Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemoenzymatic multistep retrosynthesis with transformer loops†
David
Kreutter
 and
Jean-Louis
Reymond
and
Jean-Louis
Reymond
 *
*
Department of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Freiestrasse 3, 3012 Bern, Switzerland. E-mail: david.kreutter@unibe.ch; jean-louis.reymond@unibe.ch
First published on 8th October 2024
Abstract
Integrating enzymatic reactions into computer-aided synthesis planning (CASP) should help devise more selective, economical, and greener synthetic routes. Herein we report the triple-transformer loop algorithm with biocatalysis (TTLAB) as a new CASP tool for chemo-enzymatic multistep retrosynthesis. Single-step retrosyntheses are performed using two triple transformer loops (TTL), one trained with chemical reactions from the US Patent Office (USPTO-TTL), the second one obtained by multitask transfer learning combining the USPTO dataset with preparative biotransformations from the literature (ENZR-TTL). Each TTL performs single-step retrosynthesis independently by tagging potential reactive sites in the product, predicting for each site possible starting materials (T1) and reagents or enzymes (T2), and validating the predictions via a forward transformer (T3). TTLAB combines predictions from both TTLs to explore multistep sequences using a heuristic best-first tree search and propose short routes from commercial building blocks including enantioselective biocatalytic steps. TTLAB can be used to assist chemoenzymatic route design.
Introduction
Computer-aided synthesis planning (CASP), originally proposed by E. J. Corey in the 1960's, uses computational approaches, including rule-based systems as well as various types of neural networks, to exploit synthetic methodology as recorded in the scientific literature to propose multistep syntheses of target molecules from commercial precursors.1–27 Integrating enzyme-catalyzed reactions would enable CASP to participate in the global effort towards more selective, economical, and greener chemical manufacturing processes. However, the task is challenging due to the sparsity and very different nature of biotransformations compared to chemical reactions.28–33 Both template-based and transformer-based CASP tools for biocatalysis were recently reported,34–36 which make use of biochemical reaction data describing mostly metabolic pathways as collected in databases such as BRENDA, KEGG, MetaCyc, Rhea, PathBank, MetaNetX or EzCatDB.37–43 However, these biochemical pathway datasets only partly reflect the use of enzymes in organic synthesis, where enzymes or enzyme preparations (extracts, whole cells, etc.) are used under non-natural conditions, such as in immobilized form and at very high substrate concentrations, and to convert molecules often quite different from the natural substrate.31 The CASP tool ASKCOS,44,45 on the other hand, proposes chemo-enzymatic route finding using a template-based strategy based on literature data collected from the Reaxys database46 for both chemistry and biocatalysis, which for the case of enzymes represent more relevant examples for the practice of organic synthesis compared to data from biochemical pathways.We recently showed that CASP tools based on transformer models,17,18 trained on SMILES descriptions47,48 of chemical reactions of starting materials (SM) with a set of reagents (R) to form a product (P) as collected in the public USPTO dataset,49,50 can be adapted to specific reaction subclasses by transfer learning.51 Extending on this opportunity, we then showed that literature information on a few ten thousand biotransformations extracted from Reaxys,46 for which the reagent set R is substituted with a text description of the enzyme or enzyme preparation, can be combined with the USPTO dataset to train a transformer model by multi-task transfer learning (MTL).52 The resulting enzymatic transformer performed forward predictions of enzymatic reactions as used in typical preparative biotransformations, including enantioselective processes such as kinetic resolution with lipases or enantioselective ketone reduction and reductive aminations with 71% top-2 accuracy, approaching the typical performance of forward transformer models. The key difference between our enzymatic transformer model and the other approaches for enzymatic reactions mentioned above was the use of a text description of the enzyme rather than its E.C classification or a link to literature references.
Herein we report the integration of our enzymatic transformer model into our recently reported triple transformer loop algorithm (TTLA) for multistep chemical retrosynthesis,53 to obtain a triple transformer loop algorithm with biocatalysis (TTLAB, Fig. 1). Our previously reported triple transformer loop algorithm (TTLA) performed single-step retrosynthesis prediction using a triple transformer loop (TTL) operating on products P with tagged reactive sites to explore diverse bond disconnections. In detail, potential reactive sites in P were first tagged to produce a series of P*,54 and for each P* a first transformer T1 was used to predict SM, a second transformer T2 to predict a suitable R for the proposed transformation SM → P, and finally a third transformer T3 to predict P from the predicted SM and R, thereby potentially validating the retrosynthetic step. The TTLAB presented here combines our original triple transformer loop trained on USPTO,53 here called USPTO-TTL, with ENZR-TTL, which is a new TTL trained on an updated version of our previously reported ENZR dataset containing biotransformations from the literature and originally used only for forward predictions,52 which now comprises 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 reactions.
176 reactions.
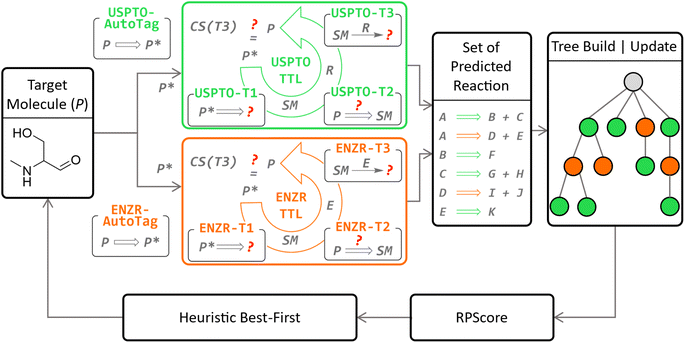 | ||
| Fig. 1 Concept of the TTLAB multistep search operating organic (USPTO-TTL, green panel)53 and enzymatic (ENZR-TTL, orange panel) catalysis in parallel. In the new enzymatic retrosynthesis, potential reactive sites in a product molecule P are first labelled by the new model ENZR-Autotag, and each labelled product P* is then passed through ENZR-TTL consisting of the new models ENZR-T1 predicting starting materials (SM) from P*, ENZR-T2 predicting the enzyme name E from SM → P, and the previously reported forward model ENZR-T3 predicting P from SM + E. The retrosynthetic step is validated if the correct product P is predicted by ENZR-T3 with a confidence score CS(T3) above 95%. The confidence scores CS(T3) are used to compute the RPScore53 to prioritize steps in the retrosynthetic tree via a heuristic best-first sorting. | ||
To predict enzymatic disconnections, ENZR-TTL tags potential reactive sites in P to produce various tagged P* by using a new tagging model for enzymatic disconnections trained on ENZR, called ENZR-AutoTag. ENZR-TTL then predicts possible SM from each possible tagged P* using a new ENZR-T1 model, and possible enzymes (E) for the predicted reaction SM → P by a new ENZR-T2 model. Both transformer models are obtained by MTL of the USPTO dataset with the ENZR dataset. Finally, ENZR-TTL validates the predicted SM + E → P reaction with the previously reported forward transformer ENZR-T3 (retrained on a more recent and larger ENZR dataset) based on the identity of the predicted and original P and the confidence score.52
To explore multistep chemo-enzymatic retrosyntheses, TTLAB considers single-step predictions from both USPTO-TTL and ENZR-TTL using the approach developed previously for TTLA. In this approach, possible routes are ranked with the route penalty score (RPScore),53 combining the simplicity of all SM along the route,55,56 with the confidence score of each retrosynthetic step, as well as route length, and the various routes are ranked and iteratively extended using a heuristic best-first tree search. TTLAB can be used to assist chemoenzymatic route design.
Methods
Chemical reaction dataset
The same United States Patent and Trademark Office (USPTO) chemical reaction dataset as in our previous report was used.53 It is a version curated by Thakkar et al.54 derived from the data mining work of Lowe.49,50Triple transformer loop models for chemical reactions (USPTO-TTL)
The models trained on the USPTO dataset are identical as in our previous study and available on Zenodo,53 and herein named USPTO-TTL. AutoTag is a tagging model predicting tagged product P* from the target product P. T1 is a disconnection-aware retrosynthesis model predicting starting materials SM from the target tagged product P*. T2 is a reaction condition model predicting reagents R, including catalyst and solvent, from the reaction SM → P. T3 is a forward validation model predicting P from SM + R.57Enzymatic dataset
The enzymatic reaction dataset, herein named ENZR, was extracted from Reaxys using the API accessible under a commercial license.46 We first isolated reactions labelled as “enzymatic reaction” in the “other conditions” field (“RXD.COND”). Next, we compiled a list of reagents, catalysts, and solvents typically associated with enzymatic reactions. This involved identifying components with the “ase” suffix in the text fields “RXD.RGT,” “RXD.CAT,” and “RXD.SOL,”. Additionally, we manually selected keywords corresponding to enzymatic transformations, such as “P450,” “NADP,” “CAL-B,” “flavin mononucleotide,” and others, from the most frequently occurring reagents and catalysts in the initial data retrieval. Finally, we queried these enzymatic components individually in the Reaxys database and retrieved the associated reactions. This process resulted in a raw dataset consisting of 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 enzymatic reactions.
865 enzymatic reactions.
Enzymatic dataset: cleaning
The process of cleaning the ENZR dataset involved several steps, wherein the RDKit library was used across various stages.58 Initially, multistep reactions and those lacking any reactant or product were excluded, leaving 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 389 reactions. Next, reactions were mapped using RxnMapper,59 for which 1333 reactions failed and were removed. Reactions with unspecified atomic symbols (“*”) were also removed. Unmapped reactant molecules were removed for each reaction. A significant number of reactions (32
389 reactions. Next, reactions were mapped using RxnMapper,59 for which 1333 reactions failed and were removed. Reactions with unspecified atomic symbols (“*”) were also removed. Unmapped reactant molecules were removed for each reaction. A significant number of reactions (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 527) with more than one product were removed. The remaining reactions were tagged with reactive atoms as described previously,53 and reactions with no tagged atoms, or with more than 10 tagged atoms, were removed. This cleaning process results in a final enzymatic dataset of 57
527) with more than one product were removed. The remaining reactions were tagged with reactive atoms as described previously,53 and reactions with no tagged atoms, or with more than 10 tagged atoms, were removed. This cleaning process results in a final enzymatic dataset of 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 unique reactions SMILES47,48 associated with textual descriptions of each reagent, including cofactors, enzymes, and solvent.
176 unique reactions SMILES47,48 associated with textual descriptions of each reagent, including cofactors, enzymes, and solvent.
