Open Access Article
Open Access ArticleCovalent-organic framework nanobionics for robust cytoprotection†
Jieying
Liang‡
*a,
Qianfan
Chen‡
b,
Joel
Yong
a,
Hiroki
Suyama
c,
Joanna
Biazik
d,
Bosiljka
Njegic
e,
Aditya
Rawal
 e and
Kang
Liang
e and
Kang
Liang
 *ab
*ab
aSchool of Chemical Engineering and Australian Centre for NanoMedicine, The University of New South Wales, Sydney, NSW 2052, Australia. E-mail: jieying.liang@unsw.edu.au; kang.liang@unsw.edu.au
bGraduate School of Biomedical Engineering, The University of New South Wales, Sydney, NSW 2052, Australia
cUNSW RNA Institute, The University of New South Wales, Sydney, NSW 2052, Australia
dElectron Microscope Unit, Mark Wainwright Analytical Centre, The University of New South Wales, Sydney, NSW 2052, Australia
eNuclear Magnetic Resonance Facility, Mark Wainwright Analytical Centre, University of New South Wales, Sydney, NSW 2052, Australia
First published on 13th December 2023
Abstract
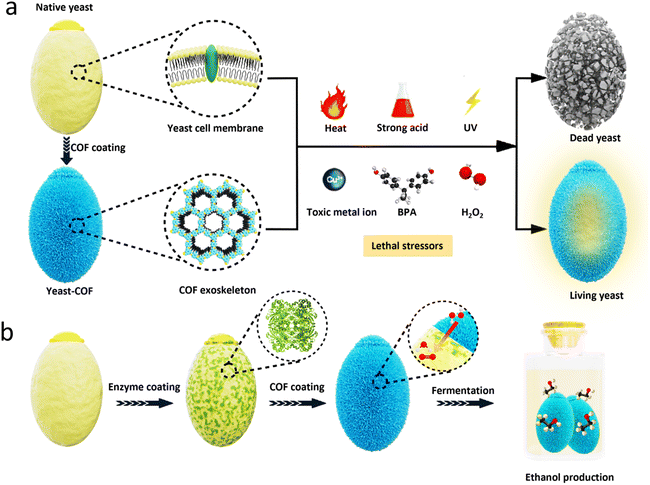
We present a novel study introducing a durable and robust covalent-organic framework (COF) nanocoating, developed in situ on living cells. This COF nanocoating demonstrates remarkable resistance against a diverse range of lethal stressors, including high temperature, extreme pH, ultraviolet radiation, toxic metal ions, organic pollutants, and strong oxidative stress. Notably, the nanocoating exhibits exceptional cell survival enhancement under high temperature and strongly acidic conditions, an aspect yet unexplored in the case of metal–organic framework nanocoatings and other nanomaterials. Moreover, functionalization of the nanocoating with an exogenous enzyme catalase enables yeast fermentation and ethanol production even under strong oxidative stress. Our findings establish the durable and robust COF nanocoating as a reliable platform for safeguarding vulnerable microorganisms to allow their utilisation in a wide range of adverse environments.
Introduction
Microorganisms play a crucial role in various aspects of human life, including medicine, food, and biotechnology.1–8 However, their survival under sudden and stressful environmental changes, such as high temperature, irradiation, high ionic strength, oxidizing agents, and toxic chemicals (e.g., heavy metals, organic pollutants), remains challenging. While natural cell membranes and/or cell walls offer limited protection, artificial shells composed of organic, inorganic, or hybrid materials have been developed to enhance their robustness at the individual cell level.9–13Among these materials, metal–organic frameworks (MOFs) have shown promise as candidate materials for artificial cell exoskeletons.14 MOFs, constructed from metal ions or metal-containing clusters and organic linkers, possess versatile functionality, tunable pore size, well-defined pore aperture, tailorable composition, and structure.15–19 These properties are particularly advantageous for controlling molecular transport through coatings.20–24 MOF coatings can be formed around living cells through in situ or post-loading strategies. For instance, previous work demonstrated the coating of living microorganisms, Saccharomyces cerevisiae and Micrococcus luteus, with thin zeolitic imidazolate framework-8 (ZIF-8) nanocoatings, inducing their hibernation state.25 Functionalization of the nanocoating with a bioactive exogenous enzyme layer, such as β-galactosidase, allowed yeast cells to survive in nutrient-depleted environments by converting lactose to glucose.26 MOFs can also be post-functionalized around bacteria through coordination interactions between teichoic acids on the cell wall and zirconium clusters on certain MOFs.27 Furthermore, tannic acid has been used as a linker between inorganic nanoparticles and the cell surface.28,29 Additionally, artificial shells have been successfully coated onto cell-like structures, including chloroplasts and viruses, using versatile MOF-based nanocoating strategies.30,31
However, the limited stability of most MOFs in biological buffers, cell medium, and serum can lead to MOF dissolution and metal ion release.32 This compromises their protective capability and can have harmful effects on living cells.33 While this feature allows for on-demand nanocoating degradation,25,34 the development of a functional and robust nanocoating that protects living cells from harsh environments, such as extreme pH and oxidative stress, would greatly expand the potential of microorganisms in biotechnology and food industries. Covalent-organic frameworks (COFs), constructed from organic building units linked by strong covalent bonds, present an ideal solution as nanocoating materials with highly uniform porosity similar to MOFs.35 Recent advancements in biofriendly COF synthesis strategies for enzyme encapsulation have further sparked interest in exploring COFs as durable and robust nanocoatings for living cells.36–38 The appealing features of COFs, including acidic stability, metal-free composition, precisely tunable functionalities and pore geometries, topological diversity, and well-defined structures, hold the potential to confer living cells with unprecedented properties.
Yeast, the most widely used single-cell research organism,39 was selected as the primary model for proof of concept. In this study, we successfully constructed COF nanocoatings on living Saccharomyces cerevisiae (baker's yeast) using an in situ strategy. The resulting COF nanocoating imparts exceptional protection to yeast cells against various adverse external environments, including high temperatures, extreme pH levels, ultraviolet radiation, toxic metal ions, organic pollutants, and strong oxidative stress. Notably, the bare yeast cells are unable to survive under such conditions. Remarkably, the COF nanocoating demonstrates exceptional resistance to high temperature and strongly acidic conditions, surpassing the capabilities of MOFs and other nanomaterials typically employed as artificial coatings. Furthermore, by incorporating the exogenous enzyme catalase (CAT) within the COF nanocoating, we enable the decomposition of hydrogen peroxide (H2O2), allowing the yeast cells to sustain continuous fermentation even in the presence of strong oxidative stress. This simple COF nanocoating strategy could find use in biotechnology, food industries, pharmaceuticals, and environmental remediation, among others (Scheme 1).
