Open Access Article
Open Access ArticleA fluorescent sensor array based on carbon dots for the accurate determination of pH†
Haobo
Guo
 ab,
Pooria
Lesani
ab,
Pooria
Lesani
 acde,
Hala
Zreiqat
ac and
Elizabeth J.
New
acde,
Hala
Zreiqat
ac and
Elizabeth J.
New
 *bcf
*bcf
aSchool of Biomedical Engineering, The University of Sydney, Sydney, NSW 2006, Australia
bSchool of Chemistry, The University of Sydney, Sydney, NSW 2006, Australia. E-mail: elizabeth.new@sydney.edu.au
cThe University of Sydney Nano Institute, Sydney, NSW 2006, Australia
dSchool of Science, STEM College, RMIT University, Melbourne, VIC 3000, Australia
eKoch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
fAustralian Research Council Centre of Excellence for Innovations in Peptide and Protein Science, The University of Sydney, Sydney, NSW 2006, Australia
First published on 12th November 2024
Abstract
In this study, we present a sensor array for precise pH monitoring, based on carbon dots (CDs) synthesised from fructose and p-phenylenediamine through a one-step hydrothermal method. The CDs exhibited significant photostability and different fluorescence emissions in different pH conditions, making them suitable for real-time pH sensing across a wide pH range of 3 to 10. Our mechanistic studies revealed how surface functionalisation affects the pH response. For statistical analysis of our array data, we employed Gaussian process regression to create a predictive model for determining pH levels of test samples based on the array spectral data, and linear discriminant analysis for high-precision classification of pH values. This combined approach enabled a comprehensive analysis of the CDs' pH-sensitive fluorescence, demonstrating a significant methodological advancement in pH sensor technology. Our research advances the understanding of the fluorescence mechanisms of CDs in response to pH variations. Furthermore, our study demonstrates the power of integrating machine learning techniques to improve the performance and application scope of fluorescent nanomaterials. These findings have important implications for chemical sensing across a range of fields, including environmental monitoring and biomedical diagnostics.
Introduction
Determination of pH is a crucial analytical challenge across a range of industrial challenges, from food spoilage1 to environmental monitoring2 to a broad range of biological questions. For example, pH sensing is crucial for biomedical applications due to its significant role in drug delivery, disease diagnosis, and physiological condition monitoring.3–8 The development of pH-responsive amphiphilic polymer nano-assemblies enhances drug delivery efficiency while reducing side effects.5 Additionally, flexible potentiometric pH sensors for wearable technologies highlight the importance of real-time health monitoring.9 These sensors are particularly valuable for continuous pH monitoring in bodily fluids, facilitating the early detection of health condition changes or abnormalities.10,11 Moreover, pH determination is vital in clinically relevant diagnoses, where sensors are utilised in drug control-released carriers and other diagnostic tools.12A broad range of analytical tools has been developed to measure pH within complex environments, including H+ permeable microelectrodes,13 advanced nuclear magnetic resonance (NMR) techniques,14 and fluorescent sensors.15 The latter are advantageous as they do not require physical breaching of cell membranes, and they can be coupled to inexpensive and readily available instruments for quantifying and imaging fluorescence. A broad range of pH-responsive fluorescent sensors have been reported to date, including those based on fluorescent proteins,16 organic fluorophores,17 and fluorescent nanomaterials.18 The protonatable groups typically have a working range within ±1 pH unit of the pKα, therefore limiting the working range of the final assay to less than 2 pH units.
One approach to address this limitation is to simultaneously use multiple sensors with different pKα values, and therefore, different pH working ranges. This can be achieved through the use of a sensor array, built by combining multiple sensors, each with specific properties, into a single platform.19 Array-based sensing offers several advantages, including increased selectivity and sensitivity, and reduced interference from extraneous substances.6,20,21 This versatility enables the detection of a wide range of pH values and the ability to tailor the sensor array for specific applications or environments. The use of array sensing technologies in pH sensing applications can lead to enhanced accuracy compared to traditional methods. This improvement in accuracy is crucial for various fields where precise pH measurements are essential, such as environmental monitoring and biomedical applications. This approach has been elegantly demonstrated by Kim and co-workers, who used a library of 30 fluorescent sensors to achieve pH determination within 0.2 pH units.22
We were interested in developing an array with fewer sensor elements, in order to increase efficiency of synthesis and array application. We therefore aimed to develop a new sensor array based on carbon dots (CDs) due to their previously reported pH-responsive properties.23–26 Traditional pH indicators, such as dyes and organic fluorophores, face limitations, including reduced stability, susceptibility to photobleaching, and potential cytotoxicity.27 Conversely, CDs have emerged as a superior alternative, renowned for their robust fluorescence, exceptional photostability, low toxicity, and facile synthesis.28–30 As small carbon nanoparticles, CDs display unique optical properties due to quantum confinement effects, surface modifications, and diverse carbonaceous compositions.31–33 These features render CDs ideal for pH sensing applications, as they can significantly alter their emission wavelength and intensity in response to environmental pH changes, thus serving as effective fluorescent probes for pH differentiation across a spectrum of applications.23,34 Recent research has focused on exploring the fluorescence mechanisms of CDs.35–37 Understanding these mechanisms is crucial for several reasons: it paves the way for enhancing their fluorescent properties, facilitates the creation of novel CDs with tailored features, and aids in developing standardised methodologies for their characterisation. In this paper, we focused on a mechanistic study of the surface functionality impact on CDs pH response.
Here we synthesised CDs from fructose and p-phenylenediamine precursors. After determining the surface groups contributing to pH sensing, we meticulously analysed the pH sensing capabilities exhibited by the CDs array using two advanced statistical methodologies: Gaussian process regression (GPR) and linear discriminant analysis (LDA).