Enzymatic AutoTag and triple transformer loop (ENZR-TTL) models
Enzymatic transformer models for the ENZR-TTL, including the AutoTag to tag reactive sites, and T1, T2 and T3 in the TTL itself, were trained using the USPTO and the ENZR dataset through MTL, similar to our previous Enzymatic Transformer model with identical training hyperparameters.52 The split ratio 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 was applied as in the USPTO dataset resulting in 51
5 was applied as in the USPTO dataset resulting in 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 459
459![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2859
2859![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2858 reactions in the training, validation, and test set respectively. The dataset split was done such that reactions resulting in identical products belong to the same splitting set.
2858 reactions in the training, validation, and test set respectively. The dataset split was done such that reactions resulting in identical products belong to the same splitting set.
During the MTL processes detailed below for all ENZR models, we incorporated instruction tokens. These tokens, “ENZYME” for the ENZR dataset and “USPTO” for the USPTO dataset, were inserted at both the start and end of the SMILES inputs. This addition aimed to provide additional context to the model and enable it to focus on specific prediction types as needed.
The ENZR-AutoTag model was trained to predict the tagged SMILES of the product (P*) from the product SMILES (P), in a similar manner to the USPTO-AutoTag model. The ENZR-T1 was trained to predict SM from P* for enzymatic retrosynthesis. In contrast, the ENZR-T2 model differs significantly from its USPTO-T2 counterpart by predicting a textual description of the enzyme (TDE) rather than reagents (R) in SMILES format from the theoretical reaction SMILES (SM → P). The ENZR-T3, previously reported as the Enzymatic Transformer,52 serves as forward validation, it was trained from SM + TDE to predict P, now retrained using the new ENZR dataset.
Disconnection-aware automatic tagging strategy
In our previous study,53 the USPTO-TTL employed a combination of three tagging strategies: (1) a systematic tagging procedure, tagging 1 to 3 neighbouring atoms, (2) tagging templates of reactive sites with a conditional structure radius of 2 atoms, and (3) the AutoTag Transformer model with a beam size of 50.The ENZR-TTL uses a specific tagging strategy combining only an AutoTag model54 and templates, excluding the systematic tagging approach. The dedicated ENZR-AutoTag was trained from the ENZR dataset and USPTO by MTL. ENZR reactive site templates were extracted from ENZR exclusively with a radius of 2 atoms.
Chemoenzymatic multistep tree search algorithm
In parallel to the existing single-step USPTO-TTL, we added the ENZR-TTL, which the multistep algorithm uses systematically and independently. The prediction outcomes of both TTLs are provided to the heuristic best-first tree search, elaborating routes mixing the predictions of both TTLs. Confidence scores of both TTLs behaving differently, the confidence scores of ENZR-T3 were adapted by polynomial fit to the USPTO-T3 distribution (Fig. S1†) to ensure a fair scoring across TTLs. The RPScore, based on molecular simplicity55,56 and confidence scores of T3 distinguishes which routes are the best to explore further, and functions the same as reported in our previous study.53Our previous report of the Enzymatic Transformer model, herein named ENZR-T3, demonstrated that a confidence score threshold was required to filter unreasonable enzymatic reactions. A similar evaluation using the round-trip evaluation of the ENZR-TTL was performed and a threshold of 90% confidence of ENZR-T3 was defined for considering ENZR-TTL predictions for multistep retrosynthesis search.
Building block (BB) set
We combined MolPort (https://www.molport.com) and Enamine (https://www.enamine.net) databases to build a database of 534![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 commercially available compounds as the building block (BB) set.
058 commercially available compounds as the building block (BB) set.
Results and discussion
Realizing the triple transformer loop algorithm with biocatalysis (TTLAB) for chemoenzymatic retrosynthesis required first to select a suitable dataset of enzymatic reactions, second to adapt our previous chemical reaction TTL to these enzymatic reactions, and finally to combine the enzymatic reaction TTL with the chemical reaction TTL in a multistep search algorithm. These steps are described in the following subsections.Chemical and enzymatic reaction datasets and their comparison
We used the USPTO reaction dataset, which lists one million chemical reactions taken from the patent literature, as a broadly accepted selection of chemical reactions used in organic synthesis.49,50 In terms of enzymatic reactions, we selected 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 enzymatic reactions from the scientific literature using the Reaxys API,46 forming an enlarged version of our earlier enzymatic reaction dataset (ENZR, see methods for details).52 The composition of this enlarged ENZR dataset is comparable to its smaller version and reflects the practice of biocatalysis in preparative organic chemistry as reported in the scientific literature, with lipases and dehydrogenases forming the largest class of enzymes (Fig. S2†).
176 enzymatic reactions from the scientific literature using the Reaxys API,46 forming an enlarged version of our earlier enzymatic reaction dataset (ENZR, see methods for details).52 The composition of this enlarged ENZR dataset is comparable to its smaller version and reflects the practice of biocatalysis in preparative organic chemistry as reported in the scientific literature, with lipases and dehydrogenases forming the largest class of enzymes (Fig. S2†).
In view of training transformer models for a combined chemoenzymatic retrosynthesis, we analyzed whether the 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 enzyme-catalyzed reactions in our ENZR dataset contained starting materials and products comparable to those in USPTO. We also analyzed the ECREACT data,36 which lists 62
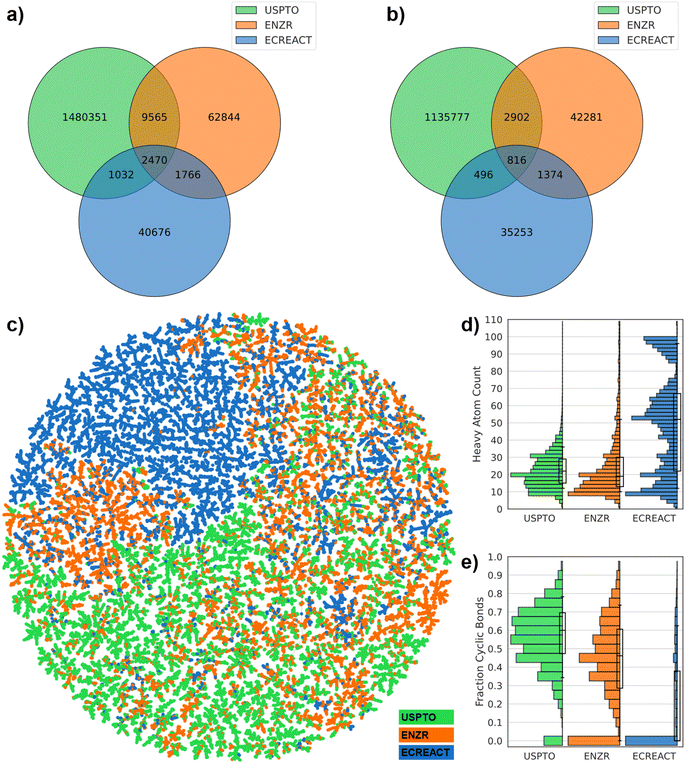
176 enzyme-catalyzed reactions in our ENZR dataset contained starting materials and products comparable to those in USPTO. We also analyzed the ECREACT data,36 which lists 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 222 enzyme-catalyzed reactions associated with their respective enzyme commission (EC) number, aggregated from the biochemical reaction pathways datasets Rhea, BRENDA, PathBank, and MetaNetX (Table 1).37,41–43 ENZR listed fewer reactions than ECREACT but more molecules, indicating a larger diversity of molecules tested in preparative biocatalysis compared to biochemical intermediates. Furthermore, ENZR shared a larger number of molecules with USPTO than ECREACT, and only shared a small number of molecules with ECREACT. A similar distribution was observed when focusing only on reaction products, with only 2470 molecules and 816 product molecules being shared between all three datasets (Fig. 2a and b).
222 enzyme-catalyzed reactions associated with their respective enzyme commission (EC) number, aggregated from the biochemical reaction pathways datasets Rhea, BRENDA, PathBank, and MetaNetX (Table 1).37,41–43 ENZR listed fewer reactions than ECREACT but more molecules, indicating a larger diversity of molecules tested in preparative biocatalysis compared to biochemical intermediates. Furthermore, ENZR shared a larger number of molecules with USPTO than ECREACT, and only shared a small number of molecules with ECREACT. A similar distribution was observed when focusing only on reaction products, with only 2470 molecules and 816 product molecules being shared between all three datasets (Fig. 2a and b).
| USPTO | ENZR | ECREACT | |
|---|---|---|---|
| Number of reactions | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 266 266![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734 734 |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 176 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 222 222 |
| Number of unique molecules | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 493![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 418 418 |
76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 645 645 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944 944 |
| Number (%) of molecules shared with USPTO | — | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 035 (15.7%) 035 (15.7%) |
3502 (7.6%) |
| Number (%) of molecules shared with ECREACT | 3502 (0.27%) | 4236 (7.4%) | — |
| Number of chiral molecules | 271![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 504 504 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 277 277 |
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 177 177 |
To compare the three datasets in terms of molecule types, we selected 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules randomly across starting materials and products in each dataset and constructed a TMAP,60 employing the MinHashed atom-pair fingerprint MAP4 as similarity measure, which considers substructures and their relative position in molecules.61 Areas of the TMAP covered by molecules from USPTO (green) also contained molecules from ENZR (orange), and to a lesser extent from ECREACT (blue), showing a certain level of overlap in structural types between the three datasets (Fig. 2c). Nevertheless, parts of the map were dominated by one of three datasets. Predominantly green areas (USPTO) contained drug-like heteroaromatic molecules, while predominantly orange areas (ENZR) featured glycosides and peptides. Furthermore, one fourth of the TMAP was standing out because it was entirely blue (ECREACT) and was populated by phospholipids and triglycerides apparently completely absent from the other two datasets, probably reflecting the difficulty to work with such molecules in terms of preparative organic synthesis.
000 molecules randomly across starting materials and products in each dataset and constructed a TMAP,60 employing the MinHashed atom-pair fingerprint MAP4 as similarity measure, which considers substructures and their relative position in molecules.61 Areas of the TMAP covered by molecules from USPTO (green) also contained molecules from ENZR (orange), and to a lesser extent from ECREACT (blue), showing a certain level of overlap in structural types between the three datasets (Fig. 2c). Nevertheless, parts of the map were dominated by one of three datasets. Predominantly green areas (USPTO) contained drug-like heteroaromatic molecules, while predominantly orange areas (ENZR) featured glycosides and peptides. Furthermore, one fourth of the TMAP was standing out because it was entirely blue (ECREACT) and was populated by phospholipids and triglycerides apparently completely absent from the other two datasets, probably reflecting the difficulty to work with such molecules in terms of preparative organic synthesis.
