Open Access Article
Open Access ArticleA cysteine-specific solubilizing tag strategy enables efficient chemical protein synthesis of difficult targets†
Wenchao
Li
a,
Michael T.
Jacobsen‡
a,
Claire
Park
a,
Jae Un
Jung
a,
Nai-Pin
Lin
 a,
Po-Ssu
Huang
b,
Rayhan A.
Lal
c and
Danny Hung-Chieh
Chou
a,
Po-Ssu
Huang
b,
Rayhan A.
Lal
c and
Danny Hung-Chieh
Chou
 *a
*a
aDivisión of Endocrinology and Diabetes, Department of Pediatrics, School of Medicine, Stanford University, Palo Alto, CA 94305, USA. E-mail: dannychou@stanford.edu
bDepartment of Bioengineering, Stanford University, Palo Alto, CA 94305, USA
cDivision of Endocrinology, Department of Medicine, School of Medicine, Stanford University, Palo Alto, CA 94305, USA
First published on 19th January 2024
Abstract
We developed a new cysteine-specific solubilizing tag strategy via a cysteine-conjugated succinimide. This solubilizing tag remains stable under common native chemical ligation conditions and can be efficiently removed with palladium-based catalysts. Utilizing this approach, we synthesized two proteins containing notably difficult peptide segments: interleukin-2 (IL-2) and insulin. This IL-2 chemical synthesis represents the simplest and most efficient approach to date, which is enabled by the cysteine-specific solubilizing tag to synthesize and ligate long peptide segments. Additionally, we synthesized a T8P insulin variant, previously identified in an infant with neonatal diabetes. We show that T8P insulin exhibits reduced bioactivity (a 30-fold decrease compared to standard insulin), potentially contributing to the onset of diabetes in these patients. In summary, our work provides an efficient tool to synthesize challenging proteins and opens new avenues for exploring research directions in understanding their biological functions.
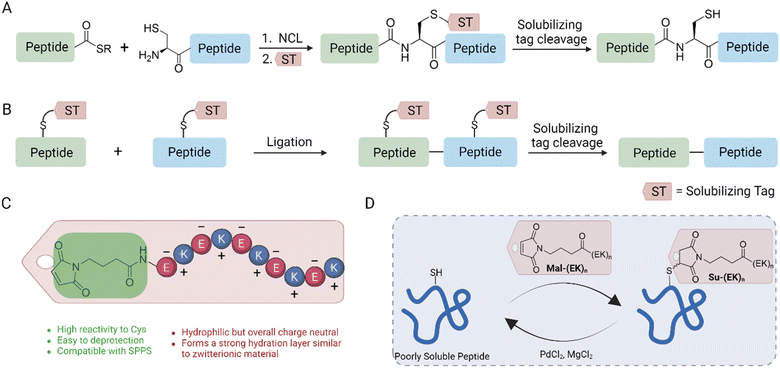
Chemical synthesis of peptides and proteins has benefited tremendously from the advance of solid-phase peptide synthesis (SPPS),1 the development of chemical ligation methods,2–5 and orthogonal protecting groups.6 From synthesizing D-proteins to insulin analogs with noncanonical amino acids, chemical protein synthesis provides access to analogs that are challenging to achieve using recombinant expression.7–9 During the course of peptide synthesis and native chemical ligation (NCL), purified peptide segments and ligation products may have poor solubility in reaction buffers, which prevents additional ligations or other handling steps.10 Therefore, hydrophilic, semipermanent group linked to peptide segments is often used as an effective strategy to enhance solubility, especially in streamlining subsequent chemical ligation reactions.11 Numerous solubilizing tags and removal strategies have been reported, providing invaluable tools for chemical protein synthesis.12–16 On the other hand, synthesizing and implementing these solubilizing tags create unique applicability constraints and limitations depending on the sequence, reactive groups, and removal conditions. These challenges have motivated us to develop a new and straightforward solubilization method that can be applied to challenging proteins and peptides (e.g., insulin).
To achieve this goal, an ideal solubilizing tag strategy should (1) be easily attached to improve the solubility of peptide segments and facilitate subsequent HPLC purifications, (2) have a stable linkage to the peptide under various conditions of chemical protein synthesis, and (3) be detached using facile conditions. Ligated products can be selectively modified after (Fig. 1A) or before NCL (Fig. 1B). Furthermore, the solubilizing tag can stay retained to ensure suitable solubility in longer intermediates, which can be globally removed at the final step. In this article, we develop a maleimide (Mal)-based solubilizing tag with a zwitterionic peptide chain, which selectively reacts with cysteine side chains17 (Fig. 1C). Mal is compatible not only with the chemical environment required for Fmoc-SPPS, but also with trifluoroacetic acid (TFA)-based cleavage conditions, making it an excellent candidate for solubilizing tags. Zwitterionic peptides (EK)n not only facilitate the formation of a hydrated layer on their surface to enhance their hydrophilicity, but also minimally impact the isoelectric point of their coupled peptides.18,19 We utilized a palladium-based detachment strategy reported by Brik et al. to remove the succinimide (Su) tag20 (Fig. 1D). We applied this method to chemically synthesize interleukin-2 (IL-2), a protein well known for its poorly soluble, hydrophobic C-terminal region.21–23 Furthermore, we extend the applicability of this method to disulfide-rich peptides and synthesize a clinically identified insulin mutant.24 These examples showcase the broad application of this methodology, enabling access to mutant proteins to answer biological questions.