Results and discussion
Synthesis of and characterisation of carbon dots
A series of CDs was synthesised via a one-pot hydrothermal method. Two carbon sources were selected as precursors: fructose, which can also introduce hydroxyl groups; and p-phenylenediamine (PPD), which can also introduce amino groups. The heating time and temperature were selected based on our previous work.38 In this study, we focused on altering the ratio of precursors to further investigate its effect on the surface functionality of the synthesised CDs. In total, five different fructose![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PPD ratios were used (100
PPD ratios were used (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0; 92
0; 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8; 69
8; 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31; 43
31; 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57; and 25
57; and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75) to give five CDs, named CD100, CD92, CD69, CD43 and CD25, respectively. The hydrothermal reactions were carried out in an oven preheated to 180 °C for 8 h to ensure consistent synthetic conditions. The synthesised CDs were purified by filtration through a 0.22 μm cellulose acetate filter to remove large and aggregated particles (TEM figures of the synthesised CDs are shown in Fig. S1 with size distribution in Table S1†).
75) to give five CDs, named CD100, CD92, CD69, CD43 and CD25, respectively. The hydrothermal reactions were carried out in an oven preheated to 180 °C for 8 h to ensure consistent synthetic conditions. The synthesised CDs were purified by filtration through a 0.22 μm cellulose acetate filter to remove large and aggregated particles (TEM figures of the synthesised CDs are shown in Fig. S1 with size distribution in Table S1†).
Attenuated total reflectance (ATR) spectroscopy provided structural insights into the five synthesised CDs. CD100, synthesised from fructose alone, displayed bands at 3300, 2970, 1660, 1405, and 1050 cm−1, corresponding to the stretching vibrations of O–H, C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, COO–, and C–O respectively (Fig. S2†). Introduction of some nitrogen doping, in CD92, and CD69, gave rise to an additional peak at approximately 1500 cm−1, corresponding to N–H. CD43 and CD25 had the highest N-doping levels, and therefore exhibited strong ATR peaks located at N–H (1510 cm−1), as well as peaks corresponding to C–N/C–O (1390, 1260, and 1070 cm−1).
O, COO–, and C–O respectively (Fig. S2†). Introduction of some nitrogen doping, in CD92, and CD69, gave rise to an additional peak at approximately 1500 cm−1, corresponding to N–H. CD43 and CD25 had the highest N-doping levels, and therefore exhibited strong ATR peaks located at N–H (1510 cm−1), as well as peaks corresponding to C–N/C–O (1390, 1260, and 1070 cm−1).
X-ray photoelectron spectroscopic (XPS) analysis of CDs (Fig. 1 and S3 and Tables S2–S5†) gave insight into the surface functionality of the particles. CD100 showed 67.5% carbon (C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–O, and O–C
C, C–O, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 32.5% oxygen (–OH and
O) and 32.5% oxygen (–OH and ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), in agreement with the ATR results (Fig. S2†). As the degree of N-doping increased from CD92 to CD25, the nitrogen surface content determined by XPS increased from 5.5% to 17.13%, with a concomitant decrease in the oxygen atomic percentage (from 26.28% to 5.81%). This corresponds to an increase in the proportion of –OH groups and a corresponding increase in the proportion of amino groups.
O), in agreement with the ATR results (Fig. S2†). As the degree of N-doping increased from CD92 to CD25, the nitrogen surface content determined by XPS increased from 5.5% to 17.13%, with a concomitant decrease in the oxygen atomic percentage (from 26.28% to 5.81%). This corresponds to an increase in the proportion of –OH groups and a corresponding increase in the proportion of amino groups.
UV-visible spectroscopy gave further insight into the composition of the CDs (Fig. 2). CD100 shows a predominance of oxygen-containing functional groups, such as hydroxyl and carboxyl groups, evidenced by strong absorption between 290–310 nm, corresponding to aromatic or graphitic π–π* transitions,39,40 and a secondary peak between 320–350 nm, corresponding to n–π* transitions of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups,41 suggesting significant oxidation.42 As the N-doping level increased from CD92 to CD25, there was an absorption increase around 350–400 nm, which was related to the π–π* transitions of the aromatic C
O groups,41 suggesting significant oxidation.42 As the N-doping level increased from CD92 to CD25, there was an absorption increase around 350–400 nm, which was related to the π–π* transitions of the aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds and n–π* transitions of C
C bonds and n–π* transitions of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N heterocycles. This shift was evidenced by a lower intensity in the 290–310 nm range and diminished n–π* transitions in the 320–350 nm range.
N heterocycles. This shift was evidenced by a lower intensity in the 290–310 nm range and diminished n–π* transitions in the 320–350 nm range.
Fluorescence characterisation and pH response of carbon dots
The fluorescence excitation and emission spectra of the synthesised CDs are shown in Fig. 3 and S7.† The excitation peaks for these CDs range from 354 nm to 393 nm, and the emission peaks span from 510 nm to 545 nm (Table 1). CD100 exhibits a Stokes shift of 150 nm, whereas the introduction of N-doping led to an initial decrease to 117 nm (CD92), then a steady increase to 186 nm (CD25). This ascending trend in Stokes shift, concomitant with an increase in amino groups on the surface of CDs, is consistent with an augmented separation between the electronic energy levels, leading to enhanced red emission.43,44 Furthermore, the increase in nitrogen doping influenced the solubility of CDs, as the proliferation of –NH2 groups led to the formation of hydrophobic CDs. These hydrophobic CDs were prone to aggregation, resulting in variations in fluorescence.44,45 Additionally, PPD, characterised by its conjugated structure, contributed to the enlargement of sp2 domains, which was advantageous for the red shift in the emission wavelength of CDs.44,46| CD | Excitation maximum (nm) | Emission maximum (nm) | Stokes shift (nm) |
|---|---|---|---|
| CD100 | 360 | 510 | 150 |
| CD92 | 393 | 510 | 117 |
| CD69 | 369 | 530 | 161 |
| CD43 | 354 | 525 | 171 |
| CD25 | 359 | 545 | 186 |
In addition to variation in fluorescence wavelengths, total emission intensity also varies across the series of CDs (Fig. 3(f)). As the proportion of the nitrogen source PPD increased during hydrothermal synthesis, the fluorescence intensity of the CDs initially rose and then declined, with CD43 exhibiting the highest fluorescence intensity.
