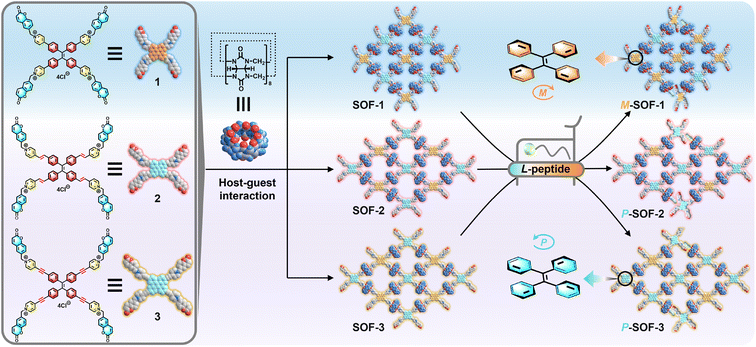
Open Access Article
Open Access Article“Gear-driven”-type chirality transfer of tetraphenylethene-based supramolecular organic frameworks for peptides in water†
Chaochao
Yan‡
a,
Qingfang
Li‡
a,
Kaige
Wang
a,
Wanni
Yang
a,
Jingyu
Han
a,
Yawen
Li
a,
Yunhong
Dong
a,
Dake
Chu
b,
Lin
Cheng
a and
Liping
Cao
 *a
*a
aCollege of Chemistry and Materials Science, Northwest University, Xi'an 710069, China. E-mail: chcaoliping@nwu.edu.cn
bDepartment of Gastroenterology, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710061, China
First published on 1st February 2024
Abstract
Chirality transfer for natural chiral biomolecules can reveal the indispensable role of chiral structures in life and can be used to develop the chirality-sensing biomolecular recognition. Here, we report the synthesis and characterization of a series of achiral supramolecular organic frameworks (SOF-1, SOF-2, and SOF-3), constructed from cucurbit[8]uril (CB[8]) and tetraphenylethene (TPE) derivatives (1, 2, and 3), respectively, as chirality-sensing platforms to explore their chirality transfer mechanism for peptides in water. Given the right-handed (P) and left-handed (M) rotational conformation of TPE units and the selective binding of CB[8] to aromatic amino acids, these achiral SOFs can be selectively triggered in water by peptides containing N-terminal tryptophan (W) and phenylalanine (F) residues into their P- or M-rotational conformation, exhibiting significantly different circular dichroism (CD) spectra. Although various peptides have the same L-type chiral configuration, they can induce positive CD signals of SOF-1 and negative CD signals of SOF-2 and SOF-3, respectively. Based on the structural analysis of the linkage units between CB[8] and TPE units in these SOFs, a “gear-driven”-type chirality transfer mechanism has been proposed to visually illustrate the multiple-step chirality transfer process from the recognition site in the CB[8]'s cavity to TPE units. Furthermore, by utilizing the characteristic CD signals generated through the “gear-driven”-type chirality transfer, these SOFs can serve as chiroptical sensor arrays to effectively recognize and distinguish various peptides based on their distinctive CD spectra.
Introduction
Chirality transfer is one of the most intriguing features in living assembly systems, playing a vital role in the formation of chiral biological structures.1 The chirality transfer of molecules to supramolecules through the self-assembly process is widely present in biological systems.2 For instance, during the DNA transcription process, through the chirality transfer facilitated by RNA polymerase (RNAP), left-handed Z-DNA can form transiently immediately upstream of RNAP.3 Chirality transfer from natural chiral biomolecules to artificial achiral assembly systems is considered one of the most significant processes in self-assembly,4 as it is not only closely associated with the origin of homochirality in nature, but also shows wide applications in chiral catalysis,5 chiral recognition,6 and chiroptical materials.7 Inspired from nature, an increasing number of synthetic supramolecular systems with controlled chirality transfer have been designed and developed over the past few decades.8 Through appropriate interactions, one-step chirality transfer from small guests (e.g., chiral biomolecules) to macrocyclic/cage hosts has been realized.9 However, it is interesting and challenging to subtly regulate chirality within a supramolecular assembly system through multiple-step chirality transfer.10Benefiting from the multivalent supramolecular assembly, supramolecular organic frameworks (SOFs) could be fabricated from cucurbit[8]uril (CB[8]) and building blocks through the head-to-tail host–guest interactions in the aqueous phase.11 These CB[8]-based SOFs could easily realize various functions by utilizing different functional units as building blocks and encapsulating different guest molecules.12 In particular, as a unique class of supramolecular polymers, employing SOFs as a class of tools for specifically recognizing natural chiral biomolecules (e.g., amino acids and peptides) for broader biological applications is worth being explored.13 Tetraphenylethylene (TPE) molecules possess the propeller-like left-handed (M) and right-handed (P) rotational chiral conformations,14 endowing the ability of chirality sensing. Consequently, TPE molecules are the ideal building blocks for SOFs to explore the conformational chirality with dynamic properties, which can be utilized to achieve chirality transfer for chiral biomolecules. We attempted to construct varied TPE-based SOFs through the precise regulation of the building block structure, and to realize the differentiated chirality transfer for varied peptides, thereby finally establishing a kind of new chirality-sensing recognition system using SOFs.