Results & discussion
Development of a cysteine-selective solubilizing tag for hydrophobic peptide segments
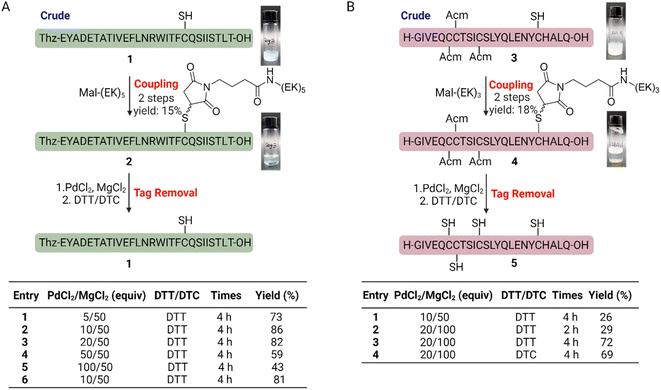
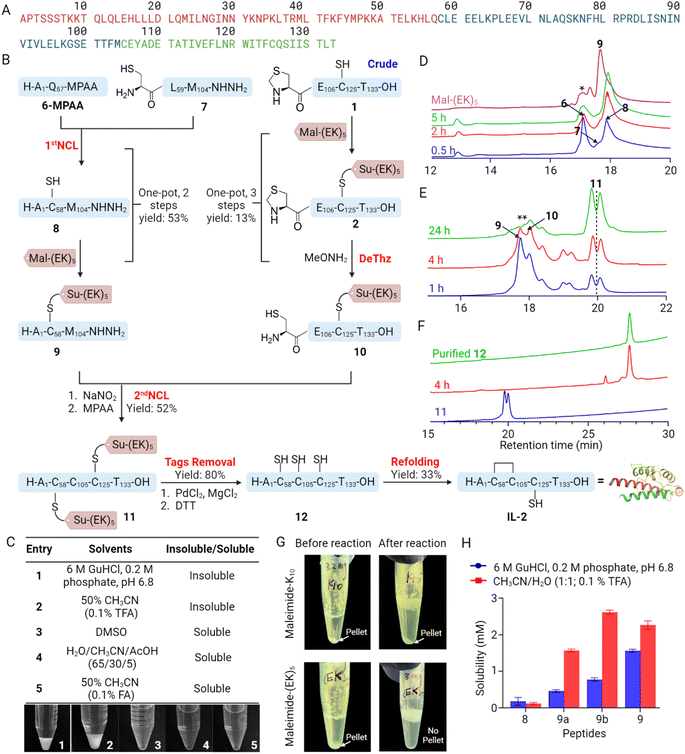
We selected two well-known hydrophobic peptide segments: the C-terminal segment of IL-2 (residue 105–133) 1 and mini-insulin A chain 3 (ref. 25) as model peptides to evaluate the performance of our solubilizing tags. The Mal-(EK)n tag can be easily prepared using SPPS-compatible approaches (Scheme S1†). We then react crude peptides 1 and 3 with Mal-(EK)n to obtain the modified products in high yields (Fig. 2). Visible changes were observed as the reactions progressed: the initially turbid solutions (top) became clear (middle, with colorful stir bars) (Fig. 2A and B). Additionally, a significant reduction in retention times was evident in the HPLC traces for both 2 and 4 (compared to 1 and 3, respectively), suggesting improved solubility in the HPLC buffer (Fig. S1†). We then attempted to cleave the linker using PdCl2 and MgCl2 at 37 °C, followed by dithiothreitol (DTT) or diethyldithiocarbamate (DTC) treatment.20 We screened multiple conditions with different PdCl2 amounts and demonstrated that effective removal of Su-(EK)n can be achieved with >80% yield (Fig. 2A). Similarly, we efficiently removed the Su-(EK)n and Acm protecting groups with >70% yield under similar conditions to obtain 5 (Fig. 2B). In summary, Mal-(EK)n solubilizing tags can be utilized to conjugate with insoluble peptides containing free thiol groups and enhance their solubility.Chemical protein synthesis of IL-2 using Mal-(EK)5 solubilizing tag
Mature IL-2, a 15.4 kDa globular protein, contains 133 amino acid residues, including three cysteine residues (Cys58, Cys105, Cys125), of which Cys58–Cys105 forms a disulfide bond that is essential for the overall tertiary structure.26 By carefully analyzing the sequence, we advanced a three segment, two NCL strategy through sites Gln57-Cys58 and Met104-Cys105, which are kinetically feasible ligation junctions27,28 (Fig. 3A). Compared to previous IL-2 synthesis described by the Hojo (Thioester ligation),21 Bode (KAHA ligation)22 and Li (STL)23 group with at least 4 segments, our method provides a more efficient and accessible route. Both IL-2(1–57)-NHNH2 (6) and IL-2(58–105)-NHNH2 (7) are relatively long peptides to synthesize by Fmoc-SPPS. To improve the synthesis, two pseudoprolines dipeptides26 were introduced into each peptide loaded on hydrazine-2-chlorotrityl resins to produce C-terminal hydrazides for subsequent NCL.29,30 The pseudoproline-containing peptides were then