Histograms further highlighted similarities and differences between molecules composing the three datasets. A histogram of molecular size as heavy atom count (HAC) showed that ENZR and USPTO contained molecules of comparable size (10 ≤ HAC ≤ 40), while more than half of ECREACT contained larger molecules (HAC > 40) (Fig. 2d). Furthermore, a histogram of the fraction of cyclic bonds showed that USPTO contained mostly cyclic molecules, while ENZR contained similarly cyclic molecules but also a sizable fraction of entirely acyclic molecules, and ECREACT was almost entirely composed of acyclic molecules (Fig. 2e). The difference in molecule properties between the three datasets was also visible in scatter plots using molecular weight, the fraction of carbon atoms and the fraction of cyclic bonds as molecular descriptors (Fig. S3†). Note that 47.9% of ECREACT molecules contained a phosphate functional group, compared to 8.2% in ENZR molecules and only 0.5% in USPTO molecules, further highlighting the different nature of molecules involved in biochemical reaction pathways compared to those in use for synthetic chemistry.
Taken together, these comparisons showed that molecules in ENZR and USPTO datasets showed a significant level of overlap and might be useful for a transformer model approach for combined chemoenzymatic retrosynthesis. By contrast, the differences between ECREACT and USPTO were more pronounced and suggested that these two datasets were almost incompatible with each other.
Enzymatic triple transformer loop (ENZR-TTL)
Our TTL approach for single-step retrosynthesis consists of tagging potential reactive sites in the product molecule P to form a series of tagged P*, and for each P* to apply three subsequent transformer models predicting SM from P* (T1), reagents R from SM → P (T2), and finally product P from SM + R (T3). T3 validates the retrosynthetic step if the predicted P is identical to the input P, and the confidence score of the T3 prediction is used to compute the route penalty score (RPScore) for the multistep search.53In our approach, potential reactive sites in the product molecule are first tagged to mark potential reactive sites. Our chemical reaction TTL used a combination of a transformer model, templates and systematic tagging. Due to the much higher substrate specificity of enzymes compared to chemical reagents, we removed the systematic tagging approach for our enzymatic TTL and only considered tagging with a transformer model and with templates. Reactive sites in product molecules of the ENZR dataset were identified from atom-mapping and labelled as previously described for the USPTO.53 An ENZR-AutoTag transformer was then trained by MTL combining the tagged and untagged datasets of ENZR and USPTO. Enzymatic templates were extracted from the atom-mapped ENZR dataset considering only templates with a radius of two bonds around reacting atoms to take enzyme specificity into account, an aspect which was also reflected by the much smaller number of ENZR templates (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 083) compared to the number of USPTO templates (281
083) compared to the number of USPTO templates (281![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 153).
153).
To complement the transformer models for the chemical TTL trained with the USPTO dataset (here named USPTO-TTL), we used MTL of USPTO with the ENZR dataset using the previously described parameters52 to obtain models for the enzymatic TTL (here named ENZR-TTL). To help the transformers to learn the differences between chemical and enzymatic reactions, all entries for MTL were labelled before and after the SMILES with “ENZYME” for ENZR data, and with “USPTO” for USPTO data. These labels helped to avoid task ambiguity between USPTO vs. ENZR caused by the substitution of reagent SMILES with enzyme names in text format for T2 (SMILES → SMILES vs. SMILES → text) and T3 (SMILES → SMILES vs. SMILES + text → SMILES). The influence of the instruction tokens “ENZYME” and “USPTO” added before and after each input was well visible in the case of ENZR-T2, for which the fraction of textual enzyme description produced increased from 85.3% for an uninstructed model to 99.7% for the instructed model.
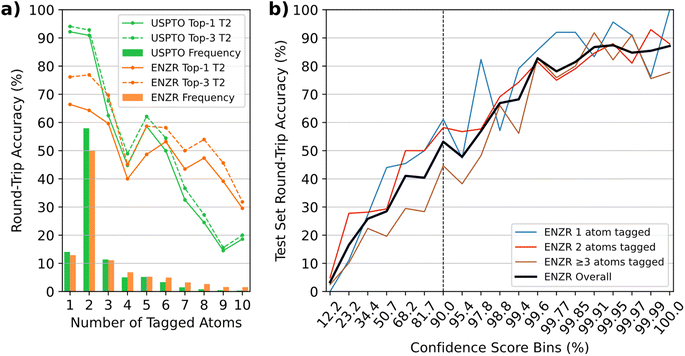
In terms of single-step round-trip accuracy,57 the ENZR-TTL achieved 59.0% top-1 accuracy on the ENZR test set, somewhat below the 81.3% top-1 accuracy of the USPTO-TTL on the USPTO test set. In both cases, the top-1 round-trip accuracy measured the percentage of cases where P predicted by T3 matched the input P, which also included cases with different SM and R compared to the ground truth in the test sets (see details in Tables S1 and S2†). In both TTLs, the round-trip accuracy decreased as function of the increasing number of tagged atoms, suggesting that the decreasing number of training examples and the increasing reaction complexity caused more difficult learning in the different transformers involved in producing the TTL predictions (Fig. 3a). ENZR-TTL top-3 round-trip accuracies were as high as 76.2% and 76.9% for single and double atom tags, compared to 94.1% and 92.8% in the case of USPTO-TTL. The lower performance of ENZR-TTL compared to USPTO-TTL probably reflects the smaller training set of enzymatic reactions learned by transfer learning, and a more difficult task associated with the prediction of enzyme names in T2. A similar analysis on the round-trip accuracy as function of the heavy atom count showed no significant higher accuracies on smaller molecules, but rather a dependence on the number of molecules per molecular size bin, emphasizing again a dependence on training set size rather than on molecular size (Fig. S4†). As for the USPTO-T3, the confidence score of ENZR-T3 was correlated with the round-trip accuracy (Fig. 3b). Analysis of test cases showed that a cut-off value of 90% had to be applied to select meaningful validated enzymatic retrosynthetic steps.
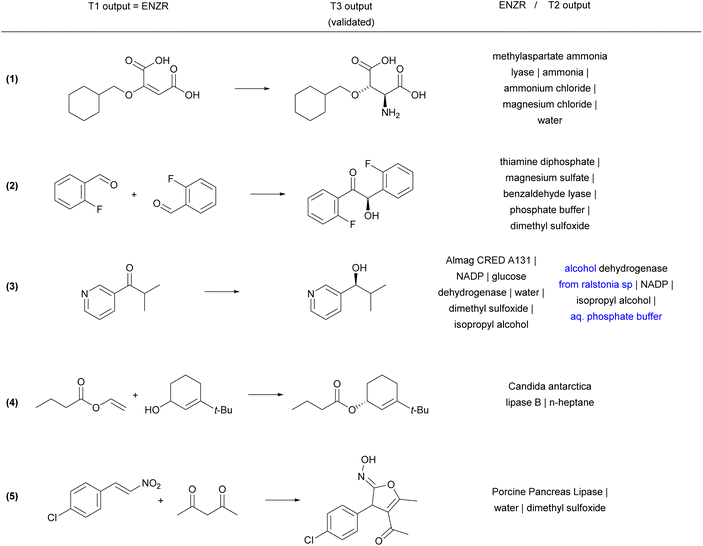
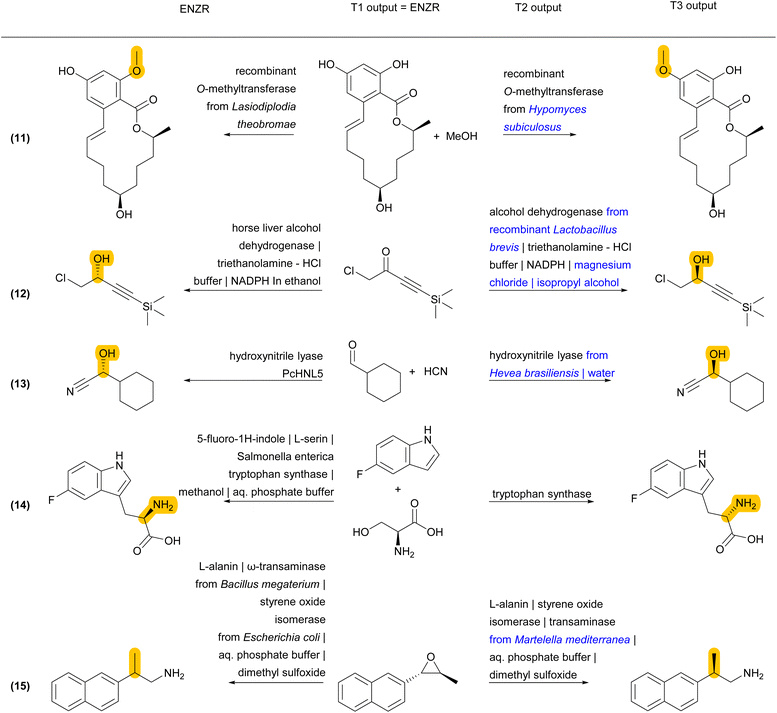
Reaction examples from the ENZR test set illustrate the performance of ENZR-TTL in terms of single-step retrosynthesis. In many cases, T1 predicts the same SM as recorded in the ENZR dataset, T2 predicts the identical or almost identical enzyme description (with enzyme name, additive and solvents), and T3 predicts the correct P (Fig. 4 and S5†). These include enantioselective reactions with non-biochemical substrates (reaction (1)),62 cofactors (reaction (2))63 and cofactor regeneration systems (reaction (3),64 here with a different T2 output), as well as lipase-catalyzed reactions such as kinetic resolutions by acylation (reaction (4))65 and heterocycle formations exploiting the catalytic promiscuity of lipases (reaction (5)).66
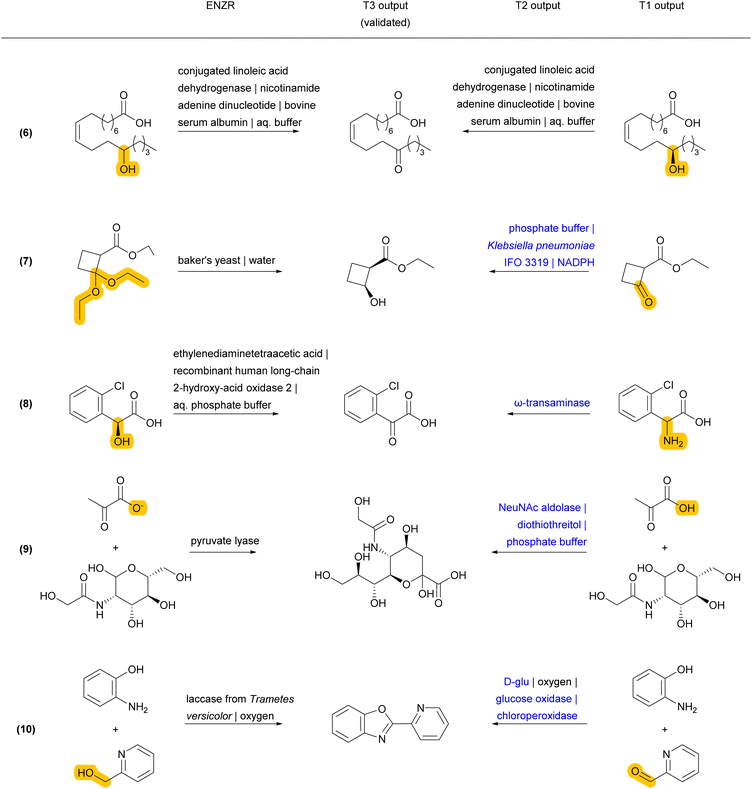
Validated retrosyntheses by ENZR-TTL include cases where the SM output by T1 and sometimes the enzyme name output by T2 are different from those recorded in ENZR, with interesting cases of reactions involving ketones and aldehydes as SM or P (Fig. 5 and S6†). In one case, the T1 output specifies alcohol chirality for a fatty acid alcohol dehydrogenase reported to be non-enantioselective (although without providing primary data, reaction (6)),67 whereby T1 probably infers alcohol chirality from other alcohol dehydrogenases. In another case, a chiral cyclobutanol is proposed by ENZR-TTL to be obtained by reduction of the parent ketone by a microbial dehydrogenase, while the database case involves baker's yeast and a ketal precursor of the cyclobutanone in aqueous pH 2, under which conditions the ketal spontaneously hydrolyzes to give the ketone (reaction (7)).68 Furthermore, a (2-chlorophenyl)-ketoacid recorded in ENZR to be formed by enzymatic oxidation of the corresponding mandelic acid,69 is predicted by ENZR-TTL to stem from a transaminase reaction from the parent phenylglycine, a known type of biotransformation (reaction (8)).70
