Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic measurement of intracellular mRNA with a molecular beacon probe towards point-of-care radiation triage†
Xin
Meng
 a,
Kechun
Wen
a,
Jingyang
Zhao
a,
Yaru
Han
a,
Shanaz A.
Ghandhi
b,
Salan P.
Kaur
b,
David J.
Brenner
b,
Helen C.
Turner
b,
Sally A.
Amundson
*b and
Qiao
Lin
*a
a,
Kechun
Wen
a,
Jingyang
Zhao
a,
Yaru
Han
a,
Shanaz A.
Ghandhi
b,
Salan P.
Kaur
b,
David J.
Brenner
b,
Helen C.
Turner
b,
Sally A.
Amundson
*b and
Qiao
Lin
*a
aDepartment of Mechanical Engineering, Columbia University, New York, NY 10027, USA. E-mail: qlin@columbia.edu
bCenter for Radiological Research, Columbia University Irving Medical Center, New York, New York 10032, USA. E-mail: saa2108@cumc.columbia.edu
First published on 2nd July 2024
Abstract
In large-scale radiation exposure events, the ability to triage potential victims by the received radiation dosage is crucial. This can be evaluated by radiation-induced biological changes. Radiation-responsive mRNA is a class of biomarkers that has been explored for dose-dependency with methods such as RT-qPCR. However, these methods are challenging to implement for point-of-care devices. We have designed and used molecular beacons as probes for the measurement of radiation-induced changes of intracellular mRNA in a microfluidic device towards determining radiation dosage. Our experiments, in which fixed TK6 cells labeled with a molecular beacon specific to BAX mRNA exhibited dose-dependent fluorescence in a manner consistent with RT-qPCR analysis, demonstrate that such intracellular molecular probes can potentially be used in point-of-care radiation biodosimetry. This proof of concept could readily be extended to any RNA-based test to provide direct measurements at the bedside.
1. Introduction
In a large-scale radiation exposure event, hundreds of people may be exposed to varying doses of radiation. It will be crucial to identify the doses received by each individual in a timely manner to triage victims of radiation and guide medical decisions. Biodosimetry refers to the determination of radiation dosage received by an individual based on observable biological changes that occur inside the biological system. These changes are reflected in specific biomarkers, which can be used to assess the magnitude of the biological absorbed dose and inform potential consequences of the radiation exposure to the individual, thereby allowing for the administration of suitable medical therapy.1 To date, the dicentric chromosome assay (DCA)2–4 has been most used and is considered the “gold standard” for dose reconstruction accuracy. However, DCA, like all cytogenetics-based assays, is time-consuming and laborious, and does not allow timely measurement of radiation exposure in a large-scale emergency.5 Thus, there is a strong need for biodosimetry assays that are more rapid and of higher throughput in events of mass radiation exposure.As an alternative method for radiation biodosimetry, additional methods such as γ-H2AX,6,7 micronuclei,5,8 microRNA,9,10 lncRNA,11,12 and protein13–15 and gene expression signatures16–18 in peripheral blood cells are being developed for this purpose.19 Gene expression assays have been particularly promising, and typically measure the mRNA content in a certain simulated scenario with reference to normal expression levels. Unlike DCA, gene expression can be easily assessed with advanced molecular assays and does not require cell division. Various techniques, such as real-time polymerase chain reaction (PCR)1 and sequencing,20 have been proposed for gene expression analysis. However, these methods, which require RNA to be purified from the cells for both real-time PCR and sequencing, are not suitable for translation to a rapid point-of-care (POC) assay.21 In comparison, analysis of gene expression at the mRNA level in whole blood cells can be highly promising for biodosimetry measurements.22,23
When implemented in field triage or POC scenarios, biodosimetry has the potential to guide medical decisions. POC devices have been widely reported for applications in disease diagnosis and monitoring24–27 based on the detection and measurement of diverse types of analytes, such as proteins,28–30 bacteria,31,32 and, in particular, nucleic acids.33,34 For POC analysis of nucleic acids, loop-mediated isothermal amplification (LAMP),35 surface acoustic wave (SAW),36 or CRISPR/Cas12a-based electrochemical DNA detection37 methods have been employed to evaluate extracellular or cell lysate samples. However, these nucleic acid analysis methods are not amenable to implementation for intracellular biodosimetry in situ, i.e., examining the analytes at the exact location where they reside. For POC biodosimetry, Balog et al. reported a protein panel-based assay in non-human primate plasma samples,38 Brengues et al. reported an RNA signature using a quantitative nuclease protection assay (qNPA),21 and Huang et al. reported an approach for quantifying mRNAs using integrated CMOS detectors.39 Studies on POC biodosimetry methods have otherwise been scarce.40