Very recently, we reported a water-soluble TPE-based SOF (SOF-1), which was self-assembled from CB[8] and 1, and it could initially distinguish dipeptide sequences based on CD responses through chirality transfer.15 Based on this result, we further designed and synthesized TPE derivatives (2) with pyridinium-4-vinyl arms and (3) with pyridinium-4-ynyl arms, which could be self-assembled with CB[8] to form SOFs (SOF-2 and SOF-3) in aqueous solutions, respectively (Scheme 1). These achiral SOFs have the hydrophobic cavity of CB[8], which is the recognition site for aromatic amino acids,16 and possess the M- and P-rotational chiral conformations of TPE units, which are chirality-responsive groups for chirality transfer. Therefore, SOFs could be ideal hosts for aromatic amino acids and peptides. Interestingly, peptides with the same L-type chiral configuration induced completely opposite CD spectra: aromatic amino acids such as tryptophan (W) and phenylalanine (F), as well as peptides containing different numbers of aromatic residues (e.g., WW and FF), induced chiral M-SOF-1 with positive CD signals, while the same guests induced chiral P-SOF-2 and P-SOF-3 with negative CD signals (Scheme 1). Furthermore, the M- or P-rotational conformation chirality of SOFs can be selectively induced by N-terminal W-/F-containing tripeptides not C-terminal ones based on the selective recognition of CB[8]. Therefore, according to different SOFs having characteristic chiral response signals for same chiral guests, SOFs could serve as chiroptical sensor arrays, which can effectively enhance the accuracy of molecular recognition.
Results and discussion
Design and formation of the SOFs
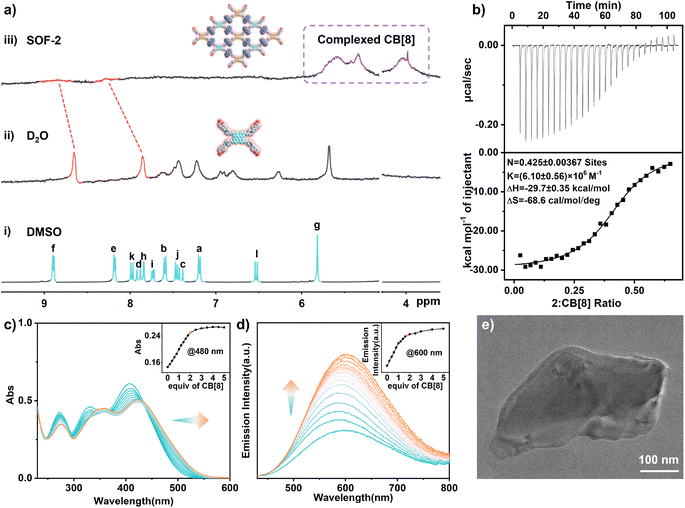
In our previous work,15 we designed and synthesized a water-soluble and fluorescent tetra(6-coumarinylmethyl-1-pyridinium) TPE derivative (1), which contains a central TPE core as the chromophore and dynamic chirality-sensing unit, and four coumarin arms at the terminal position as connecting units for the dimerization binding within CB[8], resulting in the successful construction of SOF-1 in aqueous solution. Based on the construction of SOF-1, we further designed and synthesized two water-soluble fluorescence building blocks (2 and 3) as the basic units of the SOF skeleton (synthetic details in ESI, Fig. S1–S8†). In the molecular structure of 2, double bonds were introduced between TPE and pyridine units, while triple bonds were introduced into 3. The proton resonances of 2 and 3 are sharp and identifiable in DMSO (Fig. 1a(i)), but broad and ill-defined in D2O due to their aggregation behaviour (Fig. 1a(ii)). To understand the host–guest interactions between 2 or 3 and CB[8], 1H NMR titration experiments were first performed in D2O. When 0–2.0 equiv. of CB[8] was added to a solution of 2 in D2O, all the proton resonances corresponding to 2 broadened, indicating that the host–guest polymeric species (SOF-2) was formed (Fig. 1a(iii)). Next, after the addition of excess CB[8] (3.0 equiv.), free CB[8] signals appeared and the excess CB[8] did not induce obvious chemical shifts of 2, indicating the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry of 2 and CB[8] in SOF-2 (Fig. S9†). And the 1H NMR titration experiment of 3 and CB[8] also showed similar results for proton resonance broadening, indicating the formation of SOF-3 in solution (Fig. S10†). The isothermal calorimetric titration (ITC) experiments also confirmed a 1
2 stoichiometry of 2 and CB[8] in SOF-2 (Fig. S9†). And the 1H NMR titration experiment of 3 and CB[8] also showed similar results for proton resonance broadening, indicating the formation of SOF-3 in solution (Fig. S10†). The isothermal calorimetric titration (ITC) experiments also confirmed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry with a strong ternary binding constant (Ka) of ≈ 1013 M−2, indicating that building blocks 2 or 3 can effectively bind with CB[8] to form a supramolecular assembly (Fig. 1b and S11†). Meanwhile, the Job's plot analysis also indicated a 1
2 stoichiometry with a strong ternary binding constant (Ka) of ≈ 1013 M−2, indicating that building blocks 2 or 3 can effectively bind with CB[8] to form a supramolecular assembly (Fig. 1b and S11†). Meanwhile, the Job's plot analysis also indicated a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry in SOF-2 and SOF-3 (Fig. S12 and S13†).
2 stoichiometry in SOF-2 and SOF-3 (Fig. S12 and S13†).
Additionally, dynamic light scattering (DLS) experiments showed that the average hydrodynamic diameter (DH) values of SOF-2 and SOF-3 are significantly larger than those of the monomer molecules, which clearly indicates the formation of large supramolecular assemblies. Concentration-dependent DLS showed that SOF-2 possessed various DH of ≈ 414 nm–1190 nm in the 10 μM–50 μM range owing to the reversible host–guest complexation (Fig. S14a†). Similarly, concentration-depended DLS showed that SOF-3 was stable with DH of ≈ 516 nm–1238 nm in the 10–50 μM range (Fig. S14b†). The ζ-potential experiments showed that SOF-2 (43.7 ± 0.6 mV) has a higher positive ζ-potential than 2 (17.9 ± 0.5 mV), and SOF-3 (39.8 ± 0.6 mV) also has a higher positive ζ-potential than 3 (16.3 ± 0.6 mV), indicating that SOF-2 and SOF-3 have high stability due to the repulsive forces among these large multiple-cationic supramolecular assemblies (Table S1†). These results clearly indicate the formation of large and stable supramolecular assemblies from monomer molecules (2 or 3) and CB[8] in the aqueous solution.
