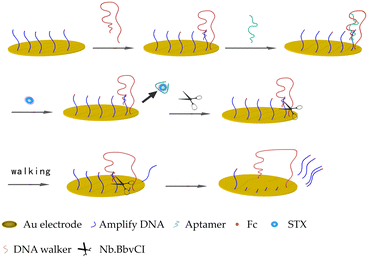
Open Access Article
Open Access ArticleDNA walker coupled with nicking endonuclease for sensitive electrochemical detection of saxitoxin†
Yiwei
Liu
,
Shumin
Feng
,
Ruoxi
Zhong
,
Yuanchang
Peng
,
Guoyuan
Mu
,
Jiayi
Bai
,
Wei
Chen
 * and
Zhan
Qu
* and
Zhan
Qu
 *
*
Department of Biophysics, Institute of Medical Engineering, School of Basic Medical Sciences, Health Science Center, Xi'an Jiaotong University, Xi'an 710061, China. E-mail: weiwcchen@xjtu.edu.cn; zhan.qu@xjtu.edu.cn
First published on 24th July 2024
Abstract
Saxitoxin (STX) as one of the paralytic shellfish toxins has become a serious public health and environmental issue. In this regards, developing highly sensitive and selective biosensors may help find a solution. Herein, a ferrocene (Fc)-labeled DNA walker coupled with nicking endonuclease Nb.BbvCI was used to construct a sensitive electrochemical aptasensor for STX detection. First, an amplified DNA, aptamer and DNA walker formed a sandwich structure on a gold electrode. This structure was disintegrated when STX was added, resulting in the hybridization of the amplified DNA and DNA walker. Thereafter, the DNA walker was activated by Nb.BbvCI to achieve stepwise cleavage of the hybridized amplified DNA. The released Fc-amplified DNA generated an electrochemical signal that decreased linearly with the logarithm value of STX concentration in the range of 1 pM–100 nM with a detection limit of 0.58 pM. Meanwhile, the proposed aptasensor exhibited good selectivity and recovery rate. The DNA walker coupled with the nicking endonuclease provides effective signal amplification for the detection of toxins and fabrication of sensitive aptasensors.
Introduction
Saxitoxin (STX) produced by certain dinoflagellate species is one of the paralytic shellfish poisoning toxins.1 STX as a sodium channel blocker disrupts the normal physiological process, resulting in paralysis or even respiratory failure.2 Thus, the STX content in seafood has a strict regulatory limit in certain countries.3 Accordingly, the detection of STX in shellfish is crucial to public health and environmental monitoring. Conventional detection methods include mouse bioassay, enzyme-linked immunosorbent assay, liquid chromatography-mass spectrometry, high-performance liquid chromatography-mass spectrometry, and capillary electrophoresis.4,5 While these methods offer high sensitivity, specificity, and accuracy, they often fall short in terms of ease of use, real-time analysis, on-site measurements, and cost-effectiveness. Electrochemical methods, however, can meet these demands and offer high specificity, sensitivity, and rapid response. To acquire higher selectivity and stability, the selection of biorecognition elements against the analyte is crucial.6,7 Aptamers, evolved by the exponential enrichment (SELEX) method, present many advantages over other biorecognition elements, especially antibodies.8 Therefore, electrochemical aptasensors have been extensively used for detecting various biomolecules,9 including STX.10,11To achieve a low detection limit and high sensitivity, signal amplification is required in electrochemical aptasensors. DNA-based signal amplification strategies include polymerase chain reaction, rolling circle amplification, hybridization chain reaction, and catalytic hairpin assembly.12 Nevertheless, these strategies have the drawbacks of strict reaction conditions, complex operations, slow electron transfer of amplified DNA structures and low turnover of catalytic cycles.13 DNA walkers exhibit advantages such as high controllability, mild reaction conditions, easy operation, precise spatial positioning, and programmability.14 DNA walkers move autonomously along programmed oligonucleotide tracks such as