Theme
We report the design and use of molecular beacons for microfluidic in situ measurement of radiation-responsive intracellular mRNA with the goal of ultimately enabling POC radiation biodosimetry.In situ measurement enabled by molecular beacons (MB)
Gene expression-based methods that require isolation and purification of target sequences, while quantitatively accurate, are generally time-consuming and labor-intensive. In contrast, measurements using molecular beacons (MBs)41 do not require the purification of the target sequence and can be more readily performed in situ.42 MBs are hairpin-shaped oligonucleotide-based probes with a fluorescent reporter and a quencher on each end, which would be strongly fluorescent only when complementary target sequences are present.43,44 The fluorescence resulting from the hairpin structure possesses a low background, which is ideal for in situ measurements.45 We for the first time use MBs for in situ measurement of radio-induced changes of mRNA level by introducing the MB into fixed and permeabilized cells. This method would enable low-background in situ measurement of mRNA in TK6 cells, and for biodosimetry, can be used for in situ hybridization of target sequences in peripheral blood cells without isolation or purification.POC potential by microfluidic technology
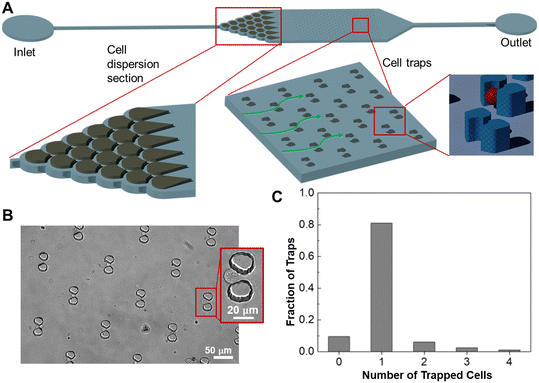
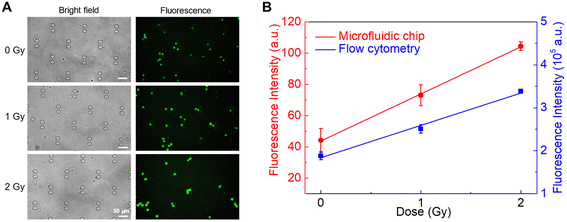
Microfluidic technology can enable POC processing and analysis of large numbers of samples.46 Toward POC biodosimetry, we have designed a microfluidic device for single-cell fluorescence measurements. This device can isolate and retain fixed and labeled cells as single cells in microscale traps to quantify the intracellular fluorescence of individual cells. The single-cell device can resolve complex fluorescent signals instigated by multilayered and overlapping cells to improve the accuracy of fluorescence quantification and be used as the measurement module in POC biodosimetry.As a proof of principle for a biodosimetry POC, we have developed MBs for a known radiation responsive biomarker BAX that will be integrated into the microfluidics system. Using the MBs, fluorescence intensities measured for TK6 cells 6 h post-exposure to radiation doses at 1 Gy and 2 Gy were respectively, 1.33-fold and 1.79-fold when compared with those of unirradiated samples. In 24 h post-exposure samples, the comparisons correspondingly became 1.45-fold for radiation at 1 Gy and 2.32-fold for radiation at 2 Gy, respectively. These changes were confirmed by experiments with NH32 p53-null control groups and found to be consistent with results from RT-qPCR quantification. Fluorescent quantification from the microfluidic device was also consistent with results obtained off-chip, suggesting the potential of the MB-based microfluidic approach for POC biodosimetry.
2. Results and discussion
2.1. Design and characterization of BAX MBs
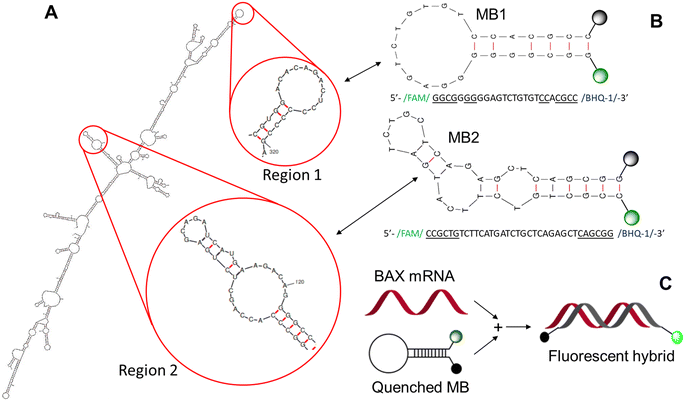 | ||
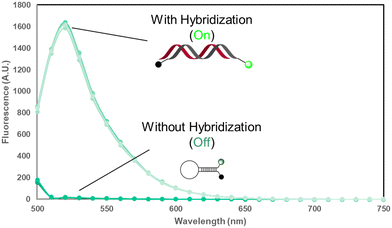
| Fig. 1 Design of BAX mRNA molecular beacons. (A) BAX mRNA secondary structure predicted by mFOLD47 and the two stem–loop regions selected for MB design. (B) BAX MB designs and sequences. (C) BAX MB working principle. The fluorescence of folded BAX MB is unquenched when it binds with the target complementary sequence. | ||
2.2. Intracellular studies of BAX MB
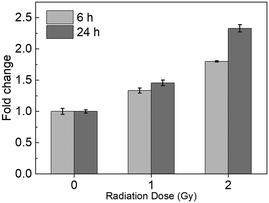 | ||
| Fig. 3 Dose-dependence of MB1 labeling with 6 h or 24 h post-exposure incubation time. All measurements were performed in triplicate and the data were shown as the mean ± SD. | ||
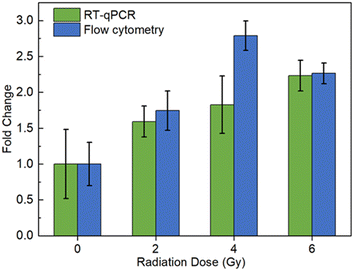
Similarly, flow cytometry analysis of BAX mRNA levels in TK6 cells was then performed in triplicate at 24 h post-exposure to X-ray, and the same three irradiation doses (0, 1, and 2 Gy) were used (Fig. 3). Compared to unirradiated samples, the 1 Gy group showed increased fluorescence of 1.45-fold, while the 2 Gy group had increased fluorescence of 2.32-fold. The 24 h post exposure fluorescence changes were consistent with the 6 h post exposure samples at 1 Gy, and slightly higher at 2 Gy. Irradiation at 0.5 Gy was tested in a separate batch of samples (Fig. S1†). Significant changes in fluorescence were detected (1.23-fold compared to that for unirradiated cells) in TK6 cells 24 h post-exposure, suggesting that this method can detect doses as low as 0.5 Gy.
The observed upregulated BAX gene response may be mainly due to activation of the p53 signal pathway, as this is one of the main responses to radiation, and BAX is a known p53-regulated gene.50 BAX is considered an apoptosis regulator, and the upregulation of BAX might indicate possible apoptosis response to radiation. To further investigate the source of the increase, MB1 labeling in NH32 cells was performed. NH32 is a TK6-derived, p53 double-knockout cell line.51 If the radiation-induced intracellular p53 increase is the main contributor for the increased levels of BAX, then the irradiated group of NH32 cells would not have such a significant increase in the fluorescence. To this end, NH32 cells were cultured and irradiated at 0.5 and 2 Gy, and cultured for 24 h before cell fixation.52 After permeabilization, the cells were stained with MB1 and imaging flow cytometry was performed on the stained cells. The results with NH32 show that compared with the control group, the 0.5 Gy-irradiated cells have a signal of 1.01-fold, while the 2 Gy group has a signal of 1.25-fold indicating that our hypothesis regarding p53 is valid. The signal in 0.5 Gy group is essentially unchanged, suggesting that the changes in TK6 cells at 0.5 Gy are coming from the p53 pathway. The 2 Gy has a slight increase in the signal. However, comparing to the 1.73-fold change in TK6 cells, it can still be concluded that the majority of the increase in TK6 cells results from the p53 pathway.
2.3. Microfluidic measurement of intracellular BAX mRNA radiation response
The ability of the microfluidic device to trap individual cells was then tested. A suspension of live cells from the CCRF-CEM leukemia cell line at a concentration of 1 × 106 cells per mL was flowed through the device at 5 μL min−1 for 5 min. The device was then washed with PBS buffer at 10 μL min−1 for 2 min. Images of the cell-trapping unit were finally taken to examine the trapped cells (Fig. 5B) and used to determine the fraction of cups trapping n cells, with n = 1 corresponding to trapped single cells (Fig. 5C). It was observed that the cups each trapped n = 0–4 cells with 80% of the cups each trapping a single cell (n = 1). Meanwhile, we observed that multiplying trapped cells allowed reliable quantification of fluorescence per cell (below), making our measurement method less dependent on single-cell trapping.
3. Experimental
TK6 and NH32 cells are gifts from Dr. Sally Amundson. CCRF-CEM cells are purchased from ATCC. Nucleic acids are purchased from Integrated DNA Technologies. All the other reagents unless otherwise specified are purchased from BD Biosciences.In vitro folding assay
The assay was performed in 96-well plates and detected with Molecular Devices SpectraMax M5 plate reader. In 250 μL microtubes, the molecular beacons (2 μM in PBSM, 50 μL) were mixed with their intended target sequence in DNA form (2 μM in PBSM, 50 μL) to reach a final concentration of 1 μM of each species. The mixture was then conditioned (10 min at 95 °C, 10 min at 0 °C and 10 min at RT) before being transferred into a 96-well plate and analyzed in a plate reader.Cell culture
TK6, NH32 and CCRF-CEM cell lines were maintained using culture media consisting of RPMI 1640 1× medium, 10% fetal bovine serum, 1% MEM non-essential amino acids, 1 mM sodium pyruvate and 1% penicillin–streptomycin–glutamine in a humidified 37 °C incubator with 5% CO2. Cell density was kept below 1 × 106 cells per mL at all times. Once the cells approached 1 × 106 cells per mL, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution was performed. All cell-related experiments were performed using cells within 25 passages.
50 dilution was performed. All cell-related experiments were performed using cells within 25 passages.
Irradiation
Mock and the irradiation of the cell line samples was performed on an X-RAD 320 Biological Irradiator (Precision X-Ray Inc.) at 320 mV, 12.5 mA, with a custom filter13 and a 40 cm sample distance to achieve 1 Gy min−1 of irradiation rate. All samples were irradiated under the same conditions with various irradiation times.Cell fixation
Cell fixation was performed according to our previously published method.15 TK6 cell samples (<15 mL each) were centrifuged at 200 × g for 10 min and the supernatants were discarded. The pellets were resuspended in 250 μL of ice-cold Fixation/Permeabilization Solution (Cytofix/Cytoperm™ Fixation/Permeabilization Solution kit, BD Biosciences; #554714) and incubated at 4 °C for 20 min to allow cell fixation. After the addition of 700 μL of 1 × Perm/Wash buffer (BD Biosciences; #554723), the suspensions were centrifuged at 300 × g for 4 min and the supernatant removed. The pellet was then washed twice with 950 μL of 1 × Perm/Wash buffer before being resuspended with 1 mL of 1% (m/v) bovine serum albumin solution in 1 × DPBS and preserved at 4 °C.Cell labeling
Cell labeling with MB1 was adapted from our earlier work.15,52 Briefly, fixed TK6 cells preserved in 1% BSA buffer were first centrifuged at 300 × g for 4 min and the supernatant was discarded. The pellet was washed with 950 μL of 1 × Perm/Wash buffer The MBs were diluted in 1 × Perm/Wash buffer and conditioned in the following order: 10 min at 95 °C; 10 min on ice; 10 min at RT. The pellet is then resuspended in 50 μL of the conditioned aptamer solution and incubated for 1 h at RT away from light. After incubation, 900 μL of 1 × Perm/Wash buffer was added to the suspension, and the sample was then centrifuged at 300 × g for 4 min. After discarding the supernatant, the pellet was washed once again with 950 μL of 1 × Perm/Wash buffer and twice with 950 μL of 1 × PBS. If not directly used in the following fluorescence analysis, the pellet was then resuspended in 1 mL of 1 × PBS and stored at 4 °C.Imaging flow cytometry
Samples were concentrated to 50 μL and acquired on ImageStreamX MkII at Columbia University Center for Radiological Research with 40× objective and 488 nm excitation laser set to 200 mW. Similar to previously described,52 a uniform analysis template on Image Data Exploration and Analysis Software (IDEAS®, Luminex ver. 6.2) was used to measure Mean Fluorescence Intensity detected on the 480–560 nm channel in focused, single, healthy cells.RNA isolation and real-time PCR
At 24 hours after irradiation, whole-cell RNA was isolated from aliquots of at least 1 × 106 exponentially growing cells per point using the Qiagen RNeasy Mini kit (cat#74106), and quantified using a Nanodrop One spectrophotometer (Thermofisher). The A260/280 ratios for all samples ranged from 1.97–2.03. For each sample, cDNA was prepared from 1 mg of total mRNA using the High-Capacity® cDNA kit (Life Technologies, Foster City, CA). Quantitative real-time RT-PCR (qRT-PCR) was then performed using 15 ng of cDNA per reaction and Taqman® assays (Life Technologies) for BAX (Hs00180269_m1) and the housekeeping gene ACTB (Hs99999903_m1). All reactions were performed in duplicate on a QuantStudio™ 7 Flex System, using standard PCR conditions. Expression Suite software ver. 1.3 (Thermofisher) was used to calculate relative fold-induction with the 2−ΔΔCT method.Microfluidic device fabrication and packaging
The PDMS mold was fabricated with various SU-8 photoresists. 33 mL of PDMS (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 base
1 base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) curing agent by weight) was poured onto the flow layer wafer and placed in a vacuum desiccator for 30 minutes to remove bubbles. Following bubble removal, the flow layer wafer was placed on a hotplate set at 72 °C for 30 minutes. The PDMS was then peeled off, cut into separate microfluidic chambers and had inlets and outlets punched with an autopsy punch. The PDMS microfluidic chambers and glass slides were then placed in an oxygen plasma etcher (Diener Plasma Etch) and exposed to oxygen plasma for 45 seconds at 100% power. Immediately after exposure, the PDMS devices were bonded with the glass slide on the reactive interface and gently pressed to put their surfaces in contact.
curing agent by weight) was poured onto the flow layer wafer and placed in a vacuum desiccator for 30 minutes to remove bubbles. Following bubble removal, the flow layer wafer was placed on a hotplate set at 72 °C for 30 minutes. The PDMS was then peeled off, cut into separate microfluidic chambers and had inlets and outlets punched with an autopsy punch. The PDMS microfluidic chambers and glass slides were then placed in an oxygen plasma etcher (Diener Plasma Etch) and exposed to oxygen plasma for 45 seconds at 100% power. Immediately after exposure, the PDMS devices were bonded with the glass slide on the reactive interface and gently pressed to put their surfaces in contact.
4. Conclusion
In summary, we described the development of a DNA molecular beacon that can bind specifically with intracellular human BAX mRNA towards POC radiation triage application. The design is based on a step-loop region of the predicted secondary structure of human BAX mRNA, and the sequence specificity is verified through BLAST. We demonstrated the unfolding of the MB probe in the presence of the target sequence in vitro. The MB fluorescence signal shows dose-dependence at both 6 h and 24 h post-exposure time, and up to 6 Gy of irradiation in fixed TK6 cells. The MB labeling is comparable with RT-qPCR quantification in terms of fold change. A microfluidic intracellular biodosimetry device was designed and fabricated to trap single fixed cells and we compared the on-chip quantification with flow cytometry analysis and the results are comparable. It can be concluded that the MB probe for BAX mRNA is suitable for microfluidic intracellular biodosimetry.We envision the prototype device developed in this manuscript as a pivotal component of a prospective point-of-care device for integrated intracellular biodosimetry. The integration of the MB probe into a point-of-care device would enable real-time, on-site assessment of radiation exposure, thereby making biodosimetric assessments more accessible and timelier. This could be especially invaluable in emergency scenarios where rapid evaluation and immediate medical intervention are critical for optimizing patient outcomes.
Data availability
Data for the article cannot be made publicly available due to legal requirements.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Institutes of Health, including a pilot grant from the Opportunity Funds Management Core of the Centers for Medical Countermeasures against Radiation, National Institute of Allergy and Infectious Diseases (grant number U19-AI067773), and grants from the National Cancer Institute (R21-CA261775) and the National Institute of Allergy and Infectious Diseases (U01-AI148309). We are also grateful to Bezalel Bacon, Leah Nemzow, Thomas Boehringer and Michelle Phillippi for discussions and experimental support related to cell culture and x-ray irradiation and imaging flow cytometry.References
- K. Brzóska and M. Kruszewski, Toward the Development of Transcriptional Biodosimetry for the Identification of Irradiated Individuals and Assessment of Absorbed Radiation Dose, Radiat. Environ. Biophys., 2015, 54, 353–363 CrossRef PubMed.
- J. K. Lee, E. A. Han, S. S. Lee, W. H. Ha, J. F. Barquinero, H. R. Lee and M. S. Cho, Cytogenetic Biodosimetry for Fukushima Travelers after the Nuclear Power Plant Accident: No Evidence of Enhanced Yield of Dicentrics, J. Radiat. Res., 2012, 53, 876–881 CrossRef PubMed.
- Y. Suto, M. Hirai, M. Akiyama, G. Kobashi, M. Itokawa, M. Akashi and N. Sugiura, Biodosimetry of Restoration Workers for the Tokyo Electric Power Company (TEPCO) Fukushima Daiichi Nuclear Power Station Accident, Health Phys., 2013, 105, 366–373 CrossRef CAS PubMed.
- A. Vaurijoux, E. Gregoire, S. Roch-Lefevre, P. P. Voisin, C. Martin, P. P. Voisin, L. Roy and G. Gruel, Detection of Partial-Body Exposure to Ionizing Radiation by the Automatic Detection of Dicentrics, Radiat. Res., 2012, 178, 357–364 CrossRef CAS.
- J. M. Sullivan, P. G. S. Prasanna, M. B. Grace, L. K. Wathen, R. L. Wallace, J. F. Koerner and C. N. Coleman, Assessment of Biodosimetry Methods for a Mass-Casualty Radiological Incident: Medical Response and Management Considerations, Health Phys., 2013, 105, 540–554 CrossRef CAS PubMed.
- L. Hernández, M. Terradas, M. Martín, L. Tusell and A. Genescà, Highly Sensitive Automated Method for DNA Damage Assessment: Gamma-H2AX Foci Counting and Cell Cycle Sorting, Int. J. Mol. Sci., 2013, 14, 15810–15826 CrossRef.
- Y. Lee, Q. Wang, I. Shuryak, D. J. Brenner and H. C. Turner, Development of a High-Throughput γ-H2AX Assay Based on Imaging Flow Cytometry, Radiat. Oncol., 2019, 14, 150 CrossRef PubMed.
- Q. Z. Qian, X. K. Cao, F. H. Shen and Q. Wang, Effects of Ionising Radiation on Micronucleus Formation and Chromosomal Aberrations in Chinese Radiation Workers, Radiat. Prot. Dosim., 2016, 168, 197–203 CAS.
- S. S. Acharya, W. Fendler, J. Watson, A. Hamilton, Y. Pan, E. Gaudiano, P. Moskwa, P. Bhanja, S. Saha, C. Guha, K. Parmar and D. Chowdhury, Serum MicroRNAs Are Early Indicators of Survival after Radiation-Induced Hematopoietic Injury, Sci. Transl. Med., 2015, 7(287), 287ra69 Search PubMed.
- B. Tomasik, W. Fendler and D. Chowdhury, Serum MicroRNAs - Potent Biomarkers for Radiation Biodosimetry, Onco Targets Ther, 2018, 9, 14038–14039 Search PubMed.
- M. J. Aryankalayil, S. Chopra, J. Levin, I. Eke, A. Makinde, S. Das, U. Shankavaram, C. Vanpouille-Box, S. Demaria and C. N. Coleman, Radiation-Induced Long Noncoding RNAs in a Mouse Model after Whole-Body Irradiation, Radiat. Res., 2018, 189, 251–263 CAS.
- Q. Lu, W. Gong, J. Wang, K. Ji, X. Sun, C. Xu, L. Du, Y. Wang and Q. Liu, Analysis of Changes to LncRNAs and Their Target MRNAs in Murine Jejunum after Radiation Treatment, J. Cell. Mol. Med., 2018, 22, 6357–6367 CrossRef CAS PubMed.
- Y. Lee, M. Pujol Canadell, I. Shuryak, J. R. Perrier, M. Taveras, P. Patel, A. Koller, L. B. Smilenov, D. J. Brenner, E. I. Chen and H. C. Turner, Candidate Protein Markers for Radiation Biodosimetry in the Hematopoietically Humanized Mouse Model, Sci. Rep., 2018, 8, 13557 CrossRef PubMed.
- S. A. Ghandhi, S. R. Morton, I. Shuryak, Y. Lee, R. K. Soni, J. R. Perrier, J. Bakke, J. Gahagan, K. Bujold, S. Authier, S. A. Amundson, D. J. Brenner, D. Nishita, P. Chang and H. C. Turner, Longitudinal Multi-Omic Changes in the Transcriptome and Proteome of Peripheral Blood Cells after a 4 Gy Total Body Radiation Dose to Rhesus Macaques, BMC Genomics, 2023, 24, 139 CrossRef CAS PubMed.
- Q. Wang, Y. Lee, I. Shuryak, M. Pujol Canadell, M. Taveras, J. R. Perrier, B. A. Bacon, M. A. Rodrigues, R. Kowalski, C. Capaccio, D. J. Brenner and H. C. Turner, Development of the FAST-DOSE Assay System for High-Throughput Biodosimetry and Radiation Triage, Sci. Rep., 2020, 10, 12716 CrossRef CAS PubMed.
- S. A. Amundson, K. T. Do, S. Shahab, M. Bittner, P. Meltzer, J. Trent and J. Fornace, Identification of Potential MRNA Biomarkers in Peripheral Blood Lymphocytes for Human Exposure to Ionizing Radiation, Radiat. Res., 2000, 154, 342–346 CrossRef CAS.
- M. Abend, S. A. Amundson, C. Badie, K. Brzoska, R. Hargitai, R. Kriehuber, S. Schüle, E. Kis, S. A. Ghandhi, K. Lumniczky, S. R. Morton, G. O'Brien, D. Oskamp, P. Ostheim, C. Siebenwirth, I. Shuryak, T. Szatmári, M. Unverricht-Yeboah, E. Ainsbury, C. Bassinet, U. Kulka, U. Oestreicher, Y. Ristic, F. Trompier, A. Wojcik, L. Waldner and M. Port, Inter-Laboratory Comparison of Gene Expression Biodosimetry for Protracted Radiation Exposures as Part of the RENEB and EURADOS WG10 2019 Exercise, Sci. Rep., 2021, 11, 9756 CrossRef CAS.
- S. A. Ghandhi, I. Shuryak, S. R. Morton, S. A. Amundson and D. J. Brenner, New Approaches for Quantitative Reconstruction of Radiation Dose in Human Blood Cells, Sci. Rep., 2019, 9, 18441 CrossRef CAS.
- C. Xiao, N. He, Y. Liu, Y. Wang and Q. Liu, Research Progress on Biodosimeters of Ionizing Radiation Damage, Radiation Medicine and Protection, 2020, 1, 127–132 CrossRef.
- M. M. Satyamitra, A. L. Dicarlo, B. A. Hollingsworth, T. A. Winters and L. P. Taliaferro, Development of Biomarkers for Radiation Biodosimetry and Medical Countermeasures Research: Current Status, Utility, and Regulatory Pathways, Radiat. Res., 2022, 197, 514–532 CAS.
- M. Brengues, B. Paap, M. Bittner, S. Amundson, B. Seligmann, R. Korn, R. Lenigk and F. Zenhausern, Biodosimetry on Small Blood Volume Using Gene Expression Assay, Health Phys., 2010, 98, 179–185 CrossRef CAS PubMed.
- C. Badie, S. Kabacik, Y. Balagurunathan, N. Bernard, M. Brengues, G. Faggioni, R. Greither, F. Lista, A. Peinnequin, T. Poyot, F. Herodin, A. Missel, B. Terbrueggen, F. Zenhausern, K. Rothkamm, V. Meineke, H. Braselmann, C. Beinke and M. Abend, Laboratory Intercomparison of Gene Expression Assays, Radiat. Res., 2013, 180, 138–148 CrossRef CAS PubMed.
- H. Ishihara, I. Tanaka, H. Yakumaru, M. Tanaka, K. Yokochi, K. Fukutsu, K. Tajima, M. Nishimura, Y. Shimada and M. Akashi, Quantification of Damage Due to Low-Dose Radiation Exposure in Mice: Construction and Application of a Biodosimetric Model Using MRNA Indicators in Circulating White Blood Cells, J. Radiat. Res., 2016, 57, 25–34 CrossRef CAS PubMed.
- S. M. Yang, S. Lv, W. Zhang and Y. Cui, Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges, Sensors, 2022, 22, 1620 CrossRef CAS PubMed.
- C. Wang, M. Liu, Z. Wang, S. Li, Y. Deng and N. He, Point-of-Care Diagnostics for Infectious Diseases: From Methods to Devices, Nano Today, 2021, 37, 101092 CrossRef CAS PubMed.
- A. N. Konwar and V. Borse, Current Status of Point-of-Care Diagnostic Devices in the Indian Healthcare System with an Update on COVID-19 Pandemic, Sensors International, 2020, 1, 100015 CrossRef.
- C. M. Pandey, S. Augustine, S. Kumar, S. Kumar, S. Nara, S. Srivastava and B. D. Malhotra, Microfluidics Based Point-of-Care Diagnostics, Biotechnol. J., 2018, 13, 1700047 CrossRef.
- L. Zhang, X. Fang, X. Liu, H. Ou, H. Zhang, J. Wang, Q. Li, H. Cheng, W. Zhang and Z. Luo, Discovery of Sandwich Type COVID-19 Nucleocapsid Protein DNA Aptamers, Chem. Commun., 2020, 56, 10235–10238 RSC.
- C. E. Anderson, C. A. Holstein, E.-M. Strauch, S. Bennett, A. Chevalier, J. Nelson, E. Fu, D. Baker and P. Yager, Rapid Diagnostic Assay for Intact Influenza Virus Using a High Affinity Hemagglutinin Binding Protein, Anal. Chem., 2017, 89, 6608–6615 CrossRef CAS.
- B. Lin, Z. Guan, Y. Song, E. Song, Z. Lu, D. Liu, Y. An, Z. Zhu, L. Zhou and C. Yang, Lateral Flow Assay with Pressure Meter Readout for Rapid Point-of-Care Detection of Disease-Associated Protein, Lab Chip, 2018, 18, 965–970 RSC.
- P. Dey, N. Fabri-Faja, O. Calvo-Lozano, R. A. Terborg, A. Belushkin, F. Yesilkoy, A. Fàbrega, J. C. Ruiz-Rodriguez, R. Ferrer, J. J. González-López, M. C. Estévez, H. Altug, V. Pruneri and L. M. Lechuga, Label-Free Bacteria Quantification in Blood Plasma by a Bioprinted Microarray Based Interferometric Point-of-Care Device, ACS Sens., 2019, 4, 52–60 CrossRef CAS PubMed.
- X. Z. Mou, X. Y. Chen, J. Wang, Z. Zhang, Y. Yang, Z. X. Shou, Y. X. Tu, X. Du, C. Wu, Y. Zhao, L. Qiu, P. Jiang, C. Chen, D. S. Huang and Y. Q. Li, Bacteria-Instructed Click Chemistry between Functionalized Gold Nanoparticles for Point-of-Care Microbial Detection, ACS Appl. Mater. Interfaces, 2019, 11, 23093–23101 CrossRef CAS PubMed.
- D. J. Heyes, S. J. O. Hardman, M. N. Pedersen, J. Woodhouse, E. De La Mora, M. Wulff, M. Weik, M. Cammarata, N. S. Scrutton and G. Schirò, Light-Induced Structural Changes in a Full-Length Cyanobacterial Phytochrome Probed by Time-Resolved X-Ray Scattering, Commun. Biol., 2019, 2, 1 CrossRef PubMed.
- R. Hajian, S. Balderston, T. Tran, T. deBoer, J. Etienne, M. Sandhu, N. A. Wauford, J. Y. Chung, J. Nokes, M. Athaiya, J. Paredes, R. Peytavi, B. Goldsmith, N. Murthy, I. M. Conboy and K. Aran, Detection of Unamplified Target Genes via CRISPR–Cas9 Immobilized on a Graphene Field-Effect Transistor, Nat. Biomed. Eng., 2019, 3, 427–437 CrossRef CAS.
- Y.-D. Ma, K. Luo, W.-H. Chang and G.-B. Lee, A Microfluidic Chip Capable of Generating and Trapping Emulsion Droplets for Digital Loop-Mediated Isothermal Amplification Analysis, Lab Chip, 2018, 18, 296–303 RSC.
- Z. Ramshani, C. Zhang, K. Richards, L. Chen, G. Xu, B. L. Stiles, R. Hill, S. Senapati, D. B. Go and H. C. Chang, Extracellular Vesicle MicroRNA Quantification from Plasma Using an Integrated Microfluidic Device, Commun. Biol., 2019, 2, 189 CrossRef.
- D. Zhang, Y. Yan, H. Que, T. Yang, X. Cheng, S. Ding, X. Zhang and W. Cheng, CRISPR/Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing, ACS Sens., 2020, 5, 557–562 CrossRef CAS PubMed.
- R. P. Balog, P. Chang, H. S. Javitz, S. Lee, H. Lin, T. Shaler and D. E. Cooper, Development of a Point-of-Care Radiation Biodosimeter: Studies Using Novel Protein Biomarker Panels in Non-Human Primates, Int. J. Radiat. Biol., 2020, 96, 35–46 CrossRef CAS PubMed.
- T.-c. D. Huang, S. Paul, P. Gong, R. Levicky, J. Kymissis, S. A. Amundson and K. L. Shepard, Gene Expression Analysis with an Integrated CMOS Microarray by Time-Resolved Fluorescence Detection, Biosens. Bioelectron., 2011, 26, 2660–2665 CrossRef CAS PubMed.
- P. Ostheim, S. A. Amundson, C. Badie, D. Bazyka, A. C. Evans, S. A. Ghandhi, M. Gomolka, M. López Riego, P. K. Rogan, R. Terbrueggen, G. E. Woloschak, F. Zenhausern, H. L. Kaatsch, S. Schüle, R. Ullmann, M. Port and M. Abend, Gene Expression for Biodosimetry and Effect Prediction Purposes: Promises, Pitfalls and Future Directions–Key Session ConRad 2021, Int. J. Radiat. Biol., 2022, 98, 843–854 CrossRef CAS PubMed.
- K. Wang, Z. Tang, C. J. Yang, Y. Kim, X. Fang, W. Li, Y. Wu, C. D. Medley, Z. Cao, J. Li, P. Colon, H. Lin and W. Tan, Molecular Engineering of DNA: Molecular Beacons, Angew. Chem., Int. Ed., 2009, 48, 856–870 CrossRef CAS PubMed.
- J. Lenaerts, H. M. Lappin-Scott and J. Porter, Improved Fluorescent in Situ Hybridization Method for Detection of Bacteria from Activated Sludge and River Water by Using DNA Molecular Beacons and Flow Cytometry, Appl. Environ. Microbiol., 2007, 73, 2020–2023 CrossRef CAS.
- S. Mao, Y. Ying, R. Wu and A. K. Chen, Recent Advances in the Molecular Beacon Technology for Live-Cell Single-Molecule Imaging, iScience, 2020, 23, 101801 CrossRef CAS PubMed.
- N. Bidar, M. Amini, F. Oroojalian, B. Baradaran, S. S. Hosseini, M. A. Shahbazi, M. Hashemzaei, A. Mokhtarzadeh, M. R. Hamblin and M. de la Guardia, Molecular Beacon Strategies for Sensing Purpose, TrAC, Trends Anal. Chem., 2021, 134, 116143 CrossRef CAS.
- S. X. Han, X. Jia, J. L. Ma and Q. Zhu, Molecular Beacons: A Novel Optical Diagnostic Tool, Arch. Immunol. Ther. Exp., 2013, 61, 139–148 CrossRef PubMed.
- J. Lacombe, S. L. Phillips and F. Zenhausern, Microfluidics as a New Tool in Radiation Biology, Cancer Lett., 2016, 371, 292–300 CrossRef CAS PubMed.
- M. Zuker, Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS PubMed.
- E. W. Sayers, R. Agarwala, E. E. Bolton, J. R. Brister, K. Canese, K. Clark, R. Connor, N. Fiorini, K. Funk, T. Hefferon, J. B. Holmes, S. Kim, A. Kimchi, P. A. Kitts, S. Lathrop, Z. Lu, T. L. Madden, A. Marchler-Bauer, L. Phan, V. A. Schneider, C. L. Schoch, K. D. Pruitt and J. Ostell, Database Resources of the National Center for Biotechnology Information, Nucleic Acids Res., 2019, 47, D23–D28 CrossRef CAS PubMed.
- S. Tyagi, D. P. Bratu and F. R. Kramer, Multicolor Molecular Beacons for Allele Discrimination, Nat. Biotechnol., 1998, 16, 49–53 CrossRef CAS PubMed.
- T. Miyashita, S. Krajewski, M. Krajewska, H. G. Wang, H. K. Lin, D. A. Liebermann, B. Hoffman and J. C. Reed, Tumor Suppressor P53 Is a Regulator of Bcl-2 and Bax Gene Expression in Vitro and in Vivo, Oncogene, 1994, 9, 1799–1805 CAS.
- Y. Y. E. Chuang, Q. Chen and H. L. Liber, Radiation-Induced Mutations at the Autosomal Thymidine Kinase Locus Are Not Elevated in P53-Null Cells, Cancer Res., 1999, 59, 3073–3076 CAS.
- L. Nemzow, T. Boehringer, B. Bacon and H. C. Turner, Development of a Human Peripheral Blood Ex Vivo Model for Rapid Protein Biomarker Detection and Applications to Radiation Biodosimetry, PLoS One, 2023, 18, e0289634 CrossRef CAS PubMed.
- A. Ho-Pun-Cheung, C. Bascoul-Mollevi, E. Assenat, F. Boissière-Michot, F. Bibeau, D. Cellier, M. Ychou and E. Lopez-Crapez, Reverse Transcription-Quantitative Polymerase Chain Reaction: Description of a RIN-Based Algorithm for Accurate Data Normalization, BMC Mol. Biol., 2009, 10, 31 CrossRef PubMed.
- J. Lacombe, C. Sima, S. A. Amundson and F. Zenhausern, Candidate Gene Biodosimetry Markers of Exposure to External Ionizing Radiation in Human Blood: A Systematic Review, PLoS One, 2018, 13, e0198851 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00079j |
| This journal is © The Royal Society of Chemistry 2024 |