The photophysical properties of the supramolecular systems of 2 or 3 with CB[8] in water were studied by UV-vis and fluorescence experiments. Initially, UV-vis titration experiments between 2 and CB[8] showed that upon increasing the amount of CB[8], the intensities of the three absorbance peaks at 273 nm, 331 nm, and 407 nm decreased in intensity, and were red shifted to 275 nm, 357 nm, and 421 nm, respectively, with three isosbestic points at 343 nm, 367 nm, and 436 nm (Fig. 1c). These results show that through the host–guest interaction between 2 and CB[8], the flexible rotation of the alkene bonds in monomer 2 is effectively limited, and the intermolecular conjugation and charge-transfer (CT) between 2 molecules are enhanced through the CB[8]'s dimerization. As illustrated in Fig. 1d, the solution of free 2 in water displayed a weak orange emission with low absolute quantum yield (ΦF = 0.77%) and short fluorescence lifetime (τ = 0.88 ns). When 0–2.0 equiv. of CB[8] was added to the solution of 2 to form SOF-2 in water, the maximum emission peak with higher absolute quantum yield and fluorescence lifetime (ΦF = 3.31%, τ = 1.32 ns) was red shifted from 580 nm to 600 nm (Fig. S15 and Table S2†). Similarly, the formation of SOF-3 was confirmed by UV-vis and fluorescence titrations in aqueous solution. In the case of 3, the addition of CB[8] (0–2.0 equiv.) resulted in absorbance peaks at 275 nm, 317 nm, and 386 nm which decrease slightly in intensity, giving two isosbestic points at 340 nm and 359 nm. Furthermore, the absorbance maximum at 386 nm, which belongs to the π–π* transition of the TPE unit, was slightly blue-shifted to 379 nm, indicating that the formation of SOF-3 restricts the rotation of TPEs to decrease the conjugation of TPE units (Fig. S17a†). The fluorescence spectrum of free 3 in water displayed a yellow emission (λmax = 640 nm) upon excitation at 395 nm. When 0–2.0 equiv. of CB[8] was added to the solution of 3 to form SOF-3 in water, the emission wavelength was blue-shifted to 608 nm, because of the decrease in the degree of conjugation of the TPE core, as a result of the restriction of rotation (RIR) of TPEs in SOF-3 (Fig. S17b†).17
Finally, the solid structure of SOFs was investigated by scanning electron microscopy (SEM), transmission electron microscopy (TEM), and synchrotron small-angle X-ray-scattering (SAXS). SEM images of the solid samples obtained from aqueous solutions of SOF-2 and SOF-3 revealed the presence of large supramolecular assemblies compared to small particles of 2 and 3 in water (Fig. S18 and S19†). TEM images showed distinct sheet-like structures and the sizes of such nanosheets were estimated to range from approximately one-hundred to several-hundred nanometers (Fig. 1e and S20†). In addition, SAXS analysis of solid samples of SOF-2 and SOF-3 displayed peaks centered at ∼1.5 nm, confirming the regularity of the framework structure (Fig. S21†). All these results strongly suggested the formation of supramolecular organic frameworks.
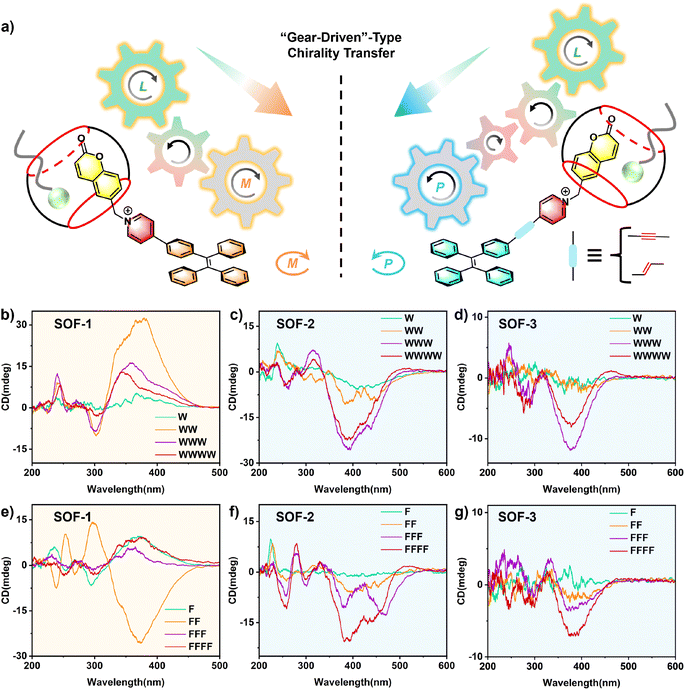
“Gear-driven”-type chirality transfer
Most chiral biomolecules, such as amino acids and peptides, are difficult to analyze due to their absorption in the far-UV region (<250 nm) and the negligible Cotton effect on the CD spectra.18 The M- and P-rotational chiral conformations of TPE compounds can show positive and negative Cotton effects in the region of 350 nm–500 nm, where CD spectra can be more identifiable and undisturbed. SOFs can bind with chiral guests through the cavities/pores of the framework to achieve chirality transfer from chiral molecules to the TPE units in the framework, inducing the excessive M- or P-rotational chiral conformation of the TPE units with CD signals in the long-wavelength region. Therefore, we expect to utilize dynamic rotational chiral conformations of TPEs to gain characteristic CD signals generated by chirality transfer from chiral biomolecules.19Initially, W and its peptides (e.g., WW, WWW, and WWWW) were selected as chiral guests to investigate the chirality transfer of the SOF system. Interestingly, these guests with the same L-type chiral configuration induced diametrically opposite chiroptical responses of different SOFs (Fig. 2). In SOF-1, W and its peptides could almost induce obvious positive Cotton effects in the long-wavelength (315 nm–470 nm) region with different intensities, respectively, which is attributed to the excess of M-conformational TPE units of SOF-1 (Fig. 2b and S22†), whereas the same amino acid and peptides induced the formation of chiral P-SOF-2 and P-SOF-3, characterized by an abundance of P-conformational TPE units, which in turn results in negative CD signals (Fig. 2c, d and S23–S24†).