double-stranded DNA, DNA origami, and DNA monolayers.13 Typically, a DNA walker is composed of DNA strands, including driving force, legs, tracks and cargo.15 Protein enzymes such as endonuclease, exonuclease and DNAzyme are usually used to power DNA walkers where nicking endonuclease is widely used. Driven by enzymatic reactions, strand displacements or conformational transitions and walking bodies migrate along the tracks, generating large numbers of irreversible and responsive new DNA strands to implement signal transduction and amplification.16 To date, DNA walkers have been applied in a variety of fields, including biosensors, bioimaging and molecule delivery.17 For example, bipedal and multipedal DNA walking nanomachines demonstrate ultrahigh sensitivity for detecting nucleic acids.18–20 Self-powered cancer biomarker biosensors based on DNA walker and graphdiyne exhibit high sensitivity.21–24
Inspired by these, we constructed a Nb.BbvCI powered DNA walker machine-based STX aptasensor (Fig. 1). First, thiol-labeled amplified DNA was self-assembled on a gold electrode. Then, the thiol-labelled DNA walker was added and a hybridization occurred between the DNA walker and amplified DNA. Thereafter, aptamers (sequence followed Zheng X., et al.) with complementary nucleotides to part of amplified DNA and DNA walker were introduced, forming a sandwich structure of DNA walker, aptamers and amplified DNA.25 Upon the addition of STX, the sandwich structure was decomposed due to the high affinity of the aptamer to STX. The amplified DNA and DNA walker were released to form the hybridized anchor again. Then, the nicking endonuclease Nb.BbvCI specifically cut the hybridized anchor, releasing amplified DNA to the solution and liberating the walking strand as a free end again.13 This free-walking strand would bind to another amplified DNA and form another anchor, repeating the previous process. Over time, dozens of DNA walkers were released to obtain the amplified signal. Therefore, a highly sensitive and selective biosensor was constructed for the detection of STX. The detection limit (LOD) was calculated using the formula LOD = 3σ/k where σ is the standard deviation and k is the slope of the curve corresponding to calibration. The convenient operation process, highly sensitive results and rapid response indicate DNA walker coupled with Nb.BbvCI is a promising tool for constructing a variety of aptasensors.
Experimental
Materials
Tris(2-carboxyethyl) phosphine hydrochloride (TCEP), potassium hexacyanoferrate(III) and potassium hexacyanoferrate(II) were purchased from Sigma-Aldrich. 6-Mercapto-1-hexanol (MCH), sulfuric acid (H2SO4), potassium chloride (KCl) and hydrogen peroxide (H2O2), PBS and Tris-HCl buffer were provided by Sinopharm Chemical Reagent Co. Ltd (Shanghai, China). The α-Al2O3 polishing powder with various diameters was bought from Tianjin Aida Hengsheng Co. Ltd (Tianjin, China). Nb.BbvCI and CutSmart Buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 100 μg mL−1 BSA) were ordered from New England Biolabs Inc. (USA). STX toxin was bought from the National Research Council of Canada (NRC). Its natural derivatives, okadaic acid (OA), dinophysistoxins (DTX), pectenotoxins (PTX), and yessotoxin (YTX) were brought from Cell Signalling Technology.Oligonucleotides were synthesized by Sangon Biotechnology Co. Ltd (Shanghai, China). The sequences of the oligonucleotides were as follows from 5′ end to 3′ end:
| Amplify DNA: TTTGCTGAAGG |
| Aptamer: TGAGGCACTGACGGTATTGAGGGTCGCATCCCGTGGAAACATGTTCATTGGGCGCACTCCGCTTTCTGTAGATGGCTCTAACTCTCCTCT |
| DNA walker: (T)47GTCAGTGCCTCAGC |
The 5′ end of the amplified DNA and DNA walker were labelled with a thiol group. While the 3′ end of the amplified DNA was labelled with an Fc molecule.