Some discrepancies between ENZR data and ENZR-TTL output are caused by database entry mistakes and illustrate the self-correcting ability of the transformer model approach. For example, N-acetylneuraminic acid is incorrectly recorded in ENZR as involving a “pyruvate lyase” due to an enzyme naming mistake in the corresponding publication (reaction (9)).71 For this reaction ENZR-TTL correctly predicts that the enzymatic conversion of SM (N-acetyl-mannosamine and pyruvic acid) is carried out by the enzyme NeuNAc aldolase.72 Similarly, the oxidative condensation of 2-pyridylmethanol with 2-aminophenol listed in Reaxys as an enzymatic process and recorded in ENZR (reaction (10)) actually involves TEMPO (2,2,6,6-tetramethylpiperidine-1-oxyl) as a chemical oxidant, which is recycled by air oxidation using laccase as enzyme but was not recorded in Reaxys.73 Here, ENZR-TTL proposes pincolinaldehyde and 2-aminophenol as SM and a true enzymatic process using glucose oxidase and chloroperoxidase. This bi-enzymatic process has been reported for the related oxidative condensation of benzaldehyde and several para-substituted benzaldehydes with 2-aminophenol to form benzoxazoles.74
Finally, some incorrect cases involve a correct SM prediction by T1, but a different choice of enzyme by T2, resulting in a valid biotransformation but a different product P predicted by T3, and a non-validated reaction in terms of round-trip accuracy of ENZR-TTL (Fig. 6 and S7†). For example, the correct phenolic SM is predicted by T1 for the formation of an O-methylated macrolactone (reaction (11)). However, T2 selects a different O-methyl transferase enzyme with a different regioselectivity, and therefore T3 predicts a different regioselectivity for the methylation. Note however that the proposed product is the correct one for the selected enzyme, as recorded in the same original publication focusing on tuning O-methylation regioselectivity.75 In a related case of a chiral propargylic alcohol stemming from reduction of the corresponding ketone by an alcohol dehydrogenase, T1 predicts the correct SM but a change of enzyme choice by T2 results in a T3 prediction of P with the opposite enantioselectivity, which is correct for the selected enzyme but incorrect relative to database entry (reaction (12)).76 A similar different enzyme choice by T2 resulting in an enantiomeric P correctly predicted by T3 also occurs for the addition of hydrogen cyanide to cyclohexane carbaldehyde catalyzed by two different hydroxynitrile lyases (reaction (13)).77,78
In a related case involving tryptophan synthase, T1 predicts the correct SM, T2 the correct enzyme, and T3 the correct L-enantiomer, however the database entry lists the D-enantiomer, which was obtained by coupling tryptophan synthase with a stereoinversion cascade involving two enzymes that were not listed in the database entry (reaction (14)).79,80 In a similar enzymatic cascade yielding 2-(2-naphthyl)propylamine from an epoxide precursor, T1 predicts the correct epoxide SM but combines styrene oxide isomerase with a different transaminase producing the (R)-enantiomeric P. By contrast, the database entry for P has an undefined stereochemistry, probably because the parent publications tested various transaminases with different enantioselectivities (reaction (15)).81,82
Taken together, the above analysis showed that biocatalytic retrosynthesis predictions by ENZR-TTL were generally relevant and sometimes even corrected inaccuracies in database entries. Encouraged by these data, we moved on to test multi-step chemoenzymatic retrosyntheses with our TTL approach.
Chemoenzymatic multistep retrosynthesis with TTLAB
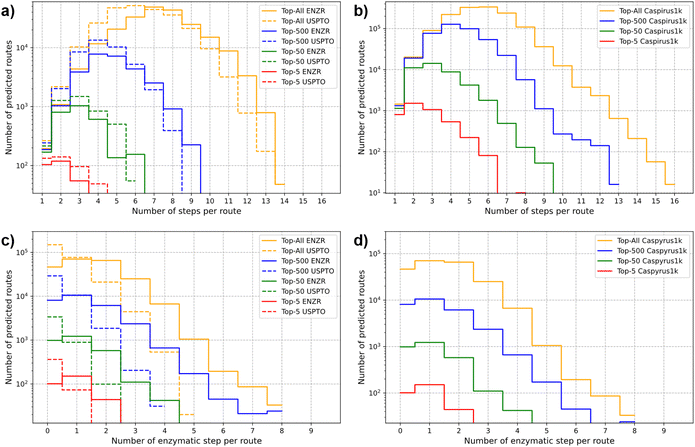
Integrating ENZR-TTL alongside the previously reported USPTO-TTL provided the chemo-enzymatic retrosynthesis prediction system, named TTLAB (Fig. 1). To ensure the reliability of the enzymatic steps selected by TTLAB, a confidence score filter of 90% was applied to ENZR-T3. This filter eliminated chemically incorrect enzymatic retrosynthetic steps which would otherwise be selected by the tree-search because they achieved a high RPScore due to a high degree of molecular simplification.We challenged TTLAB to propose retrosyntheses for 100 product molecules from the USPTO test set, 80 product molecules from the ENZR test set, and 1000 molecules from the Caspyrus dataset.83,84 A retrosynthesis was judged successful whenever the reaction sequence went back to a SM molecule available in the BB set, which consisted of 534![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 commercially available compounds (see Methods for details). TTLAB proposed synthetic routes for 88 of the 100 USPTO test set product molecules, 61 of the 80 ENZR test set product molecules, and 852 of the 1000 molecules of the Caspyrus dataset, and in almost all cases at least one of the proposed routes contained at least an enzymatic step (Table S3†). For TTLAB-predicted syntheses of USPTO and Caspyrus molecules, approximately 8% and 9% of the proposed steps were enzymatic. This percentage ranged from 17% to 50% for TTLAB predicted syntheses of ENZR molecules considering either all proposed syntheses or only top-scoring ones (Table S4†). The ability of TTLAB to identify short chemo-enzymatic synthetic routes was well visible when analyzing the number of steps per route as well as the number of enzymatic steps per route among the top-5, top-50, top-500 or all routes for USPTO, ENZR and Caspyrus molecules (Fig. 7).
058 commercially available compounds (see Methods for details). TTLAB proposed synthetic routes for 88 of the 100 USPTO test set product molecules, 61 of the 80 ENZR test set product molecules, and 852 of the 1000 molecules of the Caspyrus dataset, and in almost all cases at least one of the proposed routes contained at least an enzymatic step (Table S3†). For TTLAB-predicted syntheses of USPTO and Caspyrus molecules, approximately 8% and 9% of the proposed steps were enzymatic. This percentage ranged from 17% to 50% for TTLAB predicted syntheses of ENZR molecules considering either all proposed syntheses or only top-scoring ones (Table S4†). The ability of TTLAB to identify short chemo-enzymatic synthetic routes was well visible when analyzing the number of steps per route as well as the number of enzymatic steps per route among the top-5, top-50, top-500 or all routes for USPTO, ENZR and Caspyrus molecules (Fig. 7).
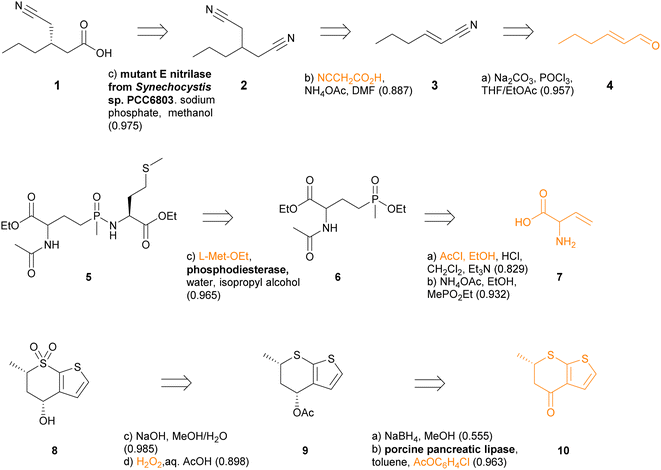
The chemoenzymatic routes predicted by TTLAB are well illustrated by three examples from the ENZR test set, for which we show in each case the best RPScoring route including at least one enzymatic step (Fig. 8). The first example is the predicted synthesis of the chiral cyanocarboxylic acid 1, which was reported as the product of the enantioselective mono-hydrolysis of the prochiral dinitrile 2 by a mutant nitrilase enzyme.85 TTLAB predicts the identical biotransformation as the first retrosynthetic operation, and proposes to assemble dinitrile 2 by Michael addition of cyanoacetic acid to unsaturated nitrile 3 and decarboxylation. Finally, TTLAB proposes to prepare nitrile 3 from the parent aldehyde 4, which is a well-known type of transformation however using different reagents.86
The second example is the predicted synthesis of the phospha-C-peptide 5, which was reported to be formed by coupling L-methionine ethyl ester with ethyl phosphinate 6 catalyzed by a phosphordiesterase.87 TTLAB proposes the identical last step using the same enzyme. Since phosphinate 6 is not present in the commercial BB set, TTLAB further proposes a synthesis from vinyl glycine 7 by N-acetylation and esterification, done as a single step, followed by addition of ethyl methylphosphinate to the double bond. The latter reaction had been reported to prepare L-phosphinothricin, a naturally occurring herbicidal amino acid, however TTLAB omits to list the required radical initiator tert-butyl per-2-ethylhexanoate.88
The third example is chiral sulfone 8, which TTLAB would prepare by deacetylation and sulfide oxidation of intermediate 9 using known chemistry.89 Intermediate 9 would be formed by diastereoselective enzymatic acetylation of the parent alcohol by porcine pancreatic lipase using p-chlorophenylacetate as acylating agent, a biotransformation reaction known from the test set.90 This parent alcohol would be formed by non-stereoselective reduction of ketone 10 using sodium borohydride. This reduction is predicted with low confidence by TTLAB because this reaction can in fact be performed stereoselectively using LiAlH4.91 Indeed, when the condition of an enzymatic step is not imposed, TTLAB readily proposes, as the second best RPScoring route, a two-step chemical synthesis of 8 from 10 by stereoselective reduction followed by thioether oxidation to the sulfone.
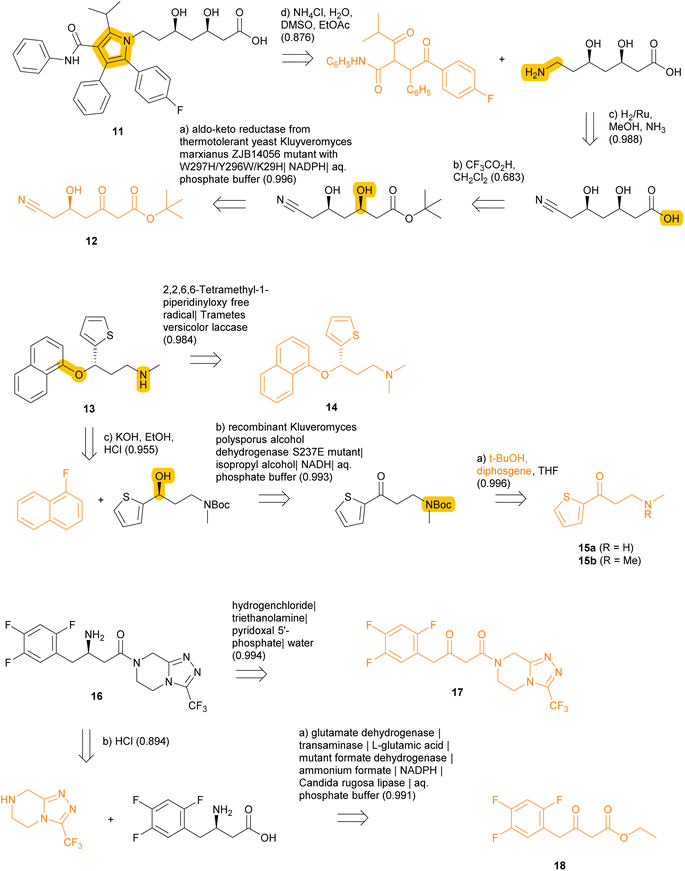
We further exemplify TTLAB in the prediction of chemoenzymatic retrosyntheses for three drugs with known chemoenzymatic routes (Fig. 9). In these cases, TTLAB often identifies steps that are part of the training sets. For the first case of the cholesterol-lowering drug atorvastatin 11, our algorithm proposes as best RPScoring route the acidic deprotection of the corresponding tert-butyl ester, which is a commercial building block. Imposing at least one enzymatic step results in a four-step sequence from a linear chiral keto-ester precursor 12, for which the first step is an enzymatic reduction by an aldo-keto reductase which was evolved precisely for this purpose and is present in the TTLAB training set.92 The overall TTLAB route design is similar to the chemoenzymatic process developed for this drug involving an enzymatic enantioselective reduction of ethyl cyanoacetoacetate as initial step.93
In the second case of the antidepressant (S)-duloxetine 13, the top-RPScoring route with at least one enzymatic step predicted by TTLAB is the single-step demethylation of the commercial N,N-dimethyl analog 14 catalyzed by a laccase, and the second best is a three-step sequence involving Boc protection of the achiral ketone precursor 15a, followed by enantioselective reduction with an alcohol dehydrogenase and arylation of the resulting alcohol with fluoronaphthalene. This route is similar to the published chemoenzymatic synthesis of this drug starting with N,N-dimethylketone 15b,94 also proposed by the ASKCOS chemical CASP tool with the help of manual intervention to introduce biocatalytic steps.95
In the third case of the DDP4 inhibitor sitagliptin (16) used to treat type II diabetes, TTLAB identifies a single-step enzymatic enantioselective retrosynthesis from the commercial β-ketoamide 17 using a transaminase. Although TTLAB only names the PLP cofactor in the reagents, this step is present in the ENZR training set using a transaminase that has been engineered for the synthesis of this drug.96 The second best RPScoring route is a similar two step sequence from the commercial ketoester 18 involving an enzymatic enantioselective reductive amination followed by amide bond formation. Note that the enzymatic step is part of the ENZR training set and uses the exact same combination of four enzymes for this biotransformation,97 illustrating that transformer model ENZR-T2 memorizes enzyme textual description with high accuracy.
The above analysis and application examples show that TTLAB can propose short chemoenzymatic retrosyntheses for various target molecules. It should be noted that enzymatic steps are selected by TTLAB only when the reaction is closely related to a training set reaction, reflecting the fact that biocatalytic reactions are often highly specific for certain types of starting materials and are intrinsically poorly generalizable.
Comparison with other chemo-enzymatic CASP tools
To compare TTLAB with other chemo-enzymatic retrosynthesis tools, we subjected the six target molecules discussed above (1, 5, 8, 11, 13 and 16, Fig. 8 and 9) to the IBM RXN for Chemistry retrosynthesis prediction tool in “Automatic mode” using the “enzymatic mode 2022-05-31” model and “high quality” tuning, which uses the reported transformer model.36 We also tested the template-based tool ASKCOS as available online using either the “reaxys_biocatalysis” and “reaxys” models combined, or just the “reaxys_biocatalysis” model alone,45 as well as the recently reported chemo-enzymatic version of BioNavi with the “Default settings” preset, allowing both “Bio-building blocks” and “Chemo-building blocks”, and combining “Enzymatic synthesis” and “Non-enzymatic synthesis”.35,98The IBM RXN for chemistry provided retrosyntheses for all six target molecules, however none of the retrosyntheses contained any enzymatic steps in the sequences and the sequences went back to chiral building blocks as source of chirality (Table 2 and Fig. S8–S13†). The difficulty of this tool in identifying biocatalytic steps might reflect the fact that it uses biochemical reaction data and very different molecule types as discussed above (Fig. 2). On the other hand, ASKCOS only predicted retrosyntheses successfully for 13 and 16. In these two cases, at least one route contained enzymatic steps and the routes were similar to those coming from TTLAB, with enzymatic steps involved in establishing stereochemistry in two cases (Table 2 and Fig. S14–S21†). This similarity might reflect the fact that ASKCOS also exploits literature data on biotransformations collected from Reaxys, although in a different manner that TTLAB. BioNavi only produced retrosyntheses for 1 and 5, however these were short and included biocatalytic steps (Fig. S22–S24†).
| Target moleculea | TTLAB | IBM RXN36 | ASKCOS45 | BioNavi98 |
|---|---|---|---|---|
| a See Fig. 8 and 9 for TTLAB retrosyntheses, Fig. S8–S13 for IBM RXN for chemistry retrosyntheses, Fig. S14–S21 for ASKCOS retrosyntheses, and Fig. S22–S24 for BioNavi retrosyntheses. b another 6 routes were proposed by BioNavi for 1 with up to 5 steps, however without enzymatic steps. | ||||
| 1 | 2C + 1B | 8C | — | 3C + 1B, 3Cb |
| 5 | 1C + 1B | 7C | — | 2C, 1B |
| 8 | 1C + 1B | 1C | — | — |
| 11 | 3C, 1B | 1C | — | — |
| 13 | 1B, 2C + 1B | 2C | 1C, 2C + 1B | — |
| 16 | 1B, 1C + 1B | 4C | 1C, 1B, 2B | — |
Although the three chemoenzymatic CASP tools tested did not perform as well than TTLAB in the examples discussed above, one cannot generalize and each retrosynthesis should be analyzed in detail for feasibility. In that respect, it must be noted that for chemical steps IBM RXN for chemistry ouputs the reagents as part of the starting materials and describes the reaction class for each transformation, thereby providing an information comparable to the output of TTLAB. For enzymatic steps however for which TTLAB provides enzyme names, IBM RXN for chemistry only provides EC numbers, which can be insufficient to choose a particular enzyme in cases such as lipases and alcohol dehydrogenases for which substrate tolerance and stereoselectivity are highly variable. On the other hand, ASKCOS does not provide reagents or enzyme names but simply links to Reaxys references, which have to be searched manually to identify the proper reaction conditions. Finally, BioNavi informs whether a given steps is enzymatic or non-enzymatic and upon request connects to a list of reagents or enzymes, which again is an output similar to TTLAB. Note that each tool uses a slightly different set of commercial building blocks, which may influence the ability to propose retrosynthetic routes as well as route length depending on the availability of advanced intermediates in the building block set.
Conclusion
In summary, our work integrates biocatalysis in a computer-assisted synthesis planning (CASP) system, going towards greener and more sustainable chemistry. We achieved this by introducing a dual multistep retrosynthesis prediction system, integrating both chemical and biocatalytic steps in the form of two triple transformer loops, namely our previously reported TTL trained on USPTO reactions for chemical steps (USPTO-TTL),53 and a related enzymatic ENZR-TTL trained on an updated version of our ENZR dataset of biotransformations extracted from Reaxys.52 ENZR-TTL makes use of a new model to mark potential biocatalytic disconnection sites (ENZR-Autotag) in product molecules and consists of three new transformers to predict and validate possible retrosynthetic biotranformations. The competitive framework, driven by the route penalty score (RPScore), drives the selection of optimal steps by our best-first tree search, incorporating both catalytic steps to generate mixed synthesis routes. In the successful routes selected by TTLAB, 8–17% of the steps (depending on the molecular dataset) are enzyme-catalyzed reactions, suggesting that our tool can valuably contribute to green process design. Our results not only showcase the tool's capabilities in proposing viable solutions for drug-like molecules but also establish it as a valuable resource for synthesis design. The continuous enrichment of data in Reaxys promises ongoing enhancements in enzymatic capabilities, progressively going towards enzymatic synthesis.Data availability
Code and instructions to compute multistep retrosynthesis as well as the code to tag reactive sites are available on our GitHub repository: https://github.com/reymond-group/MultiStepRetrosynthesisTTL. The original USPTO dataset can be found at https://doi.org/10.6084/m9.figshare.5104873.v1. The derived version of the USPTO dataset of Thakkar et al. can be found in their preprint.54 The Reaxys enzymatic dataset is a licensed commercial database that cannot be made available.Author contributions
DK designed and carried out the study and wrote the paper, JLR designed and supervised the study and wrote the paper.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported financially by Novartis. We would like to thank Dr Thierry Schlama for guidance and insightful advice, Dr Daniel Kaufmann for his support, Dr Frederic Stanger for internal testing of the retrosynthesis tool at Novartis, Dr Radka Snajdrova, Dr John Lopez, and Dr Thomas Ruch for their helpful discussions, and Reaxys for access to the API for retrieving enzymatic reactions. Calculations were performed on UBELIX (https://www.id.unibe.ch/hpc), the HPC cluster at the University of Bern.References
- E. J. Corey, General Methods for the Construction of Complex Molecules, Pure Appl. Chem., 1967, 14(1), 19–38, DOI:10.1351/pac196714010019.