cleaved from the resins to give 6 and 7 in 21% and 32% isolated yields, respectively (Fig. 3B). We next performed NCL to connect 6 and 7. The oxidation and thioesterification of 6 were completed in high yields, and the NCL reaction proceeded smoothly with the addition of 7. Monitored by analytical HPLC, the reaction was completed after 8 hours (Fig. S2†). However, we also observed white pellets in the reaction mixture after one hour. Centrifuging the mixture and analyzing the pellet and supernatant revealed that the product, peptide 8, was concentrated in the pellet. Because 8 was poorly soluble in the ligation buffer (Fig. 3C), we predicted that it would be unable to subsequently ligate with 10. The experimental results indeed confirmed this prediction. Considering that the ligation of 6 and 7 would result in a free thiol group, we used a Mal-(EK)5 solubilizing tag to improve the solubility of 8. In this approach, the acetyl acetone (acac)/3-mercaptopropionic acid (MPAA) thiolysis method31 replaced the conventional NaNO2/MPAA thiolysis, as the latter required a large excess of MPAA, which could lead to an undesired reaction with the Mal-based solubilizing tag introduced in the subsequent step, affecting the yield of the target product. Following the first NCL of peptides 6-MPAA and 7, the reaction mixture was placed in a −20 °C refrigerator for storage. Subsequently, the Mal-(EK)5 dissolved in the ligation buffer was mixed with the reaction mixture and allowed to react at −20 °C for 30 minutes. Ultimately, 9 was obtained in two steps with a 53% overall yield (Fig. 3D). We also tested two other solubilizing tags (Mal-K6, Mal-K10) on 8 and found that Mal-(EK)5 not only enhanced the solubility of 8 in HPLC buffer, but also significantly improved its solubility in NCL buffer (Fig. 3G and H).Due to the presence of a free cysteine in difficult peptide 1 (Cys125), we utilized this position to introduce another Mal-(EK)5 solubilizing tag. Initially, the N-terminal Cys105 was protected as a thiazolidine (Thz) moiety, while the central Cys125 was protected with a trityl (Trt) group. Following peptide cleavage, the crude peptide 1 reacted with Mal-(EK)5, and subsequently, the N-terminal Thz105 was converted back to a free thiol Cys105. Peptide 10 was then purified, resulting in a one-pot, three-step yield of 13% (Fig. 3B). With the purified products 9 and 10 in our possession, we effectively overcame the previous solubility challenges and accomplished the second NCL reaction, ultimately obtaining 11 with a noteworthy yield of 52% for previously poorly-soluble segments (Fig. 3E).