Inspired by the gear-driven machinery, we proposed a “gear-driven”-type chirality transfer mechanism to illustrate the process of multiple-step chirality transfer in SOFs (Fig. 2a). In SOF-1, 1 is composed of three parts: the terminal coumarin arm within CB[8], the pyridine group, and the central TPE unit, which represent the driving gear, driven gear, and output gear in a three-stage gear-driven machinery, respectively. It is worth noting that the rotational direction of adjacent gears is always opposite in gear-driven machinery. Therefore, in the three-stage gear-driven machinery, the rotational directions of the driving gear and the spaced output gear are consistent. In the context of this chirality transfer system, chiral guests with positive CD signals around 200 nm in the CD spectrum act as the “driving gear” (Fig. S25†). They induced the formation of M-SOF-1 with positive Cotton effects, characterized by TPE units as the “output gear” expressing an abundance of M-rotational conformations (Fig. 2b). In the structures of 2 and 3, alkene bonds or alkyne bonds between TPE and pyridine units are the addition of another gear in the middle of the driven gear and the output gear, thus changing the rotational direction of the output gear. As a result, as output units for multiple-step chirality transfer, there is an excess of P-rotational conformations of TPE units in SOF-2 and SOF-3 (Fig. 2c and d). In these chirality transfer systems, the cavity of CB[8] in SOFs serves as the starting point and the end point is TPE units, respectively. Compared with SOF-1, SOF-2 and SOF-3 have more “transmission shaft” units and longer distance of chirality transfer, due to the introduction of alkene bonds or alkyne bonds between TPE and pyridine units. Through this precise control of “transmission shaft” structure to achieve multiple-step chirality transfer, different SOFs exhibit completely opposite chiroptical signals for the same chiral guest.
To further expand the scope of multiple-step chirality transfer in the SOF system, another aromatic amino acid F and its peptides (e.g., FF, FFF, and FFFF) were selected as chiral guests. CD experiments showed that F and its peptides also induced M-SOF-1 with positive CD and P-SOF-2 and P-SOF-3 with negative CD, which is consistent with W and its peptides (Fig. 2e–g and S26–S28†). Notably, only FF induced P-SOF-1 with negative CD. This was probably attributed to FF undergoing a conformation change from fold to unfold state due to the binding behaviour within the cavity of CB[8], accompanied by a CD inversion from positive to negative around 200 nm in SOF-1 (Fig. S29†).20 This corresponds to the change in the rotational direction of the driving gear in the three-stage gear-driven machinery. As a result, the TPE unit acts as the “output gear” to express the opposite rich P-conformation with a negative CD signal. The CD response of SOFs to peptide guests strongly suggests that the “gear-driven”-type chirality transfer can properly elucidate the “gear-driven”-type chirality transfer mechanism of the SOF system.
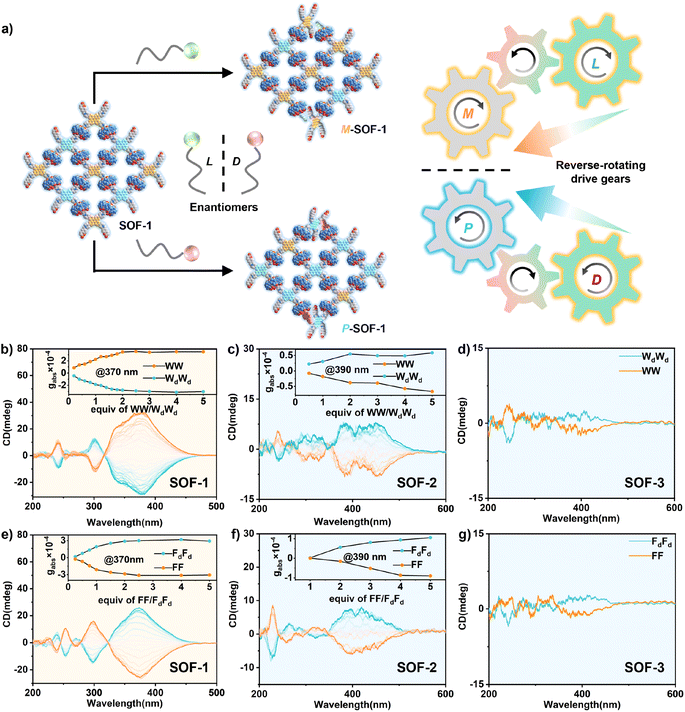
Moreover, the opposite chirality transfer of SOFs triggered by pairs of enantiomers W/Wd, WW/WdWd, F/Fd, and FF/FdFd exhibited the chiral adaptation, respectively, leading to mirror-symmetric CD spectra (Fig. 3a). CD spectra of SOF-1 titrated with WW/WdWd are shown separately: WW induced M-SOF-1 with a positive Cotton effect, whereas WdWd induced P-SOF-1 with a negative Cotton effect (Fig. 3b). In contrast, SOF-2 and SOF-3 with an added gear showed diametrically opposite results when compared to SOF-1. WW induced P-SOF-2 and P-SOF-3 with negative Cotton effects, whereas WdWd induced M-SOF-2 and M-SOF-3 with positive Cotton effects (Fig. 3c and d). Similarly, W/Wd, F/Fd, and FF/FdFd also could induce corresponding CD spectra with a mirror image, respectively (Fig. 3e–g and S30–S35†). It should be noted that SOF-3 exhibits weaker CD responses to these amino acids and dipeptides, possibly due to the lower conjugation of the triple bond and the longer chiral transfer distance in SOF-3. The results of adaptive chirality transfer further confirm that altering the absolute configuration of chiral guests in SOF systems corresponds to switching the rotational direction of the driving gear in the gear-driven chirality transfer process, further leading to the output of TPE units with opposite rotational conformations. All these results demonstrate that the “gear-driven”-type mechanism of the chirality transfer aligns perfectly with the chirality sensing observed in the SOF system.
