Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Reversible dioxygen uptake at [Cu4] clusters†
Manasseh Kusi
Osei
a,
Saber
Mirzaei
 ab,
M. Saeed
Mirzaei
a,
Agustin
Valles
ab,
M. Saeed
Mirzaei
a,
Agustin
Valles
 a and
Raúl
Hernández Sánchez
a and
Raúl
Hernández Sánchez
 *ab
*ab
aDepartment of Chemistry, Rice University, 6100 Main St., Houston, Texas, USA. E-mail: raulhs@rice.edu
bDepartment of Chemistry, University of Pittsburgh, 219 Parkman Ave., Pittsburgh, Pennsylvania 15260, USA
First published on 7th March 2024
Abstract
Dioxygen binding solely through non-covalent interactions is rare. In living systems, dioxygen transport takes place via iron or copper-containing biological cofactors. Specifically, a reversible covalent interaction is established when O2 binds to the mono or polynuclear metal center. However, O2 stabilization in the absence of covalent bond formation is challenging and rarely observed. Here, we demonstrate a unique example of reversible non-covalent binding of dioxygen within the cavity of a well-defined synthetic all-Cu(I) tetracopper cluster.
Introduction
Reversible O2 binding is the cornerstone of cellular respiration.1 Hemoglobin, myoglobin, hemerythrin, and hemocyanin all serve as O2 transporters across all living organisms.2 In the process of biological uptake, transport, and delivery of dioxygen, a covalent bond is established between O2 and Fe,3 or similarly between O2 and the Cu2 site in hemocyanin,4 resulting in charge transfer to form a superoxo or peroxo moiety, respectively. Although, non-covalent interactions have been described to be operative in stabilizing ferric–superoxo intermediates in heme proteins,5 these nominally weak contacts are generally challenging to study.Aside from biological cofactors, numerous studies have reported supramolecular complexes, porous materials, and organic cages capable of binding dioxygen. For instance, metal complexes formed within macrocyclic species, e.g., palladium-bound cyclodextrin,6 and Mn-supported calixarene,7 lead to peroxo and superoxo moieties, respectively, stabilized inside the macrocycle. Similar occurrences are observed within metal organic frameworks (MOFs), where O2 binding to embedded metal sites is used for O2/N2 separations,8 or bond activation cleaving the O–O bond.9,10 Last, macrocycles alone are also known to stabilize peroxo species.11 However, to our knowledge, non-covalent interactions alone have not been described to stabilize neutral O2 in biological metal cofactors or synthetic metal clusters. Here, we describe a polynuclear copper cluster built within a flexible supramolecular scaffold capable of creating a unique pocket binding O2 solely through non-covalent interactions.
Our group has developed modular amine-based ligands serving as templates for metal cluster formation. Recently, we reported a rigid ligand scaffold that enables the formation of square planar tetranuclear [Cu4] clusters (Fig. 1).12 Keeping some of the design principles employed before, we decided to increase the degrees of freedom of the ligand scaffold as shown in Fig. 1. Others in the field have adopted similar measures either allowing or restricting the templating ligand's rigidity to alter the cluster reactivity and composition. For example, Betley and coworkers demonstrated that increasing the template's rigidity exchanging the tame ligand backbone (tame = 1,1,1-tris(aminomethyl)ethane),13–16 for the tris-amine α-α-α-1,3,5-tris-aminocyclohexane,17–19 opens the door for substrate activation pathways at trinuclear species not available in the former tame-based clusters. Similarly, C3-symmetric 1,3,5-benzene substituted ligands developed by Holm to mimic iron sulfur clusters,20,21 inspired the synthesis of oxygen-donor congeners in work reported by Agapie and coworkers towards the creation of Mn3Ca subsite mimics of the oxygen evolving complex.22–24 Most recently, Suess et al. using a similar ligand base fragment developed a nitrogen-donor analogue capable of isolating an iron sulfur alkyl cluster which mimics elusive enzymatic intermediates.25,26 Other systems benefiting from ligand rigidification include those from the Murray group, whereupon limiting the degrees of freedom of their initial cryptand design,27 they uncovered [M3] clusters, M = Fe, Co, Cu, and Zn, capable of ligating and activating N2 and CO2.28–34 In our case, by increasing the degrees of freedom of our ligand scaffold we have created a binding pocket within an all-Cu(I) [Cu4] cluster that binds dioxygen through non-covalent interactions.
