Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploration, expansion and definition of the atropopeptide family of ribosomally synthesized and posttranslationally modified peptides†
Friederike
Biermann
 ab,
Bin
Tan
ab,
Milena
Breitenbach
ab,
Yuya
Kakumu
ab,
Bin
Tan
ab,
Milena
Breitenbach
ab,
Yuya
Kakumu
 ab,
Pakjira
Nanudorn
ab,
Yoana
Dimitrova
a,
Allison S.
Walker
ab,
Pakjira
Nanudorn
ab,
Yoana
Dimitrova
a,
Allison S.
Walker
 de,
Reiko
Ueoka
f and
Eric J. N.
Helfrich
de,
Reiko
Ueoka
f and
Eric J. N.
Helfrich
 *abc
*abc
aInstitute for Molecular Bio Science, Goethe University Frankfurt, Max-von-Laue Strasse 9, 60438 Frankfurt am Main, Germany. E-mail: eric.helfrich@bio.uni-frankfurt.de
bLOEWE Center for Translational Biodiversity Genomics (TBG), Senckenberganlage 25, 60325 Frankfurt am Main, Germany
cSenckenberg Gesellschaft für Naturforschung, Senckenberganlage 25, 60325 Frankfurt am Main, Germany
dDepartment of Chemistry, Vanderbilt University, Stevenson Center 7330, Nashville, TN 37240, USA
eDepartment of Biological Sciences, Vanderbilt University, VU Station B, Box 35-1634, Nashville, TN 37235, USA
fSchool of Marine Biosciences, Kitasato University, 1-15-1 Kitasato, Minami-ku, Sagamihara, Kanagawa 252-0373, Japan
First published on 10th September 2024
Abstract
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) constitute a diverse class of natural products. Atropopeptides are a recent addition to the class. Here we developed AtropoFinder, a genome mining algorithm to chart the biosynthetic landscape of the atropopeptides. AtropoFinder identified more than 650 atropopeptide biosynthetic gene clusters (BGCs). We pinpointed crucial motifs and residues in leader and core peptide sequences, prompting a refined definition of the atropopeptide RiPP family. Our study revealed that a substantial subset of atropopeptide BGCs harbors multiple tailoring genes, thus suggesting a broader structural diversity than previously anticipated. To verify AtropoFinder, we heterologously expressed four atropopeptide BGCs, which resulted in the identification of novel atropopeptides with varying peptide lengths, number and types of modifications. Atropopeptides serve as a proof-of-principle for the versatile genome mining approach developed in this study that can be repurposed for the identification of RiPP and other BGCs that currently evade detection.
Introduction
Peptide natural products play a crucial role in drug discovery due to their diverse structures, high potency, selectivity, and strong affinity towards drug targets, as well as favorable pharmacokinetic properties.1 Examples of pharmaceutically relevant peptides include vancomycin (antibacterial), cyclosporin (immunosuppressive), bacitracin (antibacterial), and thiostrepton (antibacterial).2–5 Historically, complex peptide natural products were believed to be solely biosynthesized by non-ribosomal peptide synthetases (NRPSs).6,7 NRPSs are giant multi-enzyme complexes that assemble complex peptides in an assembly line-like fashion independent of the ribosome. However, over the past two decades, numerous complex peptides have been discovered that are biosynthesized by ribosomally synthesized and posttranslationally modified peptide biosynthetic pathways (RiPPs).8 The array of characterized posttranslational modifications in RiPP systems is constantly increasing. RiPPs encompass a heterogenous group of natural products, which can be subdivided into more than 40 families.8 The corresponding biosynthetic gene clusters (BGCs) can be distinguished by the presence of characteristic tailoring genes (e.g., linear polyazole-containing peptides), conserved precursor genes (e.g., the Nif11-like proteusin precursors) or the intricate structure of the associated peptide (e.g., lasso peptides).9 State-of-the-art genome mining tools frequently overlook RiPP BGCs that do not belong to the well-studied RiPP families.10 Consequently, RiPP-derived products are sometimes initially misannotated as NRPS products.7,11,12 Extensive research into RiPPs over the past two decades has expanded the RiPP biosynthetic repertoire and revealed substantial overlap in the diverse array of modifications observed in peptides from RiPP and NRPS pathways (Fig. S1†).7,13 As a result, the annotation of complex peptides as nonribosomal peptides (NRPs) or RiPPs can often be challenging unless peptide class-defining modifications are present in the molecules.7 This challenge can be showcased by tryptorubin A which was initially associated with a NRPS BGC.11 The proposed BGC was the sole peptide BGC detected by state-of-the-art genome mining platforms to be present in all three reported tryptorubin A producers.11 Tryptorubin A (1) is a hexapeptide that features an unusually complex three-dimensional structure.14 The extremely rigid three-dimensional shape arises from a biaryl and a carbon–nitrogen bridge between aromatic amino acid side chains and an additional carbon–nitrogen bridge between an aromatic amino acid side chain and the peptide backbone, respectively.14 These modifications result in two possible unusual atropisomeric configurations, only one of which has been found in nature.14 Unusual atropisomerism (recently also referred to as ansamerism15) is a type of stereoisomerism in which both isomers are theoretically interconvertible by multiple nonphysical bond torsions.14 Intrigued by the structural complexity of the hexapeptide, we revisited its biosynthesis. A combination of manual genome mining and heterologous expression studies unveiled that tryptorubin A is the first member of a new RiPP-family that we named atropopeptides (also referred to as atropitides8).16 The biosynthetic blueprint for the production of characterized atropopeptides resides within small BGCs containing only two genes: one encoding a precursor peptide and another a cytochrome P450 monooxygenase.16The precursor peptide, comprising a leader and core peptide, is ribosomally synthesized. The leader peptide recruits the BGC-encoded cytochrome P450 which introduces the distinctive biaryl and carbon–nitrogen bridges in an atropospecific manner (Fig. 1A).16 In the final step, the modified core peptide is proteolytically released from the leader peptide. It is believed that an ubiquitous protease catalyzes the release of the mature hexapeptide, which can be further modified through the removal of the N-terminal alanine residue by another unknown peptidase that is not encoded in the BGC to yield the pentapeptide variant.16 A similar cytochrome P450 catalyzed biaryl-linkage was also found in biarylitides, a RiPP family described in members of the genus planomorospora.24
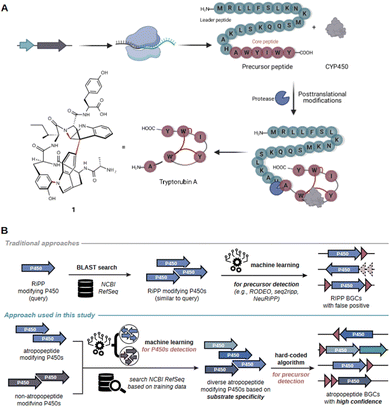 | ||
| Fig. 1 Schematic overview of the proposed model for tryptorubin A (1) biosynthesis and comparison of the genome mining concept used in AtropoFinder to state-of-the-art genome mining tools for the identification of RiPP BGCs. (A) The genes of the trp BGC are transcribed and the resulting mRNA ribosomally translated. Subsequently, the precursor peptide, consisting of a core peptide and a leader peptide, is posttranslationally modified by the BGC-encoded cytochrome P450 installing the characteristic atropopeptide crosslinks. Finally, an ubiquitous protease cleaves the leader peptide to release the hexapeptide tryptorubin A (1).16 (B) Comparison of existing machine learning approaches for RiPP discovery with the strategy developed in this study. State-of-the-art tools are frequently based on the BLAST or HMM-based search of putative RiPP-modifying tailoring enzymes.17–20 In a second step, machine learning algorithms are used to identify putative precursor genes in the genomic vicinity of the genes encoding the identified tailoring enzyme.17,18,21–23 In contrast, our approach first uses machine learning to determine atropopeptide-modifying tailoring enzymes with high precision. In a second step, a hard coded algorithm is used to identify the corresponding precursor genes with a high level of confidence. | ||
In recent genome mining efforts, the atropopeptide-modifying P450 encoded in the trp BGC was subjected to BLASTp analysis to identify homologs likely involved in atropopeptide maturation. The associated precursor peptide sequence was subsequently discerned through manual analysis of the genomic vicinity of the putative atropopeptide modifying P450s. While this approach has extended the atropopeptide family to encompass the cihunamides1,8 kitasatides25 and amyxirubins16 (also referred to as clavorubins26), it is apparent that this BLAST-based approach has limitations in capturing the full biosynthetic diversity of the atropopeptides. The BLASTp algorithm excels at detecting closely related homologs of a query sequence.10 However, to identify more distant relatives of tryptorubin A, a more flexible strategy allowing significant deviations from the original sequence is imperative. In light of these challenges, PSI-BLAST has been employed for its capability to detect more distantly related sequences through iterative searches. However, PSI-BLAST also has its limitations, as it additionally identifies a large number of P450 enzymes that are not involved in atropopeptide biosynthesis. In fact, a substantial fraction of the identified P450s are encoded in RiPP BGCs that belong to other RiPP families.27 The precise identification of exclusively atropopeptide BGCs might be achieved through the utilization of supervised machine learning (ML).28 Supervised learning entails training algorithms on a labeled dataset, which are subsequently used to classify previously unseen data by the trained algorithm.29
Herein we comprehensively explore the biosynthetic potential encoded in atropopeptide BGCs. We introduce AtropoFinder, a machine learning-based genome mining tool for the identification of atropopeptide BGCs. The AtropoFinder algorithm identified 683 putative atropopeptide BGCs across publicly accessible genome sequences, thus more than doubling the number of atropopeptide BGCs compared to BLAST analysis (683 putative BGCs found by AtropoFinder vs. 282 putative BGCs identified by Liu et al.30 Fig. S5B†). The identified atropopeptide BGCs revealed unexpected trends in core peptide composition and length, enabling us to refine the definition of atropopeptides and expand their biosynthetic and structural space. The expansion of the atropopeptide biosynthetic space is furthermore showcased by the AtropoFinder-guided identification of four new atropopeptides featuring different core peptide length, number and types of modifications. Moreover, we report the unexpected production of two atropopeptides with the same core peptide sequence but distinct non-overlapping modification patterns from a single atropopeptide BGC. Our machine learning-based approach is versatile and can be applied for the tailoring enzyme-guided discovery of BGCs associated with diverse natural product classes that currently evade unrecognized by genome mining tools.
Results and discussion
P450-guided identification of putative atropopeptide gene clusters (AtropoFinder)
Recent studies conducted by our research group and other laboratories describe the BLAST-based identification of atropopeptide biosynthetic pathways using the atropopeptide family-defining P450 as a query sequence.16,25,27,30–32 Manual analysis of the obtained results indicated that the identified core peptide sequences in the vicinity of the identified P450 genes exhibit only a moderate level of sequence diversity.16 This lack of sequence diversity, suggests that the BLASTp/PSI-BLAST approach might not comprehensively capture the entire biosynthetic diversity of atropopeptides. Furthermore, the workflow is time-intensive and requires manual search to locate the associated precursor peptide gene in the proximity of the P450 gene. The challenge of identifying the precursor sequence is exacerbated with precursor peptide sequences that display only limited similarity to known atropopeptide precursors (the leader peptides within the final dataset show only 4.5% identical sites and 54.3% pairwise identity). To address these limitations, we hypothesized that a machine learning-based approach to identify putative atropopeptide-modifying P450s is more suitable to comprehensively map the atropopeptide biosynthetic landscape. In addition, our approach aims to replace the laborious manual precursor peptide identification with a robust algorithm that detects the precursor and core peptides regardless of their level of homology to characterized atropopeptides. We decided to use the atropopeptide family-defining P450s as bait for the identification of atropopeptide BGCs. After the initial identification, a subsequent step, focused on identifying the ORF coding for the precursor peptide, serves to confirm the identified BGC (CoreFinder). This step also allows for the detailed identification of the primary amino acid sequences of both precursor and core peptides.RiPP families are frequently characterized through the BLAST or HMM-based identification of putative biosynthetic gene clusters (BGCs) based on the sequence homology of family-defining tailoring enzymes within a RiPP (sub-)family. Algorithms use sequence homology either directly, as in the cases of antiSMASH 7,19 PRISM4,20 NeuRiPP,21 RODEO2,17 or indirectly like DeepBGC,33 GECCO,34 and BiGCARP,35 which are trained on the sequences of HHM-derived Pfam domains within known gene clusters. In a subsequent step, machine learning algorithms such as RODEO2,17 seq2ripp,18 NeuRiPP,21 or DeepRiPP22 are employed to determine putative precursor sequences. Our approach markedly diverges from this strategy. We utilize machine learning to identify atropopeptide-modifying enzymes with less emphasis on their sequence homology to characterized members of the RiPP family than previous approaches. Following this step, a hard-coded algorithm is applied to pinpoint the precursor, verify the initial machine learning step, and complete the full genome mining process within one tool (Fig. 1B).
To obtain a training data set for the ML classifier, a BLAST search was conducted using the P450 encoded in the trp BGC as a query sequence (WP_007820080.1). Through manual curation, we identified 51 sequences putatively encoding tryptorubin A-like precursors (ESI File S5†). After dereplication with a 95% sequence similarity cutoff, 37 sequences remained. To train the ML-classifier to differentiate atropopeptide-modifying P450s from P450s that modify other substrates, we collected a dataset from the antiSMASH database.36 This dataset consisted of P450s encoded in BGCs involved in the biosynthesis of products that are unrelated to atropopeptides that were dereplicated at a 95% cutoff, totaling 9065 entries. We simplified the amino acid code based on the amino acids' physico-chemical properties to prevent overfitting (Fig. S2 and S3†). Next, we trained a machine-learning Random Forest classifier on a subset of the training data, which we validated using the remainder of the training dataset. The Random Forest classifier distinguishes relevant from irrelevant features during training. It uses random subsets of training data and features (four-amino-acid motifs in P450 protein functional regions) for each decision tree. Each tree independently selects features that best differentiate ‘atropopeptide-modifying P450’ from ‘non-atropopeptide-modifying P450.’ Thus, this process identifies the specific properties of atropopeptide modifying peptides from the training data.37 We then applied the classifier to a dataset of 154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 364 protein sequences from NCBI RefSeq that were annotated as “P450” (for a detailed workflow and metrics see ESI Detailed description of AtropoFinder, Fig. S2–S5 and Table S1†). In the initial round of P450 classification, the algorithm identified 202 putative atropopeptide-modifying P450s. However, the associated precursors, predicted by CoreFinder (see below), only showed moderate core peptide sequence variations. To address these limitations, we iterated the process, incorporating the 202 newly identified atropopeptide-modifying P450s into the initial training dataset to refine the algorithm. Using the refined classifier, we identified 440 additional putative atropopeptide-modifying P450s from the dataset of 154
364 protein sequences from NCBI RefSeq that were annotated as “P450” (for a detailed workflow and metrics see ESI Detailed description of AtropoFinder, Fig. S2–S5 and Table S1†). In the initial round of P450 classification, the algorithm identified 202 putative atropopeptide-modifying P450s. However, the associated precursors, predicted by CoreFinder (see below), only showed moderate core peptide sequence variations. To address these limitations, we iterated the process, incorporating the 202 newly identified atropopeptide-modifying P450s into the initial training dataset to refine the algorithm. Using the refined classifier, we identified 440 additional putative atropopeptide-modifying P450s from the dataset of 154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 364 putative P450 sequences, thus more than doubling the number of putative atropopeptide-modifying P450s in this round.
364 putative P450 sequences, thus more than doubling the number of putative atropopeptide-modifying P450s in this round.
Identification of putative atropopeptide precursor- and core peptides (CoreFinder)
More than a dozen highly sophisticated genome mining tools for the identification of putative RiPP precursor peptides have been developed.10 While these tools have led to the identification of countless putative precursor sequences and the targeted discovery of numerous RiPPs, none of the existing tools are capable of identifying atropopeptide precursors and BGCs, respectively (ESI Comparison of AtropoFinder to existing bioinformatic tools†). The manual search process for atropopeptide precursor genes is not only laborious but may also miss out on sequences that deviate significantly from known atropopeptide precursors. To address this problem, we introduced “CoreFinder” (detailed description can be found in ESI Detailed description of CoreFinder†). The command-line tool streamlines the screening for open reading frames (ORFs) in the genomic vicinity of a specified protein-encoding gene (here specifically the gene encoding the characteristic atropopeptide-modifying P450) within all GenBank nucleotide records in which the query protein is annotated. We screened the genetic neighborhood of all putative atropopeptide-modifying P450 genes from NCBI records for atropopeptide-like open reading frames. The identification criteria prioritize precursor ORFs that range between 10 to 40 amino acids in length, consisting of a leader and a core peptide, which contains two consecutive aromatic amino acids and is followed by a stop codon. The latter criterion stemmed from the observation that the atropopeptides we had previously characterized all featured at least one pair of consecutive aromatic acids that are crucial for anchoring the characteristic atropopeptide modifications. CoreFinder detected 803 potential precursor peptide-encoding genes. Out of these, 475 genes from 533 genomes exhibited overall sequence homology to the tryptorubin A precursor.Analysis of atropopeptide precursor sequences and recalculation of putative atropopeptide precursor sequences
In our endeavor to characterize conserved motifs within atropopeptide precursors, we aligned all identified putative precursor peptide sequences (ESI file S1†). We used the MEME suite38 and identified a conserved KSLK motif (or closely related motifs like ‘RSLK’, ‘ESLK’, ‘KSRK’, ‘KPLK’, or ‘PSLK’) strategically situated between residues 17–20 of the putative precursors (Fig. 2 and S6†). We thus hypothesized that the KSLK motif might be necessary for the recognition of the precursor by the P450. To verify our hypothesis we used AlphaFold2 multimer39 to investigate the predicted interaction between the precursor and corresponding P450 associated with tryptorubin A (Fig. 2). With a iptm + ptm score of 0.8395, the model seems to be a good representation of the protein interaction (for details on the pLDDT score for each residue see Fig. S7†). Our analyses underscored the putatively critical role of the KSLK motif for threading the precursor into the P450's substrate binding tunnel. The KSLK motif occupies a strategic junction, marking the transition from the N-terminal α-helix of the leader peptide—which is predicted to bind to the P450 enzyme's surface—to a second, shorter α-helix which transitions into the seemingly flexible core peptide. Based on the AlphaFold model, the KSLK motif can be regarded as a biochemical “anchor”, threading the precursor peptide into the substrate binding tunnel. This stabilization might allow the core peptide to “swirl” around within the enzyme's active site. Consequently, this flexibility likely renders the formation of different bond types at different locations within the core peptide possible. Guided by the lessons learned from the combination of sequence analysis and modeling studies, we refined the classification algorithm of CoreFinder to mandate the inclusion of one of the following motifs within the precursor: ‘KSLK’, ‘RSLK’, ‘ESLK’, ‘KSRK’, ‘KPLK’, or ‘PSLK’. The recomputation unveiled 685 putative precursor peptides dispersed across 683 nucleotide sequences, in the vicinity of 328 of the 440 putative atropopeptide p450s. Among these, 684 exhibited archetypal features of atropopeptide precursors. Only a single sequence deviates from the common precursor architecture. Although it possesses the “KSLK” motif, it significantly diverges from canonical leader peptide sequences and appeared to adopt this motif at an aberrant locus. Such deviation might represent a fusion of an atropopeptide BGC with another RiPP family. Two putative atropopeptide BGCs seemingly harbor two instead of one putative precursor gene (Fig. 3). A BiG-SCAPE40 analysis comparing all obtained gene clusters with BGCs from MIBiG41 shows that atropopeptide BGCs cluster separately from all BGCs deposited to MIBiG41 (ESI file S3 and Fig. S8†). This finding indicates atropopeptide BGCs not being related to any BGCs deposited to MIBiG. In a sequence similarity network of all identified putative precursor peptides (ESI file S2 and Fig. S9†), the precursor sequences group based on their core peptide sequence and phylogenetic origin. Surprisingly, the sequences associated with the tryptorubin A core peptide group into multiple clusters, suggesting that they might have evolved towards this sequence multiple times. A similar evolutionary pattern is observed in the phylogenesis of atropopeptide-modifying cytochrome P450s (Fig. 3).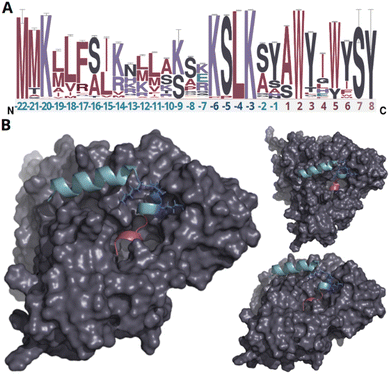 | ||
| Fig. 2 Analysis of putative precursor peptides and modeling of protein interactions of the precursor peptide and P450 encoded in the trp BGC. (A) Sequence logo of putative precursor peptide sequences. The amino acids of the leader peptide, KSLK motif and core peptide are depicted in green, blue, and red, respectively. Residues only present in very few peptides are depicted in light red (residue 7–8). The height of each letter in the sequence logo corresponds to the frequency of the respective amino acid at that position, with taller letters indicating higher frequency and conservation. The error bars indicate an approximate Bayesian 95% confidence interval. (B) AlphaFold2 multimer protein model of WP_007820080.1 and the tryptorubin A precursor peptide from different angles (a video showcasing intricate details of the protein model can be found in ESI file S4,† metrics of the modeling can be found in Fig. S7†). The leader peptide, KSLK motif, core peptide, and cytochrome P450 are depicted in green, blue, red, and gray, respectively. The leader peptide forms an α-helix that bends at the KSLK motif. The KSLK motif acts as an anchor of the precursor to the P450 that allows the core peptide to move freely within the active site. | ||
Analysis of atropopeptide core sequences
The analysis of all identified putative core peptides revealed that the majority are likely hexapeptides. Nonetheless, there are deviations from this norm. 14 are discerned as octapeptides, while three are classified as heptapeptides. Intriguingly, the octapeptides appear to incorporate additional amino acids interspersed amidst the known modified amino acids (W2, Y3, W6, Y7). In contrast, the heptapeptides exhibit an appended serine at their C-terminus (Fig. 3). Amidst the core peptide sequences (Fig. 2A), the only conserved amino acid residue is W2. We hypothesize that W2 plays a key role in one of two ways: it is either vital for forming the atropopeptide's 3D shape, especially considering all characterized atropopeptides have at least one if not multiple crosslinks anchored to this tryptophan residue. Alternatively, W2 might be crucial for binding to the P450, although our modeling studies (Fig. 2B) do not corroborate this hypothesis. A striking observation is that not all putative core peptides encompass two successive aromatic amino acids, potentially implying varied modification motifs. Moreover, in comparison to a recent report,31 not all core peptides exhibit a pair of tryptophan residues. Several core peptides originating from Xanthomonas species display an xWxxYx pattern, deviating from the conventional xWxxWx pattern reported before.31 This observation suggests that the recently coined bitryptides (xWxxW) refer to a subfamily of atropopeptides. Moreover, our in silico studies indicate that atropopeptides can be more adequately classified by the presence of conserved motifs and residue in the precursor peptide than by their complex 3-dimensional shape.Phylogenetic distribution of atropopeptides
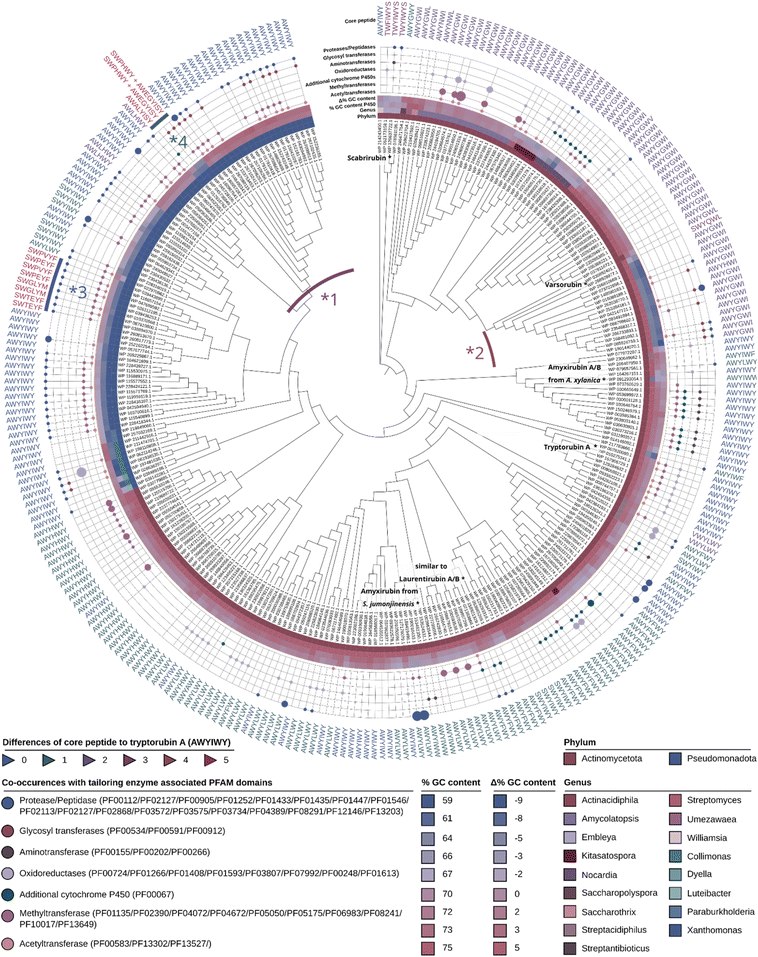
The phylogenetic distribution of putative atropopeptide BGCs revealed their abundance mainly in Streptomyces, Xanthomonas, and related genera in the phyla actinomycetota and pseudomonadota (Fig. 3 and S10†). This abundance is surprising because these phyla are not closely related within the bacterial branch of life. To delve deeper into the evolutionary history of P450s encoded in putative atropopeptide BGCs, we constructed a maximum likelihood phylogenetic tree utilizing all putative atropopeptide-modifying P450 amino acid sequences (Fig. 3). Notably, the sequences group based on the phylogeny of their producer organisms and the associated putative core peptide sequences.Upon contrasting the GC content of the P450s (serving as a proxy for the GC content of the BGC) with the mean GC content of each species' genome, we found a close correlation between the two for most BGCs. The observed pattern suggests that these BGCs have an ancient evolutionary origin and have adapted to the GC content of the producer's genomes. However, two distinct clades of P450s presented significant disparities in their GC content relative to their producer's genome GC content. Within the clade designated as *1, the P450's GC content exceeds that of the associated Pseudomonadota genomes, mirroring instead the GC profile characteristic for Actinomycetota. In contrast, the clade labeled as *2 reveals a GC content of the P450 that is markedly diminished relative to its corresponding Actinomycetota producer strains, but aligns more closely with GC contents characteristic for Pseudomonadota. Such observations might allude to recent horizontal gene transfer events between the two phyla.
Analysis of the genomic vicinity of minimal atropopeptide BGCs
For an in-depth analysis of the tailoring enzymes encoded in the putative atropopeptide BGCs, we employed RODEO's functionality to track concomitant Pfam domains encoded by a specified gene.17 Our findings underscore a frequent association between proteases and atropopeptide P450s within the genus of Xanthomonas. This is particularly noteworthy, as previously characterized atropopeptides do not harbor proteases encoded in their associated BGC. This association potentially facilitates the cleavage in instances where an external protease is absent for leader peptide excision or the proteases/peptidases are responsible for a secondary cleavage of the core peptide, reminiscent of the model for tryptorubin B biosynthesis. Other tailoring enzymes that are frequently encoded in atropopeptide BGCs are acetyltransferases, methyltransferases, oxidoreductases, additional P450s, and glycosyltransferases. It is tempting to speculate that the biosynthetic space of atropopeptides includes molecules that are more extensively modified by the identified tailoring enzymes than the previously characterized atropopeptides.AtropoFinder-based expansion of the atropopeptide chemical space
To validate the AtropoFinder algorithm and expand the biosynthetic and chemical space of characterized atropopeptides, we selected four putative atropopeptide BGCs for characterization through heterologous expression in Streptomyces albus J1074.We first selected the atropopeptide BGC from Streptomyces jumonjinensis DSM 747 (Genbank: GCF_009600885.1, NZ_VCLA01000160.1 22.964:24.216, further referred to as jum) which was predicted to harbor the same core peptide sequence as the one reported for amyxirubin A and B produced by Amycolatopsis xylanica.16 Since both organisms belong to the same class (Actinomycetes) but a different order, the identity of the P450s encoded in the jum and amyxirubin BGCs was only 59% and the P450 phylogeny suggested a distant relationship of the P450s encoded in the jum and amyxirubin BGCs (Fig. 3), we speculated that the resulting atropopeptide might show a different bridging pattern when compared to amyxirubin. Heterologous expression of the jum BGC led to the isolation of 1.37 mg of the associated atropopeptide (2) (Fig. 4A and S11†). The comparison of 1H NMR chemical shifts and high-resolution electrospray ionization mass spectrometry (HR-ESI-MS) data of 2 and an authentic standard of amyxirubin B (Fig. S12 and S13†), however, revealed that the structure of 2 is identical to amyxirubin B, suggesting that the phylogenetic placement of the atropopeptide-modifying P450 might not be a good indicator for the structural novelty of the atropopeptides.
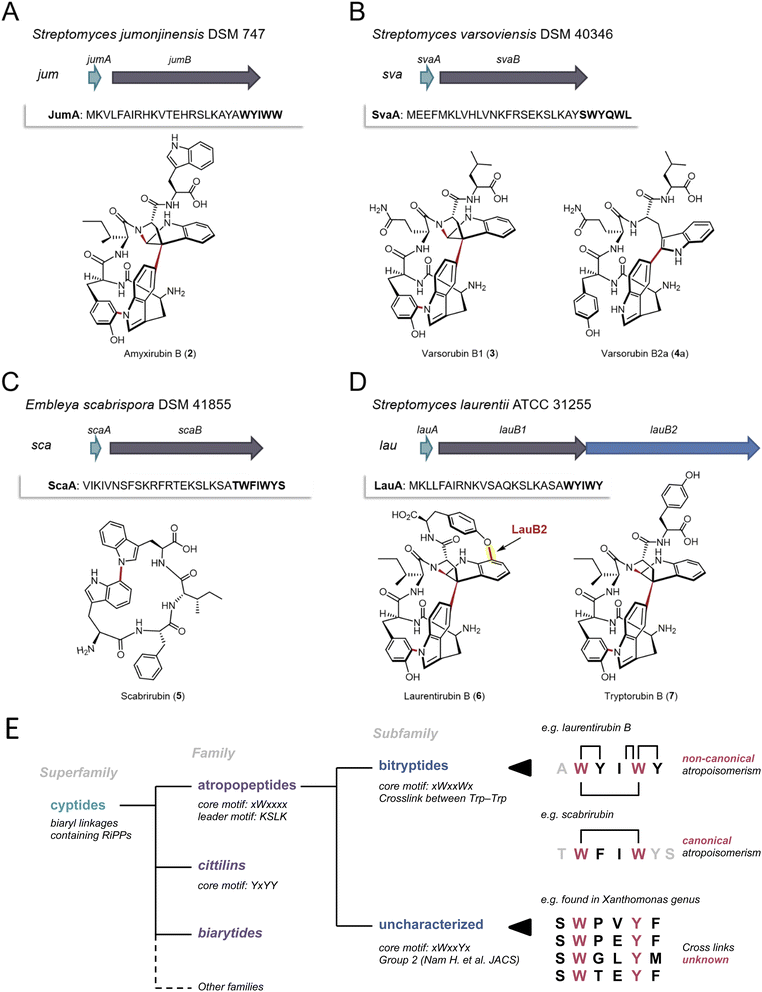 | ||
| Fig. 4 Characterization of four putative atropopeptide BGCs and overview over the landscape of RiPPs that are characterized by P450-mediated crosslinks between amino acid side chains. The precursor genes and genes encoding the atropopeptide family-defining P450s are depicted in green and gray, respectively. The additional P450 is depicted in blue. The precursor peptide sequences are shown and the putative core peptides are highlighted in bold. (A) jum BGC and structure of purified amyxirubin B (2) associated with the jum BGC. (B) sva BGC and the structure of varsorubin B1 (3), varsorubin B2a and varsorubin B2b (4a) obtained by heterologous expression of svaA and the P450 encoding gene svaB in front of which a RBS was inserted. (C) sca BGC and structure of purified scabrirubin (5) obtained by heterologous expression of the sca BGC. (D) lau BGC and structure of purified laurentirubin B (6) obtained from the heterologous expression of the full lau BGC and tryptorubin B (7), the product obtained from coexpressing lauA and lauB1. For a more detailed overview of the lau BGC, see Fig. S55.† (E) Overview over the landscape of RiPPs that are characterized by P450-mediated crosslinks between amino acid side chains and comparison of the characteristics of the individual RiPP families and subfamilies with an emphasis on the atropopeptide family. | ||
We next set out to characterize a putative atropopeptide BGC from Streptomyces varsoviensis DSM 40346 (Genbank: GCF_000718635.1, NZ_JOBF01000044.1 9.365:10.577, further referred to as sva). The predicted core peptide sequence of the putative precursor peptide is SWYQWL (Fig. 4B). The sva BGC was heterologously expressed. Extraction of HP20 resin supplemented to the culture supernatant and analysis of the crude extract by high-resolution electrospray ionization mass spectrometry (HR-ESI MS) revealed the presence of two compounds (3 and 4) detected at m/z 791.3540 [M + H]+ (calcd for m/z 791.3511, C42H47N8O8+, Δ 3.67 ppm) and m/z 793.3655 [M + H]+ (calcd for m/z 793.3668, C42H49N8O8+, Δ 1.64 ppm) (Fig. S14 and S16†) from the culture. The calculated formulae of 3 and 4 are in good agreement with molecular formulae of the predicted pentapeptide variant of the core peptide (WYQWL) that had lost four and two protons, respectively. The proton loss indicated at least one or two modifications, respectively. To increase the yield for subsequent purification, we inserted a ribosomal binding site in front of the gene encoding the P450, which resulted in a 100-fold increase in yield of the putative pentapeptides (Fig. S15†). One of the associated pentapeptides (3), named varsorubin B1 (Fig. 4B), was purified from a 16 L culture using a combination of open column chromatography and semi-preparative HPLC (yield 1.6 mg). Analysis of the 1H and 13C NMR spectra, with the aid of 2D NMR correlations (Fig. S17–S22 and Table S2†), confirmed the amino acid sequence WYQWL. Compound 3 is a pyrroloindoline-containing bicyclic macrolactam that features a C–C bond bridging two Trp residues, along with two C–N bonds between Trp-1 and Tyr-1, and between Trp-2 and the amide nitrogen. Moreover, NOESY correlations of H-11(Trp-2)/H-38(Trp-1) and H-9(Trp-2)/H-40,41(Trp-1) suggest that 3 features the “bridge-above” configuration of the bicyclic macrolactam ring as previously reported for all complex atropopeptides (Fig. S23†). Based on the recently proposed terminology for the conformational isomerism of bicyclic peptides,15 we assigned the configuration of the bridge-above macrolactam for 3 as Pansa.
Careful analysis of HPLC-HR-ESI-MS data revealed that the second peak contained 4 and an additional compound with the same molecular formula as 4 (Fig. S24 and S25†). Further chromatographic separation resulted in the isolation of 1.81 mg of 4a, which we named varsorubin B2a (Fig. 4B), as well as an additional isomer varsorubin B2b (4b). The structure of the isomer 4b was not unambiguously elucidated by NMR analysis due to its insufficient yield. Meanwhile, 1D and 2D NMR spectra of 4a indicated that the molecule no longer possesses the complex three-dimensional shape that is characteristic for 3 (Fig. S26–S33 and Table S3†). Instead, only a single modification (C–C bond between C-39 of Trp-1 and C-11 of Trp-2) was present in 4a. Interestingly, the position of this C–C bond at Trp-2 differs from the C–C bond in varsorubin B1 which is located at C-10 of Trp-2. 3 and 4a share the same amino acid sequence but are characterized by an unprecedented non-overlapping bridging pattern installed by the same P450 (Fig. 4B). Moreover, the two characterized products show a different degree of structural complexity.
Subsequently, we selected the putative atropopeptide BGC from Embleya scabrispora DSM 41855, (Genbank: GCF_000372745.1, NZ_KB889561.1, further referred to as sca) for characterization. The precursor ScaA harbors the putative heptapeptide core peptide sequence TWFIWYS (Fig. 4C). Culture extracts of S. albus harboring the sca BGC were screened for the presence of modified heptapeptides associated with the BGC. To our surprise, however, comparative HPLC-HR-ESI-MS analyses of extracts from S. albus cultures harboring the sca plasmid with an empty plasmid control revealed the presence of a new peak with an m/z 649.3132 [M + H]+ (calcd for m/z 649.3133, C37H41N6O5+, Δ 0.15 ppm) (Fig. S34 and S35†). The observed mass was significantly smaller than the expected precursor ion of the predicted heptapeptide. Nevertheless, we extracted and purified 3.5 mg of the putative atropopeptide (5) from a 10 L culture. Analysis of 1H and 13C NMR spectra, with the aid of HSQC, HMBC, and NOESY correlations (Fig. S36–S42 and Table S4†), revealed the presence of a tetrapeptide with the sequence WFIW featuring a C–N bond between C-34 of Trp-1 and N-5 of Trp-2 (Fig. S43†) that is in agreement with the N- and C-terminally truncated core peptide sequence. We named the cyclic tetrapeptide scabrirubin (5). The characterization of scabrirubin with the sequence WFIW from the putative heptapeptide core TWFIWYS suggests that peptidases in the heterologous host cleave both the N-terminal and the C-terminal amino acids of the core peptide, indicating that the C-terminus might serve as a follower peptide. While the removal of N-terminal amino acids is a common event during atropopeptide maturation, the removal of C-terminal residues is rare.
We next set out to characterize an atropopeptide BGC harboring an additional gene encoding another P450 to investigate possible additional tailoring reactions. AtropoFinder identified the putative BGC in Streptomyces sp. MMG1121, which was not available in any strain collection. We therefore conducted an NCBI BLAST42 homology search on the P450 to find a similar BGC in a commercially available strain. To our surprise, we identified a BGC from Streptomyces laurentii ATCC 31255 (Genbank: GCA_002355495.1, AP017424.1 5.439.938:5.442.374, further referred to as lau BGC) that harbors the characteristic atropopeptide-modifying P450 alongside three additional annotated P450 gene fragments that are significantly shorter than the first P450 encoded in the lau BGC (Fig. S44†). This BGC was not originally identified by AtropoFinder because we ran AtropoFinder solely on RefSeq P450s which does not include the S. laurentii P450. We initially hypothesized that the three P450 fragments might act together to form one functional P450. Alternatively, we hypothesized that the three P450 fragments are a sequencing artifact and that the three P450 gene fragments indeed encode one contiguous P450 such as in Streptomyces sp. MMG1121. To verify the latter hypothesis, we subjected the lau BGC to resequencing. Analysis of the sequencing data revealed that the three P450 gene fragments are indeed a sequencing artifact. As a consequence, the lau BGC is only predicted to harbor two genes encoding P450s, the atropopeptide-family defining P450 and a second P450 with low identity to TryB (28%) (Fig. 4D). To characterize the lau BGC, we cloned and heterologously expressed the BGC in S. albus.
HPLC-HR-ESIMS analysis of the crude extract obtained from the S. albus culture harboring the lau BGC revealed the presence of two candidate atropopeptides detected at m/z 895.3760 [M + H]+ (calc. sum formula C49H51N8O9+, Δ 1.6 ppm) and 824.3401 [M + H]+ (calc. sum formula C46H46N7O8+, Δ 0.1 ppm) to be associated with the lau BGC (Fig. S45 and S46†). The mass difference between the predicted core peptide sequence and the detected compounds are indicative of the candidate hexa- and pentapeptides with a loss of six protons which suggests the presence of at least three modifications. To pinpoint the localization of the modifications, the main product, 1.6 mg of a pentapeptide (laurentirubin B (6)) was isolated from a six liter culture. Based on the 1H NMR spectrum and 2D NMR correlations (Fig. S47–S55 and Table S5†), the structure of 6 was determined. 6 is characterized by a highly strained tricyclic macrolactam ring system that is constructed by two C–N bonds (Trp-1–Tyr-1 and Trp-2–peptide backbone), a C–C bond (Trp-1–Trp-2), and a C–O bond (Trp-2–Tyr-2) (Fig. 4). The localization of the unprecedented aryl ether linkage between C-17 of Trp-2 and C-7 of Tyr-2 is supported by density functional theory (DFT) calculations (Table S6†). Moreover, the atropisomeric configuration of Pansa for 6 was determined by NOE correlations (Fig. S55†) and 13C chemical shift calculations by the DFT method (Table S6†).
We hypothesized that the three common modifications are installed by the characteristic P450 homolog that is conserved in all atropopeptide BGCs and that the aryl ether bridge is formed via the second P450. To verify our hypothesis, we coexpressed both the precursor LauA with the first P450 LauB1 and LauA with the second P450 LauB2 in S. albus and analyzed the extracts for biosynthetic intermediates. The results showed that coexpressing lauA and lauB2 resulted in the complete abolishment of atropopeptide production. Coexpression of lauA and lauB1, on the other hand, resulted in the accumulation of tryptorubin B (7) (Fig. S56†), a biosynthetic intermediate with three modifications, indicating that the second P450 is responsible for introducing the fourth modification. Laurentirubin B is one of the most complex atropopeptides reported to date. With two exceptions,30 the lau BGC is the only characterized BGC that encodes an additional P450 that modifies the peptide beyond the characterized atropopeptide-specific modifications. Our gene coexpression studies furthermore indicate that the introduction of the ether bridge takes place after the formation of the archetypical atropopeptide modifications.
The antimicrobial activity of compounds 2, 3, 4a and 5 were investigated against all ESKAPE pathogens, Arthrobacter pascens and Candida albicans. Compounds 2, 3 and 4a did not show antibacterial or antifungal activity. Compound 5 displayed weak growth inhibition activity against A. pascens and 3 promoted the growth of P. aeruginosa and A. pascens (Fig. S57†), indicating that 3 may function as a signaling metabolite across phylum borders. This observation provides possible clues to further study the ecological role of the atropopeptides and to identify the molecular target of 3.
Conclusion
Atropopeptides are one of the recently described RiPP families that feature characteristic P450-catalyzed post-translational modifications that result in crosslinking of amino acid side chains (i.e. biaryl, aryl-nitrogen and aryl-ether linkages).8,16,25 Other families characterized by P450-mediated posttranslational modifications include the biarylitides, that feature either a P450-catalyzed biaryl or a carbon–nitrogen bridge between histidine and tyrosine residues,24 or the cittilins that harbor both a biaryl and an aryl-ether bridge between tyrosine side chains. In contrast, atropopeptides feature biaryl, carbon–nitrogen, and aryl-ether bridges, rendering them the most intricate RiPP family modified by P450s.14,16 Recently, a RiPP superfamily, named cyptides, that includes all RiPP families that feature biaryl linkages, has been proposed.27 Within this superfamily, the atropopeptides (also referred to as group 1/2 cyptides and atropitides) are the largest family with over 270 previously identified members.27 Our efforts to systematically map the biosynthetic space of the atropopeptides resulted in the significant expansion of the number of predicted BGCs associated with the atropopeptide family over previous studies that employed BLAST-based approaches, which are inherently biased towards limited sequence diversity. Moreover, our approach that is based on the unique properties of atropopeptides exclusively detects atropopeptides BGCs and thus outperforms BLAST-based approaches that have been shown to identify RiPP BGCs beyond the atropopeptide family.Our machine learning-based approach that uses the cytochrome P450 enzymes as a bait for the detection of atropopeptide BGCs, resulted in the identification of 684 putative atropopeptide BGCs. Insights from the comparison of all putative atropopeptide leader peptide sequences revealed a conserved KSLK motif in the leader that is characteristic for all atropopeptide leaders and that can be used to differentiate atropopeptides from other RiPP families that feature biaryl-linkages. Using AlphaFold2 multimer39 modeling, we were able to propose a putative role of the conserved motif in interacting with the P450, thus offering insights into its putative functional relevance as an anchor for the core peptide within the active site of the cytochrome P450. Moreover, all identified atropopeptide cores are characterized by the presence of a conserved Trp residue at the second position of the core peptide as the only conserved residue in the core. As a result, the recently proposed bitryptides31 that feature a xWxxWx core and the KSLK leader motif are a large subfamily of the atropopeptides (Fig. 4E). In contrast, atropopeptides are defined by the KSLK leader motif in conjunction with the xWxxxx core motif without a conserved W at position 5 in the precursor and core peptide, respectively. This observation parallels recent insights into biarylitide biosynthesis that likewise resulted in the expansion of the biarylitide biosynthetic space.27
Our study and recent reports from other labs25,31 suggest that not all atropopeptides are characterized by the complex 3-dimensional shape reported for tryptorubin A which results in two possible non-canonical atropisomeric configurations. While most of the characterized atropopeptides (amyxirubin B, varsorubin B1 and laurentirubin B in this study) feature this unusual type of stereoisomerism, the family members that do not (scabrirubin and varsorubin B2a in this study), at least, require canonical atropisomeric assignments.31 As a result, we propose to keep the name atropopeptides for the RiPP family. As the complex 3-dimensional shape is not a common characteristic of all atropopeptides, we propose to redefine the atropopeptide family based on their conserved biosynthetic features that include the presence of an atropopeptide class-defining P450, the conserved KSLK leader motif and a W residue at position two of the core peptide (Fig. 4E).
To validate the AtropoFinder algorithm, we characterized four atropopeptide BGCs and elucidated the structures of their associated compounds. Scabrirubin (5) features a single carbon–nitrogen bond between two tryptophan residues of the tetrapeptide core; the sva BGC produces varsorubins (3,4a and 4b) varying in complexity with three and one modifications, respectively, and laurentirubin B (6), one of the most complex atropopeptide reported to date, harbors a total of four modifications including an unprecedented ether bond between Trp-2 and Tyr-2. The presence of three different products with the same core peptide sequence and length (varsorubins) is remarkable as previous reports indicate the full conversion of the core peptide sequence into a single product. Moreover, the lau BGC one of the few characterized atropopeptide BGCs that does not harbor the minimal BGC architecture made up of precursor and P450 genes. The formation of the ether bond after the atropospecific formation of the bridge-above configuration of a tryptorubin-like intermediate by the second P450 encoded in the BGC impacts the 3-dimensional shape of the molecule. The highly rigid conformation of 6 was confirmed by the presence of asymmetric 1H and 13C NMR signals for the aromatic ring of Tyr-2 residue, along with DFT calculations (Fig. S58†).
The three conserved bridges that are present in most characterized atropopeptides result in the formation of a highly strained bimacrocyclic ring that shows non-canonical atropisomerism. In accordance with the Pansa/Mansa nomenclature for the stereochemical description of conformationally diastereomeric bismacrocyclic peptides which was recently defined by Süssmuth and co-workers,15 we determined the configuration of the bicyclic macrolactam rings for all characterized atropopeptides as Pansa. This nomenclature is applicable for the configurational description of atropopeptides in case they feature a bicyclic ring system. The only amino acid not involved in the formation of the bicyclic ring system in atropopeptides is the relatively flexible sixth amino acid of the core (e.g., Tyr6) which is not locked into place by the three characteristic modifications. In the case of laurentirubin B, the fourth modification restrains the otherwise conformationally flexible Tyr6 in the form of a cyclophane lactam, making the 3D structure more rigid.
The number of modifications installed into atropopeptides reported here ranges from one to four and the number of bond types from one (scabrirubin), to two (e.g., varsorubins) and three (laurentirubin B) that are installed by a single multi-functional P450 or alternatively four through the joint action of two P450s in the case of laurentirubin B.
In conclusion, our research expands the atropopeptide family by more than 50%, provides a comprehensive understanding of atropopeptide biosynthesis and timing of maturation. The systematic investigation of atropopeptide biosynthetic space lays the foundation for future investigations into this peptide family and showcases the potential of machine learning-based genome mining algorithms for the identification of non-canonical biosynthetic pathways that elude unrecognized by existing genome mining tools. The characterization of the atropopeptide RiPP family serves as a proof-of-concept for the versatile genome mining concept developed in this study. The machine learning-based identification of unique features in tailoring enzymes complements current genome mining strategies to chart biosynthetic dark matter. We believe that our approach can be adapted by retraining the classifier on different training data sets to chart the biosynthetic space of other RiPP families, to identify overlooked RiPP BGCs beyond family borders and other currently overlooked natural product BGCs.28,43
Experimental
Materials
All chemicals, reagents and solvents were purchased from Sigma-Aldrich or Carl Roth (Germany), unless stated otherwise. Antibiotics and medium components were purchased from Carl Roth (Germany). Oligonucleotide primers were synthesized by Microsynth AG (Germany). All restriction enzymes, Q5 High Fidelity DNA polymerase, HiFi DNA Assembly master mix, T4 DNA ligase, deoxynucleotides (dNTPs) and DNA Ladder were purchased from New England Biolabs (UK). Monarch™ DNA gel extraction kit, Monarch™ Genomic DNA Purification Kit and Monarch™ Plasmid Miniprep kit from New England Biolabs (UK) were used to purify DNA.Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 proteins annotated as ‘cytochrome p450.’ Concurrently, a positive dataset was meticulously curated by deploying the tryptorubin A cytochrome P450 WP_007820080.1 as a query for Blastp analysis against the NCBI non-redundant protein sequences (nr) database. From this analysis, 51 putative biosynthetic gene cluster (BGC) sequences were identified through a manual search for precursors situated in the genetic neighborhood of each Blast hit. To dereplicate both datasets, cdhit V4.8.1
665 proteins annotated as ‘cytochrome p450.’ Concurrently, a positive dataset was meticulously curated by deploying the tryptorubin A cytochrome P450 WP_007820080.1 as a query for Blastp analysis against the NCBI non-redundant protein sequences (nr) database. From this analysis, 51 putative biosynthetic gene cluster (BGC) sequences were identified through a manual search for precursors situated in the genetic neighborhood of each Blast hit. To dereplicate both datasets, cdhit V4.8.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44 was employed. Parameters were configured to cluster sequences with a minimum of 95% sequence similarity and to utilize a word size of 5. As a result of this dereplication process, the negative and positive datasets were refined to 9065 and 37 sequences, respectively.
44 was employed. Parameters were configured to cluster sequences with a minimum of 95% sequence similarity and to utilize a word size of 5. As a result of this dereplication process, the negative and positive datasets were refined to 9065 and 37 sequences, respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 364 protein sequences was derived from the NCBI Identical Protein Groups database (25.01.2023).45 These sequences, which encompassed the term “p450” in their descriptions and which had lengths ranging from 300 to 450 amino acids, were procured using the following query parameters: p450[All Fields] AND (refseq[filter] AND prokaryotes[filter] AND (“300”[SLEN]:“450”[SLEN])).45 The method employed to assemble the dataset mirrored that of the training dataset.
364 protein sequences was derived from the NCBI Identical Protein Groups database (25.01.2023).45 These sequences, which encompassed the term “p450” in their descriptions and which had lengths ranging from 300 to 450 amino acids, were procured using the following query parameters: p450[All Fields] AND (refseq[filter] AND prokaryotes[filter] AND (“300”[SLEN]:“450”[SLEN])).45 The method employed to assemble the dataset mirrored that of the training dataset.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46) with the default settings (clustalo-1.2.3-Ubuntu-x86_64 -i input.fasta -o clustal.aln -v --outfmt=clustal --output-order=tree-order --auto -t Protein) with a reference cytochrome P450 with known annotation of functional regions from Mycobacterium tuberculosis H2102 (GenBank: KBE51585.1) in a multiple sequence alignment. The sequences were fragmented at positions 92, 192, 275, and 395 relative to the reference sequence. The resulting fragments represent the different functional regions of the cytochrome obtained from the annotations in the Cytochrome P450 Engineering Database record 10
46) with the default settings (clustalo-1.2.3-Ubuntu-x86_64 -i input.fasta -o clustal.aln -v --outfmt=clustal --output-order=tree-order --auto -t Protein) with a reference cytochrome P450 with known annotation of functional regions from Mycobacterium tuberculosis H2102 (GenBank: KBE51585.1) in a multiple sequence alignment. The sequences were fragmented at positions 92, 192, 275, and 395 relative to the reference sequence. The resulting fragments represent the different functional regions of the cytochrome obtained from the annotations in the Cytochrome P450 Engineering Database record 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 (N-terminus, substrate binding region 1, substrate binding region 2, core region and C-terminus).47
800 (N-terminus, substrate binding region 1, substrate binding region 2, core region and C-terminus).47
To prevent overfitting, the sequences were translated into a simplified amino acid code (Fig. S3†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 for python. The dataset was balanced using the Random Over Sampler from the Python package Imbalanced-learn 0.9.0.49 A 60
48 for python. The dataset was balanced using the Random Over Sampler from the Python package Imbalanced-learn 0.9.0.49 A 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 split for the training dataset and internal validation set was used. The number of overlapping k-mers of motifs with the length of four that occurred in at least half of the 37 putative atropopeptide sequences in each segment were used as features. Different classifiers were compared and the Random Forest classifier was chosen because of its high f1 score for atropopeptide P450s (0.97). Hyperparameter tuning was performed to optimize the maximum amount of samples per leaf and the maximum tree depth assessed on the obtained maximum balanced accuracy (balanced accuracy = (recall + specificity)/2) (optimal parameters: 1 and 10, respectively). Every hit with a score >0.15 was considered “positive”. After the first run on the classification dataset, 202 putative atropopeptide cytochrome p450s were curated from the results using CoreFinder and sequence alignments. These additional putative atropopeptide cytochrome P450s were chosen to supplement the training dataset, leading to a size of 113 for the positive training data set after deduplication with cd-hit V4.8.1
40 split for the training dataset and internal validation set was used. The number of overlapping k-mers of motifs with the length of four that occurred in at least half of the 37 putative atropopeptide sequences in each segment were used as features. Different classifiers were compared and the Random Forest classifier was chosen because of its high f1 score for atropopeptide P450s (0.97). Hyperparameter tuning was performed to optimize the maximum amount of samples per leaf and the maximum tree depth assessed on the obtained maximum balanced accuracy (balanced accuracy = (recall + specificity)/2) (optimal parameters: 1 and 10, respectively). Every hit with a score >0.15 was considered “positive”. After the first run on the classification dataset, 202 putative atropopeptide cytochrome p450s were curated from the results using CoreFinder and sequence alignments. These additional putative atropopeptide cytochrome P450s were chosen to supplement the training dataset, leading to a size of 113 for the positive training data set after deduplication with cd-hit V4.8.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44 (cutoff 0.95, word size = 5). The classifiers underwent training on the newly acquired dataset, according to the previous methodology (f1 score = 0.97). Parameter optimization yielded optimal values of a singular leaf (maximum leaves = 1) and a tree depth constrained to 14 (maximum tree depth = 14).
44 (cutoff 0.95, word size = 5). The classifiers underwent training on the newly acquired dataset, according to the previous methodology (f1 score = 0.97). Parameter optimization yielded optimal values of a singular leaf (maximum leaves = 1) and a tree depth constrained to 14 (maximum tree depth = 14).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 all on default settings. The plot in Fig. S5† was created using BGCViz.50
17 all on default settings. The plot in Fig. S5† was created using BGCViz.50
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 51 from sequence alignments of core peptides and precursor peptides using CLUSTALW version 1.2.3
51 from sequence alignments of core peptides and precursor peptides using CLUSTALW version 1.2.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46 with the default settings.
46 with the default settings.
The generated sequence similarity network was then visualized using Cytoscape 3.10.1,53 and annotations were added using the AutoAnnotate 1.4 plugin.54
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46 with the default settings. The alignment was trimmed using ClipKIT version 1.4.0
46 with the default settings. The alignment was trimmed using ClipKIT version 1.4.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 using the default settings. The tree was assembled using IQ-TREE multicore version 1.6.12
55 using the default settings. The tree was assembled using IQ-TREE multicore version 1.6.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 56 with default settings and visualized using iTOL version 5.57 To perform the co-occurrence analysis, the RODEO2 web tool17 was used with all putative P450s determined by AtropoFinder as input. The GC content of the P450 was compared to the average GC content of the genome of the species queried from the NCBI taxonomy browser.58
56 with default settings and visualized using iTOL version 5.57 To perform the co-occurrence analysis, the RODEO2 web tool17 was used with all putative P450s determined by AtropoFinder as input. The GC content of the P450 was compared to the average GC content of the genome of the species queried from the NCBI taxonomy browser.58
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 in multimer mode with standard parameters. The structures were visualized in pymol.59
39 in multimer mode with standard parameters. The structures were visualized in pymol.59
To insert a ribosomal binding site in front of svaB, the plasmids pUWL201-OriT-Sva was used as template for reverse amplification by PCR using primer pairs Svar_RBS_CYP450_Fw and Svar-CYP450_Rev (Table S8†) to afford the linearized plasmid, which was then incubated with KLD mix. 1 μL of the KLD reaction mix was transformed into E. coli DH5α. The generated plasmid was named pUWL201-OriT-SvaR.
To clone lau BGC into pUWL201-OriT, the plasmid pUWL201-OriT was linearized with KpnI and BamHI. The fragment containing the lau BGC was excised from pIJ10257-lau using KpnI and BamHI. The linearized pUWL201-OriT and lau BGC fragments were ligated using T4 DNA ligase to generate pUWL201-OriT-lau after sequencing confirmation.
For heterologous expression of sva (Sva and SvaR), sca, lau, lauA + lauB1 and lauA + lauB2, the corresponding plasmids pUWL201-OriT-Sva, pUWL201-OriT-SvaR, pUWL201-OriT-sca, pUWL201-OriT-lau, pUWL201-OriT-lauA + lauB1, pUWL201-OriT-lauA + lauB2 were transformed into S. albus J1074 by conjugation as described above.
LC-MS measurements were carried out on an Ultimate 3000 LC system (Thermo Fisher) coupled to an AmaZonX (Bruker) electrospray ionization (ESI) mass spectrometer. Separation was achieved on a C18 column (ACQUITY UPLC BEH, 130 Å, 1.7 μm particle size, 2.1 × 100 mm, Waters) at a flow rate of 0.4 mL min−1 at 40 °C, using acetonitrile and Milli-Q water supplemented with 0.1% (v/v) formic acid in a gradient ranging from 5 to 95% acetonitrile over 16 min. HPLC-ESI-QTOF-MS analyses were conducted on an Ultimate 3000 LC system (Thermo Fisher) coupled to an Impact II QTOF mass spectrometer (Bruker). Separation was achieved on a C18 column (ACQUITY UPLC BEH, 130 Å, 1.7 μm particle size, 2.1 mm × 100 mm, Waters) at a flow rate of 0.4 mL min−1 at 40 °C, using acetonitrile and Milli-Q water supplemented with 0.1% (v/v) formic acid in a gradient ranging from 5 to 95% acetonitrile over 16 min. Data were acquired in positive mode at a scan range between 100 to 1200 m/z to detect atropopeptides. The software DataAnalysis 4.3 (Bruker) was used to evaluate the measurements.
The obtained crude extracts were fractionated by preparative chromatography (Büchi Pure C-850 Flash/Prep) using water and acetonitrile supplemented with 0.1% (v/v) formic acid as mobile phases A and B, respectively. Atropopeptides were purified from crude extracts using a Xbridge Prep C18 column (5 μm particle size, 250 × 19 mm, Waters) and eluted with a flow rate of 20 mL min−1 and a gradient from 10 to 45% solvent B over 40 minutes. Eluting compounds were detected with a UV-detector (254–400 nm). Fractions were screened for atropopeptides by LC-MS analysis, and the fractions containing the desired compounds were dried under reduced pressure, re-dissolved in methanol and processed further by semi-preparative HPLC on an Agilent 1260 Infinity II UV-Vis system, equipped with a phenyl–hexyl column (100 Å, 5 μm particle size, 250 × 10 mm, Phenomenex). Solvents used were Milli-Q water and acetonitrile as mobile phases A and B, respectively. Elution was achieved with a method containing three isocratic steps, 25% solvent B for 10 minutes, 28% solvent B for 20 minutes and 35% solvent B for 2 minutes using a flow rate of 5 mL min−1. After LC-MS analysis of the fractions, fractions containing the compound of interest were collected, dried under reduced pressure and re-purified on the same semi-preparative HPLC system. A gradient was set from 30 to 39% solvent B over 30 min and then 100% solvent B for 5 min, giving a total run time of 35 min, with a flow rate of 3 mL min−1. The purified compound was weighed (1.37 mg), dissolved in DMSO-d6 and then subjected to NMR analysis.
For large-scale production of varsorubin, seed cultures of Streptomyces strains carrying pUWL201-OriT-SvaR were prepared by fermentation in a culture tube containing 5 mL TSB medium with 50 μg mL−1 apramycin at 30 °C, 200 rpm, for 2–3 days. After incubation, 1 mL of the seed culture was used to inoculate (6 × 100 mL) TSB medium containing 50 μg per mL apramycin and 5% (w/v) Diaion HP-20 resin (Sigma-Aldrich) in 500 mL Erlenmeyer flasks. After 3 days, 100 mL was used to inoculate (6 × 1000 mL) mL TSB medium containing 50 μg mL−1 apramycin and 5% (w/v) Diaion HP-20 resin (Sigma-Aldrich) in a 5000 mL Erlenmeyer flask. The cultures were incubated at 30 °C, 180 rpm, for 4–6 days. The Diaion HP-20 resin in the TSB cultures was recovered by filtration with Miracloth (Milipore, MA, USA). The Diaion HP-20 resin was subsequently washed with H2O and then extracted twice with one culture volume of acetone. The crude extract was dried under reduced pressure. The obtained crude extracts were dissolved in DMSO and used to purify varsorubins by preparative HPLC on an Agilent 1260 Infinity II UV-Vis system, equipped with a XBridge BEH C18 OBD Prep Column (130 Å, 10 μm particle size, 30 mm x 250 mm, Waters). Solvents used were Milli-Q water and acetonitrile supplemented with 0.1% (v/v) formic acid as mobile phases A and B, respectively. A gradient was set from 10 to 45% solvent B over 35 min and then 100% solvent B for 5 min, giving a total run time of 40 min, with a flow rate of 20 mL min−1. The fractions containing the desired compounds were dried under reduced pressure, redissolved in DMSO and processed further by semi-preparative HPLC on an Agilent 1260 Infinity II UV-Vis system, equipped with a Luna phenyl–hexyl column (100 Å, 5 μm particle size, 250 × 4.6 mm, Phenomenex). Solvents used were Milli-Q water and acetonitrile supplemented with 0.1% (v/v) formic acid as mobile phases A and B, respectively. A gradient was set from 20 to 60% solvent B over 30 min and then 100% solvent B for 5 min, giving a total run time of 35 min, with a flow rate of 3 mL min−1. Fractions containing the desired compounds were subjected to LC-MS to check for purity. The purified compounds were weighed (1.6 mg for varsorubin B1 and 1.8 mg for varsorubin B2a) and redissolved in DMSO-d6. After that, the purified compounds were subjected to NMR analysis (1D and 2D NMR).
For the large-scale fermentation of S. albus/pUWL201-OriT-sca leading to production of scabrirubin, 6 L cultures were prepared as follows: the spores collected from one MS agar plate were incubated into a 5 L Erlenmeyer flask containing 1 L of the TSB medium with apramycin (50 μg mL−1) and incubated on a rotary shaker (180 rpm) at 30 °C for 6 days. 5% (w/v) of sterilized resin (Diaion HP-20) was added into each flask after 4 days of incubation, and the flasks were incubated for another 2 days. 4 L cultures were prepared as follows: the spores collected from one MS agar plate were inoculated into five 1 L Erlenmeyer flasks containing 200 mL of the TSB medium with apramycin (50 μg mL−1). The incubation conditions were the same as described above. The resins from the 10 L cultures were harvested by filtration through a metal sieve (40 mesh). The harvested resins were washed with water, transferred to a separatory funnel and eluted with 5 L of acetone. After evaporation of the organic solvents under reduced pressure, the crude extracts were subjected to normal phase silica gel column chromatography (230–400 mesh) and eluted with CHCl3/CH3OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 97
0, 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 95
3, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 90
5, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 8
10, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 0
1, 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, 600 mL) to yield 8 fractions (Frs.1–8). Fr.6 and Fr.7 were combined and separated by Sephadex LH-20, eluted with CHCl3/CH3OH (1
1, v/v, 600 mL) to yield 8 fractions (Frs.1–8). Fr.6 and Fr.7 were combined and separated by Sephadex LH-20, eluted with CHCl3/CH3OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to obtain 4 sub-fractions (Fr.1.1 to Fr.1.4). Fr.1.2 was further separated by semi-preparative HPLC using a reverse-phase column (Luna phenyl–hexyl, 250 × 4.6 mm, 5 μm particle size, Phenomenex) with UV detection at 300 nm to afford compound 5 (3.5 mg) using the following gradient: solvent system (solvent A, water supplementing with 0.1% formic acid; solvent B, acetonitrile); 30–39% B (0–25 min), 39–100% B (25–26 min), 100% B (26–30 min), 100–30% B (30–31 min), 30% B (31–35 min); flow rate at 2.5 mL min−1. The purified compound was weighed (3.5 mg), redissolved in DMSO-d6 and subjected to NMR analysis.
1, v/v) to obtain 4 sub-fractions (Fr.1.1 to Fr.1.4). Fr.1.2 was further separated by semi-preparative HPLC using a reverse-phase column (Luna phenyl–hexyl, 250 × 4.6 mm, 5 μm particle size, Phenomenex) with UV detection at 300 nm to afford compound 5 (3.5 mg) using the following gradient: solvent system (solvent A, water supplementing with 0.1% formic acid; solvent B, acetonitrile); 30–39% B (0–25 min), 39–100% B (25–26 min), 100% B (26–30 min), 100–30% B (30–31 min), 30% B (31–35 min); flow rate at 2.5 mL min−1. The purified compound was weighed (3.5 mg), redissolved in DMSO-d6 and subjected to NMR analysis.
For large-scale production of laurentirubin B, seed cultures of Streptomyces albus strains carrying pUWL201-OriT-Precursor-laurentii were prepared by fermentation in a culture tube containing 5 mL TSB medium with appropriate antibiotic(s) at 30 °C, 200 rpm, for 2–3 days. After incubation, 1 mL of the seed culture was used to inoculate (6 × 100 mL) TSB medium containing appropriate antibiotic(s) and 5% (w/v) Diaion HP-20 resin in 500 mL Erlenmeyer flasks. After 3 days, 100 mL was used to inoculate (6 × 1000 mL) mL TSB medium containing appropriate antibiotic(s) and 5% (w/v) Diaion HP-20 resin in a 5000 mL Erlenmeyer flask. The cultures were incubated at 30 °C, 180 rpm, for 4–6 days. The Diaion HP-20 resin in the cultures was recovered by filtration with Miracloth. The Diaion HP-20 resin was subsequently washed with H2O and then extracted twice with one culture volume of acetone. The crude extract was dried under reduced pressure. The obtained crude extracts were dissolved in DMSO and then purified by preparative HPLC on an Agilent 1260 Infinity II UV-Vis system, equipped with a XBridge BEH C18 OBD Prep Column (130 Å, 10 μm particle size, 30 mm × 250 mm, Waters). Solvents used were Milli-Q water and acetonitrile supplemented with 0.1% (v/v) formic acid as mobile phases A and B, respectively. A gradient was set from 10 to 45% solvent B over 35 min and then 100% solvent B for 5 min, giving a total run time of 40 min, with a flow rate of 20 mL min−1. The fractions containing the desired compound were dried under reduced pressure, redissolved in DMSO and processed further by semi-preparative HPLC on an Agilent 1260 Infinity II UV-Vis system, equipped with a Luna phenyl–hexyl column (100 Å, 5 μm particle size, 250 × 4.6 mm, Phenomenex). Solvents used were Milli-Q water and acetonitrile supplemented with 0.1% (v/v) formic acid as mobile phases A and B, respectively. A gradient was set from 20 to 60% solvent B over 30 min and then 100% solvent B for 5 min, giving a total run time of 35 min, with a flow rate of 3 mL min−1. Fractions containing the desired compound were dried under reduced pressure. The purified compound was weighed (1.6 mg), redissolved in DMSO-d6 and subjected to NMR analysis.
Varsorubin B2a (4): yield 1.8 mg; orange powder; 1H and 13C NMR data, see Table S3;† HRMS (ESI-QTOF) m/z [M + H]+ calcd for C42H49N8O8+ 793.3668, found 793.3655.
Scabrirubin (5): yield 3.5 mg; white powder; 1H and 13C NMR data, see Table S4;† HRMS (ESI-QTOF) m/z [M + H]+ calcd for C37H41N6O5+ 649.3133, found 649.3132.
Laurentirubin B (6): yield 1.6 mg; orangewooder; 1H and 13C NMR data, see Table S5;† HRMS (ESI-QTOF) m/z [M + H]+ calcd for C46H46N7O8+ 824.3402, found 824.3401.
Varsorubin B2a (4a) was isolated as an orange powder. Its molecular formula was determined to be C42H48N8O8 based on a protonated ion at m/z 793.3655 (calcd for C42H49N8O8+, 793.3668, Δ −1.6 ppm) in HR-ESI-QTOF-MS data (Fig. S25†). The 1H and 2D NMR spectra (Fig. S26–S28, S30 and S31†) showed similarity to those of 3 except for the presence of the AA′XX′ spin system (δH 6.95, 6.63) for a Tyr residue, a secondary amine proton signal (δH 7.24) for Trp-1, and an amide proton signal (δH 6.95) for Trp-2, and the absence of a signal for H-11 of Trp-2. These differences suggested that 4a is a peptide analog of 3 without the pyrroloindoline moiety at Trp-2 and a C–N bond between N-36 of Trp-2 and C-28 of Tyr. The HMBC correlations from H-38 and H-40 to C-11 (Fig. S29†) indicated the presence of a C–C linkage between C-39 of Trp-1 and C-11 of Trp-2. The presence of this C–C bond was further confirmed by NOESY correlations of H-8/H-38, H-8/H-40, H-9/H-38, H-9/H-40, and H-9/H-41 (Fig. S32 and S33†). The stereochemistry of the α-carbons for each amino acid was deduced as L-configuration based on the genomic data. The axial chirality between C-39 of Trp-1 and C-11 of Trp-2 was not determined in this study.
Scabrirubin (5) was obtained as a white powder. Its molecular formula was determined to be C37H40N6O5 (m/z 649.3132 [M + H]+, calcd for C37H41N6O5+, 649.3133, Δ −0.15 ppm) by HR-ESI-QTOF-MS data (Fig. S35†), suggesting the index of hydrogen deficiency to be 21. Analysis of the 1H and 13C NMR spectra, with the aid of the HSQC spectrum (Table S4 and Fig. S36–S39†), revealed the presence of 37 carbons assignable to four carbonyl carbons, eight sp2 nonprotonated carbons, 14 sp2 methines, five sp3 methines, four sp3 methylenes, and two methyls. These signals and signals for exchangeable NH protons (δH 8.04, 7.81, 7.35) are characteristic for peptides. The presence of a Phe residue was determined based on the 1H–1H COSY correlations between H-23/23′ and H-24 and the HMBC correlations of H-22/H-24, H-20/H-22, and H-20/H-21 (Fig. S43†). A continuous spin system in the aromatic region (δH 7.66, 7.52, 7.19, 7.12), a singlet signal for an aromatic proton (δH 7.85), and unequivalent methylene signals (δH 3.24, 3.15) suggested the presence of a Trp residue (Trp-2). The 1H–1H COSY spectrum (Fig. S40†) showed another constituted spin system region including resonances for two terminal methyls (δH 7.81, 4.06, 1.68, 1.66, 1.36, 0.94, 0.91), indicating the presence of an Ile residue. Furthermore, the C-34-substituted Trp (Trp-1) was assigned based on the 1H–1H COSY correlations of NH-29/H-29 and H-31/H-32 along with HMBC correlations from H-27 to C-29 and C-30, and from H-31 to C-30, C-33, and C-35, from H-32 to C-34, and from H-33 to C-35 (Fig. S43†). The amino acid sequence of 5 was determined to be NH2-Trp-Phe-Ile-Trp-CO2H based on the HMBC correlations from amino protons to carbonyl carbons (Fig. S41†). The stereochemistry of the α-carbons for each amino acid was deduced as L-configured based on the genomic data. The HMBC correlation from H-5 of Trp-2 to a quaternary carbon C-34 of Trp-1 suggested the presence of a C–N bond between C-34 of Trp-1 and N-5 of Trp-2. This bond was further confirmed by the NOESY correlations between the H-10/H-33, H-5/NH-29 (Fig. S42†).
Laurentirubin B (6) was isolated as an orange powder. Its molecular formula of C46H45N7O8 was determined by a protonated ion at m/z 824.3401 (calcd for C46H46N7O8+, 824.3402, Δ −0.1 ppm) in HR-ESI-QTOF-MS data (Fig. S46†). The 1H and 2D NMR spectra were akin to those of tryptorubin B16 except for the absence of an aromatic proton H-17 at a Trp-2 and signals of a Tyr-2 residue detected at δH 7.45, 7.10, 7.00, and 6.58 (Table S5, Fig. S47–S50 and S52†). These NMR data combined with the genomic data suggested that the amino acid sequence of 6 is identical to tryptorubin B (NH2-Trp-Tyr-Ile-Trp-Tyr-CO2H), but the modification of the amino acid sequence is different. The formation of the pyrroloindoline moiety at a Trp-2 residue was assigned from the COSY correlation between NH-12 and a sp3 methine H-12 as well as the HMBC correlations from H-12 to an α-carbon C-9, a methylene carbon C-10, and a quaternary carbon C-11. The HMBC correlation of H-10/C-41, H-12/C-41, H-40/C-11, and H-42/C-11 indicated the presence of a C–C bond between a quaternary carbon C-11 of Trp-1 and a nonprotonated sp2 carbon C-41 of Trp-1 (Fig. S52†). The presence of a C–N bond between C-30 of Tyr-1 and N-38 of Trp-1 was assigned from the 1H–15N HMBC correlations of H-28/N-38 and H-38/N-38 (Fig. S53†). This assignment was further confirmed by the NOESY cross peak between H-38 at Trp-1 and H-29 at Tyr-1 (Fig. S54†). The aryl ether bond between C-7 of Tyr-2 and C-17 of Trp-2 was deduced from the NOESY correlations of H-5′/H-10α and H-6/NH-12 (Fig. S53 and S54†), as well as asymmetric 1H and 13C signals for Tyr-2 residue (C-5, C-5′, C-6, C-6′) and the presence of a 1,2,3-trisubstituted aromatic ring in the pyrroloindoline moiety. The stereochemistry of the α-carbon for each amino acid residue was suggested as L-configuration based on the genomic data. Besides, the NOESY correlations of H-9/H-43, H-10/H-43, and H-12/H-40 (Fig. S53 and S54†) indicated the relative configuration of a quaternary carbon C-11 and an aminal carbon C-12 as (11S*,12R*) and the ansameric configuration of a bicyclic macrolactam ring as Pansa. To confirm the location of the aryl ether bridge and stereochemical configuration of the ansameric bismacrocycle, theoretical 13C NMR chemical shifts were calculated for possible conformers Pansa-6, Mansa-6, along with a possible isomer Pansa-6a that features an aryl ester bond between C-1 and C-17 instead of the aryl ether bond between C-7 and C-17 using the GIAO method. The calculated 13C NMR chemical shifts of Pansa-6a with the lowest values of the mean absolute error and mean squared error (Table S6†) were more favorable than those of Mansa-6 and Pansa-6a. Furthermore, the lowest energy conformer of Pansa-6 at the B3LYP/6-31G(d,p) level of theory (Fig. S57†) showed a rational geometry that was in agreement with the observed NOESY correlations in 6, except for the NOESY correlation of H-9/H-43. These DFT calculations supported the NMR analysis-based assignment of the ansameric configuration as Pansa, as well as the location of the aryl ether bond at C-7–C-17.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 to prepare the agar plates. Compounds were dissolved in DMSO at a concentration of 2.56 mg mL−1. Trimethoprim and ampicillin were used as positive controls for the antibacterial assays. Nystatin and cycloheximide were used as positive control for the antifungal assays. 6 mm paper disks were loaded with 5 μL of each compound, respectively, and then placed onto the agar plates. The plates were incubated at 37 °C for 16 h (ESKAPE pathogens) or for 24 h (C. albicans CAF-2), or at 30 °C for one to five days (A. pascens DSM 20545). The antimicrobial activity was determined by the size of the zone of inhibition.
100 to prepare the agar plates. Compounds were dissolved in DMSO at a concentration of 2.56 mg mL−1. Trimethoprim and ampicillin were used as positive controls for the antibacterial assays. Nystatin and cycloheximide were used as positive control for the antifungal assays. 6 mm paper disks were loaded with 5 μL of each compound, respectively, and then placed onto the agar plates. The plates were incubated at 37 °C for 16 h (ESKAPE pathogens) or for 24 h (C. albicans CAF-2), or at 30 °C for one to five days (A. pascens DSM 20545). The antimicrobial activity was determined by the size of the zone of inhibition.
Data availability
All code and raw data pertaining to the machine learning analyses presented in this study can be accessed at the following GitHub repository https://github.com/FriederikeBiermann/AtropoFinder and is publicly available as of the date of publication. Furthermore, all biosynthetic gene clusters (BGCs) and associated compounds derived from this research are available in the ESI† in the form of Genbank files and have been submitted to the MiBIG (Minimum Information about a Biosynthetic Gene Cluster) database for public access and reference. NMR spectra can be found in the ESI.† The raw data used to create Fig. 3 can be found in the ESI files (BGC_data_only_positives.csv, atropopeptide_p450s_newick.txt).† All AtropoFinder results can be found in the ESI file BGC_data.csv.†Author contributions
FB, PN, EJNH conceptualized the project, FB developed AtropoFinder and CoreFinder, performed analysis of atropopeptide BGCs, and conducted modeling studies, BT, MB, PN, YD designed and constructed plasmids, conducted heterologous expression studies, and isolated compounds, BT, YK, RU analyzed NMR data, ASW performed DFT calculations, BT conducted bioactivity assays, FB, BT, YK and EJNH wrote manuscript with contributions from all co-authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
EJNH gratefully acknowledges funding by the LOEWE Center for Translational Biodiversity Genomics (LOEWE TBG), the German Research Foundation (Emmy Noether Program), and the Funds of the Chemical Industry Germany. FB is supported by a Kekulé Fellowship of the Funds of the Chemical Industry Germany. We thank Bertholt Gust for providing strains and plasmids. We gratefully thank Dr Elke Duchardt-Ferner and Gabriele Sentis for their technical assistance with NMR measurements. Open Access funding enabled and organized by Projekt DEAL. Fig. 1A was created in BioRender.Notes and references
- T. Dang and R. D. Süssmuth, Acc. Chem. Res., 2017, 50, 1566–1576 CrossRef CAS PubMed.
- R. W. Fairbrother and B. L. Williams, Lancet, 1956, 271, 1177–1178 CrossRef CAS PubMed.
- B. A. Johnson, H. Anker and F. L. Meleney, Science, 1945, 102, 376–377 CrossRef CAS PubMed.
- R. Donovick, J. F. Pagano, H. A. Stout and M. J. Weinstein, Antibiot. Annu., 1955, 3, 554–559 Search PubMed.
- M. Dreyfuss, E. Härri, H. Hofmann, H. Kobel, W. Pache and H. Tscherter, Eur. J. Appl. Microbiol., 1976, 3, 125–133 CrossRef CAS.
- H. Kleinkauf, H. Koischwitz, J. Vater, R. Zocher, U. Keller, I. Mahmutoglu, K. Bauer, M. Altmann, R. Kittelberger, M. Marahiel and J. Salnikow, in Regulation of Secondary Product and Plant Hormone Metabolism, Elsevier, 1979, pp. 37–47 Search PubMed.
- S. L. Wenski, S. Thiengmag and E. J. N. Helfrich, Synth. Syst. Biotechnol., 2022, 7, 631–647 CrossRef CAS PubMed.
- M. Montalbán-López, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud, Y. Goto, M. J. Grande Burgos, C. Hill, S. Kim, J. Koehnke, J. A. Latham, A. J. Link, B. Martínez, S. K. Nair, Y. Nicolet, S. Rebuffat, H.-G. Sahl, D. Sareen, E. W. Schmidt, L. Schmitt, K. Severinov, R. D. Süssmuth, A. W. Truman, H. Wang, J.-K. Weng, G. P. van Wezel, Q. Zhang, J. Zhong, J. Piel, D. A. Mitchell, O. P. Kuipers and W. A. van der Donk, Nat. Prod. Rep., 2021, 38, 130–239 RSC.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K.-D. Entian, M. A. Fischbach, J. S. Garavelli, U. Göransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Müller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. T. Reaney, S. Rebuffat, R. P. Ross, H.-G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Süssmuth, J. R. Tagg, G.-L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- F. Biermann, S. L. Wenski and E. J. N. Helfrich, Beilstein J. Org. Chem., 2022, 18, 1656–1671 CrossRef CAS PubMed.
- T. P. Wyche, A. C. Ruzzini, L. Schwab, C. R. Currie and J. Clardy, J. Am. Chem. Soc., 2017, 139, 12899–12902 CrossRef CAS PubMed.
- M. C. Wilson, T. Mori, C. Rückert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. E. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. N. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crüsemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, Nature, 2014, 506, 58–62 CrossRef CAS PubMed.
- J. A. McIntosh, M. S. Donia and E. W. Schmidt, Nat. Prod. Rep., 2009, 26, 537–559 RSC.
- S. H. Reisberg, Y. Gao, A. S. Walker, E. J. N. Helfrich, J. Clardy and P. S. Baran, Science, 2020, 367, 458–463 CrossRef CAS PubMed.
- G. Yao, S. Kosol, M. T. Wenz, E. Irran, B. G. Keller, O. Trapp and R. D. Süssmuth, Nat. Commun., 2022, 13, 6488 CrossRef CAS PubMed.
- P. Nanudorn, S. Thiengmag, F. Biermann, P. Erkoc, S. D. Dirnberger, T. N. Phan, R. Fürst, R. Ueoka and E. J. N. Helfrich, Angew. Chem., Int. Ed., 2022, 61, e202208361 CrossRef CAS PubMed.
- J. I. Tietz, C. J. Schwalen, P. S. Patel, T. Maxson, P. M. Blair, H.-C. Tai, U. I. Zakai and D. A. Mitchell, Nat. Chem. Biol., 2017, 13, 470–478 CrossRef CAS PubMed.
- Y.-Y. Lee, M. Guler, D. N. Chigumba, S. Wang, N. Mittal, C. Miller, B. Krummenacher, H. Liu, L. Cao, A. Kannan, K. Narayan, S. T. Slocum, B. L. Roth, A. Gurevich, B. Behsaz, R. D. Kersten and H. Mohimani, Nat. Commun., 2023, 14, 4219 CrossRef CAS PubMed.
- K. Blin, S. Shaw, H. E. Augustijn, Z. L. Reitz, F. Biermann, M. Alanjary, A. Fetter, B. R. Terlouw, W. W. Metcalf, E. J. N. Helfrich, G. P. van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2023, 51, W46–W50 CrossRef CAS PubMed.
- M. A. Skinnider, C. W. Johnston, M. Gunabalasingam, N. J. Merwin, A. M. Kieliszek, R. J. MacLellan, H. Li, M. R. M. Ranieri, A. L. H. Webster, M. P. T. Cao, A. Pfeifle, N. Spencer, Q. H. To, D. P. Wallace, C. A. Dejong and N. A. Magarvey, Nat. Commun., 2020, 11(1), 6058 CrossRef CAS PubMed.
- E. L. C. de Los Santos, Sci. Rep., 2019, 9, 13406 CrossRef PubMed.
- N. J. Merwin, W. K. Mousa, C. A. Dejong, M. A. Skinnider, M. J. Cannon, H. Li, K. Dial, M. Gunabalasingam, C. Johnston and N. A. Magarvey, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 371–380 CrossRef CAS PubMed.
- A. M. Kloosterman, P. Cimermancic, S. S. Elsayed, C. Du, M. Hadjithomas, M. S. Donia, M. A. Fischbach, G. P. van Wezel and M. H. Medema, PLoS Biol., 2020, 18(12), e3001026 CrossRef CAS PubMed.
- M. M. Zdouc, M. M. Alanjary, G. S. Zarazúa, S. I. Maffioli, M. Crüsemann, M. H. Medema, S. Donadio and M. Sosio, Cell Chem. Biol., 2021, 28, 733–739 CrossRef CAS PubMed.
- B.-B. He, J. Liu, Z. Cheng, R. Liu, Z. Zhong, Y. Gao, H. Liu, Z.-M. Song, Y. Tian and Y.-X. Li, Angew. Chem., Int. Ed., 2023, e202311533 CAS.
- E. J. Han, S. R. Lee, S. Hoshino and M. R. Seyedsayamdost, ACS Chem. Biol., 2022, 17, 3121–3130 CrossRef CAS PubMed.
- H. Nam, J. S. An, J. Lee, Y. Yun, H. Lee, H. Park, Y. Jung, K.-B. Oh, D.-C. Oh and S. Kim, J. Am. Chem. Soc., 2024, 146(6), 3805–3815 CrossRef PubMed.
- F. Biermann and E. J. N. Helfrich, mSystems, 2021, e0084621 Search PubMed.
- C. M. Bishop, Pattern Recognition and Machine Learning, Springer, 2016 Search PubMed.
- C. L. Liu, Z. J. Wang, J. Shi, Z. Y. Yan, G. D. Zhang, R. H. Jiao, R. X. Tan and H. M. Ge, Angew. Chem., Int. Ed., 2024, 63, e202314046 CrossRef CAS PubMed.
- J. S. An, H. Lee, H. Kim, S. Woo, H. Nam, J. Lee, J. Y. Lee, S.-J. Nam, S. K. Lee, K.-B. Oh, S. Kim and D.-C. Oh, Angew. Chem., Int. Ed., 2023, e202300998 CAS.
- Y. L. Hu, F. Z. Yin, J. Shi, S. Y. Ma, Z. R. Wang, R. X. Tan, R. H. Jiao and H. M. Ge, J. Am. Chem. Soc., 2023, 145, 27325–27335 CrossRef CAS PubMed.
- G. D. Hannigan, D. Prihoda, A. Palicka, J. Soukup, O. Klempir, L. Rampula, J. Durcak, M. Wurst, J. Kotowski, D. Chang, R. Wang, G. Piizzi, G. Temesi, D. J. Hazuda, C. H. Woelk and D. A. Bitton, Nucleic Acids Res., 2019, 47, e110 CrossRef CAS PubMed.
- L. M. Carroll, M. Larralde, J. S. Fleck, R. Ponnudurai, A. Milanese, E. Cappio and G. Zeller, bioRxiv, 2021, preprint, DOI:10.1101/2021.05.03.442509.
- C. Rios-Martinez, N. Bhattacharya, A. P. Amini, L. Crawford and K. K. Yang, PLoS Comput. Biol., 2023, 19(5), e1011162 CrossRef CAS PubMed.
- K. Blin, S. Shaw, S. A. Kautsar, M. H. Medema and T. Weber, Nucleic Acids Res., 2021, 49, D639–D643 CrossRef CAS PubMed.
- A. Parmar, R. Katariya and V. Patel, in International Conference on Intelligent Data Communication Technologies and Internet of Things (ICICI) 2018, Springer International Publishing, Cham, 2019, pp. 758–763 Search PubMed.
- T. L. Bailey, J. Johnson, C. E. Grant and W. S. Noble, Nucleic Acids Res., 2015, 43, W39–W49 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- J. C. Navarro-Muñoz, N. Selem-Mojica, M. W. Mullowney, S. A. Kautsar, J. H. Tryon, E. I. Parkinson, E. L. C. De Los Santos, M. Yeong, P. Cruz-Morales, S. Abubucker, A. Roeters, W. Lokhorst, A. Fernandez-Guerra, L. T. D. Cappelini, A. W. Goering, R. J. Thomson, W. W. Metcalf, N. L. Kelleher, F. Barona-Gomez and M. H. Medema, Nat. Chem. Biol., 2020, 16, 60–68 CrossRef PubMed.
- B. R. Terlouw, K. Blin, J. C. Navarro-Muñoz, N. E. Avalon, M. G. Chevrette, S. Egbert, S. Lee, D. Meijer, M. J. J. Recchia, Z. L. Reitz, J. A. van Santen, N. Selem-Mojica, T. Tørring, L. Zaroubi, M. Alanjary, G. Aleti, C. Aguilar, S. A. A. Al-Salihi, H. E. Augustijn, J. A. Avelar-Rivas, L. A. Avitia-Domínguez, F. Barona-Gómez, J. Bernaldo-Agüero, V. A. Bielinski, F. Biermann, T. J. Booth, V. J. Carrion Bravo, R. Castelo-Branco, F. O. Chagas, P. Cruz-Morales, C. Du, K. R. Duncan, A. Gavriilidou, D. Gayrard, K. Gutiérrez-García, K. Haslinger, E. J. N. Helfrich, J. J. J. van der Hooft, A. P. Jati, E. Kalkreuter, N. Kalyvas, K. B. Kang, S. Kautsar, W. Kim, A. M. Kunjapur, Y.-X. Li, G.-M. Lin, C. Loureiro, J. J. R. Louwen, N. L. L. Louwen, G. Lund, J. Parra, B. Philmus, B. Pourmohsenin, L. J. U. Pronk, A. Rego, D. A. B. Rex, S. Robinson, L. R. Rosas-Becerra, E. T. Roxborough, M. A. Schorn, D. J. Scobie, K. S. Singh, N. Sokolova, X. Tang, D. Udwary, A. Vigneshwari, K. Vind, S. P. J. M. Vromans, V. Waschulin, S. E. Williams, J. M. Winter, T. E. Witte, H. Xie, D. Yang, J. Yu, M. Zdouc, Z. Zhong, J. Collemare, R. G. Linington, T. Weber and M. H. Medema, Nucleic Acids Res., 2023, 51, D603–D610 CrossRef CAS PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS PubMed.
- M. Baunach, J. Franke and C. Hertweck, Angew. Chem., Int. Ed., 2015, 54, 2604–2626 CrossRef CAS PubMed.
- W. Li and A. Godzik, Bioinformatics, 2006, 22(13), 1658–1659 CrossRef CAS PubMed.
- E. W. Sayers, E. E. Bolton, J. R. Brister, K. Canese, J. Chan, D. C. Comeau, R. Connor, K. Funk, C. Kelly, S. Kim, T. Madej, A. Marchler-Bauer, C. Lanczycki, S. Lathrop, Z. Lu, F. Thibaud-Nissen, T. Murphy, L. Phan, Y. Skripchenko, T. Tse, J. Wang, R. Williams, B. W. Trawick, K. D. Pruitt and S. T. Sherry, Nucleic Acids Res., 2022, 50, D20–D26 CrossRef CAS PubMed.
- J. D. Thompson, D. G. Higgins and T. J. Gibson, Nucleic Acids Res., 1994, 22, 4673–4680 CrossRef CAS PubMed.
- D. Sirim, F. Wagner, A. Lisitsa and J. Pleiss, BMC Biochem., 2009, 10, 27 CrossRef PubMed.
- R. Garreta and G. Moncecchi, Learning Scikit-Learn: Machine Learning in Python, Packt Pub Limited, 2013 Search PubMed.
- G. Lemaitre, F. Nogueira and C. K. Aridas, arXiv, 2016, preprint, arXiv:1609.06570 Search PubMed.
- P. Hrab and B. Ostash, BGCViz, 2020 Search PubMed.
- G. E. Crooks, G. Hon, J.-M. Chandonia and S. E. Brenner, Genome Res., 2004, 14, 1188–1190 CrossRef CAS PubMed.
- R. Zallot, N. Oberg and J. A. Gerlt, Biochemistry, 2019, 58(41), 4169–4182 CrossRef CAS PubMed.
- J. A. Gustavsen, S. Pai, R. Isserlin, B. Demchak and A. R. Pico, F1000Research, 2019, 8, 1774 Search PubMed.
- M. Kucera, R. Isserlin, A. Arkhangorodsky and G. D. Bader, F1000Research, 2016, 5, 1717 Search PubMed.
- J. L. Steenwyk, T. J. Buida 3rd, Y. Li, X.-X. Shen and A. Rokas, PLoS Biol., 2020, 18, e3001007 CrossRef CAS PubMed.
- J. Trifinopoulos, L.-T. Nguyen, A. von Haeseler and B. Q. Minh, Nucleic Acids Res., 2016, 44, W232–W235 CrossRef CAS PubMed.
- I. Letunic and P. Bork, Nucleic Acids Res., 2021, 49, W293–W296 CrossRef CAS PubMed.
- C. L. Schoch, S. Ciufo, M. Domrachev, C. L. Hotton, S. Kannan, R. Khovanskaya, D. Leipe, R. Mcveigh, K. O'Neill, B. Robbertse, S. Sharma, V. Soussov, J. P. Sullivan, L. Sun, S. Turner and I. Karsch-Mizrachi, Database, 2020, 2020(1), baaa062 CrossRef CAS PubMed.
- W. L. DeLano, CCP4 Newsl. Protein Crystallogr., 2002, vol. 40, pp. 82–92 Search PubMed.
- G. Yao, S. Kosol, M. T. Wenz, E. Irran, B. G. Keller, O. Trapp and R. D. Süssmuth, Nat. Commun., 2022, 13, 1–8 Search PubMed.
- I. A. Konstantinov and L. J. Broadbelt, J. Phys. Chem. A, 2011, 115, 12364–12372 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc03469d |
| This journal is © The Royal Society of Chemistry 2024 |