Results and discussion
Synthesis and characterization of COF-coated yeast cells
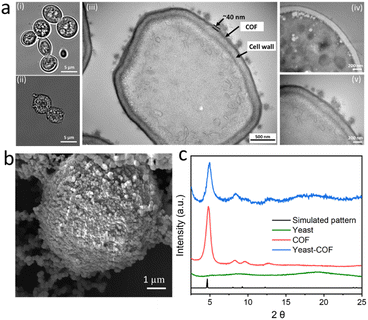
COF-LZU1, based on our previous research, was selected as the ideal nanocoating for yeast cells due to its weak host–guest interaction and substrate accessibility.36 The encapsulation of S. cerevisiae cells within COF-LZU1-based exoskeletons (referred to as yeast-COF) involved a sequential addition of 1 mL of cell suspensions (with an optical density at 600 nm (OD600) of 1.0), 1 mL of p-phenylenediamine (PPDA) (10 mg mL−1), 10 mL of benzene-1,3,5-tricarboxaldehyde (BTCA) (0.5 mg mL−1), and 1 mL of acetic acid (1.742 M). After a 5 minute reaction, 0.9 mL of NaOH (4 M) was added, followed by an additional 10 minute reaction. The yeast cells were then removed from the solution and washed with water to eliminate excess COF precursors. Optical micrographs revealed noticeable distinctions between native yeast cells and yeast-COF. Native yeast displayed a smooth edge, whereas yeast-COF exhibited a relatively rough edge (Fig. 1a, i–ii, S1, and S2i†). Fluorescent micrographs further demonstrated the homogeneous COF shell formation on each cell after infiltration with a fluorescent dye, Alexa Fluor 350, as evidenced by the continuous fluorescent signal surrounding individual cells (Fig. S2ii–iii†). Scanning electron microscopy (SEM) revealed a rough surface coating surrounding the yeast cells, while native yeast cells exhibited a smooth surface (Fig. 1b, S3, and S4†). Transmission electron microscopy (TEM) images revealed that the COF shell consisted of two components: a uniform thin film firmly adhering to the cell wall and particle aggregates (Fig. 1a, iii). The strong attachment is attributed to covalent bonding during the one-pot reaction between the COF and the amine or thiol groups of (glyco)proteins in the yeast cell wall.40 The average coating thickness was estimated to be approximately 40 nm (Fig. 1a, iii–v). X-ray diffraction (XRD) patterns confirmed the crystallinity of yeast-COF, exhibiting positions and relative intensities analogous to pure COF-LZU1, thereby verifying the structure and crystallinity of the COF nanocoating (Fig. 1c). However, the surface area of yeast-COF (22.5 m2 g−1) was lower than that of the pure COF (53 m2 g−1) (Table S1 and Fig. S5†). Fourier transform infrared (FTIR) spectra showed the emergence of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretch at 1620 cm−1, accompanied by the disappearance of aldehydic C–H and C
N stretch at 1620 cm−1, accompanied by the disappearance of aldehydic C–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of BTCA, as well as the N
O stretching vibrations of BTCA, as well as the N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) H stretching vibrations of PPDA, indicating the successful formation of the COF (Fig. S6a†). The presence of an imine C
H stretching vibrations of PPDA, indicating the successful formation of the COF (Fig. S6a†). The presence of an imine C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretch at 1620 cm−1 in yeast-COF is consistent with the pure COF (Fig. S6b†). Synchrotron terahertz/far-infrared radiation (THz FIR) spectra revealed that vibrations at ∼100 cm−1 in yeast and yeast-COF samples (Fig. S7†), which primarily originated from different lipids of the yeast cell membrane interacting through intermolecular van der Waals forces.41 Importantly, these vibrations did not arise from hydration of water, as all samples were fully freeze-dried at −60 °C for 3 days. Moreover, the yeast-COF sample displayed lower vibrations of different lipids in the cell membrane due to COF immobilization (Fig. S7†). Additionally, 1D 13C NMR spectra were recorded for yeast-COF. Yeast cells exhibited characteristic peaks, including an N–C
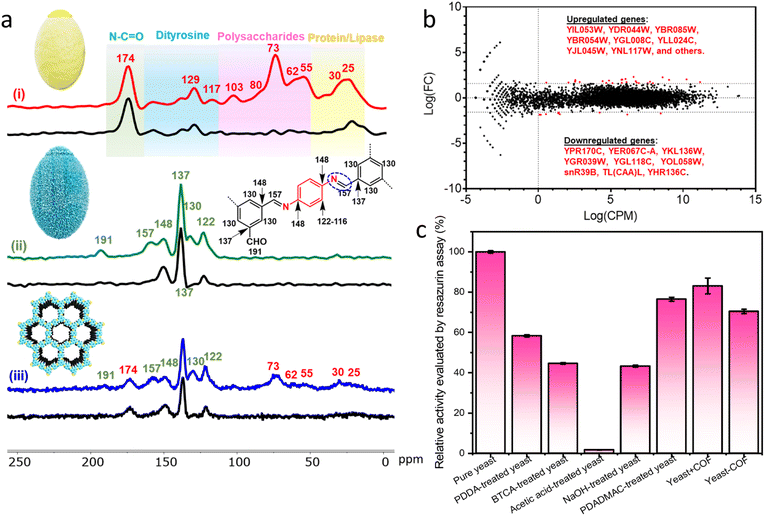
N stretch at 1620 cm−1 in yeast-COF is consistent with the pure COF (Fig. S6b†). Synchrotron terahertz/far-infrared radiation (THz FIR) spectra revealed that vibrations at ∼100 cm−1 in yeast and yeast-COF samples (Fig. S7†), which primarily originated from different lipids of the yeast cell membrane interacting through intermolecular van der Waals forces.41 Importantly, these vibrations did not arise from hydration of water, as all samples were fully freeze-dried at −60 °C for 3 days. Moreover, the yeast-COF sample displayed lower vibrations of different lipids in the cell membrane due to COF immobilization (Fig. S7†). Additionally, 1D 13C NMR spectra were recorded for yeast-COF. Yeast cells exhibited characteristic peaks, including an N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak at 174 ppm, polysaccharide peaks at 73 ppm, 62 ppm, and 55 ppm, and protein/lipase peaks at 30 ppm and 25 ppm (Fig. 2a, i). The COF-LZU1 showed a distinct 13C NMR peak at 157 ppm, corresponding to the carbon atom of the C
O peak at 174 ppm, polysaccharide peaks at 73 ppm, 62 ppm, and 55 ppm, and protein/lipase peaks at 30 ppm and 25 ppm (Fig. 2a, i). The COF-LZU1 showed a distinct 13C NMR peak at 157 ppm, corresponding to the carbon atom of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond. This peak is indicative of the condensation reaction between aldehyde BTCA and PPDA, confirming the successful integration of yeast and the COF (Fig. 2a, ii–iii). Additionally, a small amount of aldehyde groups can be observed at 191 ppm, which is attributed to the imperfect COF formation around the yeast cells despite that the unreacted COF precursors were washed away during the synthesis process. The excess aldehyde groups may have led to the formation of COF aggregates as seen on the yeast-COF surface (Fig. 1a, iii).36
N bond. This peak is indicative of the condensation reaction between aldehyde BTCA and PPDA, confirming the successful integration of yeast and the COF (Fig. 2a, ii–iii). Additionally, a small amount of aldehyde groups can be observed at 191 ppm, which is attributed to the imperfect COF formation around the yeast cells despite that the unreacted COF precursors were washed away during the synthesis process. The excess aldehyde groups may have led to the formation of COF aggregates as seen on the yeast-COF surface (Fig. 1a, iii).36
Numerous cellular activities, such as cell identification, metabolism, protein synthesis, and survival, are regulated by receptors on the cell surface. To gain insight into how the exoskeletons changed yeast cell fate, gene expression profiles of the yeast cell and yeast-COF were subjected to transcriptomics analysis. It was found that several genes associated with oxygen availability, anaerobic respiration, stress responses, energy metabolism, glucose utilization, and amino acid biosynthesis were upregulated after COF nanocoating (Fig. 2b and Table S2†). This suggests a potential increase in energy metabolism for the COF-coated yeast cells. Conversely, genes related to ribosome and amino acid biosynthesis were downregulated (Fig. 2b and Table S2†), which is unlikely to affect the yeast cell function. Out of the 7127 genes with matched reads identified in this study, only 0.49% of genes exhibited differential expression, with 9 genes potentially downregulated and 26 upregulated. The result suggests that the COF coating had negligible adverse effect on the yeast.
To evaluate the cellular viability during the COF coating process, resazurin and fluorescein diacetate (FDA) assays were conducted, assessing esterase and mitochondrial activity in metabolically intact cells, respectively.42 The relative activity of native yeast cells before and after treatment with the COF precursors was measured. The resazurin assay showed that 58.4%, 44.6%, and 43.3% of cellular activity was retained after treatment with PDDA, BTCA, and NaOH, respectively (Fig. 2c), indicating some toxicity of the precursors to the cells. Native yeast cells were completely inactive under strongly acidic conditions. However, upon immobilization within COFs, 72.5% of cellular activity was maintained, with the COF itself exhibiting negligible toxicity (90.5% cellular activity retained). Similar results were obtained with the FDA assay, confirming the minimal toxicity of the COF to yeast cells (Fig. S8†). These findings suggest that although the precursors affect cellular activity, the rapid formation of the COF during the coating process protects and minimizes the harmful effects on yeast cells. To further demonstrate the minimal toxicity of the COF to yeast growth, a control test was conducted by combining free COF particles with yeast cells in a culture medium for 48 hours. The growth curve of the yeast cells in the presence of free COF particles was essentially identical to that of native yeast cells, confirming the lack of adverse effects on yeast growth (Fig. S9†). When immobilized within the COF nanocoating, yeast cells showed negligible growth, indicating the ability of the nanocoating to prevent yeast cell division and maintain a quiescent state (Fig. S10†). Moreover, yeast cells encapsulated within the COF nanocoating remained viable in a normal cell medium for a week with minimal loss of cellular activity (Fig. S11†). XRD spectra also demonstrated the preserved crystal structure of the COF after incubation in a cell medium for a week, highlighting the durability and robustness of the COF as a protective shell for microorganisms (Fig. S12†).