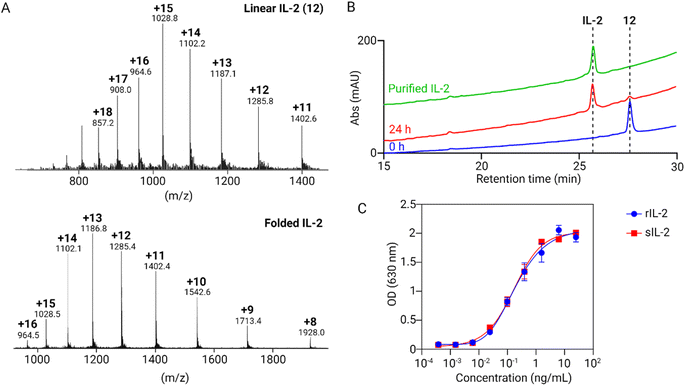
To obtain linear IL-2 (12), 11 was treated with 20 eq. of PdCl2 and 100 eq. of MgCl2 at 37 °C for 4 hours, followed by treatment with DTT. The deprotection reaction was performed efficiently and afforded fully deprotected 12 in an isolated yield of 80% following RP-HPLC (Fig. 3F). For the folding process, 12 was dissolved in 6 M GnHCl solution containing 0.1 M Tris and 30 mM reduced glutathione pH 8.0. After the protein was completely dissolved, two times the volume of oxidized glutathione buffer (0.1 M Tris and 1.5 mM oxidized glutathione, pH 8.0) was added and stored at room temperature for 24 hours.32 The shift in retention time, between linear and folded IL-2, can be seen in the analytical HPLC (Fig. 4B). The purified folded IL-2 was validated by LC-MS (Fig. 4A). The activities of recombinant and synthetic IL-2 in HEK-Blue™ IL-2 cells were comparable in cellular potency in activating STAT phosphorylation33 (Fig. 4C). Overall, our synthetic route of IL-2 led to a comparable overall yield with the work by the Bode and Li groups. Due to the use of this solubility tag, we were able to use only two native chemical ligation steps instead of three steps in previously reported methods and our method does not require any incorporation of unnatural amino acids.
Chemical protein synthesis of T8P insulin variant
The poor solubility of insulin A chain due to its hydrophobicity is well documented as a chemical synthesis challenge.34 In 2014, Liu et al. introduced an O-acyl isopeptide strategy in the A8–A9 region and increased A chain solubility; the isoacyl bond can then undergo rearrangements after A and B chain combination to produce the native amide bond.35 Recently, Lal et al. reported a case study of neonatal diabetes that occurred in a 5 months-old Hispanic male infant; the sequence panel revealed a novel heterozygous de novo insulin gene sequence variant c.289 A > C (p.T97P or T8P, the A8 residue of insulin was mutated from threonine to proline) of unknown significance.24 In the initial report, misfolding of the T8P variant proinsulin was felt to cause a pathologic ER stress response as described in several cases of INS-gene-induced diabetes of youth (MIDY). We sought to use chemical protein synthesis to access this variant and study its bioactivity. Critically, this mutation happened to be in the same residues as the isopeptide strategy,35 indicating that a different method to improve A chain solubility is needed. We deployed the Mal-(EK)n solubilization strategy to synthesize the challenging T8P insulin variant (Fig. 5A).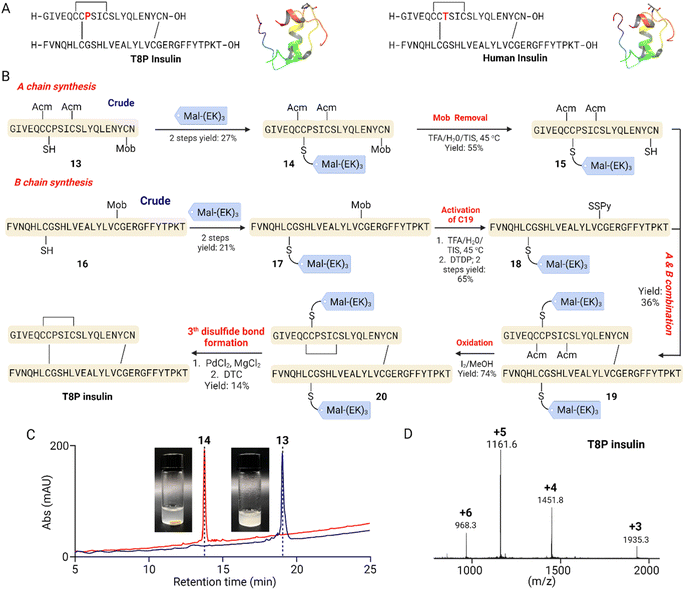 | ||
| Fig. 5 Structure and synthesis of T8P insulin. (A) Sequence and crystal structure of human insulin (PDB code 5CNY) and T8P insulin, with point mutation indicated in red. (B) Synthetic route of T8P insulin. (C) Physical states and HPLC traces of 13 before and after reaction with Mal-(EK)3, yielding 14. (D) Mass spectrum of T8P insulin. | ||
We employed a stepwise disulfide bond formation strategy to complete the synthesis of T8P insulin (Fig. 5B). First, the partially protected A-chain [C6-Acm, C7, C11-Acm, C20-Mob] 13 was synthesized using Fmoc SPPS on 2-CTC resin. The crude peptide 13, with a free thiol group at C7, was reacted with the solubilizing tag Mal-(EK)3 to yield the enhanced solubility product 14, with a two-step yield of 27%. As anticipated, the reaction solution changed from turbid to clear after the reaction (Fig. 5C). 14 was then treated with TFA/TIS/H2O cocktail at 45 °C to remove the Mob protecting group, producing 15 in 55% yield.36
Next, we proceeded to synthesize the partially protected B chain [C19-Mob] 16 using Fmoc SPPS on 2-CTC resin. Following a similar approach to the A chain, we introduced the Mal-(EK)3 solubilizing tag at the C7 position to obtain 17, with a combined yield of 21% for these two steps. The removal of the C19-Mob group from peptide 17 was achieved by treating it with a TFA/TIS/H2O cocktail at 45 °C. We then removed the Mob protecting group and activated the C19-thiol using 2,2′-dithiodipyridine (DTDP), successfully yielding product 18 with a yield of 65%.