Thus, the effect of nitrogen doping on the fluorescence intensity of CDs was not straightforwardly linear. It involved a complex interplay of factors that could either enhance or quench fluorescence, depending on the specific levels of doping and the environmental conditions.47
The emission and absorption spectra of the CDs (Fig. S5, S6 and S9 and Table S6†) demonstrate a distinct dependency on pH levels, with clear shifts in peak positions and variations in intensity. For instance, CDs with a higher oxygen content, such as CD100, consistently exhibited emission peaks around 500 nm at low pH, accompanied by Stokes shifts ranging from 85 to 160 nm. On the other hand, CDs with a higher nitrogen content, like CD25, presented higher emission peaks at lower pH levels, with Stokes shifts ranging from 130 to 190 nm. These differences implied that the fluorescence properties of CDs were significantly affected by their oxygen and nitrogen content, which in turn influenced their sensitivity and stability across various pH levels. Specifically, higher nitrogen content appears to stabilise both the emission and excitation wavelengths over a broader pH range. Conversely, CDs with a higher oxygen content displayed more pronounced shifts, particularly in their emission peaks. These findings highlight the potential use of CDs in pH sensing applications. They demonstrate how variations in oxygen and nitrogen content could be leveraged to tune the fluorescence properties of CDs for specific sensing needs.
We found a clear transition from pH turn-on to pH turn-off response with increased N-doping (Fig. 4). The fluorescence spectra of the synthesised CDs with full pH range are presented in Fig. S6 and the naked eye observation of CDs under different pH are presented in Fig. S10.† CD100 exhibits turn-on behaviour with increasing pH (Fig. 4a). We hypothesise that this pH response is dominated by surface –OH groups. In comparison, CD25, which is rich in amino groups, presents a turn-off fluorescence response with increasing pH (Fig. 4e). The intermediate CDs, CD92, CD69 and CD43, show a turn-on pH response in acidic environments and a turn-off response in basic environments (Fig. 4b–d). This can be explained by the competition between hydroxyl groups, which dominate in acidic environments, and amino groups, which take over control in basic environments.
The CDs retained their pH sensitivity even after three months in solution at room temperature without protection from sunlight (Fig. S7†). Furthermore, the reversibility of response was confirmed through alternating addition of sodium hydroxide and hydrochloric acid to CD100 and CD25 (Fig. S8†).
The oxygen content of each CD (determined both by precursor ratio and by XPS characterisation) is well-correlated with the pH value at which the highest fluorescence intensity was observed (Fig. 5). This is consistent with our hypothesis that the surface functional groups influence the pH responsiveness of CDs.
To validate our hypothesis, we masked the hydroxyl groups on CD100 and CD69 using TBS protection (Fig. 5(a)), and the amino groups on CD69 and CD25 using Boc protection (Fig. 5(b)). This approach enabled us to investigate the specific impact of hydroxyl and amino groups on the fluorescence and pH response of the CDs. For CD100, masking the –OH groups using TBS protection resulted in significant fluorescence quenching, and very little pH sensitivity (Fig. 5(c)). This result supports our hypothesis that the pH turn-on response of CD100 can largely be attributed to the presence of –OH groups.
For CD25, masking the –NH2 groups led to a significant enhancement in fluorescence, but a dampening of the pH-responsive behaviour (Fig. 5(d)). The fluorescence enhancement may be due to the fact that amino groups themselves are fluorescence quenchers through surface passivation, and through non-radiative pathways for energy dissipation.48,49 The turn-off performance of CD25 decreased by 55%, confirming our hypothesis that –NH2 groups were responsible for the CDs' pH turn-off response and limited the CDs' fluorescence.
CD69 exhibited a pH response pattern characterised by fluorescence turn-on in the pH range of 3–7 and turn-off in the pH range of 7–10, as displayed in Fig. 5(e). This behaviour was due to its surface functionalisation, which contained considerable amounts of both –OH and –NH2 groups. For CD69, the original sample had a peak fluorescence value at pH 6.525, as determined from plot fitting results. After masking the –OH groups, the peak shifted to the acidic range at pH 4.816, and the fluorescence was clearly quenched compared to the original CD69. Conversely, after masking the –NH2 groups, the peak shifted to the basic range at pH 8.163, and the fluorescence was enhanced.
These results confirmed that hydroxyl groups promoted the transformation of CDs into pH-sensitive ‘turn-on’ sensors, while amino groups drove CDs to function as pH-sensitive ‘turn-off’ sensors. The masking experiments validated our hypothesis and provided a deeper understanding of the mechanisms underlying the pH responsiveness of CDs.
Application of CDs to a pH sensor array
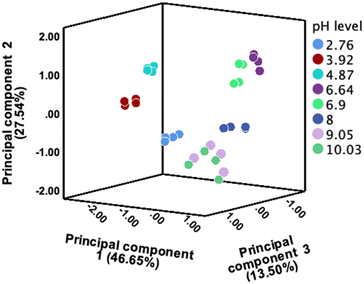
With the original 5 CDs and the 4 masked CDs (CD100–OH protected; CD25–NH2 protected; CD69–OH protected; and CD69–NH2 protected) to hand, we investigated their potential use in an array to distinguish and determine different pH values. We collected the fluorescence emission spectra and calculated integrated emission intensity for each CD at each pH value, in quadruplicate. These data were then subjected to principal component analysis (PCA) and linear discriminant analysis (LDA) using SPSS (IBM, 29.0.1.0 (171)).PCA was utilised to analyse feature sets, such as those obtained by adding sensors to a standard configuration, and to understand how the different features compared and contributed to the overall sensor array performance. From Fig. 6, PCA revealed that the combined features of the 9 CDs contributed significantly to the overall variance, indicating their collective importance in accurately sensing pH levels. This comprehensive approach leverages the strengths of each individual CD, leading to an enhanced and reliable pH sensing capability. The classification accuracy of LDA classification by using this 9 CDs array was confirmed to be 100% for both classification accuracy and cross-validation accuracy, as presented in Fig. S11 and Table S7.†
We had therefore demonstrated that these 9 CDs were able to correctly classify these pH values, over a greater than 7 pH range. However, the application of 9 elements to a sensor array is still labour-intensive, and so we sought to reduce the number of sensor elements required to still give 100% correct classification. The process of reducing the number of array elements can be achieved by an iterative trial-and-error approach,50 or by examination of the correlation statistics derived from PCA,51 as we have previously described. However, both techniques are cumbersome, and do not enable investigation of all permutations of the full sensor set. We therefore sought to develop an algorithm that would let us rapidly and comprehensively perform array reduction.