Cytoprotective role of the COF nanocoating
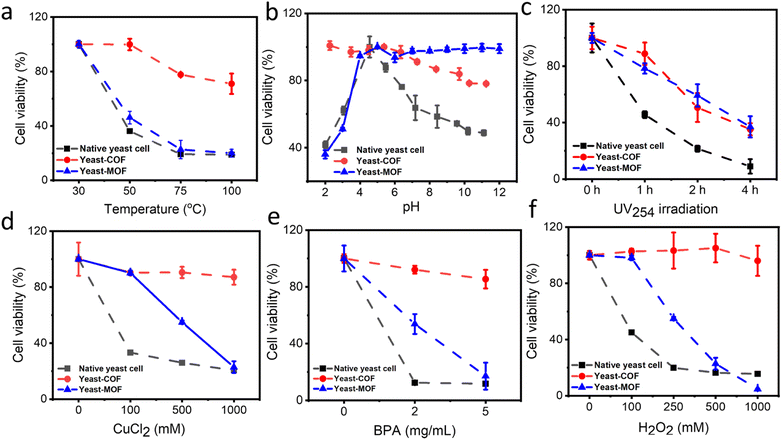
The cytoprotective properties of the COF nanocoating were investigated to evaluate its ability to protect living cells against various external cellular stressors. Industrially relevant stress factors encompass high temperatures, pH fluctuations, oxidative stress (i.e., H2O2), and high metal ion contents (e.g., Cu2+).43 In addition, other environmental conditions, including bisphenol A (BPA) and UV radiation, were investigated. BPA, known as an endocrine-disrupting chemical, is globally produced in significant quantities and continually leaches into the environment, potentially impacting the microbial community health.44 Meanwhile, UV radiation has adverse effects on living organisms, inducing cytotoxic and mutagenic DNA lesions.45First, the COF nanocoating exhibited excellent cell protection against high temperatures. Native yeast cells showed a decrease in viability with increasing temperature, with only about 20% residual viability after treatment at 100 °C for 30 minutes. In contrast, approximately 70% of COF-yeast cells remained viable under the same conditions (Fig. 3a and S13†). To understand the unexpected effects of COF on cellular viability during heating, the impact of heat exposure on heat flow and water loss was investigated. Native yeast cells and yeast-COF were first dried at 30 °C for 48 hours, and then heat flow and cell weight loss were measured using differential scanning calorimetry (DSC) and thermogravimetric analysis (TGA), respectively. The DSC results clearly demonstrated that yeast-COF exhibited higher heat flow compared to native yeast cells at temperatures of 50 °C, 75 °C, and 100 °C (Fig. S14†). This phenomenon suggests that yeast-COF undergoes a more effective endothermic process than bare yeast cells, indicating that COF shells absorb more heat due to the temperature difference between the cell samples and a reference.46 Upon increasing the temperature to 50 °C, natural yeast cells experienced a rapid loss of approximately 56% of their water content. After 30 minutes at 50 °C, the water loss increased to 67% (Fig. S15†). As the temperature increased to 70 °C and 100 °C and was maintained for 30 minutes, the water content of natural yeast cells decreased further by 2% and 2.5%, respectively. The significant reduction in water content observed in natural yeast cells is attributed to excessive evaporation at high temperatures, leading to increased osmotic stress and subsequent water loss from the cells. In contrast, when yeast-COF was exposed to a temperature increase of 50 °C, only 12% of water was lost, and after holding at 70 °C and 100 °C for 30 minutes, an additional water loss of only 2% was observed. This enhanced water retention capability is attributed to the COF coating, which enables yeast cells to remain in a fluid and water-rich microenvironment, shielding them from the detrimental effects of high temperatures. Remarkably, the temperature of yeast-COF when exposed to various ambient temperatures (∼30 °C, 50 °C, 75 °C, and 100 °C) was 0.3 °C, 0.7 °C, 1.2 °C, and 1.1 °C lower than that of native yeast cells, respectively (Fig. S16†). The lower surface temperature of the COF shells can be attributed to the low thermal conductivity of the COF shell and aggregates,47 which reduce the heat load of yeast-COF. This unique property of the COF has not been explored in MOF artificial shells. Moreover, the temperature tolerance of yeast-COF (up to 100 °C) is significantly higher than that of MOF (Fig. 3a), silica (∼50 °C)46 and SiO2–TiO2 nanoshells (∼45 °C).48
Second, we investigated the viability of yeast-COF across a pH range of 2–11, as cell metabolic processes can be disrupted by extreme pH conditions, leading to irreversible cell damage.49 As depicted in Fig. 3b and S17,† pH 4 was identified as the optimal condition for yeast cell survival, consistent with previous findings.50,51 The buffer salts demonstrate negligible effect on the cell viability (Fig. S18†). Deviating from this optimal pH range, both lower and higher pH levels were observed to detrimentally affect yeast viability. However, yeast-COF exhibited remarkable resistance to pH variations, displaying 2.5-fold and 1.6-fold higher viability than native yeast cells at pH 2 and 11, respectively. This enhanced pH resistance can be attributed to the ion chelating effect facilitated by the porous exoskeleton framework of yeast-COF.52–54 In contrast, the yeast-MOF is more stable under alkaline conditions (Fig. 3b). Notably, the exceptional performance of yeast-COF under strongly acidic conditions has not been explored in MOFs and other nanomaterial cell coatings, thereby showcasing its potential to significantly expand the applications in biotechnology and food industries.
Third, we assessed the resistance of yeast-COF to UV irradiation (λ = 254 nm, 4 W) in comparison to native yeast cells. As anticipated, native cells experienced a sharp decline in survival, with only 10% viability remaining after four hours of UV exposure. In contrast, yeast-COF exhibited 3.5 times higher viability than native yeast cells under the same exposure conditions (Fig. 3c and S19†). The enhanced UV protection of yeast-COF can be attributed to the absorbance of imine-COF in the UV region,55 which effectively shields the underlying yeast cells from the harmful effects of UV radiation. The UV protection performance of the COF is similar to that of the MOF (Fig. 3c).
Fourth, we exposed both native yeast cells and yeast-COF to toxic inorganic and organic environmental pollutants, namely copper ions (Cu2+) and bisphenol A (BPA), at various concentrations. As depicted in Fig. 3d, e and S20,† native yeast cells demonstrated high sensitivity to Cu2+ and BPA. After one hour of treatment with 1 M Cu2+ and 5 mg mL−1 BPA, only 20% and 11% of yeast cells remained viable, respectively. In contrast, yeast-COF exhibited remarkable protection, with 87% and 85% viability observed under the same treatment conditions. The yeast-MOF demonstrated a significantly decreased cell activity at higher Cu2+ and BPA concentrations (Fig. 3d and e). This substantial enhancement in protection conferred by the COF nanocoating can be attributed to specific mechanisms. The incorporation of Cu2+ into the COF skeleton through coordination interactions with imine linking groups of the COF contributes to the excellent protection against Cu2+.56 Additionally, the COF exhibits strong retention and rapid adsorption of BPA in aqueous media due to π–π interactions and hydrogen bond interactions,57 thereby effectively shielding the yeast cells from the toxic effects of BPA exposure.
Finally, we investigated the effect of excessive oxidative stress, a process known to induce oxidative damage and harmful alterations to cellular components such as lipids and DNA.58 As depicted in Fig. 3f and S21,† the viabilities of yeast-COF exhibited negligible change even with increasing H2O2 dosage, and were significantly higher than those of native yeast cells (with only 15% cell viability remaining) in the presence of escalating H2O2 concentrations. Native yeast cells possess a native oxidative stress scavenging capability, allowing them to rapidly decompose H2O2 and generate O2 (Fig. S22†). However, their tolerance is limited to small amounts of oxidative stress (up to 2 mM H2O2 (ref. 59)). When exposed to excessive levels of H2O2 beyond their tolerance capacity, native yeast cells suffer protein, DNA, and membrane injuries, ultimately leading to cell death (Fig. S21†). Compared to the native yeast, the increased resistance of yeast-COF to H2O2 can be attributed to the physical barrier provided by the COF frameworks (Table S1†), which restricts the diffusion of H2O2 molecules (Fig. S22†).36 This barrier effectively prevents the entry of excessive H2O2 into the yeast cells, thereby reducing oxidative damage and preserving cell viability. Whereas yeast-MOF demonstrated a notable reduction in cell activity as the H2O2 concentration increased (Fig. 3f). Therefore, the COF nanocoating proves to be an effective artificial shield, enabling yeast cells to withstand a wide range of lethal stressors, including oxidative stress.
