Open Access Article
Open Access Article57Fe nuclear resonance vibrational spectroscopic studies of tetranuclear iron clusters bearing terminal iron(III)–oxido/hydroxido moieties†
Jin
Xiong
 a,
Christopher
Reed
b,
Barbara
Lavina
a,
Christopher
Reed
b,
Barbara
Lavina
 cd,
Michael Y.
Hu
cd,
Michael Y.
Hu
 c,
Jiyong
Zhao
c,
Jiyong
Zhao
 c,
Esen E.
Alp
c,
Esen E.
Alp
 c,
Theodor
Agapie
c,
Theodor
Agapie
 *b and
Yisong
Guo
*b and
Yisong
Guo
 *a
*a
aDepartment of Chemistry, Carnegie Mellon University, Pittsburgh, PA 15213, USA. E-mail: ysguo@andrew.cmu.edu
bDivision of Chemistry and Chemical Engineering, California Institute of Technology, CA 91125, USA. E-mail: agapie@caltech.edu
cAdvanced Photon Source, Argonne National Laboratory, Argonne, Illinois 60439, USA
dCenter for Advanced Radiation Source, University of Chicago, Chicago, Illinois 60439, USA
First published on 9th September 2024
Abstract
57Fe nuclear resonance vibrational spectroscopy (NRVS) has been applied to study a series of tetranuclear iron ([Fe4]) clusters based on a multidentate ligand platform (L3−) anchored by a 1,3,5-triarylbenzene linker and pyrazolate or (tertbutylamino)pyrazolate ligand (PzNHtBu−). These clusters bear a terminal Fe(III)–O/OH moiety at the apical position and three additional iron centers forming the basal positions. The three basal irons are connected with the apical iron center via a μ4-oxido ligand. Detailed vibrational analysis via density functional theory calculations revealed that strong NRVS spectral features below 400 cm−1 can be used as an oxidation state marker for the overall [Fe4] cluster core. The terminal Fe(III)–O/OH stretching frequencies, which were observed in the range of 500–700 cm−1, can be strongly modulated (energy shifts of 20–40 cm−1 were observed) upon redox events at the three remote basal iron centers of the [Fe4] cluster without the change of the terminal Fe(III) oxidation state and its coordination environment. Therefore, the current study provides a quantitative vibrational analysis of how the remote iron centers within the same iron cluster exert exquisite control of the chemical reactivities and thermodynamic properties of the specific iron site that is responsible for small molecule activation.
Introduction
Metal–oxido/hydroxido (M–O/OH) species are key reactive intermediates involved in challenging oxidation reactions found in numerous biological pathways and organic synthesis.1–18 Prominent examples include selective carbon–hydrogen (C–H) bond functionalization and water oxidation in biological photosynthesis where metal–oxido/hydroxido species enable the key steps of the reactions, e.g. C–H bond cleavage5,16–19 and O–O bond formation.8,10,20–23 Given the importance of these M–O/OH species, extensive research efforts have been devoted to reveal the geometric/electronic structural features that define their chemical reactivities. There is no doubt that the metal primary coordination sphere constituted by the direct metal-binding ligand set establishes the geometric and electronic structures of M–O/OH species and further determines their reactivities.1,16,24–28 It is also increasingly clear in recent years that the secondary coordination sphere critically tunes the chemical/physical properties of metal centers.29,30 For example, Lu and coworkers have shown that by changing the composition of amino acids surrounding the copper centers in azurin, the copper redox potentials could be altered to cover the entire physiological range (>1 V).31 Among the heme-containing proteins, hydrogen bonding interactions surrounding the heme-iron centers have shown to dictate protein function. For instance, in hemoglobins and myoglobins, the proximal protein residues to the heme center stabilize the O2 binding,32–37 but in cytochrome P450 dependent enzymes, a different set of proximal protein residues facilitate O–O bond cleavage instead.7,38–41 In non-heme iron enzymes, H-bonding networks surrounding the iron center have also been shown to lower the energetic barriers for O2 activation15 and facilitate organic substrate oxidation.42,43 For nonheme M–O/OH synthetic complexes, Borovik and coworkers have demonstrated that the basicity of Mn/Fe–O/OH moieties, and subsequently their C–H activation reactivity, can be modulated by tuning the surrounding H-bonding interactions.16,29,44,45 The influence of the H-bonding interactions to the electronic properties and reactivities of Mn/Fe–O/OH moieties has also been reported by Fout and coworkers,46–50 Chang and coworkers,51 and Goldberg and coworkers.52 In addition, Lewis acidic, redox inactive metal ions, have also been demonstrated to influence the reactivities of (Fe/Mn/Co)–O moieties.53–58Compared to our understanding on the structure and reactivity modulation of M–O/OH species via changes from the first and the second coordination spheres, the understanding of the influence of remote redox active metal centers to M–O/OH moieties is much less developed. Yet, in nature, multi-nuclear metal centers are commonly used to enable essential multi-electron/proton chemistry, such as water oxidation by the oxygen evolving complex (OEC) in photosystem II,8,59 proton reduction/H2 oxidation by the H-cluster and the Ni–Fe center in hydrogenases,60–63 CO2 reduction by the C-cluster of CODH,64 O2 reduction by the copper–iron-heme center in cytochrome c oxidase,65,66 and N2 reduction by iron-molybdenum cofactor (FeMco) in nitrogenase.67–69 Some of these active sites display complex metal clusters with distal metal centers not directly involved in substrate binding. The impact of remote metal centers on substrate activation has been studied by some of us.70–74 A multidentate ligand platform (L3−) anchored by a 1,3,5-triarylbenzene linker and pyrazolate or (tertbutylamino)pyrazolate ligand (PzNHtBu−) were applied to support a [Fe3Mn] cluster and a [Fe4] cluster bearing a μ4-O ligand and a terminal MnII/III–OHx or FeIII–O/OH moiety at the apical position of the cluster, respectively (Fig. 1).73,74 Detailed thermodynamics and reactivity studies demonstrated that by only changing the redox state on the three remote base iron sites of the cluster, the bond dissociation free energy (BDE) of the O–H bond of the apical MnII–OH2 or FeIII–OH moieties can be tuned by a total of ∼16 or 12 kcal mol−1, respectively.73,74 It has also been estimated that the BDE of the O–H bond of the terminal MnIII–OH moiety could reach >100 kcal mol−1 by the same remote iron site redox controlling strategy.73 Clearly, the distal metal centers within the same metal cluster exert exquisite control of the chemical properties of the metal site that is responsible for small molecule binding and activation.
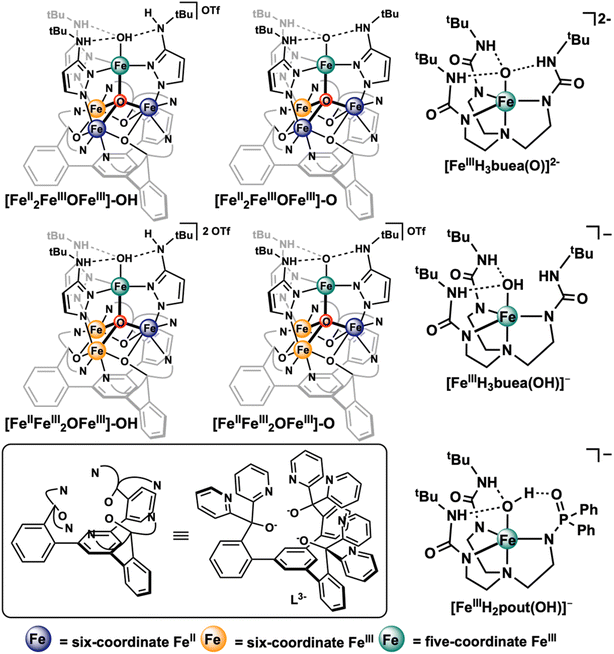 | ||
| Fig. 1 Chemical structures of the [Fe4] clusters studied in this work and the selected mononuclear iron(III)–oxido/hydroxide complexes from Borovik and coworkers75–77 that have been previously studied by 57Fe NRVS.77 | ||
Herein, to further elucidate the impact of the remote iron site redox events towards the geometric and electronic structures of the terminal M–O/OH moiety in these multi-nuclear iron clusters, we have carried out a detailed spectroscopic and computational study on a series of [LFe3O(PzNHtBu)3Fe(O/OH)]n+ clusters.7457Fe nuclear resonant vibrational spectroscopy (NRVS) has been used to explore the vibrational features of [LFeIIFeIII2O(PzNHtBu)3FeIII(O)]+ ([FeIIFeIII2OFeIII]–O) and [LFeIIFeIII2O(PzNHtBu)3FeIII(OH)]2+ ([FeIIFeIII2OFeIII]–OH), and their corresponding one electron reduced clusters, [LFeII2FeIIIO(PzNHtBu)3FeIII(O)] ([FeII2FeIIIOFeIII]–O) and [LFeII2FeIIIO(PzNHtBu)3FeIII(O)]+ ([FeII2FeIIIOFeIII]–OH). The subsequent spectral analysis with the assistance of density functional theory (DFT) calculations not only revealed the unique vibrational features of these [Fe4] clusters, but also pointed out the critical role of the Fe–μ4O bonding interactions among the four iron centers of the cluster in translating the impact of the redox event on the three remote basal iron sites to the apical terminal FeIII–O/OH moiety. Such a redox modulation effect generated by remote iron site redox state change was also compared with the modulation effect of the hydrogen-bonding interactions exerted by the secondary coordination sphere towards Fe–O/OH moiety in mononuclear iron complexes to provide a broader perspective on the influence of the chemical environment of Fe–O/OH moieties in modulating their chemical reactivity.
Results and discussions
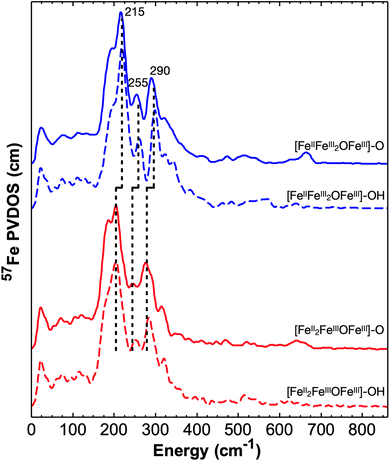
We have recorded 57Fe NRVS spectra on both [Fe4]–oxido ([Fe4]–O) and [Fe4]–hydroxido ([Fe4]–OH) complexes in two oxidation states ([FeII2FeIII2] and [FeIIFeIII3]). All the NRVS derived 57Fe PVDOS spectra are shown in Fig. 2, which exhibit strong spectral features in the region between 150 cm−1 and 400 cm−1 and a series of weak features in the region between 400 cm−1 and 800 cm−1. Additionally, the strong spectral features show systematic red shifts (Δν ∼ 10 cm−1) in going from the complexes with the oxidized [Fe4] core ([FeIIFeIII2OFeIII]) to the complexes with the one-electron reduced [Fe4] core ([FeII2FeIIIOFeIII]). We start our detailed discussion on the spectroscopic features from the [FeIIFeIII2OFeIII]–O complex (Fig. 3). The most intense 57Fe PVDOS feature is centered at 215 cm−1 with a low energy shoulder at 190 cm−1 and a high energy shoulder at 250 cm−1. The next strongest feature is observed at 290 cm−1 with a broad high energy shoulder distributed between 320 cm−1 and 400 cm−1. In the energy range above 400 cm−1, only relatively weak 57Fe PVDOS features are observed. To identify the terminal Fe–O stretching vibrations, the measurements for the analogous complex with 18O-labeling (at both the terminal oxido ligand and the bridging μ4-oxido ligand, see the ESI† for synthesis) have also been performed (Fig. 3A). A clear 18O-sensitive feature is observed at 665 cm−1 in [FeIIFeIII216OFeIII]–16O, which is red shifted to 635 cm−1 in [FeIIFeIII218OFeIII]–18O. This feature must belong to the stretching vibration of the terminal FeIII–O moiety (ν(FeIII–O)), and the isotope shift (Δν (16O/18O) = 30 cm−1) is consistent with the predicted isotope shift of an isolated Fe–O stretching vibration based on Hooke's law (Δν = 30 cm−1). The residual 665 cm−1 peak in the spectrum of [FeIIFeIII218OFeIII]–18O is most likely from the 16O labeled complex due to incomplete 18O labeling (see more discussion below). Interestingly, ν(FeIII–O) observed in [FeIIFeIII2OFeIII]–O shows nearly identical energy with the ν(FeIII–O) observed in a mononuclear ferric–oxido complex, [FeIII{H3buea}(O)]2− (ν(FeIII–O) = 660 cm−1),77 suggesting that ν(FeIII–O) is a relatively isolated vibration even in this tetranuclear iron cluster, such as [FeIIFeIII2OFeIII]–O, and the vibrational frequency is essentially determined by the FeIII–O bond length (d[Fe–O] = 1.795(8) Å in [FeIIFeIII2OFeIII]–O and 1.813(3) Å in [FeIII{H3buea}(O)]2− determined by single crystal X-ray diffraction (XRD)74,75). We note here that the stabilization of an FeIII–O moiety is achieved by three hydrogen bonds (H-bonds) provided by the NH groups of the supporting ligand in both complexes. A similar hydrogen bonded FeIII–O moiety with an FeIII–O bond length of d[Fe–O] = 1.808(1) Å has also been reported by Fout and coworker on an FeIII–O complex supported by a tripodal ligand, tris(5-cyclohexyl-amineazafulvene-2-methyl)amine (N(afaCy)3), with three H-bonds.46 Roithová and coworkers have reported the ν(FeIII–O) in gas phase for [FeIII(O)N4Py]+ (N4Py = N,N-bis(2-pyridylmethyl)-N-bis(2-pyridyl)methylamine) and [FeIII(O)TPA]+ (TPA = tris(2-pyridylmethyl)amine) with only a single H-bond provided by a water molecular to the FeIII–O moieties, which gave ν(FeIII–O) = 787 cm−1, much higher than the ν(FeIII–O) observed here.78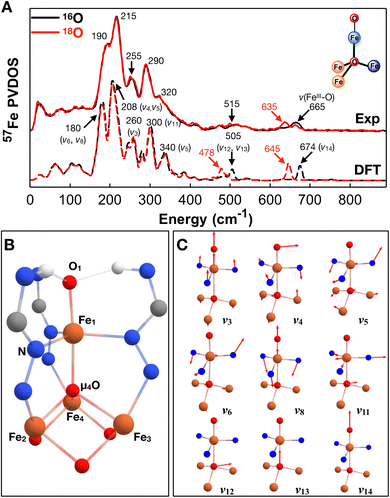 | ||
| Fig. 3 57Fe PVDOS experimental and DFT calculated spectra of [FeIIFeIII2OFeIII]–O and the associated vibrational mode assignments. (A) 57Fe PVDOS spectra (top) and DFT calculated spectra (bottom) of [FeIIFeIII2OFeIII]–O (black traces) and 18O-labeled complex, [FeIIFeIII218OFeIII]–18O (red traces); the simplified structure of this complex is also shown (see Fig. 1 for atom color code definition). See Fig. S2† for additional comparison between the experimental and DFT calculated spectra. (B) The Fe4 cluster core geometry with selected atom labeling used in this study (C) the selected normal modes of vibration of a OFe(X3)(YZ3) type molecule. | ||
To better understand the 57Fe PVDOS features exhibited by [FeIIFeIII2OFeIII]–O, we turned to DFT analysis. For the calculations, a broken symmetry state was used, where the three iron atoms (Fe2,3,4) at the base of the [Fe4] cluster are ferromagnetically coupled and the apical iron atom (Fe1) is antiferromagnetically coupled with all three base iron atoms to form an MS = 9/2 BS state. The selection of this BS state is supported by the SQUID measurements of this complex, which showed that [FeIIFeIII2OFeIII]–O exhibits an S = 9/2 ground spin state.79 By using the functional-basis set combination of B3LYP/TZVP (see the ESI† for the calculation details), the DFT optimized structure using the MS = 9/2 BS state nicely reproduced the corresponding structure determined by XRD. The comparison of the selected iron–ligand bond lengths and the atom labeling used in this study are shown in Fig. 3B (see Table S1† for additional selected bond lengths based on the XRD results and the DFT results), which demonstrates a good overall agreement between the DFT optimized and the XRD determined structures. Similar with the crystal structure, the DFT optimized structure shows that the three base irons are not structurally equivalent. One of the irons (Fe2) shows longer bond lengths with the coordinating ligands than the other two irons (davg(Fe2–L) = 2.136 (2.186, DFT) Å, davg(Fe3–L) = 2.068 (2.117, DFT) Å, and davg(Fe4–L) = 2.051 (2.114, DFT) Å), suggesting that Fe2 is the ferrous center while the other two irons (Fe3 and Fe4) are in the ferric state. Specifically, the Fe2–μ4O bond length is 2.155(7) Å in crystal structure and 2.196 Å in the DFT optimized structure while the Fe3–μ4O and Fe4–μ4O bond lengths are ∼0.2 Å shorter. Indeed, the DFT calculated spin population also support this assignment (Table S2†). Fe2 shows a Mulliken spin population of 3.81, which is smaller than those of Fe3 and Fe4 (4.15) and is more consistent with a high-spin ferrous ion. Also, the spin population of the apical iron (Fe1) is 4.01, similar with those of Fe3 and Fe4, thus confirming that Fe1, Fe3, and Fe4 are all in the ferric state. Based on this optimized structure, we carried out frequency calculations and reconstructed 57Fe PVDOS spectrum of [FeIIFeIII2OFeIII]–O. The calculated spectrum compares well with the experimental data, which allows assignments of the spectral features to specific vibrational modes. The DFT calculated ν(FeIII–O) is located at 675 cm−1, which is red shifted to 646 cm−1 with 18O-labeling on the terminal oxido ligand to give Δν (16O/18O) = 29 cm−1. Thus, this result supports the assignment of the 665 cm−1 57Fe PVDOS feature in the experimental data to the same ν(FeIII–O), which shows Δν (16O/18O) = 30 cm−1.
The vibrational mode assignments for the low energy features of [FeIIFeIII2OFeIII]–O can be best understood by comparing with the normal modes of vibration of a simplified OFe(X3)(YZ3) type molecule with a C3v symmetry,77 where in our case X represents the equatorial ligand of the apical iron, Y represents the iron ligand trans to the oxido ligand, in this case it is the bridging oxo ligand (μ4-O) in [FeIIFeIII2OFeIII]–O, and Z represents the three base iron atoms. The full normal modes of vibration of the OFe(X3)(YZ3) type molecule are listed in Fig. S1,† and selected mode pictures are listed in Fig. 3C. The strongest 57Fe PVDOS feature in the calculated spectrum of [FeIIFeIII2OFeIII]–O is located at ∼207 cm−1 with an unresolved high energy shoulder at ∼218 cm−1, which should correspond to the 215 cm−1 peak observed experimentally. This peak is mainly originated from vibrational modes that highly resembles the ν4(E) and ν5(E) modes (the swing chair motions, ν4(E), essentially the out of plane bending motions of Neq–Fe1–Neq and Febase–μ4O–Febase, and the in-phase rocking motion, ν5(E), essentially the in-phase combination of the out of plane rocking Neq–Fe1–Neq with the Febase–μ4O–Febase bending motions) from the OFe(X3)(YZ3) model. Due to the lower symmetry of [FeIIFeIII2OFeIII]–O, these two E modes split into four major vibrations calculated at 207 cm−1, 212 cm−1, 214 cm−1, and 221 cm−1. The experimentally observed 190 cm−1 peak is calculated at ∼180 cm−1, which is mainly from vibrations resembling the ν6(E) mode (the out-of-phase rocking motion, essentially the out-of-phase combination of ν5(E)) from the OFe(X3)(YZ3) model calculated at ∼180 cm−1 and the symmetric Neq–Fe1–Neq out-of-plane bending motion calculated at 185 (ν8(A1) mode). The calculated features at 300 cm−1 with a lower energy shoulder at 280 cm−1 could be assigned to the experimentally observed 290 cm−1 feature. The calculated 300 cm−1 feature mainly originates from Neq–Fe1–Neq asymmetric stretch motion combined with an expansion motion of the base Febase–μ4O structural moiety, which is reminiscent to ν11(E) mode of the OFe(X3)(YZ3) model. The calculated 280 cm−1 feature is originated from a symmetric Febase–μ4O–Febase bending motion (the ν7(A1) mode). In addition, the calculated 260 cm−1 peak with a 244 cm−1 shoulder corresponds well with the experimentally observed 255 cm−1 feature. The calculated 260 cm−1 feature can be best described as a symmetric breathing mode involving the O1–Fe1–μ4O–Febase core moiety of the [Fe4] cluster (the ν3(A1) mode), while the 244 cm−1 feature mainly represents the asymmetric Neq–Fe1–Neq stretching mode (the ν11(E) mode). The calculated 340 cm−1 feature could be assigned to the broad 320 cm−1 feature observed experimentally, which is associated with an asymmetric Febase–μ4O–Febase bending motion involving two Fe(III) centers of the base irons (resembling the ν5(E) mode). Finally, the experimental data shows a weak broad feature centered at ∼515 cm−1. The DFT calculated 57Fe PVDOS spectrum also predicts a similar weak feature centered at 505 cm−1, which mainly involves the strong movement of μ4O either along the Fe1–O1 bond vector (calculated at 507 cm−1, the ν12(E) mode) or perpendicular to it (calculated at 502 cm−1, the ν13(E) mode). The DFT calculated spectrum of [FeIIFeIII218OFeIII]–18O shows a clear 18O isotope shift of the 505 cm−1 feature with Δν (16O/18O) ∼25 cm−1 in addition to the 18O isotope shift of the terminal FeIII–O stretching mode mentioned above (ν(FeIII–16O) = 675 cm−1 and ν(FeIII–18O) = 646 cm−1 in the calculated spectra). Such an isotope shift related to μ4O is not clearly observed for the broad 515 cm−1 feature in the experimental spectrum of [FeIIFeIII218OFeIII]–18O, which we attribute to incomplete 18O isotope labeling as evidenced by the mixed ν(FeIII–16O) and ν(FeIII–18O) observed for the terminal FeIII–O in [FeIIFeIII218OFeIII]–18O. Nevertheless, calculations to probe the effect of 16O vs.18O at the μ4O position show little effect on the terminal Fe–O (as well as Fe–OH) vibrational frequencies (see Fig. S2–S5† for all the complexes studied here), which further suggests that the stretching vibration of the terminal Fe–O moiety is well isolated from other vibrational modes. The experimentally observed and DFT calculated frequencies of selected normal modes for all four complexes studied here as well as selected bond lengths are listed in Table 1. Overall, the strong 57Fe PVDOS features in the energy range between 150 cm−1 and 400 cm−1 are mainly the results of vibrations from the Fe4 cluster core (the O1–Fe1–μ4O–Febase moiety), thus reflecting the core structure of this type of complexes.
| Normal modes | [FeIIFeIII2OFeIII]–O | [FeIIFeIII2OFeIII]–OH | [FeII2FeIIIOFeIII]–O | [FeII2FeIIIOFeIII]–OH |
|---|---|---|---|---|
| a Normal modes are based on a OFe(X3)(YZ3) type molecule (see Fig. 1 and SY). b The italic numbers are derived from DFT calculations, the frequencies are in cm−1, and the bond lengths are in Å. c The feature is too weak to be assigned in the experimental data. | ||||
| ν6 | 190 | 193 | 186 | 180 |
| 180 | 176 | 168 | 174 | |
| ν8 | 190 | 193 | 186 | 180 |
| 185 | 190 | 177 | 179 | |
| ν4 | 215 | 218 | 206 | 205 |
| 207, 212 | 206, 208 | 197, 202 | 198 | |
| ν5 | 215 | 218 | 206 | 205 |
| 214, 221 | 216, 221 | 214, 215 | 208, 216 | |
| ν3 | 255 | 260 | 246 | 252 |
| 260 | 263 | 256 | 252 | |
| ν11 | 290 | 295 | 276 | 284 |
| 300 | 290 | 272 | 275 | |
| ν13 | 515 | 525 | —c | —c |
| 507 | 520 | 555 | 580 | |
| ν14 | 665 | 570 | 640 | 520 |
| 674 | 546, 550 | 670 | 525 | |
| d(Fe1–O(H)) | 1.796(8) | 1.879(2) | 1.817(2) | 1.907(4) |
| 1.781 | 1.901 | 1.785 | 1.925 | |
| d(Fe1–μ4O) | 2.049(7) | 1.948(2) | 1.965(1) | 1.889(3) |
| 2.103 | 1.980 | 2.058 | 1.929 | |
| d avg(Fe1–Neq) | 2.090 | 2.063 | 2.098 | 2.084 |
| 2.108 | 2.097 | 2.132 | 2.114 | |
With a good understanding of the vibrational features from [FeIIFeIII2OFeIII]–O, we turn to the corresponding protonated complex, [FeIIFeIII2OFeIII]–OH. The NRVS derived 57Fe PVDOS spectrum of [FeIIFeIII2OFeIII]–OH is almost identical to that of [FeIIFeIII2OFeIII]–O (both in peak positions and relative peak intensities) in the energy range below 400 cm−1, suggesting that the replacement of a terminal oxido to a terminal hydroxido ligand has a negligible impact to the Fe4 cluster core structure (Fig. 4). Indeed, the iron–ligand bond lengths of the Fe4 cluster core in [FeIIFeIII2OFeIII]–OH exhibit minimal changes when compared with those in [FeIIFeIII2OFeIII]–O. For example, the averaged Fe1–Neq bond length is 2.06 Å in [FeIIFeIII2OFeIII]–OH and 2.09 Å in [FeIIFeIII2OFeIII]–O, and the averaged iron–ligand bond lengths of the base irons (davg(Febase–L)) are 2.118 Å, 2.076 Å, and 2.058 Å in [FeIIFeIII2OFeIII]–OH and 2.136 Å, 2.068 Å, and 2.051 Å in [FeIIFeIII2OFeIII]–O. However, changes in the energy range above 400 cm−1 can be clearly identified. Especially, the 665 cm−1 peak assigned to ν(FeIII–O) in [FeIIFeIII2OFeIII]–O is not observed in [FeIIFeIII2OFeIII]–OH, instead, a broad 16O/18O sensitive feature is observed at ∼570 cm−1 in [FeIIFeIII2OFeIII]–16OH, which is shifted to ∼535 cm−1 in [FeIIFeIII218OFeIII]–18OH (Fig. 4). This isotope sensitive feature is assigned to the vibration modes involving the terminal FeIII–OH moiety. This assignment is consistent with the bond length change in going from the Fe1–OH bond (d(Fe1–OH) = 1.879(2) Å) to the Fe1–O bond (d(Fe1–O) = 1.796(8) Å). Our DFT calculated 57Fe PVDOS spectra for [FeIIFeIII2OFeIII]–OH support such an assignment. The vibrational mode exhibiting the strongest FeIII–OH stretching motion (ν(Fe1–OH)) is calculated at 546 cm−1. In comparison, ν(Fe1–O) is calculated at 674 cm−1 for [FeIIFeIII2OFeIII]–O. This mode (ν(Fe1–OH)) is down shifted to 527 cm−1 with 18O labeling in the DFT calculated results on [FeIIFeIII218OFeIII]–18OH (Fig. 3C). It is also worth noting that ν(Fe1–μ4O) in [FeIIFeIII216OFeIII]–16OH, is calculated at 520 cm−1, which is blue shifted by 13 cm−1 when compared with the same stretching mode calculated in [FeIIFeIII216OFeIII]–16O (ν(Fe1–μ4O) = 507 cm−1). This reflects the shortening of the Fe1–μ4O bond by ∼0.1 Å in going from [FeIIFeIII2OFeIII]–O to [FeIIFeIII2OFeIII]–OH, which is most likely due to the weaker trans influence of the OH ligand that in turn strengthens the Fe1–μ4O bond. This assignment is further confirmed by the DFT calculated spectrum of [FeIIFeIII218OFeIII]–18OH, which showed an 18O isotope shift of 20 cm−1 from 520 cm−1 in [FeIIFeIII216OFeIII]–16OH to 500 cm−1 in [FeIIFeIII218OFeIII]–18OH. Compared with the reported ν(FeIII–OH) frequencies in mononuclear iron complexes, such as in [FeIII{H3buea}(OH)]− (ν(FeIII–OH) = 477 cm−1) and [FeIII{H2pout}(OH)]− (ν(FeIII–OH) = 556 cm−1), ν(FeIII–OH) observed in [FeIIFeIII2OFeIII]–OH is closer to the frequency of ν(FeIII–OH) observed in the latter mononuclear iron complex, but is much higher than that in former one. This is consistent with the FeIII–OH bond lengths found in these complexes determined by XRD. d(FeIII–OH) is 1.879(2) Å in [FeIIFeIII2OFeIII]–OH, while it is 1.893(2) Å in [FeIII{H2pout}(OH)]− and 1.931(2) Å in [FeIII{H3buea}(OH)]−. The shorter FeIII–OH bond in [FeIIFeIII2OFeIII]–OH or [FeIII{H2pout}(OH)]− is due to the effect of an additional Fe–OH⋯X H-bond (X = N in [FeIIFeIII2OFeIII]–OH and X = O in [FeIII{H2pout}(OH)]−), which weakens the hydroxide O–H bond strength since the iron-bound hydroxide is the H-bond donor, leading to a strengthening of the Fe–O bond. This additional H-bond where the Fe(III)–OH as the H-bond donor is absent in [FeIII{H3buea}(OH)]−.
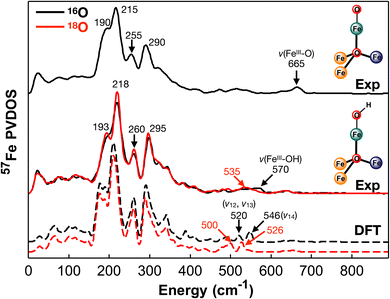 | ||
| Fig. 4 57Fe PVDOS spectra of [FeIIFeIII2OFeIII]–O and [FeIIFeIII2OFeIII]–OH. Top: The 57Fe PVDOS experimental spectra of [FeIIFeIII2OFeIII]–O with the observed frequencies of the strong spectral features highlighted; middle: the 57Fe PVDOS experimental spectra of [FeIIFeIII2OFeIII]–OH (black trace) and the 18O–labeled complex (red trace) with the key observed frequencies indicated; bottom: the DFT calculated spectra of [FeIIFeIII2OFeIII]–OH (black trace) and the 18O–labeled complex (red trace) with 18O-sensitive features indicated. The simplified structures of these two complexes are also shown (see Fig. 1 for the atom color code definition). See Fig. S3† for additional comparisons between the experimental and DFT calculated spectra. | ||
To further explore the effect of redox event of neighboring iron centers (the three base iron sites, Fe2, Fe3, and Fe4) on the vibrational properties of the terminal FeIII–O/OH moiety and the overall [Fe4] cluster, we recorded NRVS data on [FeII2FeIIIOFeIII]–O and [FeII2FeIIIOFeIII]–OH complexes, the one-electron reduced congeners of [FeIIFeIII2OFeIII]–O/OH. All the 57Fe PVDOS features observed for [FeII2FeIIIOFeIII]–O and [FeII2FeIIIOFeIII]–OH are shifted to lower energies than those of the corresponding one-electron oxidized clusters (Fig. 2, 5 and Table 1), [FeIIFeIII2OFeIII]–O and [FeIIFeIII2OFeIII]–OH respectively. Specifically, the strong features in the energy range of 150 cm−1 to 400 cm−1 are all red shifted by ∼10–20 cm−1 in going from the oxidized complexes to the reduced complexes while maintaining the relative intensities of individual peaks (Fig. 2, 5 and Table 1). This strongly suggests that the overall [Fe4] cluster core structure is maintained while the iron–ligand bond lengths, particularly related to the base irons (Fe2, Fe3, or Fe4), are elongated due to reduction, thus resulting in the overall red-shift of the vibrational features. This implication is supported by the structures of these complexes determined by XRD. In going from [FeIIFeIII2OFeIII]–O to [FeII2FeIIIOFeIII]–O, davg(Febase–L) (the averaged Fe2/3/4 – ligand bond length) is elongated, particularly for davg(Fe3–L), which changes from 2.068 Å to 2.112 Å, and for davg(Fe4–L), which changes from 2.051 Å to 2.093 Å. While the corresponding averaged bond lengths are also elongated in going from [FeIIFeIII2OFeIII]–OH to [FeII2FeIIIOFeIII]–OH, specifically davg(Fe3–L) changes from 2.058 Å to 2.124 Å. However, davg(Fe1–Neq) (the averaged Fe1–Neq bond lengths) only shows minor changes. In [FeIIFeIII2OFeIII]–O and [FeII2FeIIIOFeIII]–O, dav(Fe1–N) is 2.090 Å and 2.108 Å respectively, while in [FeIIFeIII2OFeIII]–OH and [FeII2FeIIIOFeIII]–OH, davg(Fe1–N) is 2.098 Å and 2.084 Å respectively. This implies that the reduction does not directly impact the apical iron (Fe1), which is still in the FeIII state. All these observations are consistent with the previous report indicating that reduction happens at the base iron sites (Fe2, Fe3, or Fe4).74 Although such a reduction does not impact the Fe1–Neq bond lengths, it strongly impacts the Fe1–μ4O bond. The weakening of Fe2/3/4–μ4O bonds due to reduction is in turn increasing the bonding interactions between Fe1 and μ4O. A 0.08 Å contraction of d(Fe1–μ4O) (2.049(7) Å to 1.965(1) Å) is observed in the crystal structures from [FeIIFeIII2OFeIII]–O to [FeII2FeIIIOFeIII]–O, while a 0.06 Å contraction of the same bond (1.948(2) Å to 1.889(3) Å) is observed in going from [FeIIFeIII2OFeIII]–OH to [FeII2FeIIIOFeIII]–OH. The strengthening of Fe1–μ4O generates a trans influence to weaken the Fe1–O/OH bonding interactions, which show a ∼0.02–0.03 Å elongation in the redox pair of the oxido complexes and in the redox pair of the hydroxido complexes. Accordingly, the frequencies of ν(FeIII–O) and ν(FeIII–OH) show clear red shifts upon reduction. ν(FeIII–O) is at 665 cm−1 (DFT: 674 cm−1) in [FeIIFeIII2OFeIII]–O, but at ∼640 cm−1 (DFT: 670 cm−1) in [FeII2FeIIIOFeIII]–O (Fig. 5 top), which is red shifted to 608 cm−1 (DFT: 642 cm−1) in [FeII2FeIII18OFeIII]–18O (Fig. S2†). ν(FeIII–OH) is at 570 cm−1 (DFT: 546 cm−1) in [FeIIFeIII2OFeIII]–OH, but at ∼520 cm−1 (DFT: 525 cm−1) in [FeII2FeIIIOFeIII]–OH (Fig. 5 bottom), which is red shifted to ∼500 cm−1 (DFT: 492 cm−1) in [FeII2FeIII18OFeIII]–18OH (Fig. S2†). In summary, the effect of the redox event at distal iron centers is to modulate the bonding interactions between Fe and μ4O, thus exerting a trans influence to the apical Fe center resulting in changes of the bonding interactions between Fe1 and the terminal oxido or hydroxido ligand. In addition, the 57Fe PVDOS features between 150 cm−1 and 400 cm−1 are good overall oxidation state indicators for the [Fe4]–O/OH core (Fig. 2 and 5).
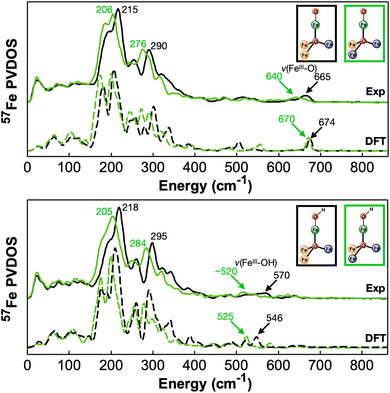 | ||
| Fig. 5 57Fe PVDOS experimental and DFT calculated spectra from different [Fe4] cluster complexes in two different oxidation states. Top: The experimental (top) and DFT calculated (bottom) spectra of [FeIIFeIII2OFeIII]–O (black) and [FeII2FeIIIOFeIII]–O (green). Bottom: The experimental (top) and DFT calculated (bottom) spectra of [FeIIFeIII2OFeIII]–OH (black) and [FeII2FeIIIOFeIII]–OH (green). The frequencies of the major spectral features, including ν(FeIII–O) and ν(FeIII–OH) in these complexes, are indicated in the figure. The simplified structures of these two complexes are also shown (see Fig. 1 for the atom color code definition). See Fig. S4 and S5† for additional comparisons between the experimental and DFT calculated spectra. | ||
To put the results of the current study into a broader context related to the iron–oxido/hydroxido moieties, we generated the correlation plot between the Fe–O(H) bond length and the NRVS-derived ν(FeIII–O(H)) frequencies from the four complexes studied here, together with the previously studied mononuclear iron complexes (Fig. 6). Since the DFT method we used in this study reproduced the experimental NRVS data well, we also extended the calculations to predict the vibrational features of the three other [Fe4] clusters previously published in this cluster series, namely the all ferric clusters, [FeIII3OFeIII]–O and [FeIII3OFeIII]–OH, and the most reduced cluster, [FeII3OFeIII]–OH. The calculated ν(FeIII–O(H)) frequencies of these three complexes are also included in Fig. 6. The present study significantly expands the data available on terminal FeIII–O(H) and FeIV![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O species. The plot clearly indicates that the Fe–O(H) bond lengths and the corresponding ν(FeIII–O(H)) frequencies of both mononuclear and tetranuclear iron–oxido/hydroxido complexes follow a similar trend and can be described by Badger's rule (Fig. 6 and S6†).77,80 This suggests that Fe–O/OH stretching vibrations in these complexes can be well described as the stretching vibration of a diatomic moiety that is relatively isolated from the rest of the molecules. But at the same time the correlation shown in Fig. 6 also suggests that Fe–O/OH bond length and stretching vibration can be strongly influenced by the surrounding environment either by the direct modulation via hydrogen bond interactions in the mononuclear Fe–O/OH complexes or by the indirect modulation via remote metal redox events in the [Fe3OFeIII]–O/OH complexes, and these two chemical strategies generate a similar effect towards the bond length and the stretching vibration of the Fe–O/OH moiety.
O species. The plot clearly indicates that the Fe–O(H) bond lengths and the corresponding ν(FeIII–O(H)) frequencies of both mononuclear and tetranuclear iron–oxido/hydroxido complexes follow a similar trend and can be described by Badger's rule (Fig. 6 and S6†).77,80 This suggests that Fe–O/OH stretching vibrations in these complexes can be well described as the stretching vibration of a diatomic moiety that is relatively isolated from the rest of the molecules. But at the same time the correlation shown in Fig. 6 also suggests that Fe–O/OH bond length and stretching vibration can be strongly influenced by the surrounding environment either by the direct modulation via hydrogen bond interactions in the mononuclear Fe–O/OH complexes or by the indirect modulation via remote metal redox events in the [Fe3OFeIII]–O/OH complexes, and these two chemical strategies generate a similar effect towards the bond length and the stretching vibration of the Fe–O/OH moiety.
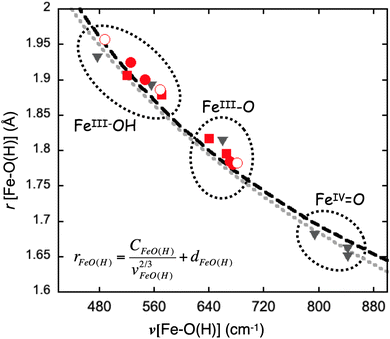 | ||
| Fig. 6 The correlation plot between the Fe–O(H) bond length and its stretching frequency. The experimental data points belonging to the [Fe3OFeIII]–O/OH complexes studied in the current work are indicated as red square boxes and the experimental data points belonging to the mononuclear Fe–O/OH complexes reported previously are indicated as black triangles, respectively. The DFT obtained data points of the [Fe3OFeIII]–O/OH complexes experimentally studied in the current work are indicated as the red dots. The DFT calculated data points of three additional [Fe4]–O/OH complexes, [FeIII3OFeIII]–O, [FeIII3OFeIII]–OH, and [FeII3OFeIII]–OH, that are not experimentally studied in this work are shown as red circles. The black curve represents a simulation using Badger's rule with the expression shown in the figure. The constants used are: CFeO(H) = 56.692, dFeO(H) = 1.038, which are taken from our previous study.77 The grey curve presents a simulation by using the results from Tolman and coworkers.80 | ||
Beyond the structural perturbation of the terminal Fe–O/OH moieties, the basal iron redox event of the [Fe3OFeIII]–O/OH complexes also impact the chemical reactivity of these moieties. Recent reactivity studies of metal–oxido (M–O) species towards C–H activation in enzymes and bioinspired model complexes strongly indicate that ground state thermodynamics of the M–O species, e.g. pKa, reduction potential, and the bond dissociation energy (BDE) of the metal bound hydroxide in the corresponding metal–hydroxido (M–OH) species, play key roles in determining M–O reactivity.81–90 Particularly, a number of model complex studies suggest that the basicity of M–O units (pKa of M–OH) is the determining factor in controlling the asynchronous proton-coupled electron transfer (PCET) process during C–H activation, where proton transfer (PT) is dominant within the transition state over electron transfer (ET).45,91–98 In a previous study by some of us, it has been shown that the C–H activation reactivity demonstrated by the [Fe3OFeIII]–O complex following a similar concerted PT-driven process. Furthermore, the redox state change of the basal iron sites that perturbs the bond length and stretching vibrational frequency of the terminal FeIII–O/OH moiety also modulates the thermodynamic properties, thus the chemical reactivity of this moiety. Specifically, the redox state change of the basal iron sites leads to modulation of the O–H BDE by as much as ∼12 kcal mol−1 in going from [FeII3OFeIII]–OH to [FeIII2FeIIOFeIII]–OH (the O–H BDE of [FeII3OFeIII]–OH was estimated to be 72 kcal mol−1 while it was estimated to be 84 kcal mol−1 for [FeIII2FeIIOFeIII]–OH).74 Thus the direct measurements of Fe–O/OH vibrational frequency by using 57Fe NRVS provide an useful tool to quantitatively evaluate the structural impact of redox event from the remote iron sites of [Fe3OFeIII]–O complexes and help construct the structure–reactivity correlation for these clusters.
Conclusion
In this study, we have carried out a detailed vibrational analysis on a series of [Fe4] clusters by using 57Fe NRVS combined with DFT calculations. These clusters bear a terminal Fe(III)–O/OH moiety at the apical position and three additional iron centers forming the base of the cluster. The three basal iron centers are connected with the apical iron center via a μ4-oxido ligand. Specifically, the complexes studied are [LFeIIFeIII2O(PzNHtBu)3FeIII(O)]+ ([FeIIFeIII2OFeIII]–O) and [LFeIIFeIII2O(PzNHtBu)3FeIII(OH)]2+ ([FeIIFeIII2OFeIII]–OH), and their corresponding one electron reduced clusters, [LFeII2FeIIIO(PzNHtBu)3FeIII(O)] ([FeII2FeIIIOFeIII]–O) and [LFeII2FeIIIO(PzNHtBu)3FeIII(O)]+ ([FeII2FeIIIOFeIII]–OH). Strong NRVS-derived 57Fe PVDOS features are observed in the frequency range of 150–400 cm−1, which reflects the vibrational features of the overall [Fe4] cluster core and can be used as a good indicator of the overall oxidation state of these clusters. In the frequency range greater than 400 cm−1, the stretching vibrations of the terminal Fe(III)–O (v[Fe(III)–O]) and Fe(III)–OH (v[Fe(III)–OH]) are also observed. For [FeIIFeIII2OFeIII]–O, v[Fe(III)–O] is observed at 660 cm−1, while for [FeIIFeIII2OFeIII]–OH, v[Fe(III)–OH] is observed at 570 cm−1. These frequency values are highly similar to v[Fe(III)–O] and v[Fe(III)–OH] observed in mononuclear Fe(III)–O/OH complexes, suggesting that v[Fe(III)–O] and v[Fe(III)–OH] are relatively isolated vibrations even in these [Fe4] clusters and are essentially determined by Fe–O or Fe–OH bond lengths via Badger's rule as shown in Fig. 6. However, the redox event in the remote basal iron centers clearly modulates the terminal Fe(III)–O/OH vibrations (as well as the bond lengths) as shown by the red shifts of v[Fe(III)–O] and v[Fe(III)–OH] (25 cm−1 and 40 cm−1 respectively) in [FeII2FeIIIOFeIII]–O/OH, the one-electron reduced complexes of [FeIIFeIII2OFeIII]–O/OH. Such a remote structural modulation is also suggested by the shifts observed in the 150–400 cm−1 region and is due to the weakening of the bonding interactions between μ4O and the three base iron sites upon reduction, which leads to the strengthening of the apical Fe–μ4O bond and the weakening of the terminal Fe–O/OH bond via a trans influence. Thus, 57Fe NRVS data provided comprehensive vibrational characterizations of the structural perturbations of the terminal Fe–O/OH moieties in these [Fe4] clusters due to the redox change of the remote basal iron sites. Overall, this indirect modulation by remote iron redox event in the [Fe4] clusters and the direct modulation via hydrogen bond interactions as reported in mononuclear Fe–O/OH complexes generate a similar effect on changing the bond length and the stretching vibration of the terminal Fe–O/OH moieties, which further modify their thermodynamic properties (such as reduction potential, pKa, and O–H BDE) and chemical reactivities as previously reported for these complexes.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization: T. A., Y. G.; methodology, J. X., C. R., B. L., M. Y. H., J. Z., E. E. A., Y. A., Y. G.; investigation: J. X.; C. R.; T. A., Y. G.; formal analysis: J. X.; C. R., T. A., Y. G.; writing – original draft: J. X., Y. G.; writing – review & editing, J. X., C. R., T. A., Y. G.; funding acquisition: T. A., Y. G.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the National Institute of Health (NIH GM125924 to Y. G., GM127588 to W. c. C.) and the NSF (CHE-1905320 to T. A.) for funding. The NRVS data were recorded at the Advanced Photon Source (APS), Argonne National Laboratory (proposal GUP-60939, GUP-68490, and GUP-73059). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. In addition, this research used Bridges-2 at Pittsburgh Supercomputing Center through allocation [CHE-200003] from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296.References
- E. I. Solomon, T. C. Brunold, M. I. Davis, J. N. Kemsley, S.-K. Lee, N. Lehnert, F. Neese, A. J. Skulan, Y.-S. Yang and J. Zhou, Geometric and Electronic Structure/Function Correlations in Non-Heme Iron Enzymes, Chem. Rev., 2000, 100, 235–350 CrossRef CAS.
- M. Costas, M. P. Mehn, M. P. Jensen and L. Que, Dioxygen Activation at Mononuclear Nonheme Iron Active Sites: Enzymes, Models, and Intermediates, Chem. Rev., 2004, 104, 939–986 CrossRef CAS.
- M. M. Abu-Omar, A. Loaiza and N. Hontzeas, Reaction Mechanisms of Mononuclear Non-heme Iron Oxygenases, Chem. Rev., 2005, 105, 2227–2252 CrossRef CAS.
- W. Nam, Dioxygen Activation by Metalloenzymes and Models, Acc. Chem. Res., 2007, 40, 465 CrossRef CAS.
- L. Que and W. B. Tolman, Biologically inspired oxidation catalysis, Nature, 2008, 455, 333–340 CrossRef CAS PubMed.
- E. G. Kovaleva and J. D. Lipscomb, Versatility of biological non-heme Fe(II) centers in oxygen activation reactions, Nat. Chem. Biol., 2008, 4, 186–193 CrossRef CAS PubMed.
- T. L. Poulos, Heme enzyme structure and function, Chem. Rev., 2014, 114, 3919–3962 CrossRef CAS.
- J. Yano and V. Yachandra, Mn4Ca Cluster in Photosynthesis: Where and How Water is Oxidized to Dioxygen, Chem. Rev., 2014, 114, 4175–4205 CrossRef CAS PubMed.
- S. Hematian, I. Garcia-Bosch and K. D. Karlin, Synthetic Heme/Copper Assemblies: Toward an Understanding of Cytochrome c Oxidase Interactions with Dioxygen and Nitrogen Oxides, Acc. Chem. Res., 2015, 48, 2462–2474 CrossRef CAS.
- J. D. Blakemore, R. H. Crabtree and G. W. Brudvig, Molecular Catalysts for Water Oxidation, Chem. Rev., 2015, 115, 12974–13005 CrossRef CAS PubMed.
- F. Nastri, M. Chino, O. Maglio, A. Bhagi-Damodaran, Y. Lu and A. Lombardi, Design and engineering of artificial oxygen-activating metalloenzymes, Chem. Soc. Rev., 2016, 45, 5020–5054 RSC.
- W. Zhang, W. Lai and R. Cao, Energy-Related Small Molecule Activation Reactions: Oxygen Reduction and Hydrogen and Oxygen Evolution Reactions Catalyzed by Porphyrin- and Corrole-Based Systems, Chem. Rev., 2017, 117, 3717–3797 CrossRef CAS PubMed.
- L. Que, 60 years of dioxygen activation, JBIC, J. Biol. Inorg. Chem., 2017, 22, 171–173 CrossRef CAS.
- X. Huang and J. T. Groves, Oxygen Activation and Radical Transformations in Heme Proteins and Metalloporphyrins, Chem. Rev., 2018, 118, 2491–2553 CrossRef CAS.
- A. J. Jasniewski and L. Que, Dioxygen Activation by Nonheme Diiron Enzymes: Diverse Dioxygen Adducts, High-Valent Intermediates, and Related Model Complexes, Chem. Rev., 2018, 118, 2554–2592 CrossRef CAS.
- J. L. Lee, D. L. Ross, S. K. Barman, J. W. Ziller and A. S. Borovik, C–H Bond Cleavage by Bioinspired Nonheme Metal Complexes, Inorg. Chem., 2021, 60, 13759–13783 CrossRef CAS.
- M. Guo, T. Corona, K. Ray and W. Nam, Heme and Nonheme High-Valent Iron and Manganese Oxo Cores in Biological and Abiological Oxidation Reactions, ACS Cent. Sci., 2019, 5, 13–28 CrossRef CAS PubMed.
- A. Gunay and K. H. Theopold, C–H Bond Activations by Metal Oxo Compounds, Chem. Rev., 2010, 110, 1060–1081 CrossRef CAS PubMed.
- A. S. Borovik, Role of metal-oxo complexes in the cleavage of C-H bonds, Chem. Soc. Rev., 2011, 40, 1870–1874 RSC.
- J. Limburg, J. S. Vrettos, L. M. Liable-Sands, A. L. Rheingold, R. H. Crabtree and G. W. Brudvig, A Functional Model for O-O Bond Formation by the O2-Evolving Complex in Photosystem II, Science, 1999, 283, 1524–1527 CrossRef CAS.
- A. I. Nguyen, M. S. Ziegler, P. Oña-Burgos, M. Sturzbecher-Hohne, W. Kim, D. E. Bellone and T. D. Tilley, Mechanistic Investigations of Water Oxidation by a Molecular Cobalt Oxide Analogue: Evidence for a Highly Oxidized Intermediate and Exclusive Terminal Oxo Participation, J. Am. Chem. Soc., 2015, 137, 12865–12872 CrossRef CAS PubMed.
- B. M. Hunter, N. B. Thompson, A. M. Müller, G. R. Rossman, M. G. Hill, J. R. Winkler and H. B. Gray, Trapping an Iron(VI) Water-Splitting Intermediate in Nonaqueous Media, Joule, 2018, 2, 747–763 CrossRef CAS.
- S. H. Kim, H. Park, M. S. Seo, M. Kubo, T. Ogura, J. Klajn, D. T. Gryko, J. S. Valentine and W. Nam, Reversible O–O Bond Cleavage and Formation between Mn(IV)-Peroxo and Mn(V)-Oxo Corroles, J. Am. Chem. Soc., 2010, 132, 14030–14032 CrossRef CAS.
- M. Y. M. Pau, J. D. Lipscomb and E. I. Solomon, Substrate activation for O2 reactions by oxidized metal centers in biology, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 18355–18362 CrossRef CAS.
- R. H. Holm and E. I. Solomon, Introduction: Bioinorganic enzymology II, Chem. Rev., 2014, 114, 4039–4040 CrossRef CAS PubMed.
- Y. Lu, S. M. Berry and T. D. Pfister, Engineering novel metalloproteins: design of metal-binding sites into native protein scaffolds, Chem. Rev., 2001, 101, 3047–3080 CrossRef CAS.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. Tian, I. Petrik, A. Bhagi and Y. Lu, Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers, Chem. Rev., 2014, 114, 4366–4369 CrossRef CAS PubMed.
- F. Yu, V. M. Cangelosi, M. L. Zastrow, M. Tegoni, J. S. Plegaria, A. G. Tebo, C. S. Mocny, L. Ruckthong, H. Qayyum and V. L. Pecoraro, Protein design: toward functional metalloenzymes, Chem. Rev., 2014, 114, 3495–3578 CrossRef CAS PubMed.
- S. A. Cook and A. S. Borovik, Molecular Designs for Controlling the Local Environments around Metal Ions, Acc. Chem. Res., 2015, 48, 2407–2414 CrossRef CAS PubMed.
- S. A. Cook, E. A. Hill and A. S. Borovik, Lessons from Nature: A Bio-Inspired Approach to Molecular Design, Biochemistry, 2015, 54, 4167–4180 CrossRef CAS PubMed.
- P. Hosseinzadeh, N. M. Marshall, K. N. Chacón, Y. Yu, M. J. Nilges, S. Y. New, S. A. Tashkov, N. J. Blackburn and Y. Lu, Design of a single protein that spans the entire 2-V range of physiological redox potentials, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 262–267 CrossRef CAS.
- B. A. Springer, S. G. Sligar, J. S. Olson and G. N. Phillips, Mechanisms of Ligand Recognition in Myoglobin, Chem. Rev., 1994, 94, 699–714 CrossRef CAS.
- J. C. Kendrew, G. Bodo, H. M. Dintzis, R. G. Parrish, H. Wyckoff and D. C. Phillips, A three-dimensional model of the myoglobin molecule obtained by X-ray analysis, Nature, 1958, 181, 662–666 CrossRef CAS PubMed.
- J. C. Kendrew, R. E. Dickerson, B. E. Strandberg, R. G. Hart, D. R. Davies, D. C. Phillips and V. C. Shore, Structure of myoglobin: a three-dimensional Fourier synthesis at 2 Å. Resolution, Nature, 1960, 185, 422–427 CrossRef CAS PubMed.
- M. F. Perutz, M. G. Rossmann, A. F. Cullis, H. Muirhead, G. Will and A. C. T. North, Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-Å. Resolution, obtained by X-ray analysis, Nature, 1960, 185, 416–422 CrossRef CAS.
- B. Shaanan, The iron–oxygen bond in human oxyhaemoglobin, Nature, 1982, 296, 683–684 CrossRef CAS.
- J. Yang, A. P. Kloek, D. E. Goldberg and F. S. Mathews, The structure of Ascaris hemoglobin domain I at 2.2 Å resolution: molecular features of oxygen avidity, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4224–4228 CrossRef CAS.
- T. L. Poulos, S. S. T. Freer, R. A. Alden, S. L. Edwards, U. Skogland, B. Eriksson, N. Xuong, T. Yonetani and J. Kraut, The Crystal Structure of Cytochrome c Peroxidase, J. Biol. Chem., 1980, 2, 575–581 CrossRef.
- T. L. Poulos and J. Kraut, The Stereochemistry of Peroxidase Catalysis, J. Biol. Chem., 1980, 255, 8199–8205 CrossRef CAS PubMed.
- I. Schlichting, J. Bernendzen, K. Chu, A. M. Stock, S. A. Maves, D. E. Benson, R. M. Sweet, D. Ringe, G. A. Petsko and S. G. Sligar, The Catalytic Pathway of Cytochrome P450cam at Atomic Resolution, Science, 2000, 287, 1615–1622 CrossRef CAS.
- S. Nagano and T. L. Poulos, Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam: implications for the dioxygen activation mechanism, J. Biol. Chem., 2005, 280, 31659–31663 CrossRef CAS.
- A. J. Fielding, J. D. Lipscomb and L. Que, A Two-Electron-Shell Game: Intermediates of the Extradiol-cleaving Catechol Dioxygenases, JBIC, J. Biol. Inorg. Chem., 2014, 19, 491–504 CrossRef CAS PubMed.
- E. G. Kovaleva, M. S. Rogers and J. D. Lipscomb, Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200, Biochemistry, 2015, 54, 5329–5339 CrossRef CAS.
- S. K. Barman, M.-Y. Yang, T. H. Parsell, M. T. Green and A. S. Borovik, Semiempirical method for examining asynchronicity in metal–oxido-mediated C–H bond activation, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2108648118 CrossRef CAS PubMed.
- S. K. Barman, J. R. Jones, C. Sun, E. A. Hill, J. W. Ziller and A. S. Borovik, Regulating the Basicity of Metal–Oxido Complexes with a Single Hydrogen Bond and Its Effect on C–H Bond Cleavage, J. Am. Chem. Soc., 2019, 141, 11142–11150 CrossRef CAS.
- E. M. Matson, Y. J. Park and A. R. Fout, Facile Nitrite Reduction in a Non-heme Iron System: Formation of an Iron(III)-Oxo, J. Am. Chem. Soc., 2014, 136, 17398–17401 CrossRef CAS.
- Y. J. Park, E. M. Matson, M. J. Nilges and A. R. Fout, Exploring Mn–O bonding in the context of an electronically flexible secondary coordination sphere: synthesis of a Mn(III)–oxo, Chem. Commun., 2015, 51, 5310–5313 RSC.
- Z. Gordon, M. J. Drummond, E. M. Matson, J. A. Bogart, E. J. Schelter, R. L. Lord and A. R. Fout, Tuning the Fe(II/III) Redox Potential in Nonheme Fe(II)-Hydroxo Complexes through Primary and Secondary Coordination Sphere Modifications, Inorg. Chem., 2017, 56, 4852–4863 CrossRef CAS.
- M. J. Drummond, C. L. Ford, D. L. Gray, C. V. Popescu and A. R. Fout, Radical Rebound Hydroxylation Versus H-Atom Transfer in Non-Heme Iron(III)-Hydroxo Complexes: Reactivity and Structural Differentiation, J. Am. Chem. Soc., 2019, 141, 6639–6650 CrossRef CAS PubMed.
- Y. J. Park, M. N. Peñas-Defrutos, M. J. Drummond, Z. Gordon, O. R. Kelly, I. J. Garvey, K. L. Gullett, M. García-Melchor and A. R. Fout, Secondary Coordination Sphere Influences the Formation of Fe(III)-O or Fe(III)-OH in Nitrite Reduction: A Synthetic and Computational Study, Inorg. Chem., 2022, 61, 8182–8192 CrossRef CAS PubMed.
- H. S. Soo, A. C. Komor, A. T. Iavarone and C. J. Chang, A Hydrogen-Bond Facilitated Cycle for Oxygen Reduction by an Acid- and Base-Compatible Iron Platform, Inorg. Chem., 2009, 48, 10024–10035 CrossRef CAS.
- V. Yadav, J. B. Gordon, M. A. Siegler and D. P. Goldberg, Dioxygen-Derived Nonheme Mononuclear Fe III (OH) Complex and Its Reactivity with Carbon Radicals, J. Am. Chem. Soc., 2019, 141, 10148–10153 CrossRef CAS.
- T. Devi, Y.-M. Lee, W. Nam and S. Fukuzumi, Metal ion-coupled electron-transfer reactions of metal-oxygen complexes, Coord. Chem. Rev., 2020, 410, 213219 CrossRef CAS.
- V. F. Oswald, J. L. Lee, S. Biswas, A. C. Weitz, K. Mittra, R. Fan, J. Li, J. Zhao, M. Y. Hu, E. E. Alp, E. L. Bominaar, Y. Guo, M. T. Green, M. P. Hendrich and A. S. Borovik, Effects of Noncovalent Interactions on High-Spin Fe(IV)–Oxido Complexes, J. Am. Chem. Soc., 2020, 142, 11804–11817 CrossRef CAS PubMed.
- S. Banerjee, A. Draksharapu, P. M. Crossland, R. Fan, Y. Guo, M. Swart and L. Que, Sc3+-Promoted O–O Bond Cleavage of a (μ-1,2-Peroxo)diiron(III) Species Formed from an Iron(II) Precursor and O2 to Generate a Complex with an FeIV2(μ-O)2 Core, J. Am. Chem. Soc., 2020, 142, 4285–4297 CrossRef CAS PubMed.
- Y. Li, S. Handunneththige, J. Xiong, Y. Guo, M. R. Talipov and D. Wang, Opening the CoIII,IV2(μ-O)2 Diamond Core by Lewis Bases Leads to Enhanced C–H Bond Cleaving Reactivity, J. Am. Chem. Soc., 2020, 142, 21670–21678 CrossRef CAS.
- D. Lionetti, S. Suseno, E. Y. Tsui, L. Lu, T. A. Stich, K. M. Carsch, R. J. Nielsen, W. A. Goddard, R. D. Britt and T. Agapie, Effects of Lewis Acidic Metal Ions (M) on Oxygen-Atom Transfer Reactivity of Heterometallic Mn3MO4 Cubane and Fe3MO(OH) and Mn3MO(OH) Clusters, Inorg. Chem., 2019, 58, 2336–2345 CrossRef CAS PubMed.
- D. B. Rice, E. N. Grotemeyer, A. M. Donovan and T. A. Jackson, Effect of Lewis Acids on the Structure and Reactivity of a Mononuclear Hydroxomanganese(III) Complex, Inorg. Chem., 2020, 59, 2689–2700 CrossRef CAS PubMed.
- J. P. McEvoy and G. W. Brudvig, Water-Splitting Chemistry of Photosystem II, Chem. Rev., 2006, 106, 4455–4483 CrossRef CAS PubMed.
- W. Lubitz, H. Ogata, O. Rüdiger and E. Reijerse, Hydrogenases, Chem. Rev., 2014, 114, 4081–4148 CrossRef CAS.
- A. Volbeda, M.-H. Charon, C. Piras, E. C. Hatchikian, M. Frey and J. C. Fontecilla-Camps, Crystal structure of the nickel–iron hydrogenase from Desulfovibrio gigas, Nature, 1995, 373, 580–587 CrossRef CAS PubMed.
- Y. Nicolet, C. Piras, P. Legrand, C. E. Hatchikian and J. C. Fontecilla-Camps, Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center, Structure, 1999, 7, 13–23 CrossRef CAS PubMed.
- J. W. Peters, W. N. Lanzilotta, B. J. Lemon and L. C. Seefeldt, X-ray Crystal Structure of the Fe-Only Hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom Resolution, Science, 1998, 282, 1853–1858 CrossRef CAS PubMed.
- H. Dobbek, V. Svetlitchnyi, L. Gremer, R. Huber and O. Meyer, Crystal Structure of a Carbon Monoxide Dehydrogenase Reveals a [Ni-4Fe-5S] Cluster, Science, 2001, 293, 1281–1285 CrossRef CAS PubMed.
- S. Ferguson-Miller and G. T. Babcock, Heme/Copper Terminal Oxidases, Chem. Rev., 1996, 96, 2889–2908 CrossRef CAS PubMed.
- T. Tsukihara, H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono and S. Yoshikawa, Structures of Metal Sites of Oxidized Bovine Heart Cytochrome c Oxidase at 2.8 Å, Science, 1995, 269, 1069–1074 CrossRef CAS PubMed.
- B. M. Hoffman, D. Lukoyanov, Z.-Y. Yang, D. R. Dean and L. C. Seefeldt, Mechanism of Nitrogen Fixation by Nitrogenase: The Next Stage, Chem. Rev., 2014, 114, 4041–4062 CrossRef CAS PubMed.
- T. Spatzal, M. Aksoyoglu, L. Zhang, S. L. a. Andrade, E. Schleicher, S. Weber, D. C. Rees and O. Einsle, Evidence for Interstitial Carbon in Nitrogenase FeMo Cofactor, Science, 2011, 334, 940 CrossRef CAS.
- O. Einsle, F. A. Tezcan, S. L. A. Andrade, B. Schmid, M. Yoshida, J. B. Howard and D. C. Rees, Nitrogenase MoFe-Protein at 1.16 Å Resolution: A Central Ligand in the FeMo-Cofactor, Science, 2002, 297, 1696–1700 CrossRef CAS.
- G. de Ruiter, N. B. Thompson, D. Lionetti and T. Agapie, Nitric Oxide Activation by Distal Redox Modulation in Tetranuclear Iron Nitrosyl Complexes, J. Am. Chem. Soc., 2015, 137, 14094–14106 CrossRef CAS PubMed.
- G. de Ruiter, K. M. Carsch, S. Gul, R. Chatterjee, N. B. Thompson, M. K. Takase, J. Yano and T. Agapie, Accelerated Oxygen Atom Transfer and C–H Bond Oxygenation by Remote Redox Changes in Fe3Mn-Iodosobenzene Adducts, Angew. Chem., Int. Ed., 2017, 56, 4772–4776 CrossRef CAS.
- C. H. Arnett, M. J. Chalkley and T. Agapie, A Thermodynamic Model for Redox-Dependent Binding of Carbon Monoxide at Site-Differentiated, High Spin Iron Clusters, J. Am. Chem. Soc., 2018, 140, 5569–5578 CrossRef CAS PubMed.
- C. J. Reed and T. Agapie, Thermodynamics of Proton and Electron Transfer in Tetranuclear Clusters with Mn–OH2/OH Motifs Relevant to H2O Activation by the Oxygen Evolving Complex in Photosystem II, J. Am. Chem. Soc., 2018, 140, 10900–10908 CrossRef CAS PubMed.
- C. J. Reed and T. Agapie, A Terminal Fe III–Oxo in a Tetranuclear Cluster: Effects of Distal Metal Centers on Structure and Reactivity, J. Am. Chem. Soc., 2019, 141, 9479–9484 CrossRef CAS.
- C. E. MacBeth, A. P. Golombek, V. G. Young, C. Yang, K. Kuczera, M. P. Hendrich and A. S. Borovik, O2 Activation by Nonheme Iron Complexes: A Monomeric Fe(III)-Oxo Complex Derived From O2, Science, 2000, 289, 938–941 CrossRef CAS PubMed.
- C. E. MacBeth, R. Gupta, K. R. Mitchell-Koch, V. G. Young, G. H. Lushington, W. H. Thompson, M. P. Hendrich and A. S. Borovik, Utilization of Hydrogen Bonds to Stabilize M–O(H) Units: Synthesis and Properties of Monomeric Iron and Manganese Complexes with Terminal Oxo and Hydroxo Ligands, J. Am. Chem. Soc., 2004, 126, 2556–2567 CrossRef CAS PubMed.
- A. C. Weitz, E. A. Hill, V. F. Oswald, E. L. Bominaar, A. S. Borovik, M. P. Hendrich and Y. Guo, Probing Hydrogen Bonding Interactions to Iron-Oxido/Hydroxido Units by 57Fe Nuclear Resonance Vibrational Spectroscopy, Angew. Chem., Int. Ed., 2018, 57, 16010–16014 CrossRef CAS PubMed.
- E. Andris, R. Navrátil, J. Jašík, M. Puri, M. Costas, L. Que and J. Roithová, Trapping Iron(III)–Oxo Species at the Boundary of the “Oxo Wall”: Insights into the Nature of the Fe(III)–O Bond, J. Am. Chem. Soc., 2018, 140, 14391–14400 CrossRef CAS PubMed.
- C. J. Reed, Activation of Nitric Oxide and Water by Transition Metal Clusters Relevant to Active Sites in Biology, California Institute of Technology, 2019 Search PubMed.
- A. D. Spaeth, N. L. Gagnon, D. Dhar, G. M. Yee and W. B. Tolman, Determination of the Cu(III)–OH Bond Distance by Resonance Raman Spectroscopy Using a Normalized Version of Badger's Rule, J. Am. Chem. Soc., 2017, 139, 4477–4485 CrossRef CAS PubMed.
- J. J. Warren, T. A. Tronic and J. M. Mayer, Thermochemistry of Proton-Coupled Electron Transfer Reagents and its Implications, Chem. Rev., 2010, 110, 6961–7001 CrossRef CAS PubMed.
- C. T. Saouma and J. M. Mayer, Do spin state and spin density affect hydrogen atom transfer reactivity?, Chem. Sci., 2014, 5, 21–31 RSC.
- D. Usharani, D. C. Lacy, A. S. Borovik and S. Shaik, Dichotomous Hydrogen Atom Transfer vs Proton-Coupled Electron Transfer during Activation of X–H Bonds (X = C, N, O) by Nonheme Iron–Oxo Complexes of Variable Basicity, J. Am. Chem. Soc., 2013, 135, 17090–17104 CrossRef CAS PubMed.
- M. S. Seo, N. H. Kim, K.-B. Cho, J. E. So, S. K. Park, M. Clémancey, R. Garcia-Serres, J.-M. Latour, S. Shaik and W. Nam, A mononuclear nonheme iron(IV)-oxo complex which is more reactive than cytochrome P450 model compound I, Chem. Sci., 2011, 2, 1039 RSC.
- J. W. Darcy, B. Koronkiewicz, G. A. Parada and J. M. Mayer, A Continuum of Proton-Coupled Electron Transfer Reactivity, Acc. Chem. Res., 2018, 51, 2391–2399 CrossRef CAS PubMed.
- J. J. D. Sacramento and D. P. Goldberg, Factors Affecting Hydrogen Atom Transfer Reactivity of Metal–Oxo Porphyrinoid Complexes, Acc. Chem. Res., 2018, 51, 2641–2652 CrossRef CAS PubMed.
- T. H. Yosca, J. Rittle, C. M. Krest, E. L. Onderko, A. Silakov, J. C. Calixto, R. K. Behan and M. T. Green, Iron(IV)hydroxide pKa and the Role of Thiolate Ligation in C-H Bond Activation by Cytochrome P450, Science, 2013, 342, 825–829 CrossRef CAS PubMed.
- C. Kupper, B. Mondal, J. Serrano-Plana, I. Klawitter, F. Neese, M. Costas, S. Ye and F. Meyer, Nonclassical Single-State Reactivity of an Oxo-Iron(IV) Complex Confined to Triplet Pathways, J. Am. Chem. Soc., 2017, 139, 8939–8949 CrossRef CAS PubMed.
- M. A. Ehudin, D. A. Quist and K. D. Karlin, Enhanced Rates of C–H Bond Cleavage by a Hydrogen-Bonded Synthetic Heme High-Valent Iron(IV) Oxo Complex, J. Am. Chem. Soc., 2019, 141, 12558–12569 CrossRef CAS PubMed.
- V. Dantignana, J. Serrano-Plana, A. Draksharapu, C. Magallón, S. Banerjee, R. Fan, I. Gamba, Y. Guo, L. Que, M. Costas and A. Company, Spectroscopic and Reactivity Comparisons between Nonheme Oxoiron(IV) and Oxoiron(V) Species Bearing the Same Ancillary Ligand, J. Am. Chem. Soc., 2019, 141, 15078–15091 CrossRef CAS.
- R. Gupta and A. S. Borovik, Monomeric Mn III/II and Fe III/II Complexes with Terminal Hydroxo and Oxo Ligands: Probing Reactivity via O–H Bond Dissociation Energies, J. Am. Chem. Soc., 2003, 125, 13234–13242 CrossRef CAS PubMed.
- T. H. Parsell, M.-Y. Yang and A. S. Borovik, C–H Bond Cleavage with Reductants: Re-Investigating the Reactivity of Monomeric Mn III/IV–Oxo Complexes and the Role of Oxo Ligand Basicity, J. Am. Chem. Soc., 2009, 131, 2762–2763 CrossRef CAS PubMed.
- J. W. Darcy, S. S. Kolmar and J. M. Mayer, Transition State Asymmetry in C–H Bond Cleavage by Proton-Coupled Electron Transfer, J. Am. Chem. Soc., 2019, 141, 10777–10787 CrossRef CAS PubMed.
- M. K. Goetz and J. S. Anderson, Experimental Evidence for pKa-Driven Asynchronicity in C–H Activation by a Terminal Co(III)–Oxo Complex, J. Am. Chem. Soc., 2019, 141, 4051–4062 CrossRef CAS PubMed.
- M. K. Goetz and J. S. Anderson, Correction to “Experimental Evidence for pKa-Driven Asynchronicity in C–H Activation by a Terminal Co(III)–Oxo Complex”, J. Am. Chem. Soc., 2020, 142, 5439–5441 CrossRef CAS.
- M. Mandal, C. E. Elwell, C. J. Bouchey, T. J. Zerk, W. B. Tolman and C. J. Cramer, Mechanisms for Hydrogen-Atom Abstraction by Mononuclear Copper(III) Cores: Hydrogen-Atom Transfer or Concerted Proton-Coupled Electron Transfer?, J. Am. Chem. Soc., 2019, 141, 17236–17244 CrossRef CAS.
- D. Bím, M. Maldonado-Domínguez, L. Rulíšek and M. Srnec, Beyond the classical thermodynamic contributions to hydrogen atom abstraction reactivity, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E10287–E10294 CrossRef.
- M. Maldonado-Domínguez and M. Srnec, H-Atom Abstraction Reactivity through the Lens of Asynchronicity and Frustration with Their Counteracting Effects on Barriers, Inorg. Chem., 2022, 61, 18811–18822 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc03396e |
| This journal is © The Royal Society of Chemistry 2024 |