Having acquired peptides 15 and 18, we proceeded to explore the formation of the three interchain and intrachain disulfide bonds. For this purpose, 15 and 18 were mixed and agitated in a buffer solution containing 6 M urea and 0.2 M NH4HCO3 (pH 8.0).35 In 20 minutes, the reaction was successfully concluded. Subsequent HPLC purification yielded a product 19 with a 36% yield. Efficient conversion of 19 to 20 was achieved through treatment with an excess of iodine in MeOH. The intrachain disulfide bond formation within the A chain occurred through an in situ process involving Acm deprotection and disulfide formation. Finally, we removed Su-(EK)3 from 20 through a sequence involving PdCl2 treatment, followed by a 30 minutes quenching period with DTC in a degassed buffer solution (6 M GnHCl and 0.2 M phosphate at pH 1). The use of a lower pH and DTC was employed to prevent the cleavage of disulfide bonds.37,38 As a result of this procedure, the mature T8P insulin was successfully formed, culminating in an isolated product yield of 14%.
Biological investigation of T8P insulin variant
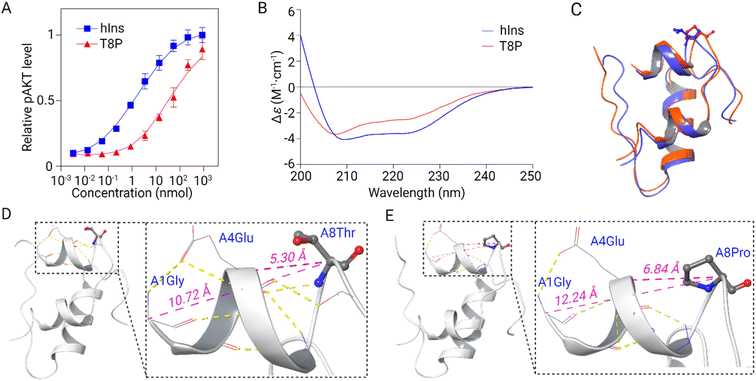
We conducted a phosphorylated AKT (Ser 473) insulin signaling stimulation assay to assess the biological activity of both human insulin and T8P insulin variant (Fig. 6A). The results indicated a 30-fold reduction in biological activity for T8P insulin (with an EC50 of 36.6 nM), as compared to native insulin (with an EC50 of 1.3 nM).39 This result indicates that the T8P mutation had a significant impact on insulin's activity and the resulting neonatal diabetes observed clinically. We then investigated T8P insulin using circular dichroism (CD) and compared it with native insulin. The helix (regular and distorted) content for native insulin and T8P insulin, calculated using the BESTSEL website,40 was 41.1 and 18.8%, respectively. This suggested that the mutation significantly perturbs the overall secondary structure of the T8P insulin, leading to reduced biological activity (Fig. 6B). Utilizing AlphaFold,41 we employed structure prediction to visualize the potential changes in the three-dimensional structure of T8P insulin and performed a comparative analysis with human insulin. We are aware that algorithms like AlphaFold biases heavily towards the native structure due to the high degree of sequence homology. Nonetheless, since the T8P insulin maintains functionality, suggesting that the mutant structure should keep the overall fold, we use AlphaFold to propose a model to investigate how the T8P mutation might have an effect. Upon alignment, notable discrepancies emerged in the N-terminal segment of the A chain between T8P insulin and human insulin, particularly in the α-helix structure located at the A chain's N-terminus in human insulin, which exhibited a more compact conformation (Fig. 6C). Detailed structural analysis was conducted to elucidate the underlying factors contributing to this phenomenon. Firstly, due to the mutated proline being the only secondary amine, its nitrogen atom cannot form hydrogen bonds between helical backbones like other amino acids. The disruption of hydrogen bonds may lead to the breaking or rotation of the α-helix.42,43 Secondly, proline is the only amino acid with a cyclic side chain, forming a five-membered ring structure with the main chain, which restricts the free rotation of its side chain.44 This unique structure leads to a fixed conformation of proline in the protein chain (Fig. 6D and E).Conclusions
In summary, we developed a new strategy utilizing a cysteine-conjugated Su-(EK)n solubilizing tag for efficient chemical protein synthesis. This approach enables on-demand coupling and removal of the tag, and the zwitterionic peptide solubilizing tag offers superior solubilization compared to traditional solubilizing tags (oligo-lysines or -arginines). With this strategy, we successfully synthesized two proteins with known challenging peptide segments. Our IL-2 chemical synthesis represents the simplest approach to date, and we intend to employ this method for synthesizing IL-2 analogs, in order to access unexplored biological activities and potential applications in cancer biology. Furthermore, the synthetic method of T8P insulin presents a convenient alternative route for preparing insulin, not dependent on specific isoacyl dipeptide motifs. Our biological studies of T8P insulin variant explain why this variant has weak bioactivity, which may play a role in the pathophysiology of the disease. This work introduced a novel solubilizing tag strategy to the field of chemical protein synthesis. Our work offers valuable prospects in synthesizing hard-to-make proteins and paves the way for deeper insights into their biological roles.Data availability
Please refer to the ESI† for extensive characterization and additional details on all experiments reported in this publication.Author contributions
W. L., M. T. J., and D. H.-C. C. conceived and designed the experiments. D. H.-C. C. directed the project. W. L., M. T. J., C. P., and J. U. J. performed the experiments. N.-P. L., P.-S. H. and R. A. L. provided analyzed experimental data along with other authors. W. L., M. T. J., and D. H.-C. C. wrote the manuscript. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Stanford Cancer Institute, an NCI-designated Comprehensive Cancer Center. We thank Prof. Amelia Fuller (Santa Clara University) for assisting with CD spectra.References
- R. Behrendt, P. White and J. Offer, Advances in Fmoc solid-phase peptide synthesis, J. Pept. Sci., 2016, 22(1), 4–27 CrossRef CAS PubMed.
- P. E. Dawson, T. W. Muir, I. Clark-Lewis and S. B. Kent, Synthesis of proteins by native chemical ligation, Science, 1994, 266(5186), 776–779 CrossRef CAS PubMed.
- V. Agouridas, O. El Mahdi, V. Diemer, M. Cargoet, J. M. Monbaliu and O. Melnyk, Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations, Chem. Rev., 2019, 119(12), 7328–7443 CrossRef CAS PubMed.
- J. S. Zheng, S. Tang, Y. K. Qi, Z. P. Wang and L. Liu, Chemical synthesis of proteins using peptide hydrazides as thioester surrogates, Nat. Protoc., 2013, 8(12), 2483–2495 CrossRef CAS PubMed.
- C. L. Lee and X. Li, Serine/threonine ligation for the chemical synthesis of proteins, Curr. Opin. Chem. Biol., 2014, 22, 108–114 CrossRef CAS PubMed.
- P. R. Hansen and A. Oddo, Fmoc Solid-Phase Peptide Synthesis, Methods Mol. Biol., 2015, 1348, 33–50 CrossRef CAS PubMed.
- A. J. Lander, Y. Jin and L. Y. P. Luk, D-Peptide and D-Protein Technology: Recent Advances, Challenges, and Opportunities, ChemBioChem, 2022, e202200537 Search PubMed.
- M. T. Weinstock, M. T. Jacobsen and M. S. Kay, Synthesis and folding of a mirror-image enzyme reveals ambidextrous chaperone activity, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(32), 11679–11684 CrossRef CAS PubMed.
- K. Mandal, M. Uppalapati, D. Ault-Riche, J. Kenney, J. Lowitz, S. S. Sidhu and S. B. Kent, Chemical synthesis and X-ray structure of a heterochiral D-protein antagonist plus vascular endothelial growth factor protein complex by racemic crystallography, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(37), 14779–14784 CrossRef CAS PubMed.
- M. Paradis-Bas, J. Tulla-Puche and F. Albericio, The road to the synthesis of “difficult peptides”, Chem. Soc. Rev., 2016, 45(3), 631–654 RSC.
- R. J. Giesler, J. M. Fulcher, M. T. Jacobsen and M. S. Kay, Controlling Segment Solubility in Large Protein Synthesis in, Total Chemical Synthesis of Proteins, 2021, pp. 185–209 Search PubMed.
- J. Liu, T. Wei, Y. Tan, H. Liu and X. Li, Enabling chemical protein (semi)synthesis via reducible solubilizing tags (RSTs), Chem. Sci., 2022, 13(5), 1367–1374 RSC.
- S. A. Abboud, E. Cisse, M. Doudeau, H. Benedetti and V. Aucagne, A straightforward methodology to overcome solubility challenges for N-terminal cysteinyl peptide segments used in native chemical ligation, Chem. Sci., 2021, 12(9), 3194–3201 RSC.