To address this challenge, we developed a MATLAB-based classifier using LDA and support vector machine (SVM) to carry out array-based classification and array optimisation for CDs based pH array development. A 4-fold cross-validation setup was implemented for robust performance evaluation. This program automatically tested all potential combinations driven by the 9 CD sensor features: a total of 511 combinations. Both SVM and LDA classifiers were trained and evaluated for each combination, individually. The performance metrics, including accuracy, precision, recall, and F1 score, were calculated and averaged over the cross-validation folds (Table S9†). The F1 score represented the mean of precision and recall, giving equal weight to both metrics. An F1 score ranges from 0 to 1, with 1 indicating a perfect model where all predictions are correct, and 0 representing the worst possible performance. In general, combinations containing our protected CDs were less accurate than those containing the original 5 CDs, consistent with the fact that masking groups led to less pH sensitivity.
The best-performing feature sets for both SVM and LDA were identified based on the highest accuracy. The SVM model used a linear kernel with a one-vs.-one approach for multi-class classification, while the LDA model employed a pseudo-linear discriminant type. This program provided a comprehensive evaluation of the sensor features, identifying the most effective combinations for pH level classification, and ensuring reliable and robust performance through meticulous data normalisation and cross-validation. Just using CD43 alone was found to achieve 100% classification accuracies for both SVM and LDA. However, since we sought to use a sensor array with at least two elements for subsequent pH quantification, we then looked for two sensor combinations that contained CD43 (Table S10†). 100% classification accuracies for both SVM and LDA classifiers were achieved by using combinations of CD43 and CD69, and CD43 and CD25. These results were verified by SPSS-based LDA, and 100% classification accuracies for both original classification and cross validation were achieved for both arrays.
The LDA classification results are shown in Fig. 7, Tables S14–S16† (CD43 and CD69), and Tables S17–S19 (CD43 and CD25).
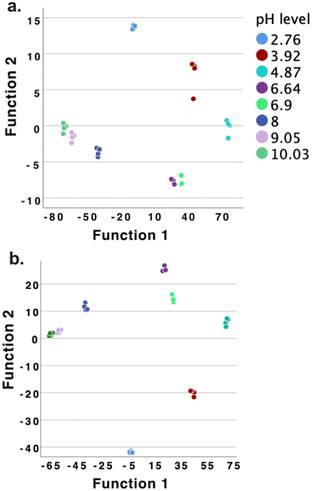 | ||
| Fig. 7 The LDA classification results by using two CDs based array (a: CD43 and CD69; b: CD43 and CD25) as pH sensor array to discriminate 8 different pH environments. | ||
Gaussian process regression-based pH prediction
With these two robust two-sensor arrays in hand, we sought to use a machine learning approach to accurately quantify pH. Gaussian process regression (GPR) is a non-parametric, Bayesian machine learning method used for regression problems, based on the Gaussian process – a collection of random variables where any finite subset follows a multivariate Gaussian distribution.52 GPR is particularly effective for modelling complex and non-linear relationships between input features and target variables.53 In our study, we developed and rigorously evaluated a MATLAB-based GPR model for predicting pH values based on a comprehensive CDs based fluorescence responses, treated as features, described in the supplementary information. Using our model, we could explore all possible combinations of CDs, but here we report the investigation of only the one-sensor CD43 and two-sensor CD43 and CD69, and CD43 and CD25, which were found to be most promising by LDA and SVM analysis. In the preliminary GPR model, we took out the pH 6.9 sample from the dataset and added it as an unknown to test the GPR. Our GPR model demonstrated superior performance, as evidenced by high coefficients of determination (R2), low mean squared errors (MSE), and minimal mean absolute errors (MAE) across all sets of arrays (Table S20†).Of the three arrays tested, the array built using CD43 and CD69 achieved the best prediction for the “unknown” pH 6.9 sample (Fig. 8(a)). For the kernel parameters, the length scale (σl) was calculated to be 427.7, indicating a significant influence of input features on predictions. The signal variance (σf) was 0.316, representing the variation of function values from their mean. The standard deviation of predicted values (n = 4) was 0.110, indicating consistent predictions close to the average value. This demonstrates that the GPR model can predict pH values close to those measured by a pH meter.
 | ||
| Fig. 8 GPR prediction by using 2 CDs array built by CD43 and CD69: (a–c) the test pH values were 6.9, 4.07, and 9.5. | ||
The R2 was 1, indicating that the model explains approximately 100% of the variance in the data. The MSE was 0, suggesting a good model fit. The MAE was 0.009, indicating that the model's predictions were accurate.