- E. J. Corey and W. T. Wipke, Computer-Assisted Design of Complex Organic Syntheses, Science, 1969, 166(3902), 178–192, DOI:10.1126/science.166.3902.178.
- D. A. Pensak and E. J. Corey, LHASA—Logic and Heuristics Applied to Synthetic Analysis, in Computer-Assisted Organic Synthesis, pp. , pp. 1–32, DOI:10.1021/bk-1977-0061.ch001.
- E. J. Corey, A. K. Long and S. D. Rubenstein, Computer-Assisted Analysis in Organic Synthesis, Science, 1985, 228(4698), 408–418, DOI:10.1126/science.3838594.
- P. Y. Johnson, I. Burnstein, J. Crary, M. Evans and T. WangDesigning an Expert System for Organic Synthesis, in Expert System Applications in Chemistry, ACS Symposium Series, American Chemical Society, 1989, vol. 408, pp. 102–123, DOI:10.1021/bk-1989-0408.ch009.
- W.-D. Ihlenfeldt and J. Gasteiger, Computer-Assisted Planning of Organic Syntheses: The Second Generation of Programs, Angew Chem. Int. Ed. Engl., 1996, 34(23–24), 2613–2633, DOI:10.1002/anie.199526131.
- J. Law, Z. Zsoldos, A. Simon, D. Reid, Y. Liu, S. Y. Khew, A. P. Johnson, S. Major, R. A. Wade and H. Y. Ando, Route Designer: A Retrosynthetic Analysis Tool Utilizing Automated Retrosynthetic Rule Generation, J. Chem. Inf. Model., 2009, 49(3), 593–602, DOI:10.1021/ci800228y.
- C. D. Christ, M. Zentgraf and J. M. Kriegl, Mining Electronic Laboratory Notebooks: Analysis, Retrosynthesis, and Reaction Based Enumeration, J. Chem. Inf. Model., 2012, 52(7), 1745–1756, DOI:10.1021/ci300116p.
- A. Bøgevig, H.-J. Federsel, F. Huerta, M. G. Hutchings, H. Kraut, T. Langer, P. Löw, C. Oppawsky, T. Rein and H. Saller, Route Design in the 21st Century: The ICSYNTH Software Tool as an Idea Generator for Synthesis Prediction, Org. Process Res. Dev., 2015, 19(2), 357–368, DOI:10.1021/op500373e.
- J. Nam and J. Kim, Linking the Neural Machine Translation and the Prediction of Organic Chemistry Reactions, arXiv, 2016, preprint, arXiv:abs/1612.09529, DOI:10.48550/arXiv.1612.09529.
- S. Szymkuć, E. P. Gajewska, T. Klucznik, K. Molga, P. Dittwald, M. Startek, M. Bajczyk and B. A. Grzybowski, Computer-Assisted Synthetic Planning: The End of the Beginning, Angew. Chem., Int. Ed., 2016, 55(20), 5904–5937, DOI:10.1002/anie.201506101.
- M. H. S. Segler and M. P. Waller, Neural-Symbolic Machine Learning for Retrosynthesis and Reaction Prediction, Chem.–Eur. J., 2017, 23(25), 5966–5971, DOI:10.1002/chem.201605499.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez and Ł. Kaiser and I. Polosukhin, Attention Is All You Need, in Advances in Neural Information Processing Systems 30, ed. Guyon, I., Luxburg, U. V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S. and Garnett, R., Curran Associates, Inc., 2017, pp. 5998–6008 Search PubMed.
- B. Liu, B. Ramsundar, P. Kawthekar, J. Shi, J. Gomes, Q. Luu Nguyen, S. Ho, J. Sloane, P. Wender and V. Pande, Retrosynthetic Reaction Prediction Using Neural Sequence-to-Sequence Models, ACS Cent. Sci., 2017, 3(10), 1103–1113, DOI:10.1021/acscentsci.7b00303.
- C. W. Coley, R. Barzilay, T. S. Jaakkola, W. H. Green and K. F. Jensen, Prediction of Organic Reaction Outcomes Using Machine Learning, ACS Cent. Sci., 2017, 3(5), 434–443, DOI:10.1021/acscentsci.7b00064.
- M. H. S. Segler, M. Preuss and M. P. Waller, Planning Chemical Syntheses with Deep Neural Networks and Symbolic AI, Nature, 2018, 555(7698), 604–610, DOI:10.1038/nature25978.
- P. Schwaller, T. Gaudin, D. Lányi, C. Bekas and T. Laino, “Found in Translation”: Predicting Outcomes of Complex Organic Chemistry Reactions Using Neural Sequence-to-Sequence Models, Chem. Sci., 2018, 9(28), 6091–6098, 10.1039/C8SC02339E.
- P. Schwaller, T. Laino, T. Gaudin, P. Bolgar, C. A. Hunter, C. Bekas and A. A. Lee, Molecular Transformer: A Model for Uncertainty-Calibrated Chemical Reaction Prediction, ACS Cent. Sci., 2019, 5(9), 1572–1583, DOI:10.1021/acscentsci.9b00576.
- A. A. Lee, Q. Yang, V. Sresht, P. Bolgar, X. Hou, J. L. Klug-McLeod and C. R. Butler, Molecular Transformer Unifies Reaction Prediction and Retrosynthesis across Pharma Chemical Space, Chem. Commun., 2019, 55(81), 12152–12155, 10.1039/C9CC05122H.
- A. Thakkar, T. Kogej, J.-L. Reymond, O. Engkvist and E. J. Bjerrum, Datasets and Their Influence on the Development of Computer Assisted Synthesis Planning Tools in the Pharmaceutical Domain, Chem. Sci., 2019, 11(1), 154–168, 10.1039/C9SC04944D.
- P. Karpov, G. Godin and I. V. Tetko, A Transformer Model for Retrosynthesis, in Artificial Neural Networks and Machine Learning – ICANN 2019: Workshop and Special Sessions, ed. Tetko, I. V., Kůrková, V., Karpov, P. and Theis, F., Lecture Notes in Computer Science, Springer International Publishing, Cham, 2019, pp. 817–830, DOI:10.1007/978-3-030-30493-5_78.
- K. Lin, Y. Xu, J. Pei and L. Lai, Automatic Retrosynthetic Route Planning Using Template-Free Models, Chem. Sci., 2020, 11(12), 3355–3364, 10.1039/C9SC03666K.
- S. Zheng, J. Rao, Z. Zhang, J. Xu and Y. Yang, Predicting Retrosynthetic Reactions Using Self-Corrected Transformer Neural Networks, J. Chem. Inf. Model., 2020, 60(1), 47–55, DOI:10.1021/acs.jcim.9b00949.
- H. Duan, L. Wang, C. Zhang, L. Guo and J. Li, Retrosynthesis with Attention-Based NMT Model and Chemical Analysis of “Wrong” Predictions, RSC Adv., 2020, 10(3), 1371–1378, 10.1039/C9RA08535A.
- F. Strieth-Kalthoff, F. Sandfort, M. H. S. Segler and F. Glorius, Machine Learning the Ropes: Principles, Applications and Directions in Synthetic Chemistry, Chem. Soc. Rev., 2020, 49(17), 6154–6168, 10.1039/C9CS00786E.
- S. Genheden, A. Thakkar, V. Chadimová, J.-L. Reymond, O. Engkvist and E. Bjerrum, AiZynthFinder: A Fast, Robust and Flexible Open-Source Software for Retrosynthetic Planning, J. Cheminf., 2020, 12(1), 70, DOI:10.1186/s13321-020-00472-1.
- A. Thakkar, S. Johansson, K. Jorner, D. Buttar, J.-L. Reymond and O. Engkvist, Artificial Intelligence and Automation in Computer Aided Synthesis Planning, React. Chem. Eng., 2021, 6(1), 27–51, 10.1039/D0RE00340A.
- N. J. Turner and E. O'Reilly, Biocatalytic Retrosynthesis, Nat. Chem. Biol., 2013, 9(5), 285–288, DOI:10.1038/nchembio.1235.
- F. H. Arnold, Directed Evolution: Bringing New Chemistry to Life, Angew. Chem., Int. Ed. Engl., 2018, 57(16), 4143–4148, DOI:10.1002/anie.201708408.
- R. A. Sheldon and J. M. Woodley, Role of Biocatalysis in Sustainable Chemistry, Chem. Rev., 2018, 118(2), 801–838, DOI:10.1021/acs.chemrev.7b00203.
- S. Wu, R. Snajdrova, J. C. Moore, K. Baldenius and U. T. Bornscheuer, Biocatalysis: Enzymatic Synthesis for Industrial Applications, Angew. Chem., Int. Ed. Engl., 2020, 59, 2–34, DOI:10.1002/anie.202006648.
- E. L. Bell, W. Finnigan, S. P. France, A. P. Green, M. A. Hayes, L. J. Hepworth, S. L. Lovelock, H. Niikura, S. Osuna, E. Romero, K. S. Ryan, N. J. Turner and S. L. Flitsch, Biocatalysis, Nat. Rev. Methods Primers, 2021, 1(1), 1–21, DOI:10.1038/s43586-021-00044-z.
- H. Gröger, F. Gallou and B. H. Lipshutz, Where Chemocatalysis Meets Biocatalysis: In Water, Chem. Rev., 2023, 123(9), 5262–5296, DOI:10.1021/acs.chemrev.2c00416.