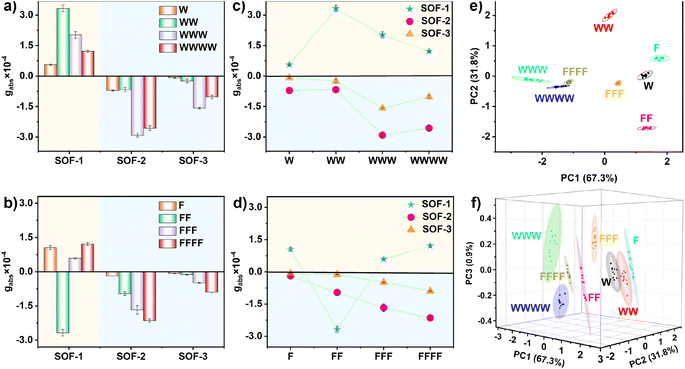
Chirality-sensing analysis
In recent years, the chiroptical sensing technology based on host–guest recognition in the CD spectrum has attracted significant attention due to its potential applications in rapid high-throughput screening.21 As SOFs exhibit excellent chirality-sensing properties, characteristic CD signals can be generated through “gear-driven”-type chirality transfer, enabling rapid differentiation of these chiral biomolecules. Peptides induce opposite M- and P-rotational conformations in the TPE unit, resulting in distinct directionally opposite signals in the CD spectrum (Fig. 4a and b). Meanwhile, in the SOF-based chirality sensing system, different peptides also induce different CD signals with varying intensities (Fig. 4c and d). Undoubtedly, these characteristic CD signals with both direction and intensity differences can be used for peptide analysis. However, in spectroscopy (e.g., fluorescence and CD), when the signals have the same direction and the intensity differences are not significant, it is often difficult to achieve truly effective differentiation based solely on visual observation of intensity differences (Fig. S36†). To improve the accuracy of recognition, sensor arrays have been proven to be effective tools for distinguishing biological molecules and cells.22Therefore, we envisioned using varied SOFs as chiroptical sensor arrays and employing the data analysis method of principal component analysis (PCA) to convert the rich differential information contained in the CD response fingerprint into interpretable classification data, thereby further effectively differentiating these chiral biomolecules.23 As shown in Fig. 4e, the PCA score plot of the top two factors of contribution (PC1, PC2) accounted for 99.1% of the total variance. Notably, the 95% confidence ellipse produced by the statistical PCA evaluation contained little overlaps between 8 distinguished peptide clusters. The 3D PCA results showed further distinction of all selected molecules absolutely (Fig. 4f). These results demonstrate that using SOFs as chiroptical sensor arrays can significantly enhance the accuracy of molecular recognition, allowing for a more precise differentiation and analysis of chiral biomolecules.
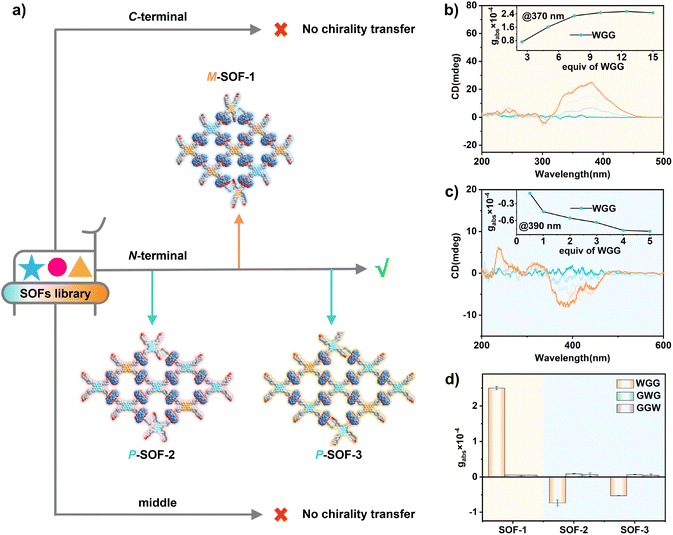
Sequence-selective chirality sensing
During this process of chirality transfer, the N-terminal aromatic amino acid residue is captured by CB[8] and the C-terminal part is located at the pore of the SOFs. Thus, the chiral microenvironment is first formed at the region around the cavity of CB[8], which shortens the distance from the chiral source of peptides (chiral donor) to the SOFs (chiral acceptor) through the cavity of CB[8]. To further uncover the key chiral source of aromatic amino acids and aromatic peptides for chirality transfer, tripeptides containing the achiral aliphatic amino acid glycine (G) coupled with the W residue (WGG, GWG, GGW) were selected as chiral guests (Fig. 5).The CD titration experiments showed that WGG with the W residue located at the N-terminal induced the M-rotational conformation of TPE units of SOF-1 with a positive Cotton effect (Fig. 5b), while inducing the P-rotational conformation of TPE units of SOF-2 and SOF-3 with negative Cotton effects (Fig. 5c and S37). This result further expands the scope of the “gear-driven”-type chirality transfer, while GWG and GGW, in which W residue is located at the middle and C-terminal, couldn't induce any obvious CD signals (Fig. 5d and S38–S40†). Moreover, ITC of SOFs titrated with peptides confirmed their energy-favourable binding behavior (Fig. S41–S49†). And DLS experiments also confirmed that the binding behavior of SOFs and peptides did not destroy the aggregation state of the SOF complexes (Fig. S50–S53†). Therefore, these results further confirmed that the selective recognition of aromatic amino acid residues located at the N-terminal with CB[8] is a crucial factor for triggering the chirality transfer process in SOFs. Strong host–guest interactions lead to the binding behavior of chiral guests within the pores of SOFs, ultimately achieving chirality transfer from the guests to the TPE units to induce the conformational chirality of achiral SOFs.