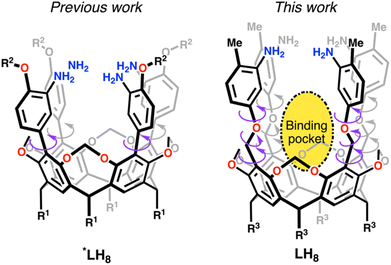 | ||
| Fig. 1 Previous ligand architecture *LH8 (R1 = n-pentyl; R2 = Me, Ph, or i-Bu) compared to LH8 (R3 = n-heptyl) reported herein with increased degrees of freedom. | ||
Results and discussion
Synthetic procedures
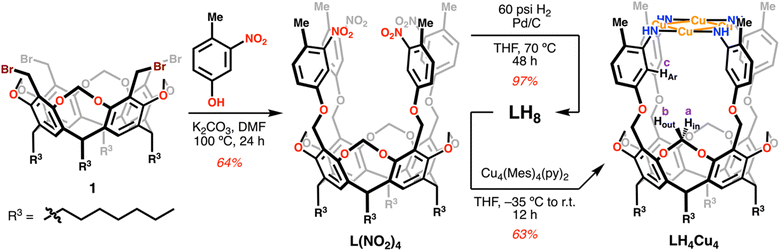
Our synthetic protocol provides square planar copper clusters in three steps from 1, a precursor obtained readily in gram-scale quantities,35 as shown in Scheme 1. First, a four-fold SN2 reaction on 1 by 4-methyl-3-nitrophenol in basic conditions using K2CO3 in DMF for 24 hours produces ligand precursor L(NO2)4 in 64% isolated yield. This tetranitro species is reduced under 60 psi of H2 over Pd/C in refluxing THF for 48 h. The reaction is quantitative by 1H NMR; however, after work up the isolated yield of LH8 is 97%. In situ deprotonation and metalation of this tetraamine ligand with Cu4(Mes)4(py)2 in THF forms the all-Cu(I) diamagnetic species LH4Cu4 in 63% yield. 1H NMR analysis of LH4Cu4 reveals its ideal C4v symmetry in solution (Fig. S11†).Cluster topology
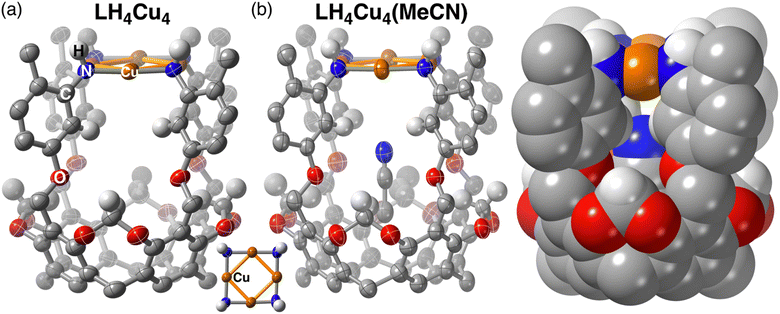
Single crystal X-ray diffraction data was collected on crystals grown by vapor diffusion of pentane into a concentrated solution of LH4Cu4 in THF. The molecular structure of LH4Cu4 confirmed the formation of a square planar arrangement of Cu atoms sitting at an average distance davg (Cu–Cu) of 2.69(2) Å (Fig. 2a). For comparison, in Cu metal the Cu–Cu distance in Cu(100) is 2.55(1) Å.36 Additionally, the [Cu4] core displays a davg (Cu–N) of 1.89(1) Å and an almost linear local Cu coordination environment with an average ∠N–Cu–N of 177.3(7) degrees. Similar square-shaped tetranuclear copper clusters mimicking CuZ in N2O reductase37 have been previously reported by Mankad and coworkers using bridging diphosphine38,39 or formamidinate40–42 ligands resulting in cluster cores with Cu–Cu distances ranging from around 2.4 to 3.5 Å across all compounds reported therein. In fact, a recent example by the same group demonstrates the formation of a [Cu4(O2)] adduct, where O2 is bound to a single metal site.43LH4Cu4 is closely related to our previously reported [Cu4] clusters;12,44 however, we hypothesized that a larger internal cavity should be created in this newly synthesized cluster located in between the [Cu4N4] fragment and the resorcinarene backbone as a consequence of the axially longer LH8 relative to *LH8. Our hypothesis was confirmed upon analyzing the molecular crystal structure obtained when LH4Cu4 is exposed to MeCN (Fig. 2b). The MeCN molecule is hosted below the [Cu4] plane establishing a [Cu4] centroid-to-NMeCN distance of 3.852 Å. Note that the structure metrics of LH4Cu4(MeCN) are relatively unchanged (davg (Cu–Cu) = 2.678(5) Å, davg (Cu–N) = 1.890(3) Å, ∠N–Cu–N = 177.4(6) degrees) from LH4Cu4.