- S. Tsuda, M. Mochizuki, H. Ishiba, K. Yoshizawa-Kumagaye, H. Nishio, S. Oishi and T. Yoshiya, Easy-to-Attach/Detach Solubilizing-Tag-Aided Chemical Synthesis of an Aggregative Capsid Protein, Angew Chem. Int. Ed. Engl., 2018, 57(8), 2105–2109 CrossRef CAS PubMed.
- M. T. Jacobsen, M. E. Petersen, X. Ye, M. Galibert, G. H. Lorimer, V. Aucagne and M. S. Kay, A Helping Hand to Overcome Solubility Challenges in Chemical Protein Synthesis, J. Am. Chem. Soc., 2016, 138(36), 11775–11782 CrossRef CAS PubMed.
- E. C. Johnson, E. Malito, Y. Shen, D. Rich, W. J. Tang and S. B. Kent, Modular total chemical synthesis of a human immunodeficiency virus type 1 protease, J. Am. Chem. Soc., 2007, 129(37), 11480–11490 CrossRef CAS PubMed.
- B. Bernardim, M. J. Matos, X. Ferhati, I. Companon, A. Guerreiro, P. Akkapeddi, A. C. B. Burtoloso, G. Jimenez-Oses, F. Corzana and G. J. L. Bernardes, Efficient and irreversible antibody-cysteine bioconjugation using carbonylacrylic reagents, Nat. Protoc., 2019, 14(1), 86–99 CrossRef CAS PubMed.
- A. K. Nowinski, F. Sun, A. D. White, A. J. Keefe and S. Y. Jiang, Sequence, Structure, and Function of Peptide Self-Assembled Monolayers, J. Am. Chem. Soc., 2012, 134(13), 6000–6005 CrossRef CAS PubMed.
- A. J. Keefe, K. B. Caldwell, A. K. Nowinski, A. D. White, A. Thakkar and S. Y. Jiang, Screening nonspecific interactions of peptides without background interference, Biomaterials, 2013, 34(8), 1871–1877 CrossRef CAS PubMed.
- G. B. Vamisetti, G. Satish, P. Sulkshane, G. Mann, M. H. Glickman and A. Brik, On-Demand Detachment of Succinimides on Cysteine to Facilitate (Semi)Synthesis of Challenging Proteins, J. Am. Chem. Soc., 2020, 142(46), 19558–19569 CrossRef CAS PubMed.
- Y. Asahina, S. Komiya, A. Ohagi, R. Fujimoto, H. Tamagaki, K. Nakagawa, T. Sato, S. Akira, T. Takao, A. Ishii, Y. Nakahara and H. Hojo, Chemical Synthesis of O-Glycosylated Human Interleukin-2 by the Reverse Polarity Protection Strategy, Angew. Chem., Int. Ed., 2015, 54(28), 8226–8230 CrossRef CAS PubMed.
- C. E. Murar, M. Ninomiya, S. Shimura, U. Karakus, O. Boyman and J. W. Bode, Chemical Synthesis of Interleukin-2 and Disulfide Stabilizing Analogues, Angew. Chem., Int. Ed., 2020, 59(22), 8425–8429 CrossRef CAS PubMed.
- H. X. Wu, Y. Tan, W. L. Ngai and X. C. Li, Total synthesis of interleukin-2 via a tunable backbone modification strategy, Chem. Sci., 2023, 14(6), 1582–1589 RSC.
- R. A. Lal, H. P. Moeller, E. A. Thomson, T. M. Horton, S. Lee, R. Freeman, P. Prahalad, A. S. Y. Poon and J. P. Annes, Novel Pathogenic De Novo INS p.T97P Variant Presenting With Severe Neonatal DKA, Endocrinology, 2022, 163(2), 1–9 CrossRef CAS PubMed.
- X. Xiong, A. Blakely, J. H. Kim, J. G. Menting, I. B. Schafer, H. L. Schubert, R. Agrawal, T. Gutmann, C. Delaine, Y. W. Zhang, G. O. Artik, A. Merriman, D. Eckert, M. C. Lawrence, U. Coskun, S. J. Fisher, B. E. Forbes, H. Safavi-Hemami, C. P. Hill and D. H. Chou, Symmetric and asymmetric receptor conformation continuum induced by a new insulin, Nat. Chem. Biol., 2022, 18, 511–519 CrossRef CAS PubMed.
- R. Hernandez, J. Poder, K. M. LaPorte and T. R. Malek, Engineering IL-2 for immunotherapy of autoimmunity and cancer, Nat. Rev. Immunol., 2022, 22, 614–628 CrossRef CAS PubMed.
- T. M. Hackeng, J. H. Griffin and P. E. Dawson, Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(18), 10068–10073 CrossRef CAS PubMed.
- B. B. Dang, T. Kubota, K. Mandal, F. Bezanilla and S. B. H. Kent, Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography, J. Am. Chem. Soc., 2013, 135(32), 11911–11919 CrossRef CAS PubMed.
- G.-M. Fang, J.-X. Wang and L. Liu, Convergent Chemical Synthesis of Proteins by Ligation of Peptide Hydrazides, 2012, vol. 51(41), pp. 10347–10350 Search PubMed.
- G.-M. Fang, Y.-M. Li, F. Shen, Y.-C. Huang, J.-B. Li, Y. Lin, H.-K. Cui and L. Liu, Protein Chemical Synthesis by Ligation of Peptide Hydrazides, 2011, vol. 50(33), pp. 7645–7649 Search PubMed.
- D. T. Flood, J. C. J. Hintzen, M. J. Bird, P. A. Cistrone, J. S. Chen and P. E. Dawson, Leveraging the Knorr Pyrazole Synthesis for the Facile Generation of Thioester Surrogates for use in Native Chemical Ligation, Angew Chem. Int. Ed. Engl., 2018, 57(36), 11634–11639 CrossRef CAS PubMed.
- P. Sengupta, K. Meena, R. Mukherjee, S. K. Jain and K. Maithal, Optimized conditions for high-level expression and purification of recombinant human interleukin-2 in E. coli, Indian J. Biochem. Biophys., 2008, 45(2), 91–97 CAS.
- E. J. Hsu, X. Cao, B. Moon, J. Bae, Z. Sun, Z. Liu and Y. X. Fu, A cytokine receptor-masked IL2 prodrug selectively activates tumor-infiltrating lymphocytes for potent antitumor therapy, Nat. Commun., 2021, 12(1), 2768 CrossRef CAS PubMed.
- J. A. Karas, J. D. Wade and M. A. Hossain, The Chemical Synthesis of Insulin: An Enduring Challenge, Chem. Rev., 2021, 121(8), 4531–4560 CrossRef CAS PubMed.
- F. Liu, E. Y. Luo, D. B. Flora and A. R. Mezo, A synthetic route to human insulin using isoacyl peptides, Angew Chem. Int. Ed. Engl., 2014, 53(15), 3983–3987 CrossRef CAS PubMed.
- N. Zheng, P. Karra, M. A. VandenBerg, J. H. Kim, M. J. Webber, W. L. Holland and D. H. C. Chou, Synthesis and Characterization of an A6-A11 Methylene Thioacetal Human Insulin Analogue with Enhanced Stability, J. Med. Chem., 2019, 62(24), 11437–11443 CrossRef CAS PubMed.
- S. Laps, F. Atamleh, G. Kamnesky, S. Uzi, M. M. Meijler and A. Brik, Insight on the Order of Regioselective Ultrafast Formation of Disulfide Bonds in (Antimicrobial) Peptides and Miniproteins, Angew Chem. Int. Ed. Engl., 2021, 60(45), 24137–24143 CrossRef CAS PubMed.
- S. Laps, H. Sun, G. Kamnesky and A. Brik, Palladium-Mediated Direct Disulfide Bond Formation in Proteins Containing S-Acetamidomethyl-cysteine under Aqueous Conditions, Angew Chem. Int. Ed. Engl., 2019, 58(17), 5729–5733 CrossRef CAS PubMed.
- M. M. Disotuar, M. E. Petersen, J. M. Nogueira, M. S. Kay and D. H. C. Chou, Synthesis of hydrophobic insulin-based peptides using a helping hand strategy, Org. Biomol. Chem., 2019, 17(7), 1703–1708 RSC.
- A. Micsonai, E. Moussong, F. Wien, E. Boros, H. Vadaszi, N. Murvai, Y. H. Lee, T. Molnar, M. Refregiers, Y. Goto, A. Tantos and J. Kardos, BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy, Nucleic Acids Res., 2022, 50, W90–W98 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Zidek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596(7873), 583–589 CrossRef CAS PubMed.
- C. M. Deber, M. Glibowicka and G. A. Woolley, Conformations of proline residues in membrane environments, Biopolymers, 1990, 29(1), 149–157 CrossRef CAS PubMed.
- J. Arnorsdottir, A. R. Sigtryggsdottir, S. H. Thorbjarnardottir and M. M. Kristjansson, Effect of proline substitutions on stability and kinetic properties of a cold adapted subtilase, J. Biochem., 2009, 145(3), 325–329 CrossRef CAS PubMed.
- F. S. Cordes, J. N. Bright and M. S. Sansom, Proline-induced distortions of transmembrane helices, J. Mol. Biol., 2002, 323(5), 951–960 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06032b |
| ‡ Current address: Novo Nordisk Research Center, Indianapolis, IN 46241, USA. |
| This journal is © The Royal Society of Chemistry 2024 |