Conclusions
In summary, we have reported a “gear-driven”-type chirality transfer of TPE-based achiral supramolecular organic frameworks for peptides in water. Given the M- and P-rotational chiral conformations of the TPE units and the specific recognition of N-terminal W/F residues by CB[8], aromatic amino acids and their peptides can successfully induce opposite chiral conformations in different SOFs to show positive and negative CD responses with different intensities, respectively. Based on the structural analysis of the linkage units between CB[8] and TPE units in these SOFs, the “gear-driven”-type chirality transfer mechanism can be used to illustrate the process of multiple-step chirality transfer in the SOF system. With the characteristic CD signals generated by chirality transfer, these SOFs can serve as supramolecular hosts and chiroptical arrays to effectively recognize and precisely analyse chiral biomolecules such as amino acids and peptides. Therefore, these SOFs with chirality-sensing recognition capabilities might further be used to distinguish other complex biomolecules, such as proteins or even chromosomes, through characteristic CD signals and array sensing patterns. On the other hand, achieving controlled chirality transfer in artificial achiral assembly systems will offer fresh inspiration for the design and application of novel chiral supramolecular systems in future.Author contributions
C. Yan (first author): investigation, data curation, formal analysis, visualization and writing – original draft; Q. Li (Co-first author): data curation, formal analysis and writing – original draft; K. Wang, W. Yang, and J. Han: synthesis of compounds; Y. Li, Y. Dong, D. Chu, and L. Cheng: edited the manuscript; L. Cao (corresponding author): conceptualization, supervision, validation, writing – review & editing and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22122108 and 22371229). D. Chu thanks the National Natural Science Foundation NSAF Joint Fund (U2230112) and the Key Laboratory Fund for Plasma Physics (6142A04210108).Notes and references
- K. Taniguchi, R. Maeda, T. Ando, T. Okumura, N. Nakazawa, R. Hatori, M. Nakamura, S. Hozumi, H. Fujiwara and K. Matsuno, Science, 2011, 333, 336–339 CrossRef PubMed.
- (a) P. Peluso and B. Chankvetadze, Chem. Rev., 2022, 122, 13235–13400 CrossRef CAS PubMed; (b) J. C. Kendrew, R. E. Dickerson, B. E. Strandberg, R. G. Hart, D. R. Davies, D. C. Phillips and V. C. Shore, Nature, 1960, 185, 422–427 CrossRef CAS PubMed; (c) M. Liu, L. Zhang and T. Wang, Chem. Rev., 2015, 115, 7304–7397 CrossRef CAS PubMed.
- D. Winogradoff, P.-Y. Li, H. Joshi, L. Quednau, C. Maffeo and A. Aksimentiev, Adv. Sci., 2021, 8, 2003113 CrossRef CAS PubMed.
- (a) G. Liu, C. Tian, X. Fan, X. Xue, L. Feng, C. Wang and Y. Liu, JACS Au, 2023, 3, 2550–2556 CrossRef CAS PubMed; (b) T. Miao, X. Cheng, Y. Guo, G. Zhang and W. Zhang, Giant, 2023, 14, 100161 CrossRef CAS.
- (a) F. Li, L. Long, Y. M. He, Z. Li, H. Chen and Q. H. Fan, Angew. Chem., Int. Ed., 2022, 61, e202202972 CrossRef CAS PubMed; (b) J. Guo, Y.-W. Xu, K. Li, L.-M. Xiao, S. Chen, K. Wu, X.-D. Chen, Y.-Z. Fan, J.-M. Liu and C.-Y. Su, Angew. Chem., Int. Ed., 2017, 56, 3852 CrossRef CAS PubMed; (c) X. Han, J. Zhang, J. Huang, X. Wu, D. Yuan, Y. Liu and Y. Cui, Nat. Commun., 2018, 9, 1294 CrossRef PubMed.