Host–guest properties
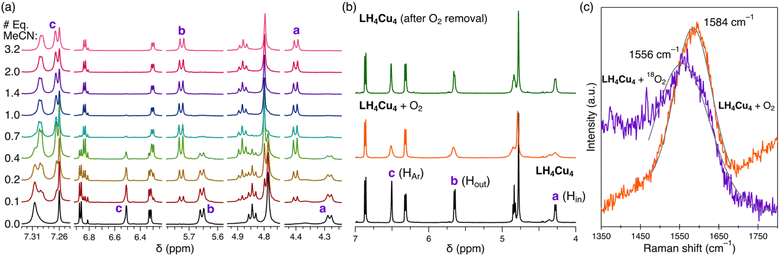
Titration of MeCN to LH4Cu4 provided insight into the hydrogen atom resonances involved in the non-covalent bonding of MeCN in the host–guest adduct LH4Cu4(MeCN). As observed in Fig. 3a, the addition of MeCN in CDCl3 leads to resonance shifts in the 1H NMR. Particularly noteworthy is that protons a, b, and c, as labeled in Scheme 1, shift downfield as equivalents of MeCN are added (Fig. S15† contains the full spectrum), a well-known effect for H atoms involved in hydrogen bonding.45,46 The 1H NMR spectrum additionally reveals the location of the MeCN signal at −1.76 ppm. Similar upfield shifts or shielding of the methyl resonance in acetonitrile has been observed in calixarene-based copper clusters.47 Moreover, the stepwise addition of MeCN to LH4Cu4 produces a new set of resonances corresponding to LH4Cu4(MeCN) while the location of those for LH4Cu4 remain unchanged, indicating a large host–guest association constant (Ka > 105 M−1 in CDCl3) that goes beyond the measurable range via NMR.48 Variable-temperature 1H NMR in C6D6 provided insight into the thermodynamics of MeCN dissociation from LH4Cu4(MeCN) (Fig. S16†). A van't Hoff analysis reveals a dissociation enthalpy and entropy of 12.7 kcal mol−1 and 25.4 cal mol−1 K−1 (Fig. S17†), respectively. The calculated binding free energy of −5.14 kcal mol−1 at 298 K is similar to solvent binding in cavitands.49 Most importantly, structural fidelity of LH4Cu4(MeCN) in combination with the 1H NMR spectroscopic signatures resulting from MeCN binding provides a roadmap to investigate binding of other guests to LH4Cu4.Copper–dioxygen chemistry is cornerstone in living systems and has inspired the realization of many synthetic compounds seeking to replicate its structure and function.50,51 Seeking to probe potential ligation and activation modes of O2 at LH4Cu4, an all-Cu(I) cluster, we dosed dry O2 to an air-free solution of LH4Cu4 in CDCl3. As the atmosphere is exchanged from N2 (Fig. 3b bottom) to O2 (Fig. 3b middle), we observe broadening in proton resonances a, b, c, and the methine at 4.85 ppm. Note that the same proton resonances a, b, and c, are affected during MeCN binding. These proton resonances sharpen back when the O2 atmosphere is removed (Fig. 3b top). Altogether, our experiments indicate that O2 is reversibly accommodated in the same binding pocket as MeCN. Note that L(NO2)4 and LH8 do not show any broadening of resonances a, b, and c under the same conditions (Fig. S19 and S20†). It is important to highlight that the vast majority of synthetic molecular [CuIn] systems, with nuclearities ranging from n = 1 to 4, engage O2 by establishing a copper–oxygen covalent bond giving rise to terminal or bridging peroxo, superoxo, or oxo complexes.52 To the best of our knowledge, this is the first formally all-Cu(I) cluster reversibly binding O2 solely via non-covalent interactions.