In summary, the COF outperforms the MOF in safeguarding yeast against high temperatures, acidic environments, highly toxic metal and organic pollutant concentrations, and strong oxidizing conditions. On the other hand, the MOF provides superior protection for yeast in alkaline environments and offers comparable protection against UV radiation. Furthermore, in comparison to previous nanocoatings (Table S3†), the COF nanocoating demonstrates remarkable enhancement in cell survival under high temperatures (up to 100 °C) and highly acidic conditions (pH 2), areas where other nanomaterials have not been explored.
Biocatalytic COF for improved cytoprotection under oxidative stress
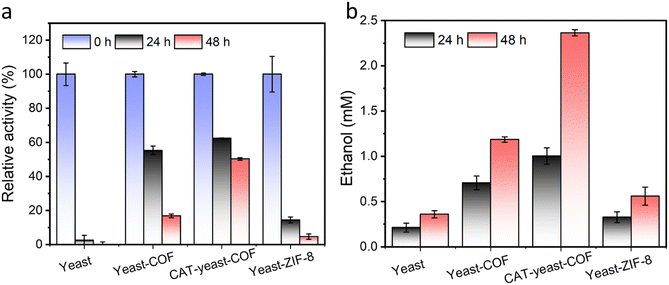
To enhance the long-term survival of yeast cells under strong oxidative stress, we developed a biocatalytic approach by immobilizing the enzyme CAT onto the yeast-COF system. To facilitate the immobilization of CAT onto the yeast cells, a poly(diallyldimethylammonium chloride) (PDADMAC) coating46 was applied to enhance the adsorption of the negatively charged CAT onto the yeast cells through electrostatic interactions. This step was necessary due to the negatively charged cell wall of S. cerevisiae (zeta potential = −36.3 mV) making it difficult for CAT to directly adsorb onto the cells. The resulting PDADMAC-yeast-CAT complex was then functionalized with COF nanocoatings through electrostatic interactions and/or covalent bonding between the COF and the amine or thiol groups of (glyco)proteins on the CAT-yeast, leading to the formation of CAT-yeast-COF. Successful immobilization of the enzyme and COF onto the yeast cells was confirmed by the positive zeta potential of PDADMAC-yeast-CAT (24.5 mV) and the negative zeta potential of CAT-yeast-COF (−10.6 mV) (Fig. S23†). Additionally, THz FIR spectra demonstrated reduced vibrations of lipids from the yeast cell membrane upon COF immobilization in CAT-yeast-COF (Fig. S24†). Optical micrographs (Fig. S25i†) and fluorescent micrographs further confirmed the successful loading of CAT within the COF structure (Fig. S25ii–iv†). Approximately 50.0% of CAT was successfully loaded onto the yeast cells, as detected using a fluorescent protein labelling method (Fig. S23 and S25†). The H2O2 decomposing rate of CAT-yeast-COF was significantly higher compared to that of yeast-COF alone (Fig. S26†), and the immobilized CAT retained 57% of its enzyme activity (Fig. S27†).When native yeast cells were incubated in 100 mM H2O2 for 24 hours, they experienced near complete cell death. In contrast, when protected by COF and CAT-COF coatings, 16.8% and 50.2% of yeast cells survived after 48 hours of incubation, respectively (Fig. 4a). These findings highlight the effectiveness of COF and enzyme coatings in preserving cellular activity over a relatively long-term period under strong oxidative stress. Alcohol fermentation is a crucial process in the production of various alcoholic beverages and bioethanol, as well as in the generation of by-products in the food industry.60S. cerevisiae is a commonly used industrial host for ethanol production, and substantial efforts have been dedicated to engineering yeast cell factories to enhance ethanol production.61 In a specific nutritional setting, native yeast reached its peak ethanol production at approximately 10 days when all the glucose was depleted. In contrast, yeast-COF demonstrated reduced ethanol production compared to their uncoated counterparts, primarily due to limited substrate diffusion (Fig. S28–S30†). Nevertheless, both types of yeast cells exhibited similar ethanol production abilities, achieving stable ethanol yields of 26.7 mM and 29.6 mM, respectively, over a 25 day period. This was a result of yeast-COF's slower but continuous nutrient consumption, with all glucose in the yeast-COF system being completely utilized by day 25 (Fig. S28–S30†). To investigate ethanol generation under strong oxidative stress, native yeast cells, yeast-COF, and CAT-yeast-COF were incubated in the presence of 100 mM H2O2 for 48 hours. The native yeast cells produced only 0.359 mM ethanol (Fig. 4b) under these conditions, as determined by the ethanol determination standard curve (Fig. S31†). In contrast, yeast-COF exhibited enhanced ethanol generation, producing 1.184 mM ethanol (Fig. 4b). This indicates that the COF coating partially protects the yeast cells from the strong oxidative environment by limiting substrate diffusion. Further improvement in ethanol generation was observed with CAT-yeast-COF, reaching 2.36 mM ethanol (Fig. 4b). These results highlight the ability of COF and CAT immobilization to enhance yeast cell fermentation under strong oxidative stress.
In addition to the comparison with native yeast cells, the performance of COF-coated yeast cells (yeast-COF) can also be compared to that of yeast cells coated with ZIF-8 (yeast-ZIF-8) using our previous methods,25,26 ZIF-8 has been extensively studied as an artificial shell for protecting living cells from adverse environments.4,5,25,26,28,30,34,62 The successful coating of ZIF-8 on yeast cells is confirmed by the SEM image and small-angle X-ray scattering (SAXS) spectra (Fig. S32 and S33†). When exposed to 100 mM H2O2 for 48 hours, only 4.7% of yeast-ZIF-8 cells remain viable (Fig. 4a). Moreover, yeast-ZIF-8 exhibits much lower ethanol generation (0.559 mM in 48 hours) compared to yeast-COF and CAT-yeast-COF, although it is slightly higher than that of native yeast cells (Fig. 4b). To investigate the reason behind the inferior protective role of ZIF-8 against oxidative stress, the stability of ZIF-8 is examined. It is observed that the pH of cell medium in native yeast cells and yeast-COF samples decreases from 5.91 to 4.96 and 4.86, respectively, upon exposure to 100 mM H2O2 for 48 hours, possibly due to the generation of organic acids during fermentation.63,64 However, the pH of the cell medium in the yeast-ZIF-8 sample (5.21) is slightly higher than that of native yeast cells and yeast-COF but lower than that of the cell medium (Fig. S34a†). Further investigation reveals that 0.43 mg L−1 of zinc is released into the supernatant after 48 hours of incubation, as demonstrated by inductively coupled plasma (ICP) analysis (Fig. S34b†). Consequently, the higher pH of the cell medium in the yeast-ZIF-8 sample is attributed to the dissolution of ZIF-8 under such conditions, where the basic 2-methylimidazole precursor of ZIF-8 is released into the supernatant. Therefore, ZIF-8 is relatively unstable when incubated in the cell medium for a relatively longer time and provides negligible protection for yeast cells under strong oxidative stress. In contrast, the COF remains stable under acidic conditions. To the best of our knowledge, this is the first study demonstrating the simultaneous co-immobilization of CAT and the COF on yeast cells to reduce strong oxidative stress and improve their metabolic products (Table S3†). These findings highlight the superior stability and protective capabilities of COF-coated yeast cells compared to ZIF-8-coated yeast cells under strong oxidative stress conditions. The stable nature of the COF in acidic environments and its ability to retain enzymatic activity contribute to its enhanced performance in cytoprotection and metabolic product generation.
Experimental section
Reagents and materials
Saccharomyces cerevisiae (Baker's yeast, domain: eukarya, kingdom: fungi), Australia, yeast extract, D-(+)-glucose, p-phenylenediamine (PPDA, 99% purity), benzene-1,3,5-tricarboxaldehyde (BTCA), dimethylformamide (DMF), catalase (CAT) from bovine liver, lyophilized powder, 2000–5000 units per mg protein, resazurin sodium salt, bisphenol A, cupric chloride (CuCl2), zinc acetate (Zn(CH3COO)2), 2-methylimidazole (99.0%), and poly(diallyldimethylammonium chloride) solution (PDADMAC) (1.04 g L−1; Mw = 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–3500
000–3500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were purchased from Sigma Aldrich (Australia). Fluorescein diacetate (FDA) was purchased from Life Technologies (Australia). Alexa Fluor 350 and Atto 647N NHS ester were purchased from Thermo Fisher Scientific (Australia), and hydrogen peroxide (27–30%, 500 mL) was purchased from Chem-Supply Pty Ltd Australia. All the other reagents were purchased from Sigma Aldrich (Australia) and used without further modification.