- (a) J. Yao, W. Wu, W. Liang, Y. Feng, D. Zhou, J. J. Chruma, G. Fukuhara, T. Mori, Y. Inoue and C. Yang, Angew. Chem., Int. Ed., 2017, 56, 6869–6873 CrossRef CAS PubMed; (b) K. Adachi, S. Fa, K. Wada, K. Kato, S. Ohtani, Y. Nagata, S. Akine and T. Ogoshi, J. Am. Chem. Soc., 2023, 145, 8114–8121 CrossRef CAS PubMed; (c) M. Quan, X. Y. Pang and W. Jiang, Angew. Chem., Int. Ed., 2022, 61, e202201258 CrossRef CAS PubMed; (d) K. Maeda, D. Hirose, N. Okoshi, K. Shimomura, Y. Wada, T. Ikai, S. Kanoh and E. Yashima, J. Am. Chem. Soc., 2018, 140, 3270–3276 CrossRef CAS PubMed.
- (a) O. Oki, C. Kulkarni, H. Yamagishi, S. C. J. Meskers, Z.-H. Lin, J.-S. Huang, E. W. Meijer and Y. Yamamoto, J. Am. Chem. Soc., 2021, 143, 8772–8779 CrossRef CAS PubMed; (b) G. Albano, G. Pescitelli and L. D. Bari, Chem. Rev., 2020, 120, 10145–10243 CrossRef CAS PubMed; (c) C. Du, Z. Li, X. Zhu, G. Ouyang and M. Liu, Nat. Nanotechnol., 2022, 17, 1294–1302 CrossRef CAS PubMed.
- (a) M. Sujith, E. K. Vishnu, S. Sappati, M. S. O. Hassan, V. Vijayan and K. G. Thomas, J. Am. Chem. Soc., 2022, 144, 5074–5086 CrossRef CAS PubMed; (b) W. Shang, X. Zhu, Y. Jiang, J. Cui, K. Liu, T. Li and M. Liu, Angew. Chem., Int. Ed., 2022, 61, e202210604 CrossRef CAS PubMed; (c) K. Harada, R. Sekiya and T. Haino, Angew. Chem., Int. Ed., 2022, 61, e202209340 CrossRef CAS PubMed; (d) H. Yan, Y. He, D. Wang, T. Han and B. Z. Tang, Aggregate, 2023, 4, e331 CrossRef CAS; (e) M. Quan, X. Y. Pang and W. Jiang, Angew. Chem., Int. Ed., 2022, 134, e202201258 CrossRef; (f) M. Fukuda, M. Morikawa, D. Hirose, T. Taniguchi, T. Nishimura, E. Yashima and K. Maeda, Angew. Chem., Int. Ed., 2023, 62, e202217020 CrossRef CAS PubMed.
- (a) Q.-P. Hu, H. Zhou, T.-Y. Huang, Y.-F. Ao, D.-X. Wang and Q.-Q. Wang, J. Am. Chem. Soc., 2022, 144, 6180–6184 CrossRef CAS PubMed; (b) H. Nian, L. Cheng, L. Wang, H. Zhang, P. Wang, Y. Li and L. Cao, Angew. Chem., Int. Ed., 2021, 60, 15354–15358 CrossRef CAS PubMed; (c) X. Song, X. Zhu, S. Qiu, W. Tian and M. Liu, Angew. Chem., Int. Ed., 2022, 61, e202208574 CrossRef CAS PubMed; (d) G. Ouyang, J. Rühe, Y. Zhang, M. J. Lin, M. Liu and F. Würthner, Angew. Chem., Int. Ed., 2022, 61, e202206706 CrossRef CAS PubMed.
- (a) J. S. Kang, S. Kang, J.-M. Suh, S. M. Park, D. K. Yoon, M. H. Lim, W. Y. Kim and M. Seo, J. Am. Chem. Soc., 2022, 144, 2657 CrossRef CAS PubMed; (b) T. Zhao, J. Yi, C. Liu, X. Liang, Y. Shen, L. Wei, X. Xie, W. Wu and C. Yang, Angew. Chem., Int. Ed., 2023, 62, e202302232 CrossRef CAS PubMed.
- (a) K.-D. Zhang, J. Tian, D. Hanifi, Y. Zhang, A. C.-H. Sue, T.-Y. Zhou, L. Zhang, X. Zhao, Y. Liu and Z.-T. Li, J. Am. Chem. Soc., 2013, 135, 17913–17918 CrossRef CAS PubMed; (b) Z.-J. Yin, S.-Y. Jiang, N. Liu, Q.-Y. Qi, Z.-Q. Wu, T.-G. Zhan and X. Zhao, CCS Chem., 2022, 4, 141–150 CrossRef CAS; (c) C. Xu, X. Lin, W. Wu and X. Ma, Chem. Commun., 2021, 57, 10178–10181 RSC.
- (a) Y. Li, Y. Dong, X. Miao, Y. Ren, B. Zhang, P. Wang, Y. Yu, B. Li, L. Isaacs and L. Cao, Angew. Chem., Int. Ed., 2018, 57, 729–733 CrossRef CAS PubMed; (b) H.-J. Wang, M.-M. Zheng, W.-W. Xing, Y.-X. Li, Y.-Y. Wang, H. Zhu, Y.-M. Zhang, Q. Yu and Y. Liu, Chem. Sci., 2023, 14, 8401–8407 RSC; (c) W. Xu, J. Y. Chao, B. Tang, Z. T. Li, J. F. Xu and X. Zhang, Chem.–Eur. J., 2022, 28, e202202200 CrossRef CAS PubMed.
- (a) F. García, R. Gómez and L. Sánchez, Chem. Soc. Rev., 2023, 52, 7524–7548 RSC; (b) A. R. A. Palmans and E. W. Meijer, Angew. Chem., Int. Ed., 2007, 46, 8948–8968 CrossRef CAS PubMed.