To further validate the performance of the pH sensor array constructed using CD43 and CD69, GPR tests were conducted at pH 4.07 and 9.05, (as presented in Fig. 8(b and c) and Table S20†). At pH 4.07, the standard deviation of the predicted values (n = 4) was 0.106, with an average predicted pH of 4.040 ± 0.106. The prediction range was 3.934 to 4.145, and the R2 was 0.987. MSE was 0.055, and the MAE was 0.165. For pH 9.05, the standard deviation of the predicted values was 0.0359, with an average predicted pH of 8.970 ± 0.0359. The prediction range was 8.934 to 9.006, and the R2 value was 0.996. The MSE was 0.019, and the MAE was 0.102. These results demonstrate that the pH sensor array provided accurate and precise pH predictions, with very high R2 values and low error metrics for both pH levels tested in acidic and basic conditions.
Overall, the CDs-based pH sensor array performed well in predicting pH values (from acidic to alkaline) based on the fluorescence response of the three sensors using the GPR model. The high R2 value, low MSE, and low MAE indicated a good fit and accurate predictions. The standard deviation of predicted values was relatively small, indicating consistency in predictions. The model's predictions for the test pH value were reasonably close to the actual value, confirming its effectiveness.
GPR-based pH prediction application with optimised CDs sensor array for pH sensing of milk
To further investigate the application of our pH sensor array for monitoring pH in food, we applied our optimised sensor array (CD69 and CD43) to determine the pH of both fresh and expired full-cream milk. The results were benchmarked against standard pH meter readings, where the fresh milk (tested 10 days before its best before date) exhibited a pH of 6.74, and the expired milk (tested 20 days after its best before date) had a pH of 5.20.The performance of the GPR model, using CD69 and CD43 as features, demonstrated a high degree of accuracy in predicting the pH of fresh milk (Fig. 9(a)). The model predicted an average pH value of 6.89 ± 0.18, with a range of predictions between 6.70 and 7.07. This closely aligns with the actual pH of 6.74, reflecting the reliability of the sensing array in accurately detecting minor pH variations in fresh milk. The model exhibits a high R2 value of 0.971, indicating a strong correlation between the predicted and observed pH values. Furthermore, the MSE (0.084) and MAE (0.234) are relatively low, supporting the model's capacity to provide precise pH predictions.
 | ||
| Fig. 9 GPR prediction by using 2 CDs array built by CD43 and CD69: a. fresh milk and b. spoiled milk. | ||
For the expired milk (Fig. 9(b)), the GPR model demonstrated strong predictive accuracy. The model predicted an average pH of 5.62 ± 0.48, with a prediction range of 5.14 to 6.09, closely approximating the actual pH value of 5.20.
The high R2 (0.9772) reflects a strong correlation between predicted and observed values, with low error rates – an MSE of 0.066 and an MAE of 0.179 – indicating precise prediction.
The relatively small standard deviation (0.48) confirms the model's robustness. These results suggest that the sensor array retained high accuracy even in expired milk, despite the potential influence of factors like microbial spoilage and chemical decomposition. The naked eye observation of pH sensor array applied for spoiled milk is shown in Fig. S12.†
Our CD-based sensor array exhibited considerable promise as a rapid and reliable method for assessing milk freshness. The high level of accuracy observed in milk pH predictions underscored its potential application in food quality monitoring systems, where real-time and non-invasive pH sensing is of significant commercial interest.
Conclusions
The research presented herein successfully demonstrated the use of CDs synthesised from fructose and p-phenylenediamine for effective pH monitoring across a broad range. We conducted a comprehensive spectroscopic analysis to assess changes in CDs' fluorescence response to pH variation. The results gleaned from this study offer deep insights into the roles hydroxyl group and amino group play in the pH sensitivity of CDs. We verified the functional group impacts by masking the functional groups respectively and established an exhaustive understanding of the pH-sensing capabilities of CDs.We were able to use this set of CDs to distinguish between pH values over a greater than 7 pH unit range, and in fact we have shown that just two carbon dots can be used to construct an array that gives 100% accuracy of classification, as well as a very high accuracy in quantification of pH. Employing Gaussian process regression and linear discriminant analysis, we developed a sophisticated analytical framework that not only predicts pH levels with remarkable accuracy but also classifies them with high precision. Importantly, we have shown that these methods can be used to rapidly and efficiently scan a full set of array data to identify the most promising set.
In our study, GPR was employed to construct a comprehensive multivariate regression model aimed at predicting pH levels in diverse test sensing environments. By analysing variations in the spectral features of the CDs' fluorescence emission in response to pH changes, GPR facilitated a nuanced understanding of the relationship between spectral attributes and pH levels, thereby enabling accurate and reliable pH estimation. The inherent flexibility of GPR, coupled with its ability to provide confidence intervals, significantly bolstered its applicability in sensor technology and analytical chemistry.
Concurrently, LDA was utilised as a powerful statistical tool for dimensionality reduction and classification within the pattern recognition domain.54,55 By maximising the ratio of between-class variance to within-class variance, LDA ensured optimal separation among different classes.56,57 In our CDs-based array, LDA harnessed the unique fluorescence-based pH responses of the CDs to create distinct fingerprints for varying pH levels. This enabled the effective reduction of data dimensionality while retaining critical distinguishing features, thereby streamlining the classification process.
This study contributes to the fields of nanotechnology and sensor development by providing detailed mechanistic insights into the fluorescence behaviour of CDs under different pH level. This study not only contributes to the fundamental understanding of CDs but also offers practical guidelines for the future design and optimisation of pH-responsive CDs, expanding their potential in various scientific and industrial applications. It also showcased the integration of nanomaterials with machine learning to create more sensitive, selective, and versatile sensing technologies. Our optimised two-CD sensor array was utilised for pH sensing in milk. During the sensor stability and fluorescence recovery tests, the CD sensors exhibited a dynamic fluorescent response to pH fluctuations, demonstrating their effectiveness in detecting pH variations. Sensors of this type could be embedded onto filter paper or a similar support for paper-based sensing, or could be used in solution, such as in a microwell plate, as we demonstrate here. While the former could provide a binary reading of high or low pH, for example for detecting food spoilage, the latter enables us to harness the full potential of intensity measurement and statistical analysis to accurately determine pH values. We are actively studying how to apply such systems to the measurement of biologically-relevant pH, both in intracellular pH mapping, and studies of clinical samples. Future research should aim to refine the synthesis and functionalization of CDs to further enhance their sensitivity and selectivity for specific ions or molecules, thereby expanding their utility in a wider range of applications such as environmental monitoring, industrial processes, and biomedical diagnostics.