Further examination of O2 binding to LH4Cu4 was carried out by collecting resonance Raman vibrational spectra on dried thin films of LH4Cu4 contained within a quartz cuvette either under vacuum or dry dioxygen atmosphere. Laser excitation at 532 nm gave a band centered at 1584 cm−1 assigned to the O–O stretching of dioxygen hosted within LH4Cu4 (Fig. 3c). Note that the symmetrical stretching band in free O2 appears at 1556.4 cm−1 (Fig. S22†).53 The same set of experiments were executed using isotopically labeled 18O2 displaying a stretching band at 1556 cm−1. The simple harmonic oscillator model predicts a shift of ∼90 cm−1, however we only observe a Δ18O of 28 cm−1.52 We hypothesize that dioxygen enters the cavity of LH4Cu4 likely through partial ligand dissociation or via the formation of a dilated aperture.54,55 Altogether, it appears that the non-covalent interactions serve to stabilize the O2 molecule within LH4Cu4 and at the same time strengthens the O–O bond by removing π* electron density.
To further investigate the electronic properties of LH4Cu4 and its adduct with O2, we employed density functional theory (DFT) methods. 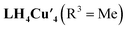 was optimized at B3LYP-D3BJ/Def2-SVP+PCM(CHCl3) level of theory. The optimized structure reproduced the experimental data with high accuracy as determined by overlaying these structures and obtaining a root mean square displacement (RMSD) of 0.3 Å and davg (Cu–Cu) of 2.67 Å (Fig. S23†), indicating the reliability of the selected method.
was optimized at B3LYP-D3BJ/Def2-SVP+PCM(CHCl3) level of theory. The optimized structure reproduced the experimental data with high accuracy as determined by overlaying these structures and obtaining a root mean square displacement (RMSD) of 0.3 Å and davg (Cu–Cu) of 2.67 Å (Fig. S23†), indicating the reliability of the selected method.
Intrigued by the non-covalent binding of O2 to LH4Cu4, the adduct 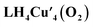 was first optimized employing B3LYP-D3BJ/Def2-SVP+PCM(CHCl3) level of theory. The binding pocket within
was first optimized employing B3LYP-D3BJ/Def2-SVP+PCM(CHCl3) level of theory. The binding pocket within 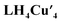 is best visualized by applying the independent gradient model based on Hirshfeld partition of molecular density (IGMH),56 which clearly reveals the contact surface of weak non-covalent interactions involved in hosting the O2 molecule. Note that the oxygen molecule positions itself in a way to maximize interactions with the C–H bonds from the bridging methylenes and top aromatic rings, protons a and c, respectively, in Scheme 1. The O2 molecule was placed at different starting positions during structure optimization, e.g., close to the resorcin[4]arene base, near the Cu4 plane, and in all cases the optimum location found is that shown in Fig. 4. Overall, the IGMH isosurface for the O2 adduct is well supported experimentally as reflected in the 1H NMR data shown in Fig. 3b.
is best visualized by applying the independent gradient model based on Hirshfeld partition of molecular density (IGMH),56 which clearly reveals the contact surface of weak non-covalent interactions involved in hosting the O2 molecule. Note that the oxygen molecule positions itself in a way to maximize interactions with the C–H bonds from the bridging methylenes and top aromatic rings, protons a and c, respectively, in Scheme 1. The O2 molecule was placed at different starting positions during structure optimization, e.g., close to the resorcin[4]arene base, near the Cu4 plane, and in all cases the optimum location found is that shown in Fig. 4. Overall, the IGMH isosurface for the O2 adduct is well supported experimentally as reflected in the 1H NMR data shown in Fig. 3b.