- (a) H. Qu, Y. Wang, Z. Li, X. Wang, H. Fang, Z. Tian and X. Cao, J. Am. Chem. Soc., 2017, 139, 18142–18145 CrossRef CAS PubMed; (b) H. Qu, X. Tang, X. Wang, Z. Li, Z. Huang, H. Zhang, Z. Tian and X. Cao, Chem. Sci., 2018, 9, 8814–8818 RSC; (c) J.-B. Xiong, H.-T. Feng, J.-P. Sun, W.-Z. Xie, D. Yang, M. Liu and Y.-S. Zheng, J. Am. Chem. Soc., 2016, 138, 11469–11472 CrossRef CAS PubMed.
- (a) Y. Li, Q. Li, X. Miao, C. Qin, D. Chu and L. Cao, Angew. Chem., Int. Ed., 2021, 60, 6744–6751 CrossRef CAS PubMed; (b) L. Cheng, P. Tian, H. Duan, Q. Li, X. Song, A. Li and L. Cao, Chem. Sci., 2023, 14, 833–842 RSC; (c) H. Duan, T. Yang, Q. Li, F. Cao, P. Wang and L. Cao, Chin. Chem. Lett., 2024, 35, 108878 CrossRef CAS.
- (a) J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS PubMed; (b) L. C. Smith, D. G. Leach, B. E. Blaylock, O. A. Ali and A. R. Urbach, J. Am. Chem. Soc., 2015, 137, 3663 CrossRef CAS PubMed; (c) M. Raeisi, K. Kotturi, I. del Valle, J. Schulz, P. Dornblut and E. Masson, J. Am. Chem. Soc., 2018, 140, 3371–3377 CrossRef CAS PubMed.
- (a) J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed; (b) Q. Peng and Z. Shuai, Aggregate, 2021, 2, e91 CrossRef CAS; (c) H.-T. Feng, Y.-X. Yuan, J.-B. Xiong, Y.-S. Zheng and B. Z. Tang, Chem. Soc. Rev., 2018, 47, 7452–7476 RSC; (d) H. Zhang, Z. Zhao, A. T. Turley, L. Wang, P. R. McGonigal, Y. Tu, Y. Li, Z. Wang, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Adv. Mater., 2020, 32, e2001457 CrossRef PubMed; (e) C. Qin, Y. Li, Q. Li, C. Yan and L. Cao, Chin. Chem. Lett., 2021, 32, 3531–3534 CrossRef CAS.
- (a) H. Zhu, Q. Li, Z. Gao, H. Wang, B. Shi, Y. Wu, L. Shangguan, X. Hong, F. Wang and F. Huang, Angew. Chem., Int. Ed., 2020, 59, 10868–10872 CrossRef CAS PubMed; (b) L. Cheng, K. Liu, Y. Duan, H. Duan, Y. Li, M. Gao and L. Cao, CCS Chem., 2021, 3, 2749–2763 CrossRef CAS; (c) L. Cheng, P. Tian, Q. Li, A. Li and L. Cao, CCS Chem., 2022, 4, 2914–2920 CrossRef CAS.
- (a) H. Zhang, L. Cheng, H. Nian, J. Du, T. Chen and L. Cao, Chem. Commun., 2021, 57, 3135–3138 RSC; (b) Y. Li, C. Yan, Q. Li and L. Cao, Sci. China: Chem., 2022, 65, 1279–1285 CrossRef CAS; (c) Y. Li, C. Qin, Q. Li, P. Wang, X. Miao, H. Jin, W. Ao and L. Cao, Adv. Opt. Mater., 2020, 8, 1902154 CrossRef CAS.
- (a) C. Yan, Q. Li, X. Miao, Y. Zhao, Y. Li, P. Wang, K. Wang, H. Duan, L. Zhang and L. Cao, Angew. Chem., Int. Ed., 2023, 62, e202308029 CrossRef CAS PubMed; (b) D. E. Clarke, G. Wu, C. Wu and O. A. Scherman, J. Am. Chem. Soc., 2021, 143, 6323–6327 CrossRef CAS PubMed.
- (a) Y.-F. Wang, H. Yao, L.-P. Yang, M. Quan and W. Jiang, Angew. Chem., Int. Ed., 2022, 61, e202211853 CrossRef CAS PubMed; (b) X. Shang, I. Song, H. Ohtsu, Y. H. Lee, T. Zhao, T. Kojima, J. H. Jung, M. Kawano and J. H. Oh, Adv. Mater., 2017, 29, 1605828 CrossRef PubMed.
- (a) H. Bai, Z. Liu, T. Zhang, J. Du, C. Zhou, W. He, J. H. C. Chau, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, ACS Nano, 2020, 14, 7552–7563 CrossRef CAS PubMed; (b) Y. Jin, N. Du, Y. Huang, W. Shen, Y. Tan, Y. Z. Chen, W.-T. Dou, X.-P. He, Z. Yang, N. Xu and C. Tan, ACS Sens., 2022, 7, 1524–1532 CrossRef CAS PubMed; (c) H. Duan, F. Cao, M. Zhang, M. Gao and L. Cao, Chin. Chem. Lett., 2022, 33, 2459–2463 CrossRef CAS.
- (a) J. Chen, B. L. Hickey, L. Wang, J. Lee, A. D. Dill, A. Favero, R. Pinalli, E. Dalcanale, R. J. Hooley and W. Zhong, Nat. Chem., 2021, 13, 488–495 CrossRef CAS PubMed; (b) J. Chen, A. D. Gill, B. L. Hickey, Z. Gao, X. Cui, R. J. Hooley and W. Zhong, J. Am. Chem. Soc., 2021, 143, 12791–12799 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details including NMR, ITC, DLS, UV/vis, fluorescence, SEM, TEM, SAXS, and CD data. See DOI: https://doi.org/10.1039/d3sc06349f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |