Improvement of DERA activity and stability in the synthesis of statin precursors by immobilization on magnetic nanoparticles†
Dino
Skendrović
a,
Anera
Švarc
b,
Tonči
Rezić
c,
Andrey
Chernev
d,
Aleksandra
Rađenović
 d and
Ana
Vrsalović Presečki
d and
Ana
Vrsalović Presečki
 *a
*a
aFaculty of Chemical Engineering and Technology, University of Zagreb, Savska c. 16, HR-10000 Zagreb, Croatia. E-mail: avrsalov@fkit.hr
bSelvita d.o.o, Prilaz Baruna Filipovića 29, HR-10000 Zagreb, Croatia
cFaculty of Food technology and Biotechnology, University of Zagreb, Pierottijeva 6, HR-10000 Zagreb, Croatia
dSchool of Engineering EPFL, Institute of Bioengineering, Ecublens 1015, CH-1007 Lausanne, Switzerland
First published on 5th September 2023
Abstract
Deoxyribose-phosphate aldolase (DERA), an enzyme applied in the synthesis of precursors for statin production, was immobilized on magnetic nanoparticles (MNPs). The silanized MNPs were functionalized/activated by several agents with different concentrations: APTMS/APTES–glutaraldehyde, APTMS/APTES–benzoquinone, APTMS/APTES–succinic anhydride and GPTMS. APTES–succinic anhydride (15%) provided the best result, and modification of the immobilization conditions was performed. The highest obtained retained activity and stability of 179% and 60%, respectively, were at pH 6.5 and 20 °C. A kinetic model for the immobilized enzyme reaction was developed and validated in a batch reactor. In reusability tests, immobilized DERA showed better stability than the free enzyme and maintained 70% enzyme activity after three cycles, while an almost complete loss of activity for the free enzyme after the second cycle was noticed.
1. Introduction
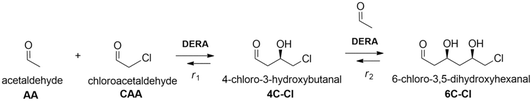
The application of aldolases in the field of organic and pharmaceutical synthesis has a long and varied history.1,2 This is mainly due to their remarkable stereoselectivity and regioselectivity features which can offer alternative paths to standard chemical syntheses, while making such processes more ecologically and economically feasible.3,4 One of the most well-known and researched reactions in this domain is the aldolase-catalyzed reaction, specifically the use of 2-deoxyribose-5-phosphate (DERA, EC 4.1.2.4) aldolase for the synthesis of statins, a class of drugs used for lowering levels of cholesterol in blood.5 By using DERA to catalyze a sequential aldol addition between acetaldehyde and chloroacetaldehyde, a valuable lactol intermediate with two chiral centers is produced, which is then further used for the synthesis of atorvastatin and rosuvastatin.6 Although this enzyme-catalyzed reaction offers many benefits, the main limitation is the enzyme inactivation due to the high concentrations of both aldehydes and reaction intermediates.5,7,8 Thus, large quantities of the enzyme are necessary to obtain industrially useful product yields.9,10 There have been many attempts in order to overcome these issues: ranging from directed evolution10–12 to different immobilization techniques.13–15 Immobilization simplifies the recovery and reuse of the enzymes, making them very suitable for continuous reactor configuration. Also, enzyme operational and storage stability is often improved, as well as tendency for denaturation towards organic solvents and high substrate and product concentrations.16 With increasing environmental concerns, enzyme immobilization that ensures high efficiency and activity is a very promising methodology, mostly because it follows current demands of biotechnological industries in providing cleaner production and sustainable operational technologies with implemented green chemistry. Among the different techniques available for enzyme immobilization, covalent immobilization is considered as one of the most efficient. That is mainly due to it not being susceptible to enzyme leaching that is often a significant problem with physical adsorption methods, simplifying carrier washing and reuse and, in some cases, it can offer additional stability and enhanced activity due to conformational changes of the immobilized enzyme.13,17,18 Materials used as carriers for covalent enzyme immobilization should be low-cost, stable, and compatible with the chosen reaction medium and desired functionalization/activation agents. Magnetic nanoparticles (MNPs) meet these requirements, while offering effortless separation from the reaction medium with the help of an external magnetic field and easily tunable particle size and morphology.19–22 Ease of work, tunability and flexibility were also the reason why we chose MNPs instead of commercial carriers. Since DERA is inactivated by aldehydes, immobilization by covalent binding via lysine amino groups on the surface seems to be a promising approach because stabilization of lipase against acetaldehyde deactivation by this method has been successfully employed for three decades.23 This type of protective covalent linking to the carrier material was also applied to stabilize DERA against high acetaldehyde concentrations. As carriers, mesoporous silica, MNPs and polymeric films were used.24–26 An acidic carbon nanotube was also used as an anionic carrier for ionic binding of lysine groups.27 Recently, DERA/polymer conjugates were covalently immobilized on a polymeric membrane carrier.28 In all mentioned cases, the immobilized DERA displayed improved stability in the presence of acetaldehyde. Generally, by studying the available literature, it is evident that publications considering DERA immobilization are very scarce,25–29 especially in terms of its application in the double aldol addition reaction of acetaldehyde and chloroacetaldehyde.27 The features of the immobilized enzyme, such as activity and stability, can be controlled to a certain extent by the choice of reaction conditions and functionalization and activation agents, and other factors.30 A number of functionalization and activation agents have been successfully used for covalent enzyme immobilization, such as organosilanes, gelatin and polyethylenimine for functionalization and glutaraldehyde (GA), carbodiimides and carbonyldiimidazoles for activation. Organosilanes and GA are without a doubt the most commonly used agents.17,31 In this work, the modification of DERA immobilization on MNPs in a batch reactor was performed, due to the simplicity of the batch process, as time is the only independent variable in all reactions. Enzyme activity and stability were monitored in the reaction of interest, i.e., in the double aldol addition of acetaldehyde and chloroacetaldehyde (Scheme 1). Several functionalization and activation agents were tested, and for the best one, different reaction conditions for the immobilization were investigated. After finding the ideal conditions, kinetic parameters were estimated and the model was validated in a batch reactor. The kinetic parameters of immobilized DERA were compared with those of the free enzyme. The application of immobilized DERA under ideal conditions was tested in a batch reactor with multiple cycles. Dynamic light scattering (DLS) and transmission electron microscopy (TEM) were performed to confirm the successful synthesis and silanization of MNPs.2. Experimental
2.1 Materials
Chemicals were mostly of reagent grade. Absolute ethanol was obtained from Scharlau (Spain). Acetaldehyde, 3-aminopropyltriethoxysilane (APTES) and (3-methylaminopropyl)trimethoxysilane (APTMS) were from Acros Organics (USA). Bovine serum albumin (BSA), succinic anhydride, chloroacetaldehyde solution (50% (w/w)) and (3-glycidyloxypropyl)trimethoxysilane (GPMTS) were from Sigma-Aldrich (Germany). Iron(III) chloride hexahydrate (FeCl3·6H2O) was from Gram-mol (Croatia). Anhydrous sodium acetate, ethylene glycol, potassium dihydrogen phosphate and dipotassium hydrogen phosphate were from Lach-Ner (Czech Republic). Ammonia solution (28% (w/w)) and tetraethyl orthosilicate were from VWR (USA). Acetonitrile and trifluoroacetic acid were from Fisher Scientific (UK). o-Benzylhydroxylamine hydrochloride was from TCI (UK). DERA024 from Thermotoga maritima was from Prozomix (UK).2.2 Synthesis of silica coated MNPs
MNPs were synthesized by a standard solvothermal method as proposed by Deng et al.32 Briefly, 3.38 g of FeCl3·6H2O was dissolved in 100 mL of ethylene glycol. After complete dissolution of FeCl3·6H2O, 6 g of sodium acetate was added to the mixture and stirred for 30 min. The solution was then placed in a Teflon-lined autoclave and heated for 8 h at 200 °C. After the solution cooled to room temperature, the Fe3O4 precipitate was separated from the solution using an external magnet, then rinsed several times with water and ethanol and dried overnight at 60 °C. For surface coating with silica, 0.93 g of obtained MNPs was dispersed in 50 mL of water, 150 mL of ethanol, and 4.5 mL of ammonia solution (28% (w/w)). This solution was then sonicated for 10 min to obtain the desired dispersion. Afterwards, 1 mL of TEOS was added to the solution dropwise and mixed for 12 h at 160 rpm. The obtained Fe3O4@SiO2 nanoparticles were then rinsed with water and ethanol and dried overnight at 60 °C. Size distribution, chemical composition and morphology of the obtained nanoparticles were checked by Dynamic Light Scattering (DLS) (Zetasizer, Malvern Panalytical, UK) and Transmission Electron Microscopy (TEM) (Talos F200S, ThermoFisher, USA) operated at 200 kV in two operating modes: TEM for collecting the bright-field TEM micrographs and scanning TEM (STEM) for obtaining the energy-dispersive X-ray (EDX) spectroscopy mappings.2.3 Functionalization and activation of Fe3O4@SiO2
The functionalization was carried out by modifying the silica surface with amine groups, to which activation groups were subsequently bound. 930 mg of Fe3O4@SiO2 was dispersed in 50 mL ethanol, to which 3 mL of either APTMS or APTES was added, and the solution was then stirred in an inert Ar atmosphere at 160 rpm and 30 °C for 24 h.The obtained Fe3O4@SiO2–NH2 precipitate was rinsed with ethanol and dried overnight at 60 °C. For the activation with benzoquinone, 25 mg of Fe3O4@SiO2–NH2 particles were stirred at 900 rpm for 2 h in 1 mL potassium phosphate buffer (0.1 M, pH 6) solution containing either 1.5, 3 or 4.5 mM benzoquinone solution. For the activation with succinic anhydride, 25 mg of Fe3O4@SiO2–NH2 particles were stirred at 900 rpm for 2 h in 1 mL potassium phosphate buffer (0.1 M, pH 6) solution containing 5/10/15% succinic anhydride, in an inert Ar atmosphere. For the activation with glutaraldehyde, 25 mg of Fe3O4@SiO2–NH2 particles were stirred at 900 rpm for 15 h in 1 mL potassium phosphate buffer (0.1 M, pH 6) solution containing 10/15/20% GA. Activation with GPTMS does not require previous functionalization, so 25 mg of Fe3O4@SiO2 particles were stirred at 900 rpm for 4 h in 1 mL potassium phosphate buffer (0.1 M, pH 6) solution containing 50% GPTMS, in an inert Ar atmosphere.