Preparation of aptasensor
The gold electrode was polished with α-Al2O3 slurry and cleaned with piranha solution (H2SO4/H2O2, 3/1) for 10 min. Then, the electrode was immersed in 0.5 M H2SO4 solution and continuously scanned in the range of −0.3 V to 1.5 V with a scan rate of 100 mV s−1 until a voltammogram characteristic of the clean gold electrode was established. 50 μL of the amplified DNA was incubated with 5.0 μL TCEP (10 mM) for 1 h to reduce disulfide bonds and diluted to total volume of 100 μL. After rinsing with distilled water and ethanol, the cleaned gold electrode was treated with 10 μL of the above solution overnight. After rinsing with PBS, 10 μL of 1 μM DNA walker (also activated by TCEP) was incubated with the electrode overnight and rinsed with PBS. Then, 10 μL of 1 μM aptamer was dropped on the electrode and incubated for 2 h. Thereafter, 5 μL of 1 mM MCH was used to block the unmodified site. Finally, the DNA walker-modified electrode was washed with PBS thoroughly to remove unfixed MCH and DNA.Measurement procedure
The electrochemical measurements were performed on an electrochemical workstation (Metrohm Dropsens) with a three-electrode system composed of a platinum wire as a counter, a saturated calomel electrode as a reference and a DNA walker-modified gold electrode as a working electrode. A certain volume of 100 nM STX was added to the surface of the electrode and incubated for 1 h. Sequentially, 5.0 μL Nb.BbvCI (0.2 U μL−1) in 1× CutSmart Buffer was added and incubated at 37 °C for 25 min. After washing with Tris-HCl buffer, the biosensor was immersed in 10 mM PBS for electrochemical impedance spectroscopy (EIS) measurement from 0.1 kHz to 100 kHz at 10 mV signal amplitude. Cyclic voltammetry (CV) test was conducted in a solution of 5 mM Fe(CN)64−/3− and 0.1 M KCl by potential scanning between −0.1 V and 0.8 V at a scan rate of 50 mV s−1. Differential plus voltammetry (DPV) analysis was recorded under the following conditions: initial voltage of −0.2 V, final voltage of 0.6 V, scan rate of 0.005 V s−1, modulation amplitude of 0.1 V, potential step of 0.005 V, and pulse time of 60 ms.The equivalent circuit model (insert image of Fig. 2c) was used to fit the impedance data, where Rct was the charge transfer resistance, Zw was the Warburg impedance, and Q represented the constant phase angle element. For the EIS results, the interaction of STX to aptamer resulted in the change of Rct due to the release of the amplified DNA. The captured STX was quantified using the following equation: ΔRct (%) = ΔRct/Rct(background) = (Rct(STX added) − Rct(background))/Rct(background). For DPV data, the STX was quantified by the following equation: ΔI (%) = ΔI/ΔI(background) = (I(peak current of STX added) − I(peak current of background))/I(peak current of background).
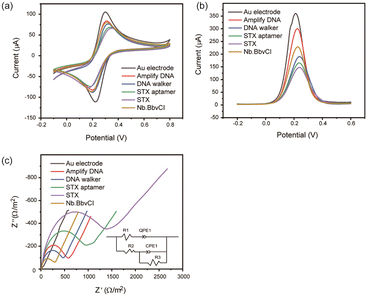 | ||
| Fig. 2 (a) CV, (b) DPV, and (c) EIS responses for bare electrode incubation with amplified DNA, DNA walker, STX aptamer, STX and Nb.BbvCI. | ||
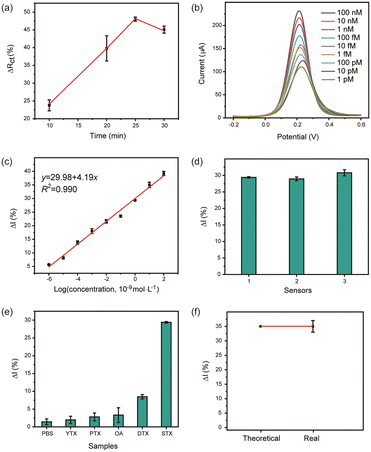
For the specificity tests, 1 nM of STX was selected as the target while 10 nM of YTX, PTX, OA, and DTX as interferences. The repeatability test was conducted in a 10 nM STX solution using three aptasensors.
Real mussel sample measurement
Fresh mussels were bought from the local market and treated following the previous method.26 Briefly, the tissues were extruded and broken using a home grinder. Then, 250 mg tissues were mixed with 1 ml of 50% methanol for 5 min. The supernatant was collected after centrifugation treatment and placed at 80 °C for 5 min. The real samples were prepared by mixing different concentrations of the STX standard sample with the previous supernatant.Results and discussion
Feasibility of the proposed aptasensor
The feasibility of a DNA walker-assisted-amplified aptasensor was verified by CV, DPV and EIS in the solution of 5 mM Fe(CN)64−/3− and 0.1 M KCl. The bare and modified electrodes displayed a typical reversible one-electron reduction/oxidation in the presence of Fe(CN)64−/3− solution (Fig. 2a). Compared with the bare electrode, electrodes modified with amplified DNA, amplified DNA/aptamer/DNA walker, and amplified DNA/aptamer/DNA walker/STX exhibited lower reduction/oxidation current and higher reduction/oxidation potentials, suggesting the electron transfer rate was reduced on the electrode surface. After amplification, the DNA was cut by Nb.BbvCI, the reduction/oxidation current increased obviously. DPV results were in accordance with the CV measurements, indicating the successful construct of the proposed aptasensor (Fig. 2b). These results suggested that the DNA walker was triggered by the toxin to release amplified DNA, accomplishing signal amplification.EIS measurements were also employed to characterize the preparation of the aptasensor (Fig. 2c). The bare gold electrode exhibited a tiny electron transfer resistance (Ret). After immobilization of amplified DNA, aptamer, DNA walker, Ret obviously increased sequentially due to the negatively charged DNA sentence repelling the Fe(CN)64−/3− to the electrode surface.13 The increase of Ret also proved the successful immobilization of DNA strands. In the presence of STX, the sandwich structure blocked the electron transfer, resulting in an enhanced Ret. Upon the addition of Nb.BbvCI, would cut and release the amplified DNA, benefiting Fe(CN)64−/3− close to the electrode surface, unblock the electron transfer and therefore lead to a decreased Ret.
Optimization and performance of proposed aptasensor
Enzymatic cleavage time of Nb.BbvCI was optimized where ΔRct (%) was used as the quantified standard (Fig. 3a). In the presence of Nb.BbvCI, EIS signal changed obviously where Rct (%) increased with incubation time at first 25 min and then decreased as time increased. At 25 min, a maximum Rct (%) was obtained. Therefore, 25 min was chosen as the optimal cleavage reaction time.The analytical performance of the aptasensor was assessed by incubating the biosensor with different concentrations of STX through the DPV method (Fig. 3b). Increasing STX concentration, the decrease of current was observed. The calibration plot showed a wide linear relationship between peak current and the logarithm value of STX concentration ranging from 1 pM to 100 nM with a correlation coefficient of 0.990 (Fig. 3c). The detection limit was calculated to be 0.58 pM. The developed sensor exhibited a wider detection range and much lower detection limit than previous reports, indicating the high sensitivity and feasibility (Table 1). The DNA walker coupled with the nicking endonuclease was the main reason for the significant signal amplification.
| Detection strategies | Active materials | Amplify technologies | Detection range (μM) | LOD (nM) | Ref. |
|---|---|---|---|---|---|
| LOD: limit of detection, NPs: nanoparticles, MWCNTs: multiwalled carbon nanotubes.a EIS: electrolyte–insulator–semiconductor.b EIS: electrochemical impedance spectroscopy. SWV: square wave voltammetry, PAH: poly(allylamine hydrochloride), AIR-SE: attenuated internal reflection-spectroscopic ellipsometry, MIP–QDs: imprinted silica layers appended to quantum dots, HCR: hybrid chain reaction, QDs: quantum dots, NMOFs: nanoscale metal–organic frameworks, 4-NTP: 4-nitrothiiophenol. | |||||
| DPV | Ag NPs | — | 0.04–0.15 | 1.0 | 27 |
| DPV | MWCNTs | — | 0.9 × 10−3–3.0 × 10−2 | 0.38 | 28 |
| EISa | PAMAM | — | 0.5–100 | 0.09 | 11 |
| EISa | PAH | — | 0.0005–0.1 | 0.05 | 29 |
| SWV/EISb | Porous Pt NPs | — | 3.0–3.0 × 105 | 1.4 | 30 |
| SWV | DNA nanotetrahedron | — | 0.001–0.4 | 0.92 | 31 |
| AIR-SE | Au | — | 2.99–1.8 × 105 | 2990 | 32 |
| Fluorescence | MIP–QDs | — | 6.0 × 103–3.0 × 104 | — | 33 |
| Fluorescence | — | HCR and CRISPR-Cas 9 | 5 × 10−9–5 × 10−5 | 1.2 × 10−6 | 34 |
| Fluorescence | Fe3O4@Au–Pt and QDs | — | 0.001–5 | 0.6 | 35 |
| Fluorescence | NMOFs | — | 0–0.19 | 1.17 | 36 |
| Colorimetry | Au NPs | — | 1.4 × 10−4–3.7 × 10−2 | 0.142 | 37 |
| Colorimetry | Au NPs | HCR | 7.8 × 10−5–2.5 × 10−3 | 0.042 | 38 |
| Raman | Au NPs@4-NTP@SiO2 | Rolling circle amplification reaction | 0.002–0.5 | 0.012 | 39 |
| Raman | Au NPs | — | 0.01–0.2 | 11.7 | 40 |
| DPV | — | DNA walker coupling with Nb.BbvCI | 0.001–100 | 0.00058 | This work |
In addition, the reproducibility of the aptasensor was manifested by executing three duplicate measurements of 1.0 nM STX with a small RSD of 3.0% (Fig. 3d). The selectivity of the proposed aptasensor was evaluated by testing 10-fold interferences including YTX, PTX, OA, and DTX (Fig. 3e). No significant current change was observed for the interfering substances due to the high affinity of aptamer to STX, showing the high specificity of this aptasensor.
To explore the feasibility of detecting a real sample, 10 nM STX was diluted into the fresh mussel samples (uncontaminated) as the target sample. The developed aptasensor exhibited good recovery of 99.97% with an RSD of 2%, suggesting that this biosensor could be used in detecting STX in real shellfish (Fig. 3f).
Conclusions
In summary, we fabricated sensitive STX biosensors using a DNA walker coupled with nicking endonuclease Nb.BbvCI amplification. The reliability and sensitivity were validated by discriminating various marine toxins and differentiating diverse concentrations of STX. The recovery ratio results indicated the practical testing feasibility of this biosensor. Although the long-term stability of nucleic acid needs consideration, the strategy of this platform provides a fast and effective method for marine toxins detection. In future work, we will proceed toward the development of integrated portable biosensors by combining our existing amplification platform with flexible printing and integrated data processing to enable direct on-site measurements.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Shaanxi Province (grant number: 2022JM-532).References
- K. Campbell, D. F. K. Rawn, B. Niedzwiadek and C. T. Elliott, Food Addit. Contam.: Part A, 2011, 28, 711–725 CrossRef CAS PubMed.
- C. H. Cho, J. H. Kim, N. S. Padalkar, Y. V. M. Reddy, T. J. Park, J. Park and J. P. Park, Biosens. Bioelectron., 2024, 255, 116269 CrossRef CAS PubMed.
- J. Li and K. M. Persson, Chemosphere, 2021, 265, 128591 CrossRef CAS PubMed.
- A. R. Humpage, V. F. Magalhaes and S. M. Froscio, Anal. Bioanal. Chem., 2010, 397, 1655–1671 CrossRef CAS PubMed.
- Y. Tian, L. Du, P. Zhu, Y. Chen, W. Chen, C. Wu and P. Wang, Biosens. Bioelectron., 2021, 176, 112899 CrossRef CAS PubMed.
- T. Mahmudiono, D. Olegovich Bokov, S. Abdalkareem Jasim, W. Kamal Abdelbasset and M. K. Dinora, Microchem. J., 2022, 179, 107460 CrossRef CAS.
- B. Noureen, N. Ullah, Y. Tian, L. Du, W. Chen, C. Wu and P. Wang, Talanta, 2022, 240, 123185 CrossRef CAS PubMed.
- R. Yuan, J. Cai, H. Ma, Y. Luo, L. Wang and S. Su, Chemosensors, 2023, 11, 488 CrossRef CAS.
- A. Villalonga, B. Mayol, R. Villalonga and D. Vilela, Sens. Actuators, B, 2022, 369, 132318 CrossRef CAS.
- N. Ullah, W. Chen, B. Noureen, Y. Tian, L. Du, C. Wu and J. Ma, Sensors, 2021, 21, 4938 CrossRef CAS PubMed.
- N. Ullah, B. Noureen, Y. Tian, L. Du, W. Chen and C. Wu, Nanomaterials, 2022, 12, 1505 CrossRef CAS PubMed.
- Y.-F. Chen, Y.-Z. Guo, S. Xiao, Y.-Q. Chai, J.-L. Liu and R. Yuan, Anal. Chem., 2024, 96, 4589–4596 CrossRef CAS PubMed.
- Y. Ji, L. Zhang, L. Zhu, J. Lei, J. Wu and H. Ju, Biosens. Bioelectron., 2017, 96, 201–205 CrossRef CAS PubMed.
- J. Shi, P. Li, Y. Huang, Y. Wu, J. Wu, K.-J. Huang, X. Tan and Y. Ya, Chem. Eng. J., 2024, 483, 149231 CrossRef CAS.
- A. Yuan, H. Xiao, F. Yang, H. Hao, X. Wang, J. Li, M. Jin, Q. Zhao, R. Sha, Z. Deng and H. Peng, TrAC, Trends Anal. Chem., 2023, 158, 116870 CrossRef CAS.
- L. Zhao, X. Yang, X. Zhong and Y. Zhuo, ChemPlusChem, 2022, 87, e202200070 CrossRef CAS PubMed.
- M. Xu and D. Tang, Anal. Chim. Acta, 2021, 1171, 338523 CrossRef CAS PubMed.
- M. Miao, H. Chai and Y. Tang, ACS Nano, 2022, 16, 4726–4733 CrossRef.
- P. Miao, Anal. Chem., 2023, 95, 6760–6764 CrossRef CAS PubMed.
- P. Miao and Y. Tang, ACS Cent. Sci., 2021, 7, 1036–1044 CrossRef CAS.
- Y.-Y. Hou, J. Xu, W. Z. Xie, K.-J. Huang, X. Tan, B. R. Zhao, S. Q. Zhang and M. T. Gao, Sens. Actuators, B, 2023, 376, 32998 CrossRef.
- J. Xu, Y. Liu, Y. Li, Y. Liu and K. J. Huang, Anal. Chem., 2023, 95, 13305–13312 CrossRef CAS PubMed.
- J. Xu, Y. Liu, K. J. Huang, R. Wang and J. Li, Chem. Eng. J., 2023, 466, 143230 CrossRef CAS.
- L. Li, Y. Zhao, X. Yan, X. Qi, L. Wang, R. Ma, S. Wang and X. Mao, Sens. Actuators, B, 2021, 344, 130320 CrossRef CAS.
- X. Zheng, B. Hu, S. X. Gao, D. J. Liu, M. J. Sun, B. H. Jiao and L. H. Wang, Toxicon, 2015, 101, 41–47 CrossRef CAS PubMed.
- Y. Chen, W. Chen, Y. Tian, P. Zhu, S. Kong, L. Du and C. Wu, J. Electrochem. Soc., 2022, 169, 057526 CrossRef CAS.
- W. Zeng, X. Tang, T. Wu, B. Han and L. Wu, Anal. Chim. Acta, 2024, 1287, 342134 CrossRef CAS PubMed.
- L. Hou, L. Jiang, Y. Song, Y. Ding, J. Zhang, X. Wu and D. Tang, Microchim. Acta, 2016, 183, 1971–1980 CrossRef CAS.
- B. Noureen, N. Ullah, Y. Tian, L. Du, W. Chen, C. Wu and P. Wang, Talanta, 2022, 240, 123185 CrossRef CAS PubMed.
- J. A. Park, N. Kwon, E. Park, Y. Kim, H. Jang, J. Min and T. Lee, Biosens. Bioelectron., 2022, 210, 114300 CrossRef CAS PubMed.
- X. Qi, X. Yan, L. Zhao, Y. Huang, S. Wang and X. Liang, J. Electroanal. Chem., 2020, 858, 113805 CrossRef CAS.
- M. O. Caglayan and Z. Üstündağ, Toxicon, 2020, 187, 255–261 CrossRef CAS PubMed.
- A. Sun, J. Chai, T. Xiao, X. Shi, X. Li, Q. Zhao, D. Li and J. Chen, Sens. Actuators, B, 2018, 258, 408–414 CrossRef CAS.
- Q. Zhao, G. Li and X. Li, Chemosensors, 2023, 11, 183 CrossRef CAS.
- L. Zhu, W. Zeng, Y. Li, Y. Han, J. Wei and L. Wu, Sci. Total Environ., 2024, 921, 171236 CrossRef CAS.
- X. Dou, S. Xu, Y. Jiang, Z. Ding and J. Xie, Spectrochim. Acta, Part A, 2023, 284, 121827 CrossRef CAS PubMed.
- L. Li, Y. Zhao, X. Yan, X. Qi, L. Wang, R. Ma, S. Wang and X. Mao, Sens. Actuators, B, 2021, 344, 130320 CrossRef CAS.
- Y. Zhao, L. Li, R. Ma, L. Wang, X. Yan, X. Qi, S. Wang and X. Mao, Anal. Chim. Acta, 2021, 1173, 338710 CrossRef CAS PubMed.
- X. Bai, W. Gong, Y. Guo, D. Zhu and X. Li, Analyst, 2023, 148, 2327–2334 RSC.
- S. Cheng, B. Zheng, D. Yao, Y. Wang, J. Tian, L. Liu, H. Liang and Y. Ding, Anal. Lett., 2019, 52, 902–918 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00167b |
| This journal is © The Royal Society of Chemistry 2024 |