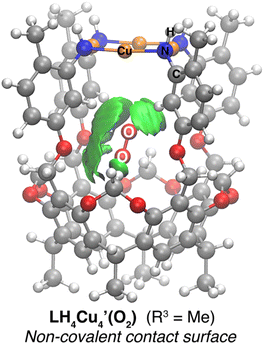 | ||
| Fig. 4 IGMH map displaying the non-covalent interactions between O2 and LH4Cu4 (isovalue = 0.0011 a.u.). | ||
Conclusions
Here, we designed a novel ligand architecture carrying four aniline moieties to obtain a rigidified square-planar [Cu4] topology despite the ligand's rotational degrees of freedom. The foregoing results show that, changing the resorcinarene backbone employed previously by our group engenders a cluster compound with a cavity to encapsulate small molecules, where we showcase a unique example of reversible non-covalent binding of O2 within the [Cu4] cluster built around a supramolecular scaffold.Data availability
All data including experimental and analytical details are in the ESI.†Author contributions
Manasseh K. Osei: conceptualization, investigation, data curation, writing – review & edition. Saber Mirzaei: conceptualization, investigation, data curation, formal analysis, writing – review & edition. M. Saeed Mirzaei: investigation, data curation, formal analysis. Agustin Valles: investigation. Raúl Hernández Sánchez: conceptualization, project administration, resources, funding acquisition, supervision, visualization, writing – original draft, writing – review & edition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by Rice University and the Robert A. Welch Foundation Young Investigator Award. Acknowledgment is made to the donors of the American Chemical Society Petroleum Research Fund for partial support of this research. The authors acknowledge the use of Raman microscope and NMR spectrometers through the Shared Equipment Authority at Rice University. Also, we thank startup funding and the Center for Research Computing from the University of Pittsburgh for their support. We thank Ying Chen for helpful discussions. We thank Dr Xu Wang for providing help with VT 1H NMR experiments.Notes and references
- G. T. Babcock, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 12971–12973 CrossRef CAS PubMed.
- J. S. Olson, Antioxid. Redox Signaling, 2019, 32, 228–246 CrossRef PubMed.
- A. Decker and E. I. Solomon, Curr. Opin. Chem. Biol., 2005, 9, 152–163 CrossRef CAS PubMed.
- D. A. Quist, D. E. Diaz, J. J. Liu and K. D. Karlin, JBIC, J. Biol. Inorg. Chem., 2017, 22, 253–288 CrossRef CAS PubMed.
- X. Huang and J. T. Groves, Chem. Rev., 2018, 118, 2491–2553 CrossRef CAS PubMed.
- R. Gramage-Doria, D. Armspach, D. Matt and L. Toupet, Chem.–Eur. J., 2012, 18, 10813–10816 CrossRef CAS PubMed.
- L.-L. Liu, H.-X. Li, L.-M. Wan, Z.-G. Ren, H.-F. Wang and J.-P. Lang, Chem. Commun., 2011, 47, 11146–11148 RSC.
- E. D. Bloch, L. J. Murray, W. L. Queen, S. Chavan, S. N. Maximoff, J. P. Bigi, R. Krishna, V. K. Peterson, F. Grandjean, G. J. Long, B. Smit, S. Bordiga, C. M. Brown and J. R. Long, J. Am. Chem. Soc., 2011, 133, 14814–14822 CrossRef CAS PubMed.
- K. Hou, J. Börgel, H. Z. H. Jiang, D. J. SantaLucia, H. Kwon, H. Zhuang, K. Chakarawet, R. C. Rohde, J. W. Taylor, C. Dun, M. V. Paley, A. B. Turkiewicz, J. G. Park, H. Mao, Z. Zhu, E. E. Alp, J. Zhao, M. Y. Hu, B. Lavina, S. Peredkov, X. Lv, J. Oktawiec, K. R. Meihaus, D. A. Pantazis, M. Vandone, V. Colombo, E. Bill, J. J. Urban, R. D. Britt, F. Grandjean, G. J. Long, S. DeBeer, F. Neese, J. A. Reimer and J. R. Long, Science, 2023, 382, 547–553 CrossRef CAS PubMed.
- X. He, A. Iliescu, T. Yang, M. Q. Arguilla, T. Chen, H. J. Kulik and M. Dincă, J. Am. Chem. Soc., 2023, 145, 16872–16878 CrossRef CAS PubMed.