Experimental section
Materials and instrumentation
All chemical reagents and solvents employed in this study were of laboratory or analytical grade, acquired from Sigma-Aldrich, Merck, or Combi-Blocks. These chemicals were used as procured, without further purification. A2 full-cream milk was purchased from a local Woolworths store and was left at room temperature for 30 days to allow it to spoil.X-ray photoelectron spectroscopy (XPS) measurements were collected using a Kratos Axis Nova spectrometer (Kratos Analytical, UK), equipped with a monochromatised aluminium X-ray source (Al Kα, 1486.6 eV), operating at 10 mA and 15 kV (150 kW). Both survey and high-resolution spectra were recorded at detector pass energies of 160 and 20 eV, respectively. The obtained XPS data were processed and analysed using Thermo Avantage software (version 5.9902). Photophysical evaluations were conducted using a PerkinElmer EnSpire Multimode Plate Reader, with experiments carried out in 300 μL, 96-well polypropylene microplates (item no.: 655209, Greiner Bio-One), ensuring standardised and reproducible measurement conditions.
Preparation of carbon dots
A one-step hydrothermal/solvothermal synthesis route was used to prepare CDs using fructose and p-phenylenediamine (PPD) as precursors. The precursors were mixed in 50 mL of Milli-Q water, then processed by ultrasonication to achieve a homogeneous mixture. The solution was then transferred into a 100 mL Teflon-lined stainless-steel autoclave chamber with a 50% filling ratio and heated at 180 °C for 8 h. The initial purification was performed by passing the solution through a 0.22 μm cellulose acetate Millipore filter (Millipore, USA) to remove large or agglomerated particles. The sample was then concentrated by rotary evaporation and dried using a freeze dryer. A dark brown powder was obtained, which was subsequently used to produce a 1 mg mL−1 CDs solution by adding Milli-Q water. The synthesis process was universal and applicable for all CDs. To prepare CDs of different varieties with varying proportions of –OH and –NH2 groups, the weight ratio of precursors and synthesis time were adjusted for each CDs species, as detailed in Table 2. This table illustrated the variations in precursor ratios and synthesis times used to achieve the different functional group compositions in the CDs, which influenced their fluorescence and pH responsiveness.| CD | Fructose/g | p-Phenylenediamine/g | Synthesis time/h | Synthesis temperature/°C |
|---|---|---|---|---|
| CD100 | 0.5 | 0 | 8 | 180 |
| CD92 | 0.43 | 0.07 | 8 | 180 |
| CD69 | 0.278 | 0.222 | 8 | 180 |
| CD43 | 0.147 | 0.353 | 8 | 180 |
| CD25 | 0.078 | 0.422 | 8 | 180 |
pH buffer preparation
A series of standard pH buffers was prepared using different buffering agents to cover a range of pH values from 3 to 10: sodium citrate dihydrate (50 mM) for pH 3–5; potassium phosphate dibasic (50 mM) for pH 6–8. and sodium bicarbonate (50 mM) for pH 9–10. Each buffer solution was meticulously adjusted using either NaOH or HCl to achieve the desired pH level. The resultant pH values were accurately determined using a SevenCompact pH meter. The measured pH values for each buffer are detailed in Table 3.| Expected pH | Actual pH | Chemical 1 | Chemical 2 | Buffer concentration/mM |
|---|---|---|---|---|
| 3 | 2.76 | Sodium citrate dihydrate | Citric acid | 50 |
| 4 | 3.92 | Sodium citrate dihydrate | Citric acid | 50 |
| 5 | 4.87 | Sodium citrate dihydrate | Citric acid | 50 |
| 6 | 6.9 | Potassium phosphate dibasic | Potassium phosphate monobasic | 50 |
| 7 | 6.64 | Potassium phosphate dibasic | Potassium phosphate monobasic | 50 |
| 8 | 8 | Potassium phosphate dibasic | Potassium phosphate monobasic | 50 |
| 9 | 9.05 | Sodium bicarbonate | Sodium carbonate (anhydrous) | 50 |
| 10 | 10.03 | Sodium bicarbonate | Sodium carbonate (anhydrous) | 50 |
TBS protection of carbon dots
CDs (50 mg) were dissolved in N,N-dimethylformamide (DMF, 10 mL). Subsequently, imidazole (18.5 μL) and tert-butyldimethylsilyl chloride (25 mg) were added to the solution at a controlled temperature of 0 °C. The reaction mixture was gradually allowed to attain room temperature and was stirred continuously for 24 h to ensure complete reaction. To terminate the reaction, an adequate amount of water was introduced to the mixture. The resulting mixture was then concentrated under reduced pressure, leading to the formation of a solid CDs product with hydroxyl groups masked.The obtained solid was redissolved in Milli-Q water and transferred to a dialysis bag (molecular weight cutoff: 2 kDa) for purification purposes. The dialysis process was conducted over 24 h to remove any unreacted reagents and by-products.
Boc protection of carbon dots
For amino group protection, 50 mg of CDs were dissolved in N,N-dimethylformamide (10 mL), followed by the addition of di-tert-butyl dicarbonate (50 mg) and triethylamine (25 μL). The reaction mixture was stirred at room temperature for 24 h. After the reaction, the mixture was concentrated under reduced pressure to obtain the solid product. The resulting solid was then purified using a similar dialysis procedure as described for hydroxyl group protection. The dialysis process, conducted over a span of 24 h with a molecular weight cutoff of 2 kDa, effectively removed any unreacted reagents and by-products.Fluorescence spectroscopy
To prepare the CDs solution, the synthesised CDs were first dispersed in Milli-Q water then mixed with pH buffer, achieving a final concentration of 1 mg mL−1 of CDs solution and 25 mM of pH buffer (50% v/v). For fluorescence measurements, precisely 200 μL of this CDs solution was pipetted into the wells of 96-well plates. Using an EnSpire Multimode Plate Reader (Revvity), the fluorescence spectra were then recorded. The settings for excitation and emission wavelengths were carefully selected based on the unique properties of the CDs under investigation. Emission spectra were diligently collected over a wavelength range of 400–700 nm. To ensure the reliability and accuracy of the measurements, each sample underwent triplicate testing. This rigorous approach not only confirmed the reproducibility of the results but also enhanced the overall precision of the fluorescence data obtained.Statistical analysis of array data
The fluorescence data obtained from the CDs array were subjected to sophisticated statistical analyses. Specifically, Gaussian process regression was used to construct predictive models for estimating pH levels. The spectral data served as inputs to the GPR model, which provided predictions accompanied by confidence intervals to indicate the reliability of these predictions. Furthermore, linear discriminant analysis was employed to categorise the fluorescence responses into distinct pH levels. The LDA model was designed to maximise the separation between different pH classes, thereby enhancing the accuracy of classification.GPR and LDA analyses were conducted using MATLAB (MathWorks, USA). Custom scripts were meticulously developed to handle the fluorescence data, train the models, and evaluate their performance comprehensively. To ensure the robustness and generalisability of the models, cross-validation techniques were employed to validate the results from both GPR and LDA.
Carbon dot sensors' fluorescence recovery test
CD100 and CD25 were synthesised and prepared as 10 mL solutions at a concentration of 1 mg mL−1 in Milli-Q water. For the fluorescence recovery test of CD100, an initial 200 μL aliquot of the solution was loaded in quadruplicate into a 96-well fluorescent sensing plate to serve as the blank control (represented by the blue columns in Fig. S8†). To create an acidic environment and quench the fluorescence of CD100, 200 μL of 5 M HCl was added to the solution, which was then mixed thoroughly using sonication. After mixing, four 200 μL aliquots were collected and transferred into the sensing plate. Next, 400 μL of 5 M NaOH was added to neutralise the acidic environment and shift the solution to a basic pH, allowing CD100’s fluorescence to be activated (“turned on”). Four additional 200 μL replicates were taken and loaded into the plate. The cycle was repeated by adding 400 μL of 5 M HCl, followed by sonication, and four replicate samples were collected. Finally, 400 μL of 5 M NaOH was added to return the solution to basic conditions, and four more replicates were transferred into the plate. Fluorescence measurements were conducted using a plate reader with an excitation wavelength of 370 nm, and emission spectra were recorded from 390 to 700 nm.For the fluorescence recovery test of CD25, a similar procedure was followed, with the order of HCl and NaOH additions reversed. In this case, 200 μL of 5 M NaOH was added first, followed by 400 μL of 5 M HCl, then 400 μL of NaOH, and finally 400 μL of HCl. After each addition, the solution was mixed thoroughly, and four 200 μL aliquots were collected. Fluorescence measurements were performed under the same conditions as for CD100.
Carbon dots-based pH sensor array for milk pH sensing
Fresh A2 full-cream milk was purchased from a local Woolworths store, and a spoiled sample was prepared by leaving the milk at room temperature. A carbon dots sensor array, consisting of CD43 and CD69, was utilised for sensing the pH of both fresh and spoiled milk samples. The pH values of the milk samples were measured using a pH meter as a reference. The synthesised CDs were first dispersed in Milli-Q water and then mixed with the milk samples and pH buffer. For the test group, Milli-Q water was used in place of the pH buffer. The final solution contained 1 mg mL−1 CDs, 25 mM pH buffer, and the milk sample, with each component contributing 33.33% (v/v) to a total volume of 200 μL per well. The procedure was consistent with the previously described fluorescence spectroscopy and statistical analysis of array data.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Haobo Guo: conceptualisation, methodology, data curation, formal analysis, investigation, validation, writing – original draft.; Pooria Lesani: conceptualisation, data curation, formal analysis, investigation, writing – review & editing; Hala Zreiqat: resources, writing – review; Elizabeth J. New: conceptualisation, investigation, resources, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Australian Research Council Training Centre in Innovative BioEngineering (IC170100022) for research funds and a postgraduate scholarship, ARC Training Centre in Innovative BioEngineering Postgraduate Research Scholarship (SC2398), and the Australian Research Council (ARC) DP funding scheme (Project DP220102876). The authors acknowledge the facilities and the scientific and technical assistance of Sydney Analytical, a core research facility at The University of Sydney.References
- M. Weston, S. Geng and R. Chandrawati, Food sensors: Challenges and opportunities, Adv. Mater. Technol., 2021, 6, 2001242 CrossRef.
- W. Xiao and Q. Dong, The recent advances in bulk and microfluidic-based pH sensing and its applications, Catalysts, 2022, 12, 1124 CrossRef.
- L. Gao, Z. Xie and M. Zheng, Highly efficient carbon dots for quantitatively visualizing pH fluctuations in cells, zebrafish, mice and tumors, J. Photochem. Photobiol., B, 2023, 238, 112620 CrossRef PubMed.
- M. Ghoneim, A. Nguyen, N. Dereje, J. Huang, G. Moore, P. Murzynowski and C. Dagdeviren, Recent progress in electrochemical pH-sensing materials and configurations for biomedical applications, Chem. Rev., 2019, 119, 5248–5297 CrossRef PubMed.
- S. R. Mane, A. Sathyan and R. Shunmugam, Biomedical applications of pH-responsive amphiphilic polymer nanoassemblies, ACS Appl. Nano Mater., 2020, 3, 2104–2117 CrossRef.
- D. G. Smith, I. L. Topolnicki, V. E. Zwicker, K. A. Jolliffe and E. J. New, Fluorescent sensing arrays for cations and anions, Analyst, 2017, 142, 3549–3563 RSC.
