral bioavailability. These results suggest that compound 29 is a promising lead for further development in the fight against MRSA.
1. Introduction
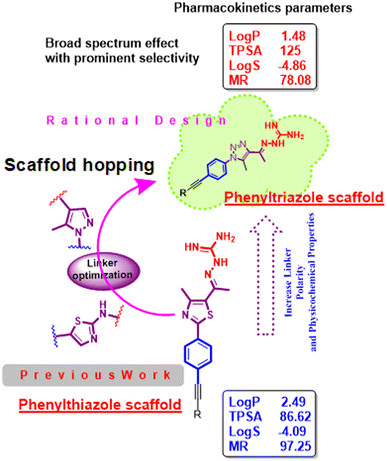
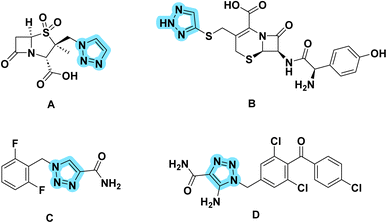
Antimicrobial resistance poses a major public health risk worldwide.1 Antibiotic-resistant infections, once considered a problem of the future, are now a familiar reality that kill tens of thousands of people every year. A prime example is methicillin-resistant Staphylococcus aureus (MRSA), which has become resistant to several antibacterial drugs.2,3 MRSA infections are associated with high mortality rates and are challenging to treat. The emergence of bacteria resistant to multiple drugs raises the possibility of a post-antibiotic era in which there are inadequate treatment options and recurrent infections that can lead to death.4,5 The consequence is enormous health and economic losses across the world, which is running out of its arsenal of antibacterial agents. According to a list published by the WHO, Gram-positive bacteria are considered a significant health risk, especially multidrug-resistant bacteria such as methicillin-resistant Staphylococcus aureus (M.R.S.A.).6,7 Additionally, the Infectious Disease Society of America (I.D.S.A.) defines the E.S.K.A.P.E. pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter cloacae) as posing a worldwide threat.8 Therefore, new clues need to be found to defeat resistant enterobacteria without affecting the microbiota. As the number of antimicrobials decreases worldwide, everyone should have the responsibility to address antimicrobial resistance. In our laboratory, the search for a good antibacterial agent began with phenylthiazole, which was originally discovered as a thiazole core with a guanidine head and an alkyl chain end.9–19 Recently, this scaffold has been intensively studied and optimized to improve its activity and metabolic profile. While the phenylthiazoles showed anti-biofilm activity, the poor permeability of phenylthiazole may play a role in its narrow spectrum. Therefore, a modification was made to the linker between the head and the scaffold. However, the solubility of the resulting product was not encouraging.20–23 Then a modification to the tail gave us the second generation, which showed intracellular effectiveness.24,25 However, the metabolic profile had to be improved by incorporation of a nitrogen atom in place of the active metabolite (–CH2) to obtain the third generation as phenyltriazole (Fig. 1).Therefore, this study attempts to modify the scaffold to explain its activity against a variety of microbial organisms and expand our knowledge on the structure–activity relationship of this new class of antimicrobial agents. The idea of the present scaffold is based on replacing the thiazole core with a 1,2,3-triazole ring, which is suggested to have antimicrobial activity against a variety of microorganisms and enhanced physicochemical properties. 1,2,3-Phenyltriazole derivatives have been extensively studied for their antibacterial and antifungal activities and DNA gyrase inhibitor has been proposed as a potential target for the development of antibacterial and antifungal agents. Many 1,2,3-triazoles have been shown to be effective and have several useful biological properties, such as antiviral,26 antiepileptic,27 antifungal,28 antibacterial,29 antimicrobial,30 antiprotozoal,31 antioxidant32 and anticancer activities.33 These 1,2,3-triazoles find their way into medicinal chemistry and form some drugs available on the markets. The most well-known structures (Fig. 2), include tazobactam (A),34 cefatrizine (B),35 rufinamide (C),36 and carboxyamidotriazole (D).37
2. Results and discussion
2.1. Chemistry
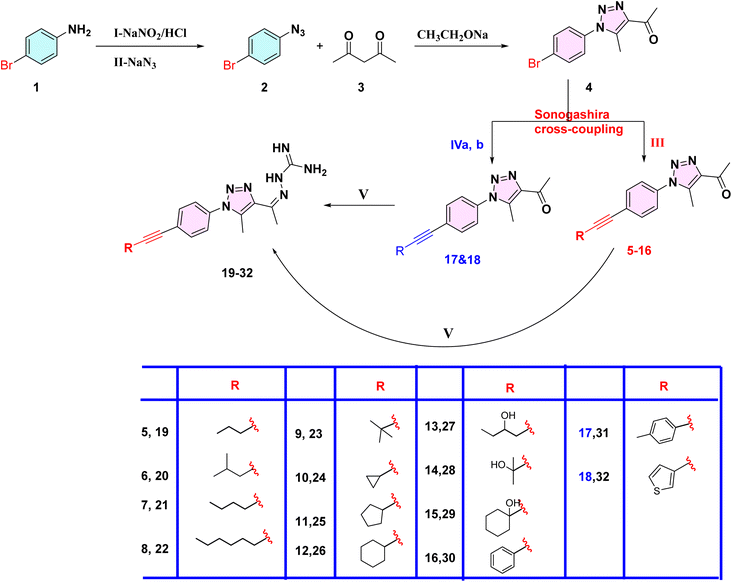
In Scheme 1, the first step is to prepare the commercially expensive 1-azido-4-bromobenzene (2) (CAS number: 2101-88-4) by oxidation of readily available p-bromoaniline via diazotization, then treating with sodium azide to afford compound 2. The latter was allowed to react with acetylacetone in the presence of sodium ethoxide to afford compound 4 as yellow crystals. According to the standard Sonogashira cross-coupling protocol, compound 4 was reacted with alkyne derivatives in the presence of copper(I) iodide as a catalyst, bis(triphenylphosphine)palladium(II) dichloride and triethylamine as a base, to afford compounds 5–16. While compounds 17 and 18 were available as 4-bromotoluene and 3-bromothiophene, we first reacted ethynyltrimethyl silane with compound 4 via Sonogashira cross-coupling without using potassium carbonate to avoid deprotection of TMS (trimethyl silane), thereby yielding a byproduct using an excess of triethylamine as a base, and then deprotected the TMS using potassium carbonate in methanol to obtain the intermediate 1-(1-(4-ethynylphenyl)-5-methyl-1H-1,2,3-triazol-4-yl)ethan-1-one, which reacts with two aryl halides (4-bromotoluene and 3-bromothiophene) via typical Sonogashira cross-coupling to afford compounds 17 and 18. Subsequently, the target compounds 19–32 were obtained by condensation of the phenyltriazole derivatives 5–18 with aminoguanidine hydrochloride (Scheme 1).2.2. Biological screening
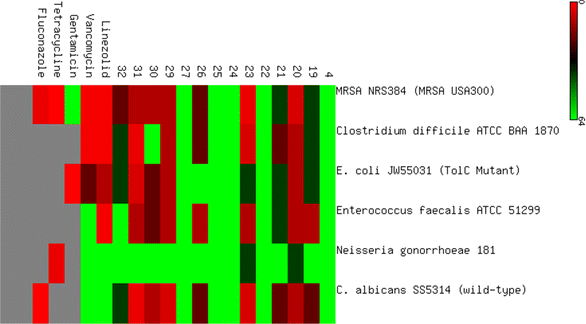
| Cp. | MRSA NRS384 (MRSA USA300) | Clostridium difficile ATCC BAA 1870 | E. coli JW55031 (TolC mutant) | Enterococcus faecalis ATCC 51299 | Neisseria gonorrhoeae 181 | C. albicans SS5314 (wild type) | Escherichia coli BW25113 (wild type) |
|---|---|---|---|---|---|---|---|
| a NT: Not tested. | |||||||
| 19 | 32 | 32 | 32 | 8 | 64 | 16 | >64 |
| 20 | 4 | 8 | 8 | 8 | 32 | 8 | >64 |
| 21 | 32 | 16 | 32 | 32 | >64 | 16 | >64 |
| 22 | 64 | 64 | 64 | 64 | >64 | 64 | >64 |
| 23 | 4 | 4 | 32 | 8 | >32 | 4 | >64 |
| 24 | 64 | 64 | 64 | >64 | >64 | >64 | >64 |
| 25 | 64 | >64 | 64 | >64 | 64 | >64 | >64 |
| 26 | 16 | 16 | 64 | 8 | >64 | 16 | >64 |
| 27 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| 28 | 16 | 16 | 64 | 16 | >64 | 16 | >64 |
| 29 | 4 | 8 | 8 | 8 | 64 | 4 | >64 |
| 30 | 4 | >64 | 16 | 16 | >64 | 8 | >64 |
| 31 | 8 | 2 | 4 | 8 | >64 | 2 | >64 |
| 32 | 16 | 32 | 32 | >64 | >64 | 32 | >64 |
| Linezolid | 1 | 1 | 8 | 1 | 64 | >64 | NT |
| Vancomycin | 1 | 1 | 16 | 64 | >64 | >64 | NT |
| Gentamicin | NT | NT | 0.25 | NT | NT | NT | NT |
| Tetracycline | NT | NT | NT | NT | 2 | NT | NT |
| Fluconazole | NT | NT | NT | NT | NT | 0.5 | NT |
The antibacterial evaluation was then expanded to include four additional Gram-positive and Gram-negative strains, including Staphylococcus aureus (ATCC 43300, clinical isolates), Klebsiella pneumoniae (ATCC 13883), and Pseudomonas aeruginosa (ATCC 10145) as Gram-negative bacteria.
In general, the synthesized compounds were more effective against Gram-positive microorganisms than against negative ones, as shown in (Fig. 3).
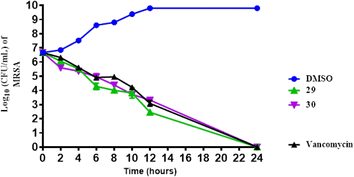
Overall, both compounds 29 and 30 exhibit potent and time-dependent bactericidal activity against MRSA, achieving complete eradication within 24 hours, with results comparable to those of vancomycin. This suggests that these phenyltriazole derivatives are promising candidates for the treatment of MRSA infections.
| Organism type | MIC (μg mL−1) | Selectivity indexa | |
|---|---|---|---|
| Caco-2 | WI38 | ||
| a Selectivity index = IC50/MIC. The IC50 values of compound 29 against both Caco-2 and W-I38 cells were 21.98 ± 1.12 and 70.2 ± 3.57 μg mL−1, respectively. The IC50 value = the concentration that reduced Caco-2 or WI38 cell viability by 75%. | |||
| MRSA NRS384 (MRSA USA300) | 4 | 5.5 | 17.5 |
| Clostridium difficile ATCC BAA 1870 | 4 | 5.5 | 17.55 |
| E. coli JW55031 (TolC mutant) | 32 | 0.69 | 2.194 |
| Enterococcus faecalis ATCC 51299 | 8 | 2.75 | 8.775 |
| Neisseria gonorrhoeae 181 | 32 | 0.69 | 2.194 |
| C. albicans SS5314 (wild type) | 4 | 5.5 | 17.55 |
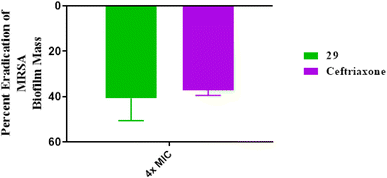 | ||
| Fig. 7 Disruption of mature MRSA USA300 biofilm by the tested phenyltriazole and ceftriaxone (at 4 × MIC). The data are presented as percent disruption of MRSA mature biofilm. | ||
| ID | Organism | IZD | MIC | MBC | T | Inhibition of biofilm formation (%) |
|---|---|---|---|---|---|---|
| a IZD: inhibition zone diameter expressed as mean number of millimeters ± SD, MIC: minimum inhibitory concentration; μg mL−1, MBC: minimum inhibitory concentration; μg mL−1, T = tolerance: MBC/MIC. | ||||||
| 29 | MRSA ATCC 43300 | 33.34 ± 0.47 | 3.9 | 15.625 | 4 | 79.39 ± 0.37 |
| K. pneumonia ATCC 13883 | 28.67 ± 0.47 | 7.8 | 31.25 | 4 | 72.11 ± 0.51 | |
| E. coli ATCC 25922 | 30.67 ± 0.47 | 3.9 | 31.25 | 8.01 | 65.43 ± 0.45 | |
| C. albicans ATCC 10231 | 37.67 ± 0.47 | 7.8 | 31.25 | 4 | 75.29 ± 0.53 | |
| P. aeruginosa ATCC 10145 | 21.67 ± 0.47 | 31.25 | 62.5 | 2 | 46.99 ± 0.66 | |
| Ceftriaxone | MRSA ATCC 43300 | 30.34 ± 1.24 | 8 | 8 | 1 | 73.262 ± 0.21 |
| K. pneumonia ATCC 13883 | 34.34 ± 0.47 | 4 | 8 | 2 | 61.23 ± 0.22 | |
| E. coli ATCC 25922 | 36 ± 1.50 | 16 | 32 | 2 | 59.14 ± 0.16 | |
| C. albicans ATCC 10231 | 37.67 ± 0.47 | 7.8 | 31.25 | 4 | 82.50 ± 0.14 | |
| P. aeruginosa ATCC 10145 | 35 ±1.85 | 8 | 16 | 2 | 74.39 ± 0.68 | |
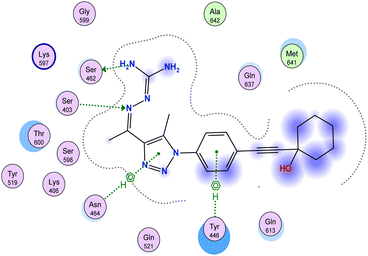
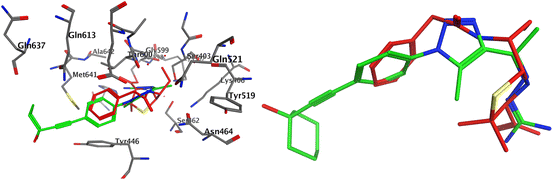
2.3. Molecular docking of the PBP2a target
Antibiotic resistance has emerged in response to the overuse of currently available antibiotics.50 Therefore, it is necessary to discover and develop novel antibacterial compounds with new formulations to combat severe infections.51 MRSA infection is one of the most common causes of hospital-acquired disease and is currently associated with poor prognosis and increased mortality/morbidity.15,22,46,47,49 The key factor in broad-spectrum β-lactam resistance in MRSA strains is penicillin-binding protein 2a (PBP2a). Due to its low affinity for β-lactams, PBP2a provides transpeptidase activity to enable cell wall synthesis at β-lactam concentrations that inhibit the β-lactam-sensitive PBPs normally produced by S. aureus. In the present study, molecular docking simulations were performed to identify the possible mechanism of such a novel phenyltriazole scaffold as a prominent cell wall disruptor of the active site of SauPBP2a. However, targeting the PBP2a allosteric site is another alternative strategy to reduce SauPBP2a activity. Therefore, we also evaluated the binding affinity of our potent analogs in PBP2a active site inhibitors including compound 29 and recorded binding data (Table 4). The crystal structure of PBP2a was considered, and the PDB code 1MWT was used for the experiments. All previous validation steps were performed, and re-docking of the bound ligand was performed. Interestingly, compound 29 exhibited high binding affinity to the active site of PBP2a with a ΔGbinding value of −7.39 kcal mol−1.2.4. Binding affinity analysis of penicillin-binding protein 2a
Penicillin-binding protein 2a (PBP2a) is an essential protein involved in the resistance of methicillin-resistant Staphylococcus aureus (MRSA) to β-lactam antibiotics and a potential antibacterial target.52 PBP2a can also perform the function of transpeptidase, complete cell wall synthesis and maintain the growth and proliferation of bacteria, thereby exhibiting multi drug resistance.53 Therefore, it is urgent to develop potential specific PBP2a inhibitors to overcome the multi drug resistance of MRSA to most current antibiotics. The binding affinity of potent analogs based on the inhibitory effect on MRSA and other microbial strains toward the PBP2a target was examined using inhibition constant data, and Ki is an effective parameter for cell wall destruction. The diagram in Fig. 8 shows the mode of action.The dock scores of potent analogs 20, 23, 29, 30, and 31 are estimated and using an equation the inhibition constants, Ki, were calculated and compared (Table 5). The equation used for Ki calculation is as follows:
| Ki = exp(ΔG/(R × T)), (T = 298 K) |
| Compound | ΔG of binding (kcal mol−1) | Inhibition constant (Ki, nM) | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
|---|---|---|---|
| 20 | −5.68 | 679.07 | 4.1 |
| 23 | −6.60 | 143.01 | 4.9 |
| 29 | −7.39 | 37.80 | 4.2 |
| 30 | −7.21 | 51.21 | 5.1 |
| 31 | −7.11 | 60.60 | 3.5 |
| PNAM | −7.89 | 16.20 | −0.86 |
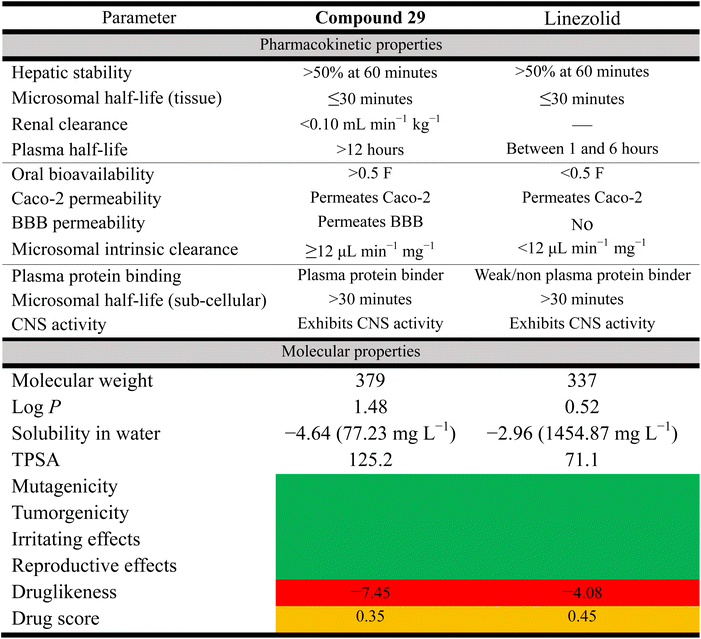
2.5. Pharmacokinetic profiling
One of the fundamental challenges of translating promising compounds from the bench to clinical trials is addressing the physicochemical limitations of a compound, with almost 90% of drugs currently in the drug discovery/development pipeline having poor physicochemical properties, such as limited aqueous solubility, poor permeability, or both.54 Our earlier generation phenylthiazoles exhibited poor physicochemical profiles including limited ability to permeate the gastrointestinal tract, as evaluated via the Caco-2 bidirectional permeability assay. This was due in large part to the lead compound being a substrate for the P-gp efflux system.55 Additionally, phenylthiazoles containing an n-alkyl chain lipophilic tail were found to be substrates for CYP450. This resulted in a very short half-life and high clearance rate (t1/2 of 15a <30 minutes, Fig. 1–8).55 Based on the data presented in (Table 6), the toxicity hazards of all top hits were as predicted by the OSIRIS Property Explorer, and the two compounds have similar pharmacokinetic properties in many respects such as hepatic stability, but differ in their distribution and CNS activity. Compound 29 likely has a longer duration of action than linezolid and the first generation of alkylphenylthiazole (t1/2 = 4.4 h) due to its longer plasma half-life and higher plasma protein binding, while linezolid has better solubility and a slightly better druglikeness and drug score (Table 6). These parameters may lead to further modifications to optimize compound 29 as a potential therapeutic agent.2.6. Conclusion
This study successfully showed the potential of phenyltriazole derivatives as effective anti-MRSA agents. Using rational design and scaffold hopping, a series of phenyltriazole compounds were synthesized and evaluated against methicillin-resistant Staphylococcus aureus (MRSA). The introduction of the 1,2,3-triazole ring, replacing the conventional thiazole core, was a strategic move aimed at enhancing the antimicrobial properties while improving the physicochemical properties of these molecules. Among the synthesized derivatives, compound 29 exhibited the most promising activity, with an MIC value comparable to or better than standard antibiotics such as vancomycin. Its rapid bactericidal action and significant disruption of MRSA biofilms make it a suitable candidate for the treatment of MRSA infections. The compound also showed a favorable selectivity index, highlighting its safety profile for therapeutic use. Molecular docking studies further supported the proposed mechanism of action and suggested a strong binding affinity to penicillin-binding protein 2a (PBP2a), which is crucial for cell wall biosynthesis in MRSA. This study not only highlights the importance of scaffold modifications but also provides valuable insights into the structure–activity relationship (SAR) of phenyltriazole compounds. The observed correlation between a terminal branched chain structure and antimicrobial efficacy highlights the critical role of molecular design in the development of effective antimicrobial agents. Future research should focus on optimizing the pharmacokinetic properties of these derivatives and exploring their efficacy in vivo. Additionally, expanding the scope to other resistant bacterial strains could further validate the potential of phenyltriazole scaffolds as broad-spectrum antibacterial agents. The promising results of compound 29, including its minimal cytotoxicity, potent anti-biofilm activity, and favorable pharmacokinetics, suggest that it could serve as a lead compound for the development of new antibiotics targeting resistant pathogens.3. Experimental
3.1. Chemistry
The chemicals used in the synthetic protocols were purchased from Sigma-Aldrich Corp., USA. The melting points of the target compounds were uncorrected and recorded using a Stuart Scientific Co. Ltd apparatus. The IR analysis was done by a Jasco FT/IR 460 Plus spectrophotometer utilizing a KBr disc unit. The 1H (400 MHz) and 13C (100 MHz) NMR spectra were measured on a BRUKER AV 850 MHz spectrometer in DMSO-d6 solvent using tetramethyl silane (TMS) as an internal standard. All compound purities were analyzed by HPLC-mass spectrometry on an Agilent 1100/ZQ MSD including a C18 column and diode array UV detector using a suitable mobile phase. In addition, elemental analysis data were recorded on an RCMP unit, Al-Azhar University, Cairo, Egypt. TLC on precoated Merck sheets was utilized for following reactions and separations.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HCl (1
HCl (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and stirred at 0–5 °C for 30 min. The aqueous NaNO2 was added dropwise to form diazotize amine hydrochloride. After that the solution was treated with aqueous NaN3 and further stirred for 1 h at room temperature. Finally, the reaction mixture was extracted with ethyl acetate, dried over anhydrous sodium sulphate and concentrated in vacuo to afford compound (2) in quantitative yield.56
1) and stirred at 0–5 °C for 30 min. The aqueous NaNO2 was added dropwise to form diazotize amine hydrochloride. After that the solution was treated with aqueous NaN3 and further stirred for 1 h at room temperature. Finally, the reaction mixture was extracted with ethyl acetate, dried over anhydrous sodium sulphate and concentrated in vacuo to afford compound (2) in quantitative yield.56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Yields, physical properties, and spectral data of isolated purified products are listed below.
1). Yields, physical properties, and spectral data of isolated purified products are listed below.3.1.4.1 General procedures for compounds 17 and 18. To dry DME (5 mL) in a 25 mL sealed tube compound 4 (1 equiv.), ethynyltrimethylsilane (2 equiv.), and triethylamine (2 mL) were added. After the reaction mixture was purged with dry nitrogen gas for 20 min, dichlorobis(triphenylphosphine)palladium(II) (5% mol) and copper(I) iodide (7.5% mol) were added. The sealed tube was then placed in an oil bath and stirred at 50 °C for 2 h. After cooling down to room temperature, the reaction mixture was passed through Celite, followed by chloroform (2 × 50 mL). The organic materials were then concentrated under reduced pressure. The crude materials were purified and washed by silica gel flash column chromatography using hexane-ethyl acetate (9
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford the intermediate compound 1-(5-methyl-1-(4-((trimethylsilyl)ethynyl)phenyl)-1H-1,2,3-triazol-4-yl)ethan-1-one.
1) to afford the intermediate compound 1-(5-methyl-1-(4-((trimethylsilyl)ethynyl)phenyl)-1H-1,2,3-triazol-4-yl)ethan-1-one.
3.1.4.2 1-(1-(4-Ethynylphenyl)-5-methyl-1H-1,2,3-triazol-4-yl)ethan-1-one. To dry methanol (10 mL) in a 25 mL round flask, the intermediate compound 1-(5-methyl-1-(4-((trimethylsilyl)ethynyl)phenyl)-1H-1,2,3-triazol-4-yl)ethan-1-one (1 equiv.) and anhydrous potassium carbonate (0.1% mol) were added. The reaction mixture was stirred at room temperature for 1 h until an off-white precipitate formed, and then concentrated under reduced pressure. The reaction was then quenched with very diluted hydrochloric acid and extracted with diethyl ether and then dried over anhydrous MgSO4. The solvent was evaporated under reduced pressure and the product was used in the next step without purification.
3.1.4.3 Compounds 17 and 18. Compounds 17 and 18 were prepared by the typical Sonogashira cross-coupling as mentioned above. Yields, physical properties, and spectral data of isolated purified products are listed below.
3.2 Synthesis of 1-(5-methyl-1-(4-substituted aryl)-1H-1,2,3-triazol-4-yl)ethan-1-one (17–18)
3.2.1.1 Synthesis of 1-(5-methyl-1-(4-((trimethylsilyl)ethynyl)phenyl)-1H-1,2,3-triazol-4-yl)ethan-1-one 4a. To dry DME (25 mL) in a sealed tube, compound 4 (1 mmol, 1 equiv.), appropriate alkynes (2 mmol, 2 equiv.), triethylamine (3 mL), and potassium carbonate (2 mmol, 2 equiv.) were added. After the reaction mixture was purged with dry nitrogen gas for 10 min, dichloro bis(triphenyl phosphine) palladium(II) (5% mole) and copper(I) iodide (7.5% mole) were added. The sealed tube was then placed in an oil bath and stirred at 110 °C for 24 h. After cooling to room temperature, the reaction mixture was extracted 3 times by ethyl acetate (20 mL). The organic materials were then concentrated under reduced pressure. The crude materials were purified by silica gel flash column chromatography using hexane-ethyl acetate (9
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield was 500 mg (94%).
1). The yield was 500 mg (94%).
3.2.1.2 Deprotection step to synthesis 1-(1-(4-ethynylphenyl)-5-methyl-1H-1,2,3-triazol-4-yl) ethan-1-one 4b. To dry methanol (10 mL) in a 25 mL round flask compound 4a (300 mg, 1.07 mmol) and anhydrous potassium carbonate (70 mg, 0.5 mmol) were added. The reaction mixture was stirred at room temperature for 1 h until an off-white precipitate formed, and then concentrated under reduced pressure. It was then quenched with very dilute hydrochloric acid and extracted with diethyl ether, and then dried over anhydrous MgSO4. The solvent was evaporated under reduced pressure. The yield was 200 mg (88%).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The yield, physical properties and spectral data of the isolated purified product are listed below.
3). The yield, physical properties and spectral data of the isolated purified product are listed below.Brown oil (190 mg, 67%); 1H NMR (DMSO-d6); δ 8.19 (d, J = 8.2 Hz, 2H), 8.16 (d, J = 8.2 Hz, 2H), 7.85–7.44 (m, 4H), 2.67 (s, 3H), 2.25 (s, 3H), 2.16 (s, 3H); 13C NMR (DMSO-d6); δ 199.03, 155.79, 152.25, 148.44, 139.26, 137.13, 129.36, 127.01, 122.72, 117.77, 117.74, 91.01, 89.79, 25.34, 23.61, 17.50; MS (m/z); 315.38 (M+, 77.51%).
3.3. Biological screening
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in a 96-well micro-titer plate of polypropylene material. The plate was covered and incubated at 37 °C for 24 h to culture the biofilms. Each experiment always had a negative control (culture media only) and positive control (culture media with inoculation 1%). All experiments were conducted in replicates of three unless otherwise mentioned.62 Crystal violet assay was performed to assess the biofilm inhibiting activity of the chemical compounds. For biofilm inhibition assay the medium and planktonic cells were discarded after 24 h of static growth at 37 °C. Each well was rinsed thrice unless and until complete removal of all media and planktonic cells. Adhered cells were stained with 125 μL of crystal violet (0.1%) for 30 min at room temperature. Then the crystal violet (CV) was washed out while using d.H2O. The CV bound to biofilm was re-solubilized in 30% acetic acid and its absorbance was measured at 550 nm.63–65 The change in the biofilm was calculated as follows:
100 in a 96-well micro-titer plate of polypropylene material. The plate was covered and incubated at 37 °C for 24 h to culture the biofilms. Each experiment always had a negative control (culture media only) and positive control (culture media with inoculation 1%). All experiments were conducted in replicates of three unless otherwise mentioned.62 Crystal violet assay was performed to assess the biofilm inhibiting activity of the chemical compounds. For biofilm inhibition assay the medium and planktonic cells were discarded after 24 h of static growth at 37 °C. Each well was rinsed thrice unless and until complete removal of all media and planktonic cells. Adhered cells were stained with 125 μL of crystal violet (0.1%) for 30 min at room temperature. Then the crystal violet (CV) was washed out while using d.H2O. The CV bound to biofilm was re-solubilized in 30% acetic acid and its absorbance was measured at 550 nm.63–65 The change in the biofilm was calculated as follows:| Biofilm change (%) = 1 − (OD of tested compound treated cells)/(OD of tested compound untreated cells) × 100.66 |
| Hemolytic activity (%) = (AbsS − AbsN/AbsP − AbsN) × 100 | (1) |
3.4. Molecular docking and binding data studies
PBP2a from methicillin-resistant Staphylococcus aureus (PDB ID: 1MWT) was used for docking simulations when covalently bound to the PNAM drug. The newly synthesized compound 29 was docked into the active site of the ATP-pocket of the 1MWT complex.71 Re-docking studies were performed for the crystal structure 1MWT as a validation method. AutoDock tools72 were used to process the enzyme. PyRx73 was used to perform the docking. Discovery Studio Visualizer74 was used to visualize and access the docking results. The AutoDock Tools package was employed to generate the docking input files and to analyze the docking results. A grid box size of 90 × 90 × 90 points with a spacing of 0.375 Å between the grid points was generated that covered almost the entire protein surface. Ligands were built by means of the AutoDock builder interface, and their geometries were optimized with the CHARMM forcefield75 and then prepared for docking calculations with the Python scripts available in the Autodock package. For each ligand 50 runs were performed, and the resulting poses were clustered with 1.8 Å tolerance. Lamarckian GA was used for the conformational space search with an initial population set to 150, and fitness function evaluations set to 25, 000, 000. The most abundant low energy clusters were selected for analysis.76–80Data availability
The data supporting this article have been included in the main manuscript and ESI.†Author contributions
I confirm that the contributory role of each author listed in the article is shown below and some authors may have contributed to multiple roles. I am responsible for ensuring that the descriptions are accurate and agreed by all authors in this work. M. A. S.: synthesis and adjustment of methodology, spectroscopic analysis, and writing original draft. H. E. A. A.: conceptualization, supervision, molecular modelling, writing paper and review, and project administration. All authors have read and agreed to the published version of the manuscript. E. H. E. and A. T. A. B.: conceptualization, supervision, methodology, and writing original draft. M. A.: Molecular modeling and contribution to biochemical data evaluation and discussion, writing, review and editing, and funding acquisition. A. K. A. and S. S.: biological evaluation of target compounds, data analysis, and partial funding acquisition. M. R. A. and A. A.: writing proposal, supervision, synthesis, and partial funding acquisition.Conflicts of interest
The authors declare that they have no competing interests. No financial support was received for this work. All authors have contributed significantly to the conception, design, execution, analysis, and interpretation of the work. All authors have reviewed and approved the final manuscript.Acknowledgements
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Large Research Project under grant number RGP2/346/45.References
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. R. Aguilar and A. Gray, et al., Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399(10325), 629–655 CrossRef CAS PubMed.
- T. Thompson, The staggering death toll of drug-resistant bacteria, Nature, 2022 DOI:10.1038/d41586-022-00228-x.
- P. Hawkey, The growing burden of antimicrobial resistance, J. Antimicrob. Chemother., 2008, 62(suppl_1), i1–i9 CrossRef CAS PubMed.
- S. W. Long, The Ongoing Threat of Methicillin-Resistant Staphylococcus aureus, Oxford University Press US, 2020, pp. 1943–1945 Search PubMed.
- G. M. Rossolini, F. Arena, P. Pecile and S. Pollini, Update on the antibiotic resistance crisis, Curr. Opin. Pharmacol., 2014, 18, 56–60 CrossRef CAS PubMed.
- W. C. Reygaert, An overview of the antimicrobial resistance mechanisms of bacteria, AIMS Microbiol., 2018, 4(3), 482 CAS.
- G. Cornaglia, Fighting Infections Due to Multidrug-Resistant Gram-Positive Pathogens, Elsevier, 2009, pp. 209–211 Search PubMed.
- J. Dai, R. Han, Y. Xu, N. Li, J. Wang and W. Dan, Recent progress of antibacterial natural products: Future antibiotics candidates, Bioorg. Chem., 2020, 101, 103922 CrossRef CAS PubMed.
- M. M. Elsebaei, H. Mohammad, M. Abouf, N. S. Abutaleb, Y. A. Hegazy and A. Ghiaty, et al., Alkynyl-containing phenylthiazoles: Systemically active antibacterial agents effective against methicillin-resistant Staphylococcus aureus (MRSA), Eur. J. Med. Chem., 2018, 148, 195–209 CrossRef CAS PubMed.
- M. M. Elsebaei, N. S. Abutaleb, A. A. Mahgoub, D. Li, M. Hagras and H. Mohammad, et al., Phenylthiazoles with nitrogenous side chain: an approach to overcome molecular obesity, Eur. J. Med. Chem., 2019, 182, 111593 CrossRef PubMed.
- M. M. Elsebaei, H. Mohammad, A. Samir, N. S. Abutaleb, A. B. Norvil and A. R. Michie, et al., Lipophilic efficient phenylthiazoles with potent undecaprenyl pyrophosphatase inhibitory activity, Eur. J. Med. Chem., 2019, 175, 49–62 CrossRef CAS PubMed.
- Y. Hosny, N. S. Abutaleb, M. Omara, M. Alhashimi, M. M. Elsebaei and H. S. Elzahabi, et al., Modifying the lipophilic part of phenylthiazole antibiotics to control their drug-likeness, Eur. J. Med. Chem., 2020, 185, 111830 CrossRef CAS PubMed.
- A. Mancy, N. S. Abutaleb, M. M. Elsebaei, A. Y. Saad, A. Kotb and A. O. Ali, et al., Balancing physicochemical properties of phenylthiazole compounds with antibacterial potency by modifying the lipophilic side chain, ACS Infect. Dis., 2019, 6(1), 80–90 CrossRef PubMed.
- I. Eid, M. M. Elsebaei, H. Mohammad, M. Hagras, C. E. Peters and Y. A. Hegazy, et al., Arylthiazole antibiotics targeting intracellular methicillin-resistant Staphylococcus aureus (MRSA) that interfere with bacterial cell wall synthesis, Eur. J. Med. Chem., 2017, 139, 665–673 CrossRef CAS PubMed.
- M. M. Elsebaie, H. T. N. El-Din, N. S. Abutaleb, A. A. Abuelkhir, H.-W. Liang and A. S. Attia, et al., Exploring the structure-activity relationships of diphenylurea as an antibacterial scaffold active against methicillin-and vancomycin-resistant Staphylococcus aureus, Eur. J. Med. Chem., 2022, 234, 114204 CrossRef CAS PubMed.
- A. M. Helal, A. M. Sayed, M. Omara, M. M. Elsebaei and A. S. Mayhoub, Peptidoglycan pathways: there are still more, RSC Adv., 2019, 9(48), 28171–28185 RSC.
- A. M. Sayed, N. S. Abutaleb, A. Kotb, H. G. Ezzat, M. N. Seleem and A. S. Mayhoub, et al., Arylpyrazole as selective anti-enterococci; synthesis and biological evaluation of novel derivatives for their antimicrobial efficacy, J. Heterocycl. Chem., 2023, 60(1), 134–144 CrossRef CAS.
- I. G. Shahin, K. O. Mohamed, A. T. Taher, M. M. Elsebaei, A. S. Mayhoub and A. E. Kassab, et al., New Phenylthiazoles: Design, Synthesis, and Biological Evaluation as Antibacterial, Antifungal, and Anti-COVID-19 Candidates, Chem. Biodiversity, 2023, 20(11), e202301143 CrossRef CAS PubMed.
- A. Mancy, Synthesis and evaluation of antimicrobial activity of arylazole derivatives, Al-Azhar J. Pharm. Sci., 2019, 60(2), 43–50 CrossRef.
- M. ElAwamy, H. Mohammad, A. Hussien, N. S. Abutaleb, M. Hagras and R. A. T. Serya, et al., Alkoxyphenylthiazoles with broad-spectrum activity against multidrug-resistant gram-positive bacterial pathogens, Eur. J. Med. Chem., 2018, 152, 318–328 CrossRef CAS PubMed.
- M. M. Elsebaei, N. S. Abutaleb, A. A. Mahgoub, D. Li, M. Hagras and H. Mohammad, et al., Phenylthiazoles with nitrogenous side chain: An approach to overcome molecular obesity, Eur. J. Med. Chem., 2019, 182, 111593 CrossRef PubMed.
- M. M. Elsebaei, H. Mohammad, M. Abouf, N. S. Abutaleb, Y. A. Hegazy and A. Ghiaty, et al., Alkynyl-containing phenylthiazoles: Systemically active antibacterial agents effective against methicillin-resistant Staphylococcus aureus (MRSA), Eur. J. Med. Chem., 2018, 148, 195–209 CrossRef CAS PubMed.
- A. Hammad, N. S. Abutaleb, M. M. Elsebaei, A. B. Norvil, M. Alswah and A. O. Ali, From phenylthiazoles to phenylpyrazoles: broadening the antibacterial spectrum toward carbapenem-resistant bacteria, J. Med. Chem., 2019, 62(17), 7998–8010 CrossRef CAS PubMed.
- I. Eid, M. M. Elsebaei, H. Mohammad, M. Hagras, C. E. Peters and Y. A. Hegazy, et al., Arylthiazole antibiotics targeting intracellular methicillin-resistant Staphylococcus aureus (MRSA) that interfere with bacterial cell wall synthesis, Eur. J. Med. Chem., 2017, 139, 665–673 CrossRef CAS PubMed.
- A. Hammad, N. S. Abutaleb, M. M. Elsebaei, A. B. Norvil, M. Alswah and A. O. Ali, et al., From phenylthiazoles to phenylpyrazoles: broadening the antibacterial spectrum toward carbapenem-resistant bacteria, J. Med. Chem., 2019, 62(17), 7998–8010 CrossRef CAS PubMed.
- Y.-W. He, C.-Z. Dong, J.-Y. Zhao, L.-L. Ma, Y.-H. Li and H. A. Aisa, 1, 2, 3-Triazole-containing derivatives of rupestonic acid: click-chemical synthesis and antiviral activities against influenza viruses, Eur. J. Med. Chem., 2014, 76, 245–255 CrossRef CAS PubMed.
- S. Ulloora, R. Shabaraya and A. V. Adhikari, Facile synthesis of new imidazo [1, 2-a] pyridines carrying 1, 2, 3-triazoles via click chemistry and their antiepileptic studies, Bioorg. Med. Chem. Lett., 2013, 23(11), 3368–3372 CrossRef CAS PubMed.
- S. N. Darandale, N. A. Mulla, D. N. Pansare, J. N. Sangshetti and D. B. Shinde, A novel amalgamation of 1, 2, 3-triazoles, piperidines and thieno pyridine rings and evaluation of their antifungal activity, Eur. J. Med. Chem., 2013, 65, 527–532 CrossRef CAS PubMed.
- A. Kamal, S. M. A. Hussaini, S. Faazil, Y. Poornachandra, G. N. Reddy and C. G. Kumar, et al., Anti-tubercular agents. Part 8: synthesis, antibacterial and antitubercular activity of 5-nitrofuran based 1, 2, 3-triazoles, Bioorg. Med. Chem. Lett., 2013, 23(24), 6842–6846 CrossRef CAS PubMed.
- C. Kaushik, K. Lal, A. Kumar and S. Kumar, Synthesis and biological evaluation of amino acid-linked 1, 2, 3-bistriazole conjugates as potential antimicrobial agents, Med. Chem. Res., 2014, 23, 2995–3004 CrossRef CAS.
- Y. Dürüst, H. Karakuş, M. Kaiser and D. Tasdemir, Synthesis and anti-protozoal activity of novel dihydropyrrolo [3, 4-d][1, 2, 3] triazoles, Eur. J. Med. Chem., 2012, 48, 296–304 CrossRef PubMed.
- M. F. Mady, G. E. Awad and K. B. Jørgensen, Ultrasound-assisted synthesis of novel 1, 2, 3-triazoles coupled diaryl sulfone moieties by the CuAAC reaction, and biological evaluation of them as antioxidant and antimicrobial agents, Eur. J. Med. Chem., 2014, 84, 433–443 CrossRef CAS PubMed.
- W. Zhang, Z. Li, M. Zhou, F. Wu, X. Hou and H. Luo, et al., Synthesis and biological evaluation of 4-(1, 2, 3-triazol-1-yl) coumarin derivatives as potential antitumor agents, Bioorg. Med. Chem. Lett., 2014, 24(3), 799–807 CrossRef CAS PubMed.
- Y. Yang, B. A. Rasmussen and D. M. Shlaes, Class A β-lactamases—enzyme-inhibitor interactions and resistance, Pharmacol. Therapeut., 1999, 83(2), 141–151 CrossRef CAS PubMed.
- A. J. Weinstein, The cephalosporins: activity and clinical use, Drugs, 1980, 20(2), 137–154 CrossRef CAS PubMed.
- E. Perucca, J. Cloyd, D. Critchley and E. Fuseau, Rufinamide: clinical pharmacokinetics and concentration–response relationships in patients with epilepsy, Epilepsia, 2008, 49(7), 1123–1141 CrossRef CAS PubMed.
- S. B. Ötvös, Á. Georgiádes, I. M. Mándity, L. Kiss and F. Fülöp, Efficient continuous-flow synthesis of novel 1, 2, 3-triazole-substituted β-aminocyclohexanecarboxylic acid derivatives with gram-scale production, Beilstein J. Org. Chem., 2013, 9(1), 1508–1516 CrossRef PubMed.
- S. Sonoda, T. Yamaguchi, K. Aoki, D. Ono, A. Sato and C. Kajiwara, et al., Evidence of latent molecular diversity determining the virulence of community-associated MRSA USA300 clones in mice, Immun., Inflammation Dis., 2018, 6(3), 402–412 CrossRef CAS PubMed.
- I. A. Tickler, R. V. Goering, J. R. Mediavilla, B. N. Kreiswirth, F. C. Tenover and H. A. I. Consortium, Continued expansion of USA300-like methicillin-resistant Staphylococcus aureus (MRSA) among hospitalized patients in the United States, Diagn. Microbiol. Infect. Dis., 2017, 88(4), 342–347 CrossRef PubMed.
- C. B. Long, R. P. Madan and B. C. Herold, Diagnosis and management of community-associated MRSA infections in children, Expert Rev. Anti., 2010, 8(2), 183–195 Search PubMed.
- B. Wang, B. Pachaiyappan, J. D. Gruber, M. G. Schmidt, Y. M. Zhang and P. M. Woster, Antibacterial Diamines Targeting Bacterial Membranes, J. Med. Chem., 2016, 59(7), 3140–3151 CrossRef CAS PubMed.
- P. S. Stewart, Mechanisms of antibiotic resistance in bacterial biofilms, Int. J. Med. Microbiol., 2002, 292(2), 107–113 CrossRef CAS PubMed.
- A. Gristina, Biomaterial-centered infection: microbial adhesion versus tissue integration. 1987, Clin. Orthop. Relat. Res., 2004, 427, 4–12 CrossRef.
- A. G. Gristina, Biomaterial-centered infection: microbial adhesion versus tissue integration, Science, 1987, 237(4822), 1588–1595 CrossRef CAS PubMed.
- European Medicines Agency, Guideline on Strategies to Identify and Mitigate Risks for First-In-Human and Early Clinical Trials with Investigational Medicinal Products; EMEA/CHMP/SWP/28367/07 Rev. 1; Committee for Medicinal Products for Human Use (CHMP), Amsterdam, The Netherlands, 2017 Search PubMed.
- H. Mohammad, H. E. Eldesouky, T. Hazbun, A. S. Mayhoub and M. N. Seleem, Identification of a Phenylthiazole Small Molecule with Dual Antifungal and Antibiofilm Activity Against Candida albicans and Candida auris, Sci. Rep., 2019, 9(1), 18941 CrossRef CAS PubMed.
- A. Hammad, N. S. Abutaleb, M. M. Elsebaei, A. B. Norvil, M. Alswah and A. O. Ali, et al., From Phenylthiazoles to Phenylpyrazoles: Broadening the Antibacterial Spectrum toward Carbapenem-Resistant Bacteria, J. Med. Chem., 2019, 62(17), 7998–8010 CrossRef CAS PubMed.
- M. A. Seleem, A. M. Disouky, H. Mohammad, T. M. Abdelghany, A. S. Mancy and S. A. Bayoumi, et al., Second-Generation Phenylthiazole Antibiotics with Enhanced Pharmacokinetic Properties, J. Med. Chem., 2016, 59(10), 4900–4912 CrossRef CAS PubMed.
- M. M. Elsebaie, H. T. Nour El-Din, N. S. Abutaleb, A. A. Abuelkhir, H.-W. Liang and A. S. Attia, et al., Exploring the structure-activity relationships of diphenylurea as an antibacterial scaffold active against methicillin- and vancomycin-resistant Staphylococcus aureus, Eur. J. Med. Chem., 2022, 234, 114204 CrossRef CAS PubMed.
- G. Muteeb, M. T. Rehman, M. Shahwan and M. Aatif, Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review, Pharmaceuticals, 2023, 16(11) CrossRef CAS PubMed.
- L. L. Silver, Challenges of antibacterial discovery, Clin. Microbiol. Rev., 2011, 24(1), 71–109 CrossRef CAS PubMed.
- N. Lv, Q. Kong, H. Zhang and J. Li, Discovery of novel Staphylococcus aureus penicillin binding protein 2a inhibitors by multistep virtual screening and biological evaluation, Bioorg. Med. Chem. Lett., 2021, 41, 128001 CrossRef CAS PubMed.
- T. A. Łeski and A. Tomasz, Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A, J. Bacteriol., 2005, 187(5), 1815–1824 CrossRef PubMed.
- R. Lipp, The Innovator Pipeline: Bioavailability Challenges and Advanced Oral Drug Delivery Opportunities, http://www.americanpharmaceuticalreview.com/Featured-Articles/135982-The-Innovator-Pipeline-Bioavailability-Challenges-and-Advanced-Oral-Drug-Delivery-Opportunities, 2013 May 11, 2017 [cited 2013.
- H. Mohammad, A. S. Mayhoub, A. Ghafoor, M. Soofi, R. A. Alajlouni and M. Cushman, et al., Discovery and characterization of potent thiazoles versus methicillin- and vancomycin-resistant Staphylococcus aureus, J. Med. Chem., 2014, 57(4), 1609–1615 CrossRef CAS PubMed.
- S. Kumar, A. S. Pathania, N. K. Satti, P. Dutt, N. Sharma and F. A. Mallik, et al., Synthetic modification of hydroxychavicol by Mannich reaction and alkyne–azide cycloaddition derivatives depicting cytotoxic potential, Eur. J. Med. Chem., 2015, 92, 236–245 CrossRef CAS PubMed.
- J. An, G. Y. Zuo, X. Y. Hao, G. C. Wang and Z. S. Li, Antibacterial and synergy of a flavanonol rhamnoside with antibiotics against clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA), Phytomedicine : international journal of phytotherapy and phytopharmacology, 2011, 18(11), 990–993 CrossRef CAS PubMed.
- N. Rezki, S. A. Al-Sodies, H. E. A. Ahmed, S. Ihmaid, M. Messali and S. Ahmed, et al., A novel dicationic ionic liquids encompassing pyridinium hydrazone-phenoxy conjugates as antimicrobial agents targeting diverse high resistant microbial strains, J. Mol. Liq., 2019, 284, 431–444 CrossRef CAS.
- I. Wiegand, K. Hilpert and R. E. Hancock, Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances, Nat. Protoc., 2008, 3(2), 163–175 CrossRef CAS PubMed.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65(1–2), 55–63 CrossRef CAS PubMed.
- F. Demirci and K. H. C. Başer, Bioassay Techniques for Drug Development By Atta-ur-Rahman, M. Iqbal Choudhary (HEJRIC, University of Karachi, Pakistan), William J. Thomsen (Areana Pharmaceuticals, San Diego, CA). Harwood Academic Publishers, Amsterdam, The Netherlands. 2001. xii + 223 pp. 15.5 × 23.5 cm. $79.00. ISBN 90-5823-051-1, J. Nat. Prod., 2002, 65(7), 1086–1087 CrossRef CAS.
- G. A. O'Toole, Microtiter dish biofilm formation assay, J. Vis. Exp., 2011, 47 Search PubMed.
- K. Alam, D. A. A. Farraj, E. F. S. Mah, M. A. Yameen, M. S. Elshikh and R. M. Alkufeidy, et al., Anti-biofilm activity of plant derived extracts against infectious pathogen-Pseudomonas aeruginosa PAO1, J. Infect. Public. Health., 2020, 13(11), 1734–1741 CrossRef PubMed.
- H. E. A. Ahmed, H. A. Abdel-Salam and S. MAJBc, Synthesis, characterization, molecular modeling, and potential antimicrobial and anticancer activities of novel 2-aminoisoindoline-1,3-Dione derivatives, Bioorg. Chem., 2016, 66, 1–11 CrossRef CAS PubMed.
- A. M. Omar, S. Ihmaid, E. E. Habib, S. S. Althagfan, S. Ahmed and H. S. Abulkhair, et al., The rational design, synthesis, and antimicrobial investigation of 2-Amino-4-Methylthiazole analogues inhibitors of GlcN-6-P synthase, Bioorg. Chem., 2020, 99, 103781 CrossRef CAS PubMed.
- A. Hend, El-sayed YAF. Correlation between biofilm formation and multidrug resistance in clinical isolates of Pseudomonas aeruginosa, Microbes and Infectious Diseases, 2021, 2(3), 541–549 Search PubMed.
- J. C. Bastos, N. S. M. Vieira, M. M. Gaspar, A. B. Pereiro and J. M. M. Araújo, Human Cytotoxicity, Hemolytic Activity, Anti-Inflammatory Activity and Aqueous Solubility of Ibuprofen-Based Ionic Liquids, Sustainable Chem., 2022, 3(3), 358–375 CrossRef CAS.
- M. M. Gaspar, S. Calado, J. Pereira, H. Ferronha, I. Correia and H. Castro, et al., Targeted delivery of paromomycin in murine infectious diseases through association to nano lipid systems, Nanomed. Nanotechnol. Biol. Med., 2015, 11(7), 1851–1860 CrossRef CAS PubMed.
- F. M. War Nongkhlaw and S. R. Joshi, Microscopic study on colonization and antimicrobial property of endophytic bacteria associated with ethnomedicinal plants of Meghalaya, J. Microsc. Ultrastruct., 2017, 5(3), 132–139 CrossRef PubMed.
- B. Sarkar, B. K. Dey, M. Sarkar and S. J. Kim, A smart production system with an autonomation technology and dual channel retailing, Computers & Industrial Engineering, 2022, 173, 108607 Search PubMed.
- S. Howard, V. Berdini, J. A. Boulstridge, M. G. Carr, D. M. Cross and J. Curry, et al., Fragment-Based Discovery of the Pyrazol-4-yl Urea (AT9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity, J. Med. Chem., 2009, 52(2), 379–388 CrossRef CAS PubMed.
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart and R. K. Belew, et al., Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function, J. Comput. Chem., 1998, 19(14), 1639–1662 CrossRef CAS.
- S. Dallakyan and A. J. Olson, Small-molecule library screening by docking with PyRx, Methods in Molecular Biology (Clifton, NJ), 2015, vol. 1263, 243–250 Search PubMed.
- M. A. Soliman, H. E. Ahmed, E. H. Eltamany, A. T. Boraei, A. Aljuhani and S. A. Salama, et al., Novel bis-benzimidazole-triazole hybrids: anticancer study, in silico approaches, and mechanistic investigation, Future Med. Chem., 2024, 1–15 Search PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong and J. Shim, et al., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J. Comput. Chem., 2010, 31(4), 671–690 CrossRef CAS PubMed.
- A. A. Awaji, W. A. Z. E. Zaloa, M. A. Seleem, M. Alswah, M. M. Elsebaei and A. H. Bayoumi, et al., N- and s-substituted Pyrazolopyrimidines: A promising new class of potent c-Src kinase inhibitors with prominent antitumor activity, Bioorg. Chem., 2024, 145, 107228 CrossRef CAS PubMed.
- A. M. E. A. Malebari, H. Ahmed, S. K. Ihmaid, A. M. Omar, Y. A. Muhammad and S. S. Althagfan, et al., Exploring the dual effect of novel 1,4-diarylpyranopyrazoles as antiviral and anti-inflammatory for the management of SARS-CoV-2 and associated inflammatory symptoms, Bioorg. Chem., 2023, 130, 106255 CrossRef CAS PubMed.
- M. T. Khayat, H. E. A. Ahmed, A. M. Omar, Y. A. Muhammad, K. A. Mohammad and A. M. Malebari, et al., A novel class of phenylpyrazolone-sulphonamides rigid synthetic anticancer molecules selectively inhibit the isoform IX of carbonic anhydrases guided by molecular docking and orbital analyses, J. Biomol. Struct. Dyn., 2023, 1–19 Search PubMed.
- M. Almaghrabi, A. Musa, A. K. B. Aljohani, H. E. A. Ahmed, M. Alsulaimany and S. F. Miski, et al., Introducing of novel class of pyrano[2,3-c]pyrazole-5-carbonitrile analogs with potent antimicrobial activity, DNA gyrase inhibition, and prominent pharmacokinetic and CNS toxicity profiles supported by molecular dynamic simulation, J. Biomol. Struct. Dyn., 2023, 1–18 Search PubMed.
- M. Alsehli, A. Aljuhani, S. K. Ihmaid, S. M. El-Messery, D. I. A. Othman and A.-A. A. A. El-Sayed, et al., Design and Synthesis of Benzene Homologues Tethered with 1,2,4-Triazole and 1,3,4-Thiadiazole Motifs Revealing Dual MCF-7/HepG2 Cytotoxic Activity with Prominent Selectivity via Histone Demethylase LSD1 Inhibitory Effect, Int. J. Mol. Sci., 2022, 23(15), 8796 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra07367c |
| This journal is © The Royal Society of Chemistry 2024 |