000) were purchased from Sigma Aldrich (Australia). Fluorescein diacetate (FDA) was purchased from Life Technologies (Australia). Alexa Fluor 350 and Atto 647N NHS ester were purchased from Thermo Fisher Scientific (Australia), and hydrogen peroxide (27–30%, 500 mL) was purchased from Chem-Supply Pty Ltd Australia. All the other reagents were purchased from Sigma Aldrich (Australia) and used without further modification.
Formation of COF coatings on living yeast cells
2 mg of dry yeast cells were cultured in 10 mL yeast culture media containing yeast extract (10 mg mL−1) and glucose (20 mg mL−1) with continuous shaking at 30 °C and 220 rpm for 18 hours. The yeast cells were then washed with deionized (DI) water three times and suspended in 1 mL aqueous solution. To the yeast cell aqueous solution, 250 μL of DMF containing 5 mg of BTCA was added, followed by dispersion in 10 mL of DI water. Then, 1 mL of aqueous solution containing 10 mg PPDA and 1 mL of acetic acid (1.742 M) were sequentially added to the yeast cell solution. The mixture was placed on a shaking stage at 300 rpm for 5 minutes. Next, 0.9 mL of NaOH (4 M) was added, and the mixture was further shaken for another 10 minutes to facilitate the formation of COF coatings. The coated cells were washed with DI water three times to remove excess COF precursors and finally suspended in 1 mL of DI water. For the CAT-yeast-COF synthesis, 1 mL of yeast cells was first coated with 1 mL of PDADMAC (0.1 g![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L−1). After removing the excess PDADMAC, 100 μL of CAT solution (10 mg mL−1) was added to the 1 mL yeast solution prior to synthesis. The same procedures as described above for yeast-COF formation were followed for CAT-yeast-COF synthesis.
L−1). After removing the excess PDADMAC, 100 μL of CAT solution (10 mg mL−1) was added to the 1 mL yeast solution prior to synthesis. The same procedures as described above for yeast-COF formation were followed for CAT-yeast-COF synthesis.
Quantification of enzyme loading in CAT-yeast-COFs
A fluorescent protein labelling method was used for quantitative analysis of the enzyme loading in the CAT-yeast-COF sample using a CLARIOstar microplate reader (BMG LABTECH Germany). 50 μL of Atto 647N NHS ester (λex 318 nm; λem 663 nm) dissolved in DMSO (2 mg mL−1) was added to 1 mL of enzyme solution (10 mg mL−1 CAT). After 2 h, excess unreacted dye molecules were removed via a NAP-25 Column (GE Healthcare). The dye-labelled CAT enzymes were then subjected to the same procedure to synthesize the CAT-yeast-COF biocomposite. Finally, 100 μL of supernatant was added into a 96-well plate. The loading of encapsulated enzymes in CAT-yeast-COFs was determined by subtracting the fluorescent signal of enzymes in the supernatant.Characterization
An FEI Nova NanoSEM 230 was used to obtain SEM images of the samples after Pt coating. Crystallinity analysis of biocatalytic enzyme-COF was performed using XRD-MPD-powder diffraction. The samples were vacuum dried and analysed using PXRD measurement. Small-angle X-ray scattering (SAXS) patterns were collected at the Australian Synchrotron using a Pilatus 1 M detector and a 20 keV beam. Fourier transform infrared (FTIR) spectra were recorded on a Bruker Alpha FTIR Spectrometer. Metal ion content in the samples was determined using Inductively coupled plasma-optical emission spectroscopy (ICP-OES) instruments (Optima7000 and Avio from PerkinElmer, USA). Synchrotron Far-infrared (FIR)/terahertz (THz) radiation was collected at the Australian Synchrotron using an attenuated total reflectance (ATR) sampling accessory in the range of 50–750 cm−1.Transmission electron microscopy (TEM)
A TEM (JEOL 1400) was utilized to characterize the COF shell. Yeast and COF-yeast samples were fixed in a 2.5% (w/v) glutaraldehyde solution in 0.2 M sodium phosphate buffer at 4 °C overnight. Subsequently, the fixed samples underwent rinsing with 0.1 M sodium phosphate buffer, followed by post-fixation in a 1% osmium tetroxide solution (ProSciTech) in 0.2 M sodium buffer using a BioWave Pro+ Microwave Tissue Processor (Ted Pella, USA). After another round of rinsing with 0.1 M sodium phosphate buffer, the samples were dehydrated using a graded series of ethanol, infiltrated with Procure 812 resin (EMS, USA), and polymerized in an oven at 60 °C for 48 hours. Ultrathin sections measuring 70 nm were then obtained using a diamond knife (Diatome, Switzerland) and collected onto carbon-coated copper slot TEM grids (Ted Pella, USA). The grids were subsequently post-stained with 2% uranyl acetate (EMS, USA) and lead citrate. For each sample, two grids were collected and imaged using a JEOL 1400 TEM (Tokyo, Japan) operating at 100 kV.Solid-state NMR
The 1D solid-state 13C NMR experiments were conducted on a Bruker AVANCE III 300 spectrometer equipped with a 7 Tesla superconducting magnet operating at frequencies of 300 MHz and 75 MHz for the 13C nuclei. The sample was packed into 4 mm zirconia rotors fitted with a Kel-F® cap and spun at magic angle (MAS) frequencies of 8 kHz and 12 kHz. For the acquisition of 1D 13C spectra, a 1 ms ramped cross-polarization from the 1H nuclei was employed, along with the Total Suppression of Spinning sidebands (TOSS) scheme to prevent overlap of the isotropic peaks by the spinning sidebands. Recycle delays of 2 s were used in the acquisition of 1D spectra, with 1H decoupling field strength set at 86 kHz using the Spinal-64 scheme. Additionally, up to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 signal transients were averaged to ensure an adequate signal-to-noise ratio.
000 signal transients were averaged to ensure an adequate signal-to-noise ratio.