- N. Lopez, D. J. Graham, R. McGuire, G. E. Alliger, Y. Shao-Horn, C. C. Cummins and D. G. Nocera, Science, 2012, 335, 450 CrossRef CAS PubMed.
- M. K. Osei, S. Mirzaei, X. Bogetti, E. Castro, M. A. Rahman, S. Saxena and R. Hernández Sánchez, Angew. Chem., Int. Ed., 2022, 61, e202209529 CrossRef CAS PubMed.
- Q. L. Zhao and T. A. Betley, Angew. Chem., Int. Ed., 2011, 50, 709–712 CrossRef CAS PubMed.
- A. R. Fout, Q. L. Zhao, D. N. J. Xiao and T. A. Betley, J. Am. Chem. Soc., 2011, 133, 16750–16753 CrossRef CAS PubMed.
- T. D. Harris and T. A. Betley, J. Am. Chem. Soc., 2011, 133, 13852–13855 CrossRef CAS PubMed.
- E. V. Eames and T. A. Betley, Inorg. Chem., 2012, 51, 10274–10278 CrossRef CAS PubMed.
- T. M. Powers, A. R. Fout, S. L. Zheng and T. A. Betley, J. Am. Chem. Soc., 2011, 133, 3336–3338 CrossRef CAS PubMed.
- A. K. Bartholomew, C. E. Juda, J. N. Nessralla, B. Lin, S. G. Wang, Y.-S. Chen and T. A. Betley, Angew. Chem., Int. Ed., 2019, 58, 5687–5691 CrossRef CAS PubMed.
- T. M. Powers and T. A. Betley, J. Am. Chem. Soc., 2013, 135, 12289–12296 CrossRef CAS PubMed.
- T. D. P. Stack and R. H. Holm, J. Am. Chem. Soc., 1987, 109, 2546–2547 CrossRef CAS.
- T. D. P. Stack, J. A. Weigel and R. H. Holm, Inorg. Chem., 1990, 29, 3745–3760 CrossRef CAS.
- E. Y. Tsui, M. W. Day and T. Agapie, Angew. Chem., Int. Ed., 2011, 50, 1668–1672 CrossRef CAS PubMed.
- E. Y. Tsui, J. S. Kanady, M. W. Day and T. Agapie, Chem. Commun., 2011, 47, 4189–4191 RSC.
- J. S. Kanady, E. Y. Tsui, M. W. Day and T. Agapie, Science, 2011, 333, 733–736 CrossRef CAS PubMed.
- A. McSkimming and D. L. M. Suess, Inorg. Chem., 2018, 57, 14904–14912 CrossRef CAS PubMed.
- M. Ye, N. B. Thompson, A. C. Brown and D. L. M. Suess, J. Am. Chem. Soc., 2019, 141, 13330–13335 CrossRef CAS PubMed.
- G. L. Guillet, F. T. Sloane, M. F. Dumont, K. A. Abboud and L. J. Murray, Dalton Trans., 2012, 41, 7866–7869 RSC.
- L. J. Murray, W. W. Weare, J. Shearer, A. D. Mitchell and K. A. Abboud, J. Am. Chem. Soc., 2014, 136, 13502–13505 CrossRef CAS PubMed.
- Y. Lee, F. T. Sloane, G. Blondin, K. A. Abboud, R. Garcia-Serres and L. J. Murray, Angew. Chem., Int. Ed., 2015, 54, 1499–1503 CrossRef CAS PubMed.
- D. M. Ermert, I. Ghiviriga, V. J. Catalano, J. Shearer and L. J. Murray, Angew. Chem., Int. Ed., 2015, 54, 7047–7050 CrossRef CAS PubMed.
- Y. Lee, K. J. Anderton, F. T. Sloane, D. M. Ermert, K. A. Abboud, R. García-Serres and L. J. Murray, J. Am. Chem. Soc., 2015, 137, 10610–10617 CrossRef CAS PubMed.
- B. J. Cook, G. N. Di Francesco, K. A. Abboud and L. J. Murray, J. Am. Chem. Soc., 2018, 140, 5696–5700 CrossRef CAS PubMed.
- R. B. Ferreira, B. J. Cook, B. J. Knight, V. J. Catalano, R. García-Serres and L. J. Murray, ACS Catal., 2018, 8, 7208–7212 CrossRef CAS PubMed.