- W. Finnigan, L. J. Hepworth, S. L. Flitsch and N. J. Turner, RetroBioCat as a Computer-Aided Synthesis Planning Tool for Biocatalytic Reactions and Cascades, Nat. Catal., 2021, 4(2), 98–104, DOI:10.1038/s41929-020-00556-z.
- S. Zheng, T. Zeng, C. Li, B. Chen, C. W. Coley, Y. Yang and R. Wu, Deep Learning Driven Biosynthetic Pathways Navigation for Natural Products with BioNavi-NP, Nat. Commun., 2022, 13(1), 3342, DOI:10.1038/s41467-022-30970-9.
- D. Probst, M. Manica, Y. G. Nana Teukam, A. Castrogiovanni, F. Paratore and T. Laino, Biocatalysed Synthesis Planning Using Data-Driven Learning, Nat. Commun., 2022, 13(1), 964, DOI:10.1038/s41467-022-28536-w.
- I. Schomburg, A. Chang and D. Schomburg, BRENDA, Enzyme Data and Metabolic Information, Nucleic Acids Res., 2002, 30(1), 47–49 CrossRef CAS.
- M. Kanehisa, The KEGG Database, in ‘In Silico’ Simulation of Biological Processes, John Wiley & Sons, Ltd, 2002, pp. 91–103, DOI:10.1002/0470857897.ch8.
- P. D. Karp, M. Riley, S. M. Paley and A. Pellegrini-Toole, The MetaCyc Database, Nucleic Acids Res., 2002, 30(1), 59–61, DOI:10.1093/nar/30.1.59.
- N. EzC. D. B. Nagano, The Enzyme Catalytic-Mechanism Database, Nucleic Acids Res., 2005, 33(suppl_1), D407–D412, DOI:10.1093/nar/gki080.
- R. Alcántara, K. B. Axelsen, A. Morgat, E. Belda, E. Coudert, A. Bridge, H. Cao, P. de Matos, M. Ennis, S. Turner, G. Owen, L. Bougueleret, I. Xenarios and C. Steinbeck, Rhea–a Manually Curated Resource of Biochemical Reactions, Nucleic Acids Res., 2012, 40(Database issue), D754–D760, DOI:10.1093/nar/gkr1126.
- M. Ganter, T. Bernard, S. Moretti, J. Stelling and M. Pagni, MetaNetX.Org: A Website and Repository for Accessing, Analysing and Manipulating Metabolic Networks, Bioinformatics, 2013, 29(6), 815–816, DOI:10.1093/bioinformatics/btt036.
- D. S. Wishart, C. Li, A. Marcu, H. Badran, A. Pon, Z. Budinski, J. Patron, D. Lipton, X. Cao, E. Oler, K. Li, M. Paccoud, C. Hong, A. C. Guo, C. Chan, W. Wei and M. Ramirez-Gaona, PathBank: A Comprehensive Pathway Database for Model Organisms, Nucleic Acids Res., 2020, 48(D1), D470–D478, DOI:10.1093/nar/gkz861.
- C. W. Coley, D. A. Thomas, J. A. M. Lummiss, J. N. Jaworski, C. P. Breen, V. Schultz, T. Hart, J. S. Fishman, L. Rogers, H. Gao, R. W. Hicklin, P. P. Plehiers, J. Byington, J. S. Piotti, W. H. Green, A. J. Hart, T. F. Jamison and K. F. Jensen, A Robotic Platform for Flow Synthesis of Organic Compounds Informed by AI Planning, Science, 2019, 365(6453), eaax1566, DOI:10.1126/science.aax1566.
- I. Levin, M. Liu, C. A. Voigt and C. W. Coley, Merging Enzymatic and Synthetic Chemistry with Computational Synthesis Planning, Nat. Commun., 2022, 13(1), 7747, DOI:10.1038/s41467-022-35422-y.
- A. J. Lawson; J. Swienty-Busch; T. Géoui and D. Evans, The Making of Reaxys—Towards Unobstructed Access to Relevant Chemistry Information, in The Future of the History of Chemical Information, ACS Symposium Series, American Chemical Society, 2014, vol. 1164, pp. 127–148, DOI:10.1021/bk-2014-1164.ch008.
- D. Weininger, SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules, J. Chem. Inf. Comput. Sci., 1988, 28(1), 31–36, DOI:10.1021/ci00057a005.
- D. Weininger, A. Weininger and J. L. Weininger, SMILES. 2. Algorithm for Generation of Unique SMILES Notation, J. Chem. Inf. Comput. Sci., 1989, 29(2), 97–101, DOI:10.1021/ci00062a008.
- D. M. Lowe Extraction of Chemical Structures and Reactions from the Literature, PhD thesis, University of Cambridge, 2012, DOI:10.17863/CAM.16293.
- D. Lowe, Chemical Reactions from US Patents (1976–Sep 2016). figshare. dataset., 2017, DOI:10.6084/m9.figshare.5104873.v1.
- G. Pesciullesi, P. Schwaller, T. Laino and J.-L. Reymond, Transfer Learning Enables the Molecular Transformer to Predict Regio- and Stereoselective Reactions on Carbohydrates, Nat. Commun., 2020, 11(1), 4874, DOI:10.1038/s41467-020-18671-7.
- D. Kreutter, P. Schwaller and J.-L. Reymond, Predicting Enzymatic Reactions with a Molecular Transformer, Chem. Sci., 2021, 12(25), 8648–8659, 10.1039/D1SC02362D.
- D. Kreutter and J.-L. Reymond, Multistep Retrosynthesis Combining a Disconnection Aware Triple Transformer Loop with a Route Penalty Score Guided Tree Search, Chem. Sci., 2023, 14(36), 9959–9969, 10.1039/D3SC01604H.
- A. Thakkar, A. C. Vaucher, A. Byekwaso, P. Schwaller, A. Toniato and T. Laino, Unbiasing Retrosynthesis Language Models with Disconnection Prompts, ACS Cent. Sci., 2023, 9(7), 1488–1498, DOI:10.1021/acscentsci.3c00372.
- C. W. Coley, L. Rogers, W. H. Green and K. F. Jensen, SCScore: Synthetic Complexity Learned from a Reaction Corpus, J. Chem. Inf. Model., 2018, 58(2), 252–261, DOI:10.1021/acs.jcim.7b00622.
- P. Schwaller, R. Petraglia, V. Zullo, H. V. Nair, R. Andreas Haeuselmann, R. Pisoni, C. Bekas, A. Iuliano and T. Laino, Predicting Retrosynthetic Pathways Using Transformer-Based Models and a Hyper-Graph Exploration Strategy, Chem. Sci., 2020, 11(12), 3316–3325, 10.1039/C9SC05704H.
- P. Schwaller, R. Petraglia, V. H. Nair and T. Laino, Evaluation Metrics for Single-Step Retrosynthetic Models, Second Workshop on Machine Learning and the Physical Sciences (NeurIPS 2019), 2019 Search PubMed.
- G. Landrum, RDKit: Open-Source Cheminformatics, 2006 Search PubMed.
- P. Schwaller, D. Probst, A. C. Vaucher, V. H. Nair, D. Kreutter, T. Laino and J.-L. Reymond, Mapping the Space of Chemical Reactions Using Attention-Based Neural Networks, Nat. Mach. Intell., 2021, 3(2), 144–152, DOI:10.1038/s42256-020-00284-w.
- D. Probst and J.-L. Reymond, Visualization of Very Large High-Dimensional Data Sets as Minimum Spanning Trees, J. Cheminf., 2020, 12(1), 12, DOI:10.1186/s13321-020-0416-x.
- A. Capecchi, D. Probst and J.-L. Reymond, One Molecular Fingerprint to Rule Them All: Drugs, Biomolecules, and the Metabolome, J. Cheminf., 2020, 12(1), 43, DOI:10.1186/s13321-020-00445-4.
- H. Fu, J. Zhang, P. G. Tepper, L. Bunch, A. A. Jensen and G. J. Poelarends, Chemoenzymatic Synthesis and Pharmacological Characterization of Functionalized Aspartate Analogues As Novel Excitatory Amino Acid Transporter Inhibitors, J. Med. Chem., 2018, 61(17), 7741–7753, DOI:10.1021/acs.jmedchem.8b00700.
- A. S. Demir, Ö. Şeşenoglu, E. Eren, B. Hosrik, M. Pohl, E. Janzen, D. Kolter, R. Feldmann, P. Dünkelmann and M. Müller, Enantioselective Synthesis of α-Hydroxy Ketones via Benzaldehyde Lyase-Catalyzed C–C Bond Formation Reaction, Adv. Synth. Catal., 2002, 344(1), 96–103, DOI:10.1002/1615-4169(200201)344:1<96::AID-ADSC96>3.0.CO;2-Z.
- A. S. Rowan, T. S. Moody, R. M. Howard, T. J. Underwood, I. R. Miskelly, Y. He and B. Wang, Preparative Access to Medicinal Chemistry Related Chiral Alcohols Using Carbonyl Reductase Technology, Tetrahedron: Asymmetry, 2013, 24(21), 1369–1381, DOI:10.1016/j.tetasy.2013.09.015.
- B. Wagner, F. P. C. Binder, X. Jiang, T. Mühlethaler, R. C. Preston, S. Rabbani, M. Smieško, O. Schwardt and B. Ernst, A Structural-Reporter Group to Determine the Core Conformation of Sialyl Lewisx Mimetics, Molecules, 2023, 28(6), 2595, DOI:10.3390/molecules28062595.
- M.-Y. Wu, K. Li, T. He, X.-W. Feng, N. Wang, X.-Y. Wang and X.-Q. Yu, A Novel Enzymatic Tandem Process: Utilization of Biocatalytic Promiscuity for High Stereoselective Synthesis of 5-Hydroxyimino-4,5-Dihydrofurans, Tetrahedron, 2011, 67(14), 2681–2688, DOI:10.1016/j.tet.2011.01.060.
- M. Takeuchi, S. Kishino, S.-B. Park, N. Kitamura and J. Ogawa, Characterization of Hydroxy Fatty Acid Dehydrogenase Involved in Polyunsaturated Fatty Acid Saturation Metabolism in Lactobacillus Plantarum AKU 1009a, J. Mol. Catal. B Enzym., 2015, 117, 7–12, DOI:10.1016/j.molcatb.2015.03.020.
- D. Buisson and R. Azerad, Preparation and Use of (S)-O-Acetyllactyl Chloride (Mosandl's Reagent) as a Chiral Derivatizing Agent, Tetrahedron: Asymmetry, 1999, 10(15), 2997–3002, DOI:10.1016/S0957-4166(99)00285-2.
- Y. Zhang, C. Su, J. Lei, L. Chen, H. Hu, S. Zeng and L. Yu, Studies on the L-2-Hydroxy-Acid Oxidase 2 Catalyzed Metabolism of S-Mandelic Acid and Its Analogues, Drug Metabol. Pharmacokinet., 2019, 34(3), 187–193, DOI:10.1016/j.dmpk.2019.02.003.
- D. Zhu and L. Hua, Biocatalytic Asymmetric Amination of Carbonyl Functional Groups – a Synthetic Biology Approach to Organic Chemistry, Biotechnol. J., 2009, 4(10), 1420–1431, DOI:10.1002/biot.200900110.
- O. M. T. Pearce and A. Varki, Chemo-Enzymatic Synthesis of the Carbohydrate Antigen N-Glycolylneuraminic Acid from Glucose, Carbohydr. Res., 2010, 345(9), 1225–1229, DOI:10.1016/j.carres.2010.04.003.
- Y. Li, H. Yu, H. Cao, K. Lau, S. Muthana, V. K. Tiwari, B. Son and X. Chen, Pasteurella multocida Sialic Acid Aldolase: A Promising Biocatalyst, Appl. Microbiol. Biotechnol., 2008, 79(6), 963–970, DOI:10.1007/s00253-008-1506-2.
- M. Mogharabi-Manzari, M. Kiani, S. Aryanejad, S. Imanparast, M. Amini and M. A. Faramarzi, A Magnetic Heterogeneous Biocatalyst Composed of Immobilized Laccase and 2,2,6,6-Tetramethylpiperidine-1-Oxyl (TEMPO) for Green One-Pot Cascade Synthesis of 2-Substituted Benzimidazole and Benzoxazole Derivatives under Mild Reaction Conditions, Adv. Synth. Catal., 2018, 360(18), 3563–3571, DOI:10.1002/adsc.201800459.
- A. Kumar, S. Sharma and R. A. Maurya, Bienzymatic Synthesis of Benzothia/(Oxa)Zoles in Aqueous Medium, Tetrahedron Lett., 2010, 51(48), 6224–6226, DOI:10.1016/j.tetlet.2010.06.012.
- X. Wang, C. Wang, L. Duan, L. Zhang, H. Liu, Y. Xu, Q. Liu, T. Mao, W. Zhang, M. Chen, M. Lin, A. A. L. Gunatilaka, Y. Xu and I. Molnár, Rational Reprogramming of O-Methylation Regioselectivity for Combinatorial Biosynthetic Tailoring of Benzenediol Lactone Scaffolds, J. Am. Chem. Soc., 2019, 141(10), 4355–4364, DOI:10.1021/jacs.8b12967.
- T. Schubert, W. Hummel and M. Müller, Highly Enantioselective Preparation of Multifunctionalized Propargylic Building Blocks, Angew. Chem., Int. Ed. Engl., 2002, 41(4), 634–637, DOI:10.1002/1521-3773(20020215)41:4<634::AID-ANIE634>3.0.CO;2-U.
- H. Griengl, N. Klempier, P. Pöchlauer, M. Schmidt, N. Shi and A. A. Zabelinskaja-Mackova, Enzyme Catalysed Formation of (S)-Cyanohydrins Derived from Aldehydes and Ketones in a Biphasic Solvent System, Tetrahedron, 1998, 54(48), 14477–14486, DOI:10.1016/S0040-4020(98)00901-6.
- Y.-C. Zheng, L.-Y. Ding, Q. Jia, Z. Lin, R. Hong, H.-L. Yu and J.-H. Xu, A High-Throughput Screening Method for the Directed Evolution of Hydroxynitrile Lyase towards Cyanohydrin Synthesis, Chembiochem, 2021, 22(6), 996–1000, DOI:10.1002/cbic.202000658.
- H. M. Ge, W. Yan, Z. K. Guo, Q. Luo, R. Feng, L. Y. Zang, Y. Shen, R. H. Jiao, Q. Xu and R. X. Tan, Precursor-Directed Fungal Generation of Novel Halogenated Chaetoglobosins with More Preferable Immunosuppressive Action, Chem. Commun., 2011, 47(8), 2321–2323, 10.1039/C0CC04183A.
- F. Parmeggiani, A. Rué Casamajo, C. J. W. Walton, J. L. Galman, N. J. Turner and R. A. Chica, One-Pot Biocatalytic Synthesis of Substituted d-Tryptophans from Indoles Enabled by an Engineered Aminotransferase, ACS Catal., 2019, 9(4), 3482–3486, DOI:10.1021/acscatal.9b00739.
- R. Xin, W. W. L. See, H. Yun, X. Li and Z. Li, Enzyme-Catalyzed Meinwald Rearrangement with an Unusual Regioselective and Stereospecific 1,2-Methyl Shift, Angew. Chem., Int. Ed. Engl., 2022, 61(28), e202204889, DOI:10.1002/anie.202204889.
- W. W. L. See, X. Li and Z. Li, Biocatalytic Cascade Conversion of Racemic Epoxides to (S)-2-Arylpropionic Acids, (R)- and (S)-2-Arylpropyl Amines, Adv. Synth. Catal., 2023, 365(1), 68–77, DOI:10.1002/adsc.202201061.
- P. Torren-Peraire, A. K. Hassen, S. Genheden, J. Verhoeven, D.-A. Clevert, M. Preuss and I. V. Tetko, Models Matter: The Impact of Single-Step Retrosynthesis on Synthesis Planning, Digital Discovery, 2024, 3(3), 558–572, 10.1039/D3DD00252G.
- O. J. M. Béquignon, B. J. Bongers, W. Jespers, A. P. IJzerman, B. van der Water, B. van Westen and G. J. P. Papyrus, A Large-Scale Curated Dataset Aimed at Bioactivity Predictions, J. Cheminf., 2023, 15(1), 3, DOI:10.1186/s13321-022-00672-x.
- S. Yu, J. Li, P. Yao, J. Feng, Y. Cui, J. Li, X. Liu, Q. Wu, J. Lin and D. Zhu, Inverting the Enantiopreference of Nitrilase-Catalyzed Desymmetric Hydrolysis of Prochiral Dinitriles by Reshaping the Binding Pocket with a Mirror-Image Strategy, Angew. Chem., Int. Ed. Engl., 2021, 60(7), 3679–3684, DOI:10.1002/anie.202012243.
- D. J. Quinn, G. J. Haun and G. Moura-Letts, Direct Synthesis of Nitriles from Aldehydes with Hydroxylamine-O-Sulfonic Acid in Acidic Water, Tetrahedron Lett., 2016, 57(34), 3844–3847, DOI:10.1016/j.tetlet.2016.07.047.
- I. A. Natchev, Organophosphorus Analogues and Derivatives of the Natural L-Aminocarboxylic Acid and Peptides Vii. Enzyme Synthesis of Phospha-c Peptides, Tetrahedron, 1991, 47(7), 1239–1248, DOI:10.1016/S0040-4020(01)86380-8.
- H.-J. Zeiss, Enantioselective Synthesis of L-Phosphinothricin from L-Methionine and L-Glutamic Acid viaL-Vinylglycine, Tetrahedron, 1992, 48(38), 8263–8270, DOI:10.1016/S0040-4020(01)80494-4.
- O. Tempkin, T. J. Blacklock, J. Andrew Burke and M. Anastasia, β-Butyrolactone as a Chiral Building Block in Organic Synthesis: A Convenient Synthesis of MK-0507 Keto Sulfone, Tetrahedron: Asymmetry, 1996, 7(9), 2721–2724, DOI:10.1016/0957-4166(96)00350-3.
- M. C. Turcu, M. Rantapaju and L. T. Kanerva, Applying Lipase Catalysis to Access the Enantiomers of Dorzolamide Intermediates, Eur. J. Org Chem., 2009, 2009(32), 5594–5600, DOI:10.1002/ejoc.200900672.
- T. J. Blacklock, P. Sohar, J. W. Butcher, T. Lamanec and E. J. J. Grabowski, An Enantioselective Synthesis of the Topically-Active Carbonic Anhydrase Inhibitor MK-0507: 5,6-Dihydro-(S)-4-(Ethylamino)-(S)-6-Methyl-4H-Thieno[2,3-b]Thiopyran-2-Sulfonamide 7,7-Dioxide Hydrochloride, J. Org. Chem., 1993, 58(7), 1672–1679, DOI:10.1021/jo00059a013.
- S. Qiu, F. Cheng, L.-J. Jin, Y. Chen, S.-F. Li, Y.-J. Wang and Y.-G. Zheng, Co-Evolution of Activity and Thermostability of an Aldo-Keto Reductase KmAKR for Asymmetric Synthesis of Statin Precursor Dichiral Diols, Bioorg. Chem., 2020, 103, 104228, DOI:10.1016/j.bioorg.2020.104228.
- S. K. Ma, J. Gruber, C. Davis, L. Newman, D. Gray, A. Wang, J. Grate, G. W. Huisman and R. A. Sheldon, A Green-by-Design Biocatalytic Process for Atorvastatin Intermediate, Green Chem., 2010, 12(1), 81–86, 10.1039/B919115C.
- X. Chen, Z.-Q. Liu, C.-P. Lin and Y.-G. Zheng, Chemoenzymatic Synthesis of (S)-Duloxetine Using Carbonyl Reductase from Rhodosporidium Toruloides, Bioorg. Chem., 2016, 65, 82–89, DOI:10.1016/j.bioorg.2016.02.002.
- K. Sankaranarayanan and K. F. Jensen, Computer-Assisted Multistep Chemoenzymatic Retrosynthesis Using a Chemical Synthesis Planner, Chem. Sci., 2023, 14(23), 6467–6475, 10.1039/D3SC01355C.
- C. K. Savile, J. M. Janey, E. C. Mundorff, J. C. Moore, S. Tam, W. R. Jarvis, J. C. Colbeck, A. Krebber, F. J. Fleitz, J. Brands, P. N. Devine, G. W. Huisman and G. J. Hughes, Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture, Science, 2010, 329(5989), 305–309, DOI:10.1126/science.1188934.
- G.-H. Kim, H. Jeon, T. P. Khobragade, M. D. Patil, S. Sung, S. Yoon, Y. Won, S. Sarak and H. Yun, Glutamate as an Efficient Amine Donor for the Synthesis of Chiral β- and γ-Amino Acids Using Transaminase, ChemCatChem, 2019, 11(5), 1437–1440, DOI:10.1002/cctc.201802048.
- T. Zeng, Z. Jin, S. Zheng, T. Yu and R. Wu, Developing BioNavi for Hybrid Retrosynthesis Planning, JACS Au, 2024, 4(7), 2492–2502, DOI:10.1021/jacsau.4c00228.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc02408g |
| This journal is © The Royal Society of Chemistry 2024 |