Viability test
Cell viability was assessed using both the FDA and resazurin assays independently. The FDA assay relies on the hydrolysis of FDA by esterases in metabolically active cells, resulting in the production of fluorescently bright fluorescein. To prepare the FDA stock solution, 5 mg of FDA was dissolved in 1 mL of acetone. Next, 1 μL of the FDA stock solution was added to each 0.1 mL of the yeast suspension and incubated at room temperature for 20 minutes. In the resazurin assay, 10 μL of a resazurin solution (0.15 mg mL−1) was added to each 0.1 mL of the yeast suspension. The mixture was then incubated at room temperature for 2 hours. During this incubation period, resazurin measures the mitochondrial activity within the cells. Fluorescent images of the samples were captured using an epifluorescent microscope (Olympus IX53) equipped with a Q-imaging OPTIMOS camera. The FDA and resazurin assay results were recorded using a CLARIOstar microplate reader (BMG LABTECH Germany) in fluorescence mode (FDA assays: excitation filters between 485 and 490 nm and emission filters between 515 and 525 nm; resazurin assays were read using excitation filters between 530 and 570 nm and emission filters between 580 and 620 nm) at room temperature.Thermal gravimetric analysis (TGA) and differential scanning calorimetry (DSC)
Before performing the TGA analysis and DSC analysis, the native yeast and yeast-COF samples underwent a washing step with distilled water, which was repeated three times. Subsequently, the samples were dried at 30 °C for 48 hours to ensure complete removal of any residual moisture. The TGA analysis was carried out using a TA Instrument Q5000, while the DSC analysis was performed using a Q20 instrument. The heating procedure involved annealing the samples in a nitrogen (N2) atmosphere. The heating rate was set at 5 °C per minute. The temperature profile during the analysis was as follows: The temperature was ramped from room temperature to 50 °C and maintained at this temperature for 30 minutes. It was then further increased to 75 °C and held for an additional 30 minutes. Finally, the temperature was raised to 100 °C and maintained for 30 minutes.Cell surface temperature measurements
The samples were carefully placed into tubes, which were subsequently inserted into a temperature-controlled thermomixer (Eppendorf 5418) known for its precise temperature control capabilities. To ensure accuracy, the sample temperatures were measured using a highly sensitive thermometer (FLIR, TG165). The temperature adjustments were made as needed until the equilibrium states were reached, guaranteeing consistent and reliable temperature conditions for the samples.Heat treatment
To initiate the experiment, 50 μL of yeast (2.97 mg mL−1), yeast-COF (10.8 mg mL−1), and COF (7.1 mg mL−1) samples were separately combined with 600 μL of water. Subsequently, these mixtures were incubated at temperatures of 30, 50, 75, and 100 °C for a duration of 30 minutes. Following the incubation period, the cellular activity of the samples was assessed using the FDA and resazurin assays.Acid or basic treatment
The acetic acid–sodium acetate buffer, phosphate buffered saline (PBS), HCl and NaOH were used to adjust the pH. 50 μL of yeast (2.97 mg mL−1), yeast-COF (10.8 mg mL−1), and COF (7.1 mg mL−1) samples were incubated with 600 μL of buffers at different pH levels for a duration of 1 hour. Following the incubation, the samples were washed three times with deionized (DI) water. The cellular activity was then assessed using the FDA and resazurin assays.UV treatment
10 μL of yeast (2.97 mg mL−1), yeast-COF (10.8 mg mL−1), and COF (7.1 mg mL−1) samples were dropped onto glass bottles. These samples were then exposed to UV light at a wavelength of 254 nm for either 2 or 4 hours. After the irradiation, the samples were dissolved in 300 μL of DI water and subsequently washed three times with DI water. The cellular activity was evaluated using the FDA and resazurin assays.Toxic metal ion treatment
10 μL of yeast (2.97 mg mL−1) and yeast-COF (10.8 mg mL−1) samples were separately incubated with 900 μL of CuCl2 solution at concentrations of 0, 100, 500, and 1000 mM for a duration of 1 hour. Following the incubation, the samples were washed three times with DI water. The cellular activity was then assessed using the FDA and resazurin assays.Bisphenol A (BPA) treatment
10 μL of yeast (2.97 mg mL−1) and yeast-COF (10.8 mg mL−1) samples were incubated in 300 μL of BPA solution at concentrations of 2 and 5 mg mL−1 for 1 hour. After the incubation, the samples were washed three times with DI water, and the cellular activity was evaluated using the FDA and resazurin assays.Strong oxidative stress treatment
For the strong oxidative stress treatment, 10 μL of yeast (2.97 mg mL−1), yeast-COF (10.8 mg mL−1), and COF (7.1 mg mL−1) samples were incubated with 300 μL of H2O2 solution at concentrations of 0, 100, 250, 500, and 1000 mM for a duration of 1 hour. Subsequently, the samples were washed three times with DI water, and the cellular activity was evaluated using the FDA and resazurin assays.In the long-term 48 hour experiment, 1 mL of yeast (2.97 mg mL−1), yeast-COF (10.8 mg mL−1), and CAT-yeast-COF (11.2 mg mL−1) samples were incubated in 5 mL of 100 mM H2O2 solution at 30 °C in a sealed tube for 24 and 48 hours. After the incubation period, the samples were washed three times with DI water. The cellular activities were evaluated using the FDA assay, and ethanol production was measured.
Ethanol measurement
To measure ethanol levels, enzymatic assay of alcohol dehydrogenase (ADH) was employed. The reactions were carried out in a total volume of 200 μL, consisting of 40 μL of 50 mM phosphate buffer, 30 μL of 15 mM NAD+, 30 μL of 20 units per mL ADH, 50 μL of H2O, and 50 μL of the sample. After a reaction time of 30 minutes, the samples were detected at 340 nm. All measurements were recorded using a CLARIOstar microplate reader (BMG LABTECH Germany) at room temperature.RNA extraction and RNA-sequence analysis
1.28 × 106 yeast cells and yeast-COF were used for the RNA extraction. The RNA sequence was extracted using a RiboPure™ RNA Purification Kit according to the manufacturer's instructions. After extraction, RNA samples were quality checked for concentration and purity using an Epoch microplate spectrophotometer, and for integrity using an Agilent TapeStation 4200. The RNA was determined to be intact with a RIN of 8.4 and 8.1 for optimal analysis. The 260 nm peaks were clear and visible, with no shift to 270 nm on spectroscopy. 260/280 values are 1.8–2.2 and 260/230 values are ideally 2.33 and 2.38 for native yeast and yeast-COF samples. Then, the yeast RNA was analysed by Ramaciotti Centre for Genomics (UNSW Sydney, Australia). Library QC was performed using the Illumina Stranded mRNA Prep Ligation kit with 500 ng input and 11 PCR cycles. Illumina IDT for Illumina RNA UD Indexes were used to uniquely identify samples. Libraries were quality checked using a ThermoFisher Qubit 4.0 fluorometer (dsDNA HS assay) and a PerkinElmer GX Touch HT (High Sensitivity DNA assay). Libraries were sequenced on NextSeq1000 P1 1 × 100 bp. The reads were trimmed using BBMap (v38.63) and aligned to the S. cerevisiae reference genome (sacCer3) using Burrows Wheeler Aligner (BWA, v0.7.17). The reads were sorted using SAMtools (v1.15.1) and counts were extracted using the R (v4.2.3) Subreads package (v2.0.6). Differential expression analysis was performed using the edgeR package (v3.40.2). Functional category analysis was performed using the information provided by the Saccharomyces Genome Database (SGD).Conclusions
In summary, our study presents a durable and robust COF nanocoating that provides effective cytoprotection in various adverse environments, such as high temperature, extreme pH, UV radiation, exposure to toxic metal ions and organic pollutants, and strong oxidative stress. Notably, the COF nanocoating exhibits remarkable effectiveness in promoting yeast cell survival under high temperature and strongly acidic conditions, surpassing the performance of MOF nanocoatings and other nanomaterials. Additionally, by incorporating exogenous enzymes into the nanocoating, stable yeast fermentation and ethanol production are achieved even under strong oxidative stress. The simplicity and compatibility of this COF coating approach make it a reliable strategy for enhancing the environmental resistance of vulnerable microorganisms under adverse conditions.Data availability
The N2 physisorption isotherms, genes analysis, optical micrographs, fluorescent images, SEM, FTIR, synchrotron THz/Far-IR, cell activity, XRD, DSC and TGA analysis, oxygen concentration, zeta potential, enzyme activity, ethanol production, glucose retention, and ICP results (Tables S1–S3 and Fig. S1–S34†) are available in the ESI.† Access to all data can be obtained from the authors upon written request.Author contributions
J. Liang and Q. Chen contributed equally in this work. J. Liang formulated the project and wrote the initial manuscript. J. Liang and Q. Chen synthesized the materials and measured the performance and analysed the data. J. Yong and J. Liang extracted the RNA. H. Suyama analysed the RNA data. J. Biazik conducted the TEM measurement and wrote the method. B. Njegic, A. Rawal and J. Liang conducted the solid-state NMR experiments. K. Liang contributed to the draft and project administration, conceptualization, writing – review and editing. All authors discussed and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has received support from the NHMRC Investigator Grants project (2026262), the Australian Research Council (ARC) through grants DP210100422 and FT220100479, and the Scientia program at UNSW. The authors would like to acknowledge the assistance provided by Microscopy Australia at the Electron Microscope Unit (EMU) within the Mark Wainwright Analytical Centre (MWAC) at UNSW Sydney and Ramaciotti Centre for Genomics (UNSW Sydney, Australia). The authors also express their gratitude to the Australian Synchrotron, part of the Australian Nuclear Science and Technology Organisation (ANSTO), for providing access to the small-/wide-angle X-ray scattering (SAXS/WAXS) beamlines and THz-Far-IR beamlines, which were instrumental in conducting parts of this research.References
- R. Franco-Duarte, L. Černáková, S. Kadam, K. S. Kaushik, B. Salehi, A. Bevilacqua, M. R. Corbo, H. Antolak, K. Dybka-Stępień and M. Leszczewicz, Advances in chemical and biological methods to identify microorganisms—from past to present, Microorganisms, 2019, 7, 130 CrossRef CAS PubMed.
- D. Endy, Foundations for engineering biology, Nature, 2005, 438, 449–453 CrossRef CAS PubMed.
- S. R. Ceccato-Antonini and E. A. Covre, From baker's yeast to genetically modified budding yeasts: the scientific evolution of bioethanol industry from sugarcane, FEMS Yeast Res., 2020, 20, foaa065 CrossRef CAS PubMed.
- W. Wang, L. Zhang, Q. Deng, Z. Liu, J. Ren and X. Qu, Yeast@MOF bioreactor as a tumor metabolic symbiosis disruptor for the potent inhibition of metabolically heterogeneous tumors, Nano Today, 2022, 42, 101331 CrossRef CAS.
- H. Yang, Y. Zhang, L. Zeng, W. Yin, Y. Xu, J. Chen, S.-Y. Liu, X. Zou, Z. He and Z. Dai, Cell-Selective Encapsulation within Metal–Organic Framework Shells via Precursor-Functionalized Aptamer Identification for Whole-Cell Cancer Vaccine, Small Methods, 2022, 6, 2101391 CrossRef CAS PubMed.
- D. Yu, H. Zhang, Z. Liu, C. Liu, X. Du, J. Ren and X. Qu, Hydrogen-Bonded Organic Framework (HOF)-Based Single-Neural Stem Cell Encapsulation and Transplantation to Remodel Impaired Neural Networks, Angew. Chem., Int. Ed., 2022, 61, e202201485 CrossRef CAS PubMed.
- S. Arslan-Tontul, H. Çetin-Babaoğlu, M. Aslan and İ. Tontul, Refractance window drying in the production of instant baker's yeast and its effect on the quality characteristics of bread, J. Food Sci., 2022, 87, 4991–5000 CrossRef CAS PubMed.
- O. A. Kamanina, E. A. Saverina, P. V. Rybochkin, V. A. Arlyapov, A. N. Vereshchagin and V. P. Ananikov, Preparation of hybrid sol-gel materials based on living cells of microorganisms and their application in nanotechnology, Nanomaterials, 2022, 12, 1086 CrossRef CAS PubMed.
- R. F. Fakhrullin and Y. M. Lvov, “Face-lifting” and “make-up” for microorganisms: layer-by-layer polyelectrolyte nanocoating, ACS Nano, 2012, 6, 4557–4564 CrossRef CAS PubMed.
- W. Geng, L. Wang, N. Jiang, J. Cao, Y.-X. Xiao, H. Wei, A. K. Yetisen, X.-Y. Yang and B.-L. Su, Single cells in nanoshells for the functionalization of living cells, Nanoscale, 2018, 10, 3112–3129 RSC.
- B. Wang, P. Liu, W. Jiang, H. Pan, X. Xu and R. Tang, Yeast cells with an artificial mineral shell: protection and modification of living cells by biomimetic mineralization, Angew. Chem., Int. Ed., 2008, 120, 3616–3620 CrossRef.
- J. C. Rooke, A. Léonard and B.-L. Su, Targeting photobioreactors: immobilisation of cyanobacteria within porous silica gel using biocompatible methods, J. Mater. Chem., 2008, 18, 1333–1341 RSC.
- N. Jiang, G.-L. Ying, A. K. Yetisen, Y. Montelongo, L. Shen, Y.-X. Xiao, H. J. Busscher, X.-Y. Yang and B.-L. Su, A bilayered nanoshell for durable protection of single yeast cells against multiple, simultaneous hostile stimuli, Chem. Sci., 2018, 9, 4730–4735 RSC.
- J. Liang and K. Liang, Nanobiohybrids: Synthesis strategies and environmental applications from micropollutants sensing and removal to global warming mitigation, Environ. Res., 2023, 116317 CrossRef CAS PubMed.
- J. Liang and K. Liang, Multi-enzyme Cascade Reactions in Metal-Organic Frameworks, Chem. Rec., 2020, 20, 1100–1116 CrossRef CAS PubMed.
- J. Liang and K. Liang, Biocatalytic Metal–Organic Frameworks: Prospects Beyond Bioprotective Porous Matrices, Adv. Funct. Mater., 2020, 30, 2001648 CrossRef CAS.
- J. Liang and K. Liang, Nano-bio-interface engineering of metal-organic frameworks, Nano Today, 2021, 40, 101256 CrossRef CAS.
- J. Liang, J. Liu, M. S. Lord, Y. Wang and K. Liang, De Novo Engineering of Metal–Organic Framework-Printed In Vitro Diagnostic Devices for Specific Capture and Release of Tumor Cells, Small, 2021, 17, 2103590 CrossRef CAS PubMed.
- J. Liang, M. Y. B. Zulkifli, S. Choy, Y. Li, M. Gao, B. Kong, J. Yun and K. Liang, Metal–Organic Framework–Plant Nanobiohybrids as Living Sensors for On-Site Environmental Pollutant Detection, Environ. Sci. Technol., 2020, 54, 11356–11364 CrossRef CAS PubMed.
- J. Liang, M. Y. Bin Zulkifli, J. Yong, Z. Du, Z. Ao, A. Rawal, J. A. Scott, J. R. Harmer, J. Wang and K. Liang, Locking the Ultrasound-Induced Active Conformation of Metalloenzymes in Metal–Organic Frameworks, J. Am. Chem. Soc., 2022, 144, 17865–17875 CrossRef CAS PubMed.
- J. Liang, Q. Chen, J. Xue and K. Liang, Reticular chemistry for improving the activity of biocatalysts: Synthesis strategies and advanced characterization techniques, Chem Catal., 2022, 2, 2515–2551 CrossRef CAS.
- J. Liang, S. Gao, J. Liu, M. Y. B. Zulkifli, J. Xu, J. Scott, V. Chen, J. Shi, A. Rawal and K. Liang, Hierarchically Porous Biocatalytic MOF Microreactor as a Versatile Platform towards Enhanced Multienzyme and Cofactor-Dependent Biocatalysis, Angew. Chem., Int. Ed., 2021, 60, 5421–5428 CrossRef CAS PubMed.
- J. Liang, B. Johannessen, Z. Wu, R. F. Webster, J. Yong, M. Y. B. Zulkifli, J. S. Harbort, Y. R. Cheok, H. Wen, Z. Ao, B. Kong, S. L. Y. Chang, J. Scott and K. Liang, Regulating the Coordination Environment of Mesopore-confined Single Atoms from Metalloprotein-MOFs for Highly Efficient Biocatalysis, Adv. Mater., 2022, 34, 2205674 CrossRef CAS PubMed.
- J. Liang, F. Mazur, C. Tang, X. Ning, R. Chandrawati and K. Liang, Peptide-induced super-assembly of biocatalytic metal–organic frameworks for programmed enzyme cascades, Chem. Sci., 2019, 10, 7852–7858 RSC.
- K. Liang, J. J. Richardson, J. Cui, F. Caruso, C. J. Doonan and P. Falcaro, Metal–Organic Framework Coatings as Cytoprotective Exoskeletons for Living Cells, Adv. Mater., 2016, 28, 7910–7914 CrossRef CAS PubMed.
- K. Liang, J. J. Richardson, C. J. Doonan, X. Mulet, Y. Ju, J. Cui, F. Caruso and P. Falcaro, An Enzyme-Coated Metal–Organic Framework Shell for Synthetically Adaptive Cell Survival, Angew. Chem., Int. Ed., 2017, 56, 8510–8515 CrossRef CAS PubMed.
- Z. Ji, H. Zhang, H. Liu, O. M. Yaghi and P. Yang, Cytoprotective metal-organic frameworks for anaerobic bacteria, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 10582–10587 CrossRef CAS PubMed.
- W. Zhu, J. Guo, S. Amini, Y. Ju, J. O. Agola, A. Zimpel, J. Shang, A. Noureddine, F. Caruso and S. Wuttke, SupraCells: living mammalian cells protected within functional modular nanoparticle-based exoskeletons, Adv. Mater., 2019, 31, 1900545 CrossRef PubMed.
- J. Guo, M. Suástegui, K. K. Sakimoto, V. M. Moody, G. Xiao, D. G. Nocera and N. S. Joshi, Light-driven fine chemical production in yeast biohybrids, Science, 2018, 362, 813–816 CrossRef CAS PubMed.
- L. Shi, A. Li, W. Zhang, H. Wu and Y. Chi, Endowing chloroplasts with artificial “cell walls” using metal–organic frameworks, Nanoscale, 2020, 12, 11582–11592 RSC.
- R. Riccò, W. Liang, S. Li, J. J. Gassensmith, F. Caruso, C. Doonan and P. Falcaro, Metal–organic frameworks for cell and virus biology: a perspective, ACS Nano, 2018, 12, 13–23 CrossRef PubMed.
- M. A. Luzuriaga, C. E. Benjamin, M. W. Gaertner, H. Lee, F. C. Herbert, S. Mallick and J. J. Gassensmith, ZIF-8 degrades in cell media, serum, and some—but not all—common laboratory buffers, Supramol. Chem., 2019, 31, 485–490 CrossRef CAS PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- C. Sun, L. Chang, K. Hou, S. Liu and Z. Tang, Encapsulation of live cells by metal-organic frameworks for viability protection, Sci. China Mater., 2019, 62, 885–891 CrossRef CAS.
- T. Ma, Y. Zhou, C. S. Diercks, J. Kwon, F. Gándara, H. Lyu, N. Hanikel, P. Pena-Sánchez, Y. Liu, N. J. Diercks, R. O. Ritchie, D. M. Proserpio, O. Terasaki and O. M. Yaghi, Catenated covalent organic frameworks constructed from polyhedra, Nat. Synth., 2023, 2, 286–295 Search PubMed.
- J. Liang, J. Ruan, B. Njegic, A. Rawal, J. Scott, J. Xu, C. Boyer and K. Liang, Insight into Bioactivity of In-situ Trapped Enzyme-Covalent-organic Frameworks, Angew. Chem., Int. Ed., 2023, e202303001 CAS.
- Y. Zheng, S. Zhang, J. Guo, R. Shi, J. Yu, K. Li, N. Li, Z. Zhang and Y. Chen, Green and Scalable Fabrication of High-Performance Biocatalysts Using Covalent Organic Frameworks as Enzyme Carriers, Angew. Chem., Int. Ed., 2022, 134, e202208744 CrossRef.
- C. Zhong, G. Li, W. Tian, D. Ouyang, Y. Ji, Z. Cai and Z. Lin, Construction of Covalent Organic Framework Capsule-Based Nanoreactor for Sensitive Glucose Detection, ACS Appl. Mater. Interfaces, 2023, 15, 10158–10165 CrossRef CAS PubMed.
- C. M. Coughlan and J. L. Brodsky, Use of yeast as a model system to investigate protein conformational diseases, Mol. Biotechnol., 2005, 30, 171–180 CrossRef CAS PubMed.
- S. H. Yang, S. M. Kang, K.-B. Lee, T. D. Chung, H. Lee and I. S. Choi, Mussel-Inspired Encapsulation and Functionalization of Individual Yeast Cells, J. Am. Chem. Soc., 2011, 133, 2795–2797 CrossRef CAS PubMed.
- G. D'Angelo, V. Conti Nibali, C. Crupi, S. Rifici, U. Wanderlingh, A. Paciaroni, F. Sacchetti and C. Branca, Probing Intermolecular Interactions in Phospholipid Bilayers by Far-Infrared Spectroscopy, J. Phys. Chem. B, 2017, 121, 1204–1210 CrossRef PubMed.
- J. H. Park, K. Kim, J. Lee, J. Y. Choi, D. Hong, S. H. Yang, F. Caruso, Y. Lee and I. S. Choi, A Cytoprotective and Degradable Metal–Polyphenol Nanoshell for Single-Cell Encapsulation, Angew. Chem., Int. Ed., 2014, 53, 12420–12425 CrossRef CAS PubMed.
- Q. Deparis, A. Claes, M. R. Foulquié-Moreno and J. M. Thevelein, Engineering tolerance to industrially relevant stress factors in yeast cell factories, FEMS Yeast Res., 2017, 17, fox036 CrossRef PubMed.
- C. Bereketoglu, K. Y. Arga, S. Eraslan and B. Mertoglu, Analysis of transcriptional profiles of Saccharomyces cerevisiae exposed to bisphenol A, Curr. Genet., 2017, 63, 253–274 CrossRef CAS PubMed.
- R. P. Sinha and D.-P. Häder, UV-induced DNA damage and repair: a review, Photochem. Photobiol. Sci., 2002, 1, 225–236 CrossRef CAS PubMed.
- G. Wang, L. Wang, P. Liu, Y. Yan, X. Xu and R. Tang, Extracellular Silica Nanocoat Confers Thermotolerance on Individual Cells: A Case Study of Material-Based Functionalization of Living Cells, ChemBioChem, 2010, 11, 2368–2373 CrossRef CAS PubMed.
- S. K. S. Freitas, R. S. Borges, C. Merlini, G. M. O. Barra and P. M. Esteves, Thermal Conductivity of Covalent Organic Frameworks as a Function of Their Pore Size, J. Phys. Chem. C, 2017, 121, 27247–27252 CrossRef CAS.
- E. H. Ko, Y. Yoon, J. H. Park, S. H. Yang, D. Hong, K.-B. Lee, H. K. Shon, T. G. Lee and I. S. Choi, Bioinspired, Cytocompatible Mineralization of Silica–Titania Composites: Thermoprotective Nanoshell Formation for Individual Chlorella Cells, Angew. Chem., Int. Ed., 2013, 52, 12279–12282 CrossRef CAS PubMed.
- S. K. Parks, J. Chiche and J. Pouysségur, Disrupting proton dynamics and energy metabolism for cancer therapy, Nat. Rev. Cancer, 2013, 13, 611–623 CrossRef CAS PubMed.
- Y. Lin, W. Zhang, C. Li, K. Sakakibara, S. Tanaka and H. Kong, Factors affecting ethanol fermentation using Saccharomyces cerevisiae BY4742, Biomass Bioenergy, 2012, 47, 395–401 CrossRef CAS.
- S. Periyasamy, S. Venkatachalam, S. Ramasamy and V. Srinivasan, Production of bio-ethanol from sugar molasses using Saccharomyces cerevisiae, Model. Appl. Sci., 2009, 3, 32–37 CAS.
- S. S. Han, H. Furukawa, O. M. Yaghi and W. A. Goddard Iii, Covalent organic frameworks as exceptional hydrogen storage materials, J. Am. Chem. Soc., 2008, 130, 11580–11581 CrossRef CAS PubMed.
- H. Fan, A. Mundstock, A. Feldhoff, A. Knebel, J. Gu, H. Meng and J. r. Caro, Covalent organic framework–covalent organic framework bilayer membranes for highly selective gas separation, J. Am. Chem. Soc., 2018, 140, 10094–10098 CrossRef CAS PubMed.
- Z. Wang, S. Zhang, Y. Chen, Z. Zhang and S. Ma, Covalent organic frameworks for separation applications, Chem. Soc. Rev., 2020, 49, 708–735 RSC.
- W. Helweh, N. C. Flanders, S. Wang, B. T. Phelan, P. Kim, M. J. Strauss, R. L. Li, M. S. Kelley, M. S. Kirschner and D. O. Edwards, Layered structures of assembled imine-linked macrocycles and two-dimensional covalent organic frameworks give rise to prolonged exciton lifetimes, J. Mater. Chem. C, 2022, 10, 3015–3026 RSC.
- M. Mu, Y. Wang, Y. Qin, X. Yan, Y. Li and L. Chen, Two-Dimensional Imine-Linked Covalent Organic Frameworks as a Platform for Selective Oxidation of Olefins, ACS Appl. Mater. Interfaces, 2017, 9, 22856–22863 CrossRef CAS PubMed.
- Z. Liu, H. Wang, J. Ou, L. Chen and M. Ye, Construction of hierarchically porous monoliths from covalent organic frameworks (COFs) and their application for bisphenol A removal, J. Hazard. Mater., 2018, 355, 145–153 CrossRef CAS PubMed.
- A. Nel, T. Xia, L. Mädler and N. Li, Toxic Potential of Materials at the Nanolevel, Science, 2006, 311, 622–627 CrossRef CAS PubMed.
- S. Izawa, Y. Inoue and A. Kimura, Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae, Biochem. J., 1996, 320, 61–67 CrossRef CAS PubMed.
- S.-S. Vamvakas and J. Kapolos, Factors affecting yeast ethanol tolerance and fermentation efficiency, World J. Microbiol. Biotechnol., 2020, 36, 114 CrossRef CAS PubMed.
- J. Sharma, V. Kumar, R. Prasad and N. A. Gaur, Engineering of Saccharomyces cerevisiae as a consolidated bioprocessing host to produce cellulosic ethanol: Recent advancements and current challenges, Biotechnol. Adv., 2022, 56, 107925 CrossRef CAS PubMed.
- W. Chen, S. Kong, M. Lu, F. Chen, W. Cai, L. Du, J. Wang and C. Wu, Comparison of different zinc precursors for the construction of zeolitic imidazolate framework-8 artificial shells on living cells, Soft Matter, 2020, 16, 270–275 RSC.
- K. Nakanishi, H. Tokuda, T. Soga, T. Yoshinaga and M. Takeda, Effect of electric current on growth and alcohol production by yeast cells, J. Ferment. Bioeng., 1998, 85, 250–253 CrossRef CAS.
- I. Borodina and J. Nielsen, Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals, Biotechnol. J., 2014, 9, 609–620 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc04973f |
| ‡ The authors contributed equally in this work. |
| This journal is © The Royal Society of Chemistry 2024 |