- M. C. Eaton, V. J. Catalano, J. Shearer and L. J. Murray, J. Am. Chem. Soc., 2021, 143, 5649–5653 CrossRef CAS PubMed.
- R. Wu, T. F. Al-Azemi and K. S. Bisht, RSC Adv., 2014, 4, 16864–16870 RSC.
- I.-K. Suh, H. Ohta and Y. Waseda, J. Mater. Sci., 1988, 23, 757–760 CrossRef CAS.
- T. Rasmussen, B. C. Berks, J. Sanders-Loehr, D. M. Dooley, W. G. Zumft and A. J. Thomson, Biochemistry, 2000, 39, 12753–12756 CrossRef CAS PubMed.
- B. J. Johnson, S. V. Lindeman and N. P. Mankad, Inorg. Chem., 2014, 53, 10611–10619 CrossRef CAS PubMed.
- C.-W. Hsu, S. C. Rathnayaka, S. M. Islam, S. N. MacMillan and N. P. Mankad, Angew. Chem., Int. Ed., 2020, 59, 627–631 CrossRef CAS PubMed.
- B. J. Johnson, W. E. Antholine, S. V. Lindeman and N. P. Mankad, Chem. Commun., 2015, 51, 11860–11863 RSC.
- B. J. Johnson, W. E. Antholine, S. V. Lindeman, M. J. Graham and N. P. Mankad, J. Am. Chem. Soc., 2016, 138, 13107–13110 CrossRef CAS PubMed.
- S. C. Rathnayaka, C. W. Hsu, B. J. Johnson, S. J. Iniguez and N. P. Mankad, Inorg. Chem., 2020, 59, 6496–6507 CrossRef CAS PubMed.
- N. P. Mankad, Chem. Sci., 2024, 15, 1820–1828 RSC.
- LH4Cu4 also displays an irreversible oxidation near 0 V vs. Fc/Fc+ (Fig. S13†).
- K. Choi and A. D. Hamilton, J. Am. Chem. Soc., 2003, 125, 10241–10249 CrossRef CAS PubMed.
- S. Mirzaei, V. M. Espinoza Castro and R. Hernández Sánchez, Chem. Sci., 2022, 13, 2026–2032 RSC.
- N. Frank, A. Dallmann, B. Braun-Cula, C. Herwig and C. Limberg, Angew. Chem., Int. Ed., 2020, 59, 6735–6739 CrossRef CAS PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- O. Dumele, N. Trapp and F. Diederich, Angew. Chem., Int. Ed., 2015, 54, 12339–12344 CrossRef CAS PubMed.
- C. Wurtele, E. Gaoutchenova, K. Harms, M. C. Holthausen, J. Sundermeyer and S. Schindler, Angew. Chem., Int. Ed., 2006, 45, 3867–3869 CrossRef PubMed.
- E. I. Solomon, D. E. Heppner, E. M. Johnston, J. W. Ginsbach, J. Cirera, M. Qayyum, M. T. Kieber-Emmons, C. H. Kjaergaard, R. G. Hadt and L. Tian, Chem. Rev., 2014, 114, 3659–3853 CrossRef CAS PubMed.
- C. E. Elwell, N. L. Gagnon, B. D. Neisen, D. Dhar, A. D. Spaeth, G. M. Yee and W. B. Tolman, Chem. Rev., 2017, 117, 2059–2107 CrossRef CAS PubMed.
- H. G. M. Edwards, D. A. Long, K. A. B. Najm and M. Thomsen, J. Raman Spectrosc., 1981, 10, 60–63 CrossRef CAS.
- A. V. Davis and K. N. Raymond, J. Am. Chem. Soc., 2005, 127, 7912–7919 CrossRef CAS PubMed.
- S. Rieth, Z. Yan, S. Xia, M. Gardlik, A. Chow, G. Fraenkel, C. M. Hadad and J. D. Badjić, J. Org. Chem., 2008, 73, 5100–5109 CrossRef CAS PubMed.
- T. Lu and Q. Chen, J. Comput. Chem., 2022, 43, 539–555 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, characterization, and spectroscopic data. CCDC 2332560 and 2333831. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3sc06390a |
| This journal is © The Royal Society of Chemistry 2024 |