2.4 Enzyme immobilization and protein concentration
After activation, enzyme immobilization was performed by mixing 25 mg of activated MNPs with 1 mL of 0.6 mg mL−1 DERA solution in potassium phosphate buffer (0.1 M, pH 6) for 2 h at 25 °C and 900 rpm. After 2 h, a supernatant sample was taken to calculate the amount of unbound enzyme via standard Bradford assay33 and from this, the immobilization yield was calculated using the following expression: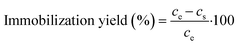 | (1) |
2.5 HPLC measurements
For measuring enzyme activity, substrate and product concentrations were analyzed by HPLC using a Phenomenex Kinetex column (5 μm, C18, 100 Å, 250 × 4.6 mm). For the samples to be UV detectable, they had to be derivatized with a solution of 0.02 g mL−1o-benzylhydroxylamine in a mixture of pyridine/methanol/water (33/15/2). 5 μL of the sample was taken and mixed with 50 μL of the derivatization solution. After 20 min of incubation at 25 °C and 900 rpm, the derivatization reaction was stopped by adding 350 μL of methanol, and the samples were centrifuged and then analyzed by high-performance liquid chromatography. The mobile phase consisted of solvent A (0,1% v/v TFA in an 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ACN
20 ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture) and solvent B (0.1% v/v TFA in water) with a gradient elution from 90 to 28.4% solvent B for the first 10 min and then from 28.4 to 90% solvent B from minute 14 to 16. The flow rate was 1.5 mL min−1, the column temperature was 30 °C and UV detection was set at 215 nm.
water mixture) and solvent B (0.1% v/v TFA in water) with a gradient elution from 90 to 28.4% solvent B for the first 10 min and then from 28.4 to 90% solvent B from minute 14 to 16. The flow rate was 1.5 mL min−1, the column temperature was 30 °C and UV detection was set at 215 nm.
2.6 Enzyme assay
DERA activity was determined in the aldol addition reaction shown in Fig. 1. 100 μL of the free or immobilized enzyme solution was mixed with 100 μL of aldehyde solution at 25 °C and 900 rpm, and the samples were taken at regular intervals during the first 20 min of the reaction and then analyzed by HPLC. The aldehyde solution consisted of 200 mM acetaldehyde and 100 mM chloroacetaldehyde in potassium phosphate buffer (0.1 M, pH 6). Specific enzyme activity was calculated using eqn (2), where Δc/Δt represents the change of product concentration with time and γe is the enzyme concentration. 1 mg mL−1 of free enzyme activity corresponds to 1.16 U mL−1.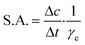 | (2) |
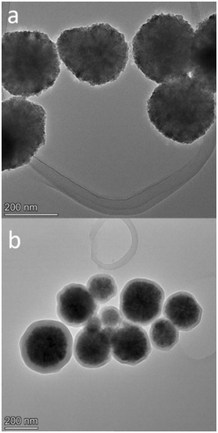 | ||
| Fig. 1 High-resolution TEM (HRTEM) images of Fe3O4 (a) and Fe3O4@SiO2 (b). Both samples were deposited and dried on a lacey-carbon copper TEM grid. HRTEM measurements were performed at 200 kV. | ||
2.7 Retained activity and stability measurements
After the activation of MNPs, retained activity was measured by comparing the free and immobilized enzyme activities with the following equation: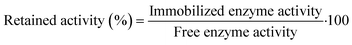 | (3) |
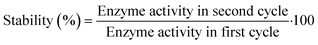 | (4) |
2.8 Influence of pH and temperature on immobilization yield and activity
Using the best performing activating agent, the ideal pH and temperature for the immobilization process were determined. The pH was varied between 6 and 8 in 0.5 steps using potassium phosphate buffer (0.1 M), and the temperature was varied between 20 and 35 °C in 5 °C steps. After each immobilization, the yield was calculated using eqn (1) and the retained activity was measured using eqn (3).2.9 Enzyme reusability measurements
The operational stability of the DERA enzyme immobilized on MNPs after recycling was investigated by measuring the relative activity of the enzyme during the process of circular reaction under the assay conditions, i.e., in the reaction of double aldol addition of acetaldehyde and chloroacetaldehyde. The reaction time was set to 10 min. The DERA activity in the first cycle was set to 100%. After each cycle, the MNPs were separated by applying an external magnetic field. The recycled MNPs were washed three times with potassium phosphate buffer (0.1 M, pH 6), and then used in the next cycle with the addition of a fresh substrate.For recycling free enzyme mixtures, the free enzyme was tested in the same reaction as the immobilized one and the activity of the enzyme in the first cycle was set to 100%. After each cycle, the free enzyme mixture was recovered by ultrafiltration using a 10 kDa ultrafiltration tube at 4 °C. The recovered free enzyme mixtures were washed three times with potassium phosphate buffer (0.1 M, pH 6), and then used in the next cycle.
2.10 Kinetic measurements
Kinetic measurements were performed for the first and second steps of the reaction shown in Scheme 1. The influence of each substrate on specific enzyme activity was tested. Measurements were performed using the initial rate method, i.e. when the substrate conversion is below 10%, and all variables except the concentration of the monitored substrate were kept constant. The used concentrations are shown in the ESI† (S4). The samples were taken for the first 5 minutes of each reaction, and measurements were performed using HPLC as described in 2.5.2.11 Data handling
The processing of experimental data was carried out by using the SCIENTIST software. Kinetic parameters (Vm, Km, Ki, n) of the Michaelis–Menten kinetics were estimated using non-linear regression methods (simplex fit and least squares fit) incorporated in the software. Non-linear regression is used for more precise fitting of experimental data considering the complexity of the Michaelis–Menten kinetics. To assess the validity and accuracy of this model, model validation was performed in a batch reactor. The set of optimum parameters used for the simulation was according to the proposed models. For simulations, the built-in Episode algorithm for the stiff system of differential equations was used. Standard deviations (σ) and coefficients of determination (R2) as measures of goodness-of-fit were calculated using the Scientist built-in statistical functions.3. Results and discussion
3.1 Synthesis and characterization of magnetic nanoparticles
To confirm the particle sizes, chemical composition and morphology, TEM and DLS were performed on both Fe3O4 and Fe3O4@SiO2. The results of TEM analysis are shown in Fig. 1 and in the ESI† (S1). Both Fe3O4 and silica-coated Fe3O4 nanoparticles exhibit spherical geometry, high polycrystallinity, and narrow size distribution. Fe3O4 had an average diameter of 197 ± 56.7 nm (based on size measurements of 31 nanoparticles), which increased to 254 ± 62.8 nm after silica coating (based on size measurements of 74 nanoparticles), indicating that we successfully applied a relatively thin silica shell of about 54 nm thickness. STEM analysis (ESI† S2) also independently confirms the application of this uniform silica coating. Size distribution by intensity as given by DLS analysis on a Zetasizer is shown in the ESI† (S3), and here we could also observe a relatively narrow size distribution with a mean diameter of 267.2 ± 70.3 nm for Fe3O4 and 310.7 ± 59.4 nm for the silica coated Fe3O4@SiO2. Table 1 shows the comparison of particle sizes for both SEM and DLS analysis and their discrepancy, which can be safely attributed to the widely observed difference between dry and hydrodynamic diameters.34,35| Particle | SEM (nm) | DLS (nm) |
|---|---|---|
| Fe3O4 | 197 ± 56.7 | 267.2 ± 70.3 |
| Fe3O4@SiO2 | 254 ± 62.8 | 310.7 ± 59.4 |
3.2 Improving the immobilization process
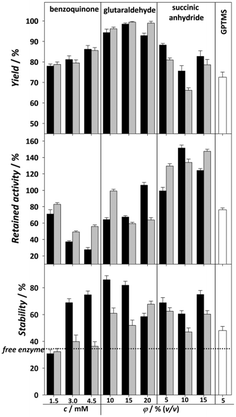
Although covalent immobilization usually results in increased stability of the enzyme, a major drawback is the significant loss of enzyme activity that occurs during immobilization. This depends on the utilized techniques in the process of immobilization. The loss of activity is usually attributed to the changes in the active sites or orientation of the enzyme, which can make the active sites inaccessible. The choice of reaction conditions in the process of enzyme immobilization such as pH, temperature, and buffer, as well as the choice of functionalization and activation agents, can have an impact on the desired features of the immobilized enzyme.30 In order to obtain an immobilized biocatalyst with the highest activity and stability, activation agents were tested and then the immobilization reaction conditions were modified (pH and temperature).The tested activating agents were as follows: APTMS/APTES–glutaraldehyde, APTMS/APTES–benzoquinone, APTMS/APTES–succinic anhydride and GPTMS (Fig. 2). APTMS and APTES are both widely used organosilanes available at low prices and have similar behavior, their only difference being the methyl group bonded to an amino group in APTMS, which theoretically could have some effect on subsequent activation group bonding.17 Glutaraldehyde and benzoquinone have already been used in the process of DERA immobilization24,25 and some preliminary measurements have been performed for succinic anhydride,36 which was promising, while immobilization on the epoxy group was chosen simply because it is a common method of carrier activation, where the two-step process of functionalization and activation can be replaced by a single process using an appropriate agent.37 The above reagents were selected to bind the carrier to the lysine amino group to stabilize the enzyme against acetaldehyde-induced deactivation.4 The agents all have different spacer lengths (glutaraldehyde – long, benzoquinone – medium, succinic anhydride – short) as they can specifically tune the catalytic properties of the immobilized protein.17 In addition, another type of covalent interaction between the enzyme and carrier is covered: succinic anhydride is strictly oriented to the ε-NH2 group of lysine,38 benzoquinone and the epoxy group, among others, react with amine, hydroxyl or thiol groups of proteins,39,40 while glutaraldehyde usually binds to the α-NH2 group of lysine.17 Moreover, the immobilization of enzymes via epoxy and glutaraldehyde activation can lead to multi-point binding of the enzymes and the carrier.17 This can lead to a significant improvement in the stability of the enzyme, but on the other hand, it can also affect its activity. The concentration of the used activation agent can have an impact on the success of the immobilization,13,41 so DERA was immobilized on carriers activated with different concentrations of all activating agents.
The results of the effects of different combinations of functionalization and activation agents on the immobilization yield, activity and stability of immobilized DERA compared to the free enzyme are shown in Fig. 2. We can see that regarding the immobilization yield, both APTES/glutaraldehyde and APTMS/glutaraldehyde have shown the best results for all three concentrations. The percentage of the bound enzyme was almost 100%, resulting in an enzyme loading capacity of about 24 mg g−1 carrier. This higher yield in comparison to the other used agents can be explained by the possibility of crosslinking happening along with covalent bonding, which is common with glutaraldehyde.42 Regarding retained activity, succinic anhydride showed by far the best results at practically every concentration and functionalization method tested, with the rare occurrence of hyperactivation, which means that the activity of the immobilized enzyme is higher than that of the free enzyme, possibly due to a structural change to a more active conformation.16 Regarding stability, almost all combinations of functionalizing and activating agents have shown better results than the free enzyme, which has a stability of 37%. In general, all MNPs functionalized with APTMS have shown higher stability, which is most prominent at lower concentrations of glutaraldehyde. If we compare all three immobilization parameters, it seems that 10% v/v APTMS–glutaraldehyde should be the ideal candidate for further modifications due to its highest stability value of 82%, because stability is generally the most desired improvement in the immobilization process. Nevertheless, the 140% retained activity obtained with 15% v/v APTES–succinic anhydride should not be underestimated considering that, in addition to the 40% increase in activity over the free enzyme, it presents a more than twofold improvement over 10% v/v APTMS–glutaraldehyde while having only 20% less stability and yield. For these reasons, 15% v/v APTES–succinic anhydride was chosen for further modifications of the pH and temperature environment during the immobilization process. These results are very promising in terms of the activity obtained compared to the other immobilization methods used, while the stability and yield are in line with previously published results. For example, in DERA immobilization by ionic exchange interactions on oxidized multi-walled carbon nanotubes, a maximal retained activity of 56% was achieved while the retained stability was approximately 80% after exposure to the aldehyde solution.27
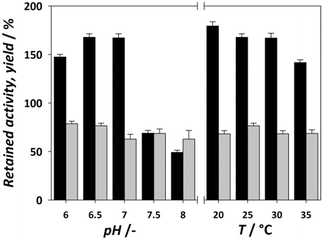
The retained activity and stability of EcoDERA immobilized on a nanomagnetic material (functionalized and activated by APTES and GA) were 75% and 85%.25 The recently applied DERA immobilization method consisting of conjugation of DERA with polymer conjugates and subsequent covalent immobilization of such conjugates in a film resulted in the retained activity of 30–60%, while the stability was also around 80%.28,43 A significantly lower stability of 50% was obtained by the physical method of immobilization on the interparticle pore type of mesoporous silica, and a retained activity of 84% was observed in this case.29 The next step in modifying the immobilization process is to find the ideal reaction conditions for the reaction between the carrier and the enzyme. Improvement of these conditions can greatly affect the retained activity of the enzyme. Fig. 3 shows the influence of the pH of immobilization (0.1 M potassium phosphate buffer) and temperature on the activity and yield obtained. For pH, we can observe an increase in activity at pH 6.5 and 7, which then drops sharply. The immobilization yield generally follows a downward trend with each pH increase. With temperature, the yield remains mostly static, but we can observe a lower retained activity the higher the temperature, suggesting that the enzyme is sensitive to both higher pH and higher temperature. Thus, the ideal conditions for the immobilization process were found to be pH 6.5 and 20 °C. In summary, the results of immobilization success after process improvement were a retained activity of 179% and stability of 60%. Compared to the data available in the literature for the immobilization of the DERA enzyme, the obtained results show an approximately equal or slightly lower stability, while the retained activity exhibits a significant step forward.24–29,43 Since all activity and stability measurements were performed in the reaction of double aldol addition of acetaldehyde and chloroacetaldehyde, the used method has potential to be applied in this process.
3.3 Kinetics of the immobilized enzyme
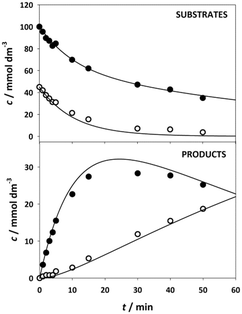
The kinetics of the immobilized enzyme (ESI,† S4) for the first and second steps of the reaction were determined from the experimental data using the initial reaction rate method described in Section 2.10.The kinetic equations used to describe the reaction kinetics and determine the parameters are listed in Table 2. Looking at the data for chloroacetaldehyde in the first step of the reaction, we can see that a strong inhibition of the substrate occurs at concentration higher than 200 mM. Švarc et al.44 performed kinetic measurements for the free enzyme in the same reaction, where no substrate inhibition occurred. However, their measurements were performed only up to 200 mM, i.e. the concentration where inhibition starts to occur. For that reason, we have repeated the measurements for the first reaction and modeled it again (ESI,† S5), and it is clear that the same case of strong substrate inhibition exists with the free enzyme. The standard Michaelis–Menten equation used for substrate inhibition did not fit the experimental data for both the free and immobilized enzymes in this case, but we found the occurrence of the same kinetic phenomena in the work of Bapiro et al.,45 who solved this problem with the addition of an extra parameter n representing the number of molecules bound to the inhibition sites on the enzyme (Table 2, eqn (5)). If we now compare all the kinetic parameters of the free and the immobilized enzymes, we can see that although the Km (Michaelis constant) values were higher for the immobilized enzyme, implying that the affinity of the enzyme for the two substrates is lower after immobilization, the maximum reaction rate Vm was more than threefold higher than that of the free enzyme, which also correlates with a much higher relative activity, i.e. hyperactivation of our immobilized enzyme. The hyperactivation possibly occurs because the DERA enzyme belongs to class 1 of aldolases, which form a Schiff base between the donor substrate and the NH2 group of a lysine residue in the active site4 and succinic anhydride reacts specifically with the ε-NH2 group of lysine, likely making it more accessible to the substrate.38 The second part of the reaction seems to be a bit slower with the immobilized enzyme, along with slight substrate inhibition present with acetaldehyde, but both Km and Ki (inhibition constant) values for 4-chloro-3-hydroxybutanal were improved, implying higher affinity of the enzyme towards the intermediate along with less pronounced inhibition. To assess the accuracy of the kinetic model, mathematical model validation (Table 2, kinetic and batch reactor models) was performed in a batch reactor (Fig. 4). According to the previous findings for the free DERA, enzyme deactivation was described using second-order kinetics.8,44 We can see that there is a very good overlap of the experimental and model data which was also proved by statistic goodness-of-fit (σ = 1.92, R2 = 0.99). This indicates that the model could serve for accurate reaction predictions and further reaction improvements. Operation stability decay was described by the second order kinetics (Table 2, eqn (11))44 and the estimated deactivation constant was kd = 0.001345 min−1. Fig. 5 shows a comparison of the simulation of the decrease in enzyme activity for the immobilized enzyme and for the free enzyme. The simulation for the free enzyme was performed using the previously published parameters under the same initial conditions in which the experiment, shown in Fig. 4, was performed.
| Parameter | Unit | Free DERA024 | Immobilised DERA | Kinetic model | |
|---|---|---|---|---|---|
| a Data taken from Švarc et al.44 | |||||
| First aldol addition | |||||
| V m1 | U mg−1 | 4.31 ± 0.63 | 13.64 ± 0.33 |
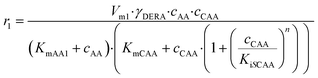 (5) (5) |
|
| K mAA1 | mM | 11.10 ± 2.54 | 28.34 ± 2.82 | ||
| K mCAA | mM | 73.51 ± 9.11 | 188.20 ± 8.11 | ||
| K iSCAA | mM | 260.96 ± 26.31 | 204.88 ± 3.27 | ||
| n | mM | 20.00 ± 4.21 | 15.43 ± 3.75 | ||
| Second aldol addition | |||||
| V m2 | U mg−1 | 1.53 ± 0.26a | 1.06 ± 0.02 |
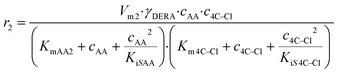 (6) (6) |
|
| K mAA2 | mM | 2.03 ± 0.28a | 13.11 ± 1.52 | ||
| K m4C–Cl | mM | 86.26 ± 7.62a | 47.90 ± 3.15 | ||
| K iSAA | mM | — | 115.19 ± 9.70 | ||
| K iS4C–Cl | mM | 72.44 ± 7.54a | 103.42 ± 8.03 | ||
| Batch reactor model | |||||
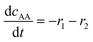 (7) (7) |
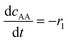 (8) (8) |
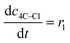 (9) (9) |
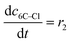 (10) (10) |
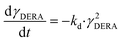 (11) (11) |
|
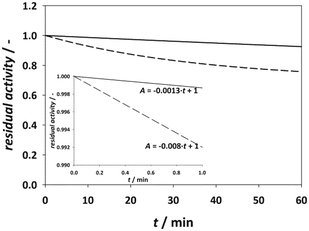 | ||
| Fig. 5 Comparison of model simulation for immobilized (solid line) and free enzyme* (dash line) (cacetaldehyde = 100 mM, cchloroacetaldehyde = 45 mM, 0.1 M phosphate buffer pH 6, 25 °C, γDERA = 2 mg cm−3). *Simulation provided by using parameters from Švarc et al.44 | ||
According to the model simulation, the remaining activity of the isolated enzyme is 92.53%, which is higher than the result of the measurements during the experimental modifications to the immobilization method (Fig. 2). This is likely due to the process of centrifugation and stirring during washing of the enzyme in our stability tests, which can lead to mechanical denaturation.46 A comparison with the free enzyme shows that it lost about 24% of its initial activity, which is about three times more than the immobilized enzyme whose initial activity decreased by 7.5%. This indicates that the modified immobilization method contributes significantly to the stability of the enzyme. In addition, the rate of initial activity loss was also extracted from these results (Fig. 5, inner graph).
According to the data, the loss of activity in the initial phase of the reaction at high aldehyde concentrations is more than six times higher for the free enzyme than for the immobilized enzyme, proving that the immobilized enzyme is less sensitive to high aldehyde concentrations.
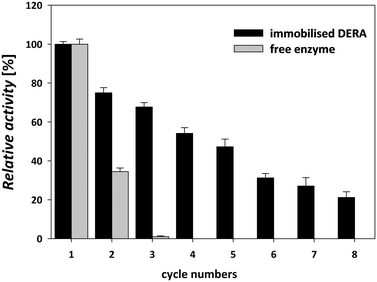
3.4 Reusability of immobilized DERA
DERA immobilization on MNPs enabled simple enzyme recycling in a sequential batch-wise process, thereby lowering the enzyme costs. The magnetic nature of the carrier facilitated easy separation and recovery of the immobilized enzyme from the reaction mixture.47 Reusability was tested in the sequential aldol addition reaction of acetaldehyde and chloroacetaldehyde (Scheme 1). This type of reusability assay for the immobilized DERA enzyme was not tested in previous reports.24–29,43 The immobilized enzyme was stable for up to five consecutive cycles of statin side-chain synthesis and retained approximately 50% of its initial activity; thereafter, the activity decreased significantly (Fig. 6).The immobilized enzyme maintained about 70% activity until the third cycle, while the free enzyme did not retain any activity after the second cycle. The gradual loss of the enzyme activity after only a few cycles occurs due to factors such as structural modification of the enzyme and protein denaturation and/or enzyme inactivation.48 The results of the catalytic activity and stability of the immobilized DERA described above indicated that the used method for immobilization is suitable for the construction of a biocatalyst that can be applied in the synthetic reaction for the production of statin side chain precursors.
4. Conclusion
Magnetic nanoparticles were used as carriers for DERA immobilization to improve the reusability of the enzyme in the double aldol addition reaction. Modified MNPs were prepared by direct silanization of Fe3O4 nanoparticles. The combination of APTES–succinic anhydride (15%) for functionalization and activation of the carrier gave the best results in terms of activity and stability of the immobilized enzyme. By modifying the immobilization conditions, significant hyperactivation of the immobilized enzyme was achieved, and the stability of the enzyme was improved almost threefold compared to the free enzyme. A mathematical model for the reaction of the immobilized enzyme was established, and the parameters were compared with those of the free enzyme, with a significant improvement in the maximal reaction rate Vm in the first step of the reaction. The model was successfully validated in a batch reactor. In the reusability test performed on the double aldol addition reaction of acetaldehyde and chloroacetaldehyde, the immobilized DERA showed higher stability than the free enzyme, indicating that it is suitable for the synthesis of statin side chain precursors on an industrial scale.Author contributions
Dino Skendrović: conceptualization, investigation, methodology, validation, visualization, and writing – original draft. Anera Švarc: validation, methodology, and writing – review & editing. Tonči Rezić: methodology and writing – review & editing. Andrey Chernev: investigation and writing – review & editing. Aleksandra Rađenović: investigation and writing – review and editing. Ana Vrsalović Presečki: conceptualization, methodology, validation, visualization, supervision, funding acquisition, project administration, resources, and writing – original draft.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was supported by the University of Zagreb short term financial scientific research support entitled “Enzyme immobilization for green and sustainable pharmaceutical production”. The authors would like to thank Prozomix for the supply of DERA024.Notes and references
- P. Clapés, W. D. Fessner, G. A. Sprenger and A. K. Samland, Curr. Opin. Chem. Biol., 2010, 14, 154–167 CrossRef.
- S. M. Dean, W. A. Greenberg and C.-H. Wong, Adv. Synth. Catal., 2007, 349, 1308–1320 CrossRef CAS.
- P. Clapés and X. Garrabou, Adv. Synth. Catal., 2011, 353, 2263–2283 CrossRef.
- M. Haridas, E. M. M. Abdelraheem and U. Hanefeld, Appl. Microbiol. Biotechnol., 2018, 102, 9959–9971 CrossRef CAS.
- M. Schürmann, in Industrial Enzyme Applications, ed. A. Vogel and O. May, Wiley-VCH, Weinheim, Germany, 2019, ch. 5.2, pp. 385–403 Search PubMed.
- W. A. Greenberg, A. Varvak, S. R. Hanson, K. Wong, H. Huang, P. Chen and M. J. Burk, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 5788–5793 CrossRef CAS PubMed.
- A. Švarc, M. Fekete, K. Hernandez, P. Clapés, Z. Findrik Blažević, A. Szekrenyi, D. Skendrović, Đ. Vasić-Rački, S. J. Charnock and A. V. Presečki, Chem. Eng. Sci., 2021, 231, 116312 CrossRef.
- A. Švarc, Z. Findrik, D. Vasic-Racki, A. Szekrenyi, W. Fessner, S. Charnock and A. Presečki, J. Chem. Technol. Biotechnol., 2019, 94, 1832–1842 CrossRef.
- M. Ošlaj, J. Cluzeau, D. Orkić, G. Kopitar, P. Mrak and Z. Časar, PLoS One, 2013, 8, e62250 CrossRef.
- X.-C. Jiao, J. Pan, G.-C. Xu, X.-D. Kong, Q. Chen, Z.-J. Zhang and J.-H. Xu, Catal. Sci. Technol., 2015, 5, 4048–4054 RSC.
- S. Jennewein, M. Schürmann, M. Wolberg, I. Hilker, R. Luiten, M. Wubbolts and D. Mink, Biotechnol. J., 2006, 1, 537–548 CrossRef CAS.
- I. Kullartz and J. Pietruszka, J. Biotechnol., 2011, 161, 174–180 CrossRef.
- A. Wang, M. Wang, Q. Wang, F. Chen, F. Zhang, H. Li, Z. Zeng and T. Xie, Bioresour. Technol., 2011, 102, 469–474 CrossRef CAS PubMed.
- H. Fei, C. Zheng, X. Liu and Q. Li, Process Biochem., 2017, 63, 55–59 CrossRef CAS.
- B. Grabner, Y. Pokhilchuk and H. Gruber-Woelfler, Catalyst, 2020, 10, 137 CrossRef CAS.
- R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
- P. Zucca and E. Sanjust, Mol., 2014, 19, 14139–14194 CrossRef.
- Y. T. Zhu, X. Y. Ren, Y. M. Liu, Y. Wei, L. Sen Qing and X. Liao, Mater. Sci. Eng., C, 2014, 38, 278–285 CrossRef CAS PubMed.
- J. Xu, J. Sun, Y. Wang, J. Sheng, F. Wang and M. Sun, Mol., 2014, 19, 11465–11486 CrossRef.
- S. Hasan, Res. J. Recent Sci., 2015, 4, 9–11 Search PubMed.
- S. F. Hasany, I. Ahmed, R. Jose and A. Rehman, Nanosci. Nanotechnol., 2012, 2, 148–158 CrossRef.
- M. Bilal, Y. Zhao, T. Rasheed and H. M. N. Iqbal, Int. J. Biol. Macromol., 2018, 120, 2530–2544 CrossRef CAS PubMed.
- M. Mischitz, U. Pöschl and K. Faber, Biotechnol. Lett., 1991, 13, 653–656 CrossRef CAS.
- A. Wang, W. Gao, F. Zhang, F. Chen, F. Du and X. Yin, Bioprocess Biosyst. Eng., 2012, 35, 857–863 CrossRef CAS PubMed.
- H. Fei, G. Xu, J. P. Wu and L. R. Yang, J. Mol. Catal. B: Enzym., 2014, 101, 87–91 CrossRef CAS.
- S. Reinicke, H. C. Rees, P. Espeel, N. Vanparijs, C. Bisterfeld, M. Dick, R. R. Rosencrantz, G. Brezesinski, B. G. de Geest, F. E. Du Prez, J. Pietruszka and A. Böker, ACS Appl. Mater. Interfaces, 2017, 9, 8317–8326 CrossRef CAS.
- F. Subrizi, M. Crucianelli, V. Grossi, M. Passacantando, G. Botta, R. Antiochia and R. Saladino, ACS Catal., 2014, 4, 3059–3068 CrossRef CAS.
- S. Zhang, J. Bramski, M. Tutus, J. Pietruszka, A. Böker and S. Reinicke, ACS Appl. Mater. Interfaces, 2019, 11, 34441–34453 CrossRef CAS.
- T. Y. Nara, H. Togashi, S. Ono, M. Egami, C. Sekikawa, Y. H. Suzuki, I. Masuda, J. Ogawa, N. Horinouchi, S. Shimizu, F. Mizukami and T. Tsunoda, J. Mol. Catal. B: Enzym., 2011, 68, 181–186 CrossRef CAS.
- S. B. Sigurdardóttir, J. Lehmann, S. Ovtar, J.-C. Grivel, M. Della Negra, A. Kaiser and M. Pinelo, Adv. Synth. Catal., 2018, 360, 2578–2607 CrossRef.
- L. Treccani, T. Yvonne Klein, F. Meder, K. Pardun and K. Rezwan, Acta Biomater., 2013, 9, 7115–7150 CrossRef CAS.
- H. Deng, X. Li, Q. Peng, X. Wang, J. Chen and Y. Li, Angew. Chem., Int. Ed., 2005, 44, 2782–2785 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- J. Lim, S. P. Yeap, H. X. Che and S. C. Low, Nanoscale Res. Lett., 2013, 8, 381 CrossRef.
- M. Kaasalainen, V. Aseyev, E. von Haartman, D. Ş. Karaman, E. Mäkilä, H. Tenhu, J. Rosenholm and J. Salonen, Nanoscale Res. Lett., 2017, 12, 74 CrossRef.
- H. Guo, Y. Tang, Y. Yu, L. Xue and J. Q. Qian, Int. J. Biol. Macromol., 2016, 87, 537–544 CrossRef CAS.
- Y.-B. Shao, T. Jing, J.-Z. Tian, Y.-J. Zheng and M.-H. Shang, Chem. Pap., 2015, 69, 1298–1311 CAS.
- S. Danait-Nabar and R. S. Singhal, Process Biochem., 2022, 122, 181–195 CrossRef CAS.
- N. S. Rios, D. M. A. Neto, J. C. S. dos Santos, P. B. A. Fechine, R. Fernández-Lafuente and L. R. B. Gonçalves, Int. J. Biol. Macromol., 2019, 134, 936–945 CrossRef CAS.
- K. Banjanac, M. Mihailovic, N. Prlainović, M. Ćorović, M. Carević, A. Marinkovic and D. Bezbradica, J. Chem. Technol. Biotechnol., 2016, 91, 2654–2663 CrossRef CAS.
- R. Chen, Q. Wei, X. Wei, Y. Liu, X. Zhang, X. Chen, X. Yin and T. Xie, PLoS One, 2020, 15, e0242564 CrossRef CAS PubMed.
- I. Migneault, C. Dartiguenave, M. J. Bertrand and K. C. Waldron, BioTechniques, 2004, 37, 790–802 CrossRef CAS PubMed.
- S. Zhang, C. Bisterfeld, J. Bramski, N. Vanparijs, B. G. De Geest, J. Pietruszka, A. Böker and S. Reinicke, Bioconjugate Chem., 2018, 29, 104–116 CrossRef CAS.
- A. Švarc, Z. Findrik Blažević, Đ. Vasić-Rački, S. J. Charnock and A. Vrsalović Presečki, Chem. Eng. Res. Des., 2020, 164, 35–45 CrossRef.
- T. E. Bapiro, A. Sykes, S. Martin, M. Davies, J. W. T. Yates, M. Hoch, H. E. Rollison and B. Jones, Drug Metab. Dispos., 2018, 46, 1268–1276 CrossRef CAS.
- C. B. Elias and J. B. Joshi, Adv. Biochem. Eng./Biotechnol., 1998, 59, 47–71 CAS.
- R. E. Abraham, M. L. Verma, C. J. Barrow and M. Puri, Biotechnol. Biofuels, 2014, 7, 90 CrossRef PubMed.
- J. Jordan, C. S. S. R. Kumar and C. Theegala, J. Mol. Catal. B: Enzym., 2011, 68, 139–146 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3re00388d |
| This journal is © The Royal Society of Chemistry 2024 |