- N. K. Tan, J. Zhu and E. J. New, Fluorescent Sensors of the Cellular Environment, Mol. Fluoresc. Sens. Cell. Stud., 2022, 173–200 CAS.
- N. K. Tan and J. Zhu, in Molecular Fluorescent Sensors for Cellular Studies, ed. E. J. New, 2022, pp. 173–200 Search PubMed.
- L. Manjakkal, S. Dervin and R. Dahiya, Flexible potentiometric pH sensors for wearable systems, RSC Adv., 2020, 10, 8594–8617 RSC.
- J. Kim, A. S. Campbell, B. E.-F. de Ávila and J. Wang, Wearable biosensors for healthcare monitoring, Nat. Biotechnol., 2019, 37, 389–406 CrossRef CAS PubMed.
- J. H. Yoon, S. M. Kim, H. J. Park, Y. K. Kim, D. X. Oh, H. W. Cho, K. G. Lee, S. Y. Hwang, J. Park and B. G. Choi, Highly self-healable and flexible cable-type pH sensors for real-time monitoring of human fluids, Biosens. Bioelectron., 2020, 150, 111946 CrossRef CAS.
- S.-H. Kuo, C.-J. Shen, C.-F. Shen and C.-M. Cheng, Role of pH value in clinically relevant diagnosis, Diagnostics, 2020, 10, 107 CrossRef PubMed.
- F. Mariani, I. Gualandi, W. Schuhmann and E. Scavetta, Micro-and nano-devices for electrochemical sensing, Microchim. Acta, 2022, 189, 459 CrossRef PubMed.
- Y.-W. Chen, H.-Q. Liu, Q.-X. Wu, Y.-H. Huang, Y.-Y. Tung, M.-H. Lin, C.-H. Lin, T.-C. Chen, E. C. Lin and D. W. Hwang, pH mapping of skeletal muscle by chemical exchange saturation transfer (CEST) imaging, Cell, 2020, 9, 2610 CrossRef PubMed.
- J. Han and K. Burgess, Fluorescent indicators for intracellular pH, Chem. Rev., 2010, 110, 2709–2728 CrossRef PubMed.
- G. Miesenböck, D. A. De Angelis and J. E. Rothman, Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins, Nature, 1998, 394, 192–195 CrossRef.
- Y. Liu, D. Zhang, Y. Qu, F. Tang, H. Wang, A. Ding and L. Li, Advances in Small-Molecule Fluorescent pH Probes for Monitoring Mitophagy, Chem. Biomed. Imaging, 2023, 2, 81–97 CrossRef PubMed.
- M. Shamsipur, A. Barati and Z. Nematifar, Fluorescent pH nanosensors: Design strategies and applications, J. Photochem. Photobiol., 2019, 39, 76–141 CrossRef.
- H. Xu, W. Wu, Y. Chen, T. Qiu and L.-J. Fan, Construction of response patterns for metal cations by using a fluorescent conjugated polymer sensor array from parallel combinatorial synthesis, ACS Appl. Mater. Interfaces, 2014, 6, 5041–5049 CrossRef PubMed.
- Y. Wu, Y. Tan, J. Wu, S. Chen, Y. Z. Chen, X. Zhou, Y. Jiang and C. Tan, Fluorescence array-based sensing of metal ions using conjugated polyelectrolytes, ACS Appl. Mater. Interfaces, 2015, 7, 6882–6888 CrossRef.
- L. Mitchell, E. J. New and C. S. Mahon, Macromolecular optical sensor arrays, ACS Appl. Polym. Mater., 2021, 3, 506–530 CrossRef.
- H. Kim, S. Lee, J. S. Min, E. Kim, J. Choi, J. Ko and E. Kim, Fluorescent sensor array for high-precision pH classification with machine learning-supported mobile devices, Dyes Pigm., 2021, 193, 109492 CrossRef.
- P. Lesani, G. Singh, Z. Lu, M. Mirkhalaf, E. J. New and H. Zreiqat, Two-photon ratiometric carbon dot-based probe for real-time intracellular pH monitoring in 3D environment, Chem. Eng. J., 2022, 433, 133668 CrossRef.
- C. Wang, Z. Xu, H. Cheng, H. Lin, M. G. Humphrey and C. Zhang, A hydrothermal route to water-stable luminescent carbon dots as nanosensors for pH and temperature, Carbon, 2015, 82, 87–95 CrossRef.
- M. Zhang, R. Su, J. Zhong, L. Fei, W. Cai, Q. Guan, W. Li, N. Li, Y. Chen and L. Cai, Red/orange dual-emissive carbon dots for pH sensing and cell imaging, Nano Res., 2019, 12, 815–821 CrossRef.
- N. K. Tan, H. Chan, Z. Lu, H. Zreiqat, G. Lakhwani, P. Lesani and E. J. New, Ultrasensitive Dual Fluorophore-Conjugated Carbon Dots for Intracellular pH Sensing in 3D Tumor Models, ACS Appl. Mater. Interfaces, 2024, 16, 47303–47313 CrossRef PubMed.
- K. McCarthy, B. Braun and I. Hunter, Importance of pH monitoring in the care of long-term catheters, Br. J. Community Nurs., 2001, 10, 1240–1247 CrossRef.
- F. Yan, Y. Zou, M. Wang, X. Mu, N. Yang and L. Chen, Highly photoluminescent carbon dots-based fluorescent chemosensors for sensitive and selective detection of mercury ions and application of imaging in living cells, Sens. Actuators, B, 2014, 192, 488–495 CrossRef.
- P. Lesani, A. H. Mohamad Hadi, M. Khetarpaul, Z. Lu, E. J. New and H. Zreiqat, The Facile Synthesis of Efficient Red-Emissive Two-Photon Carbon Dots for Real-Time Cellular Imaging and High-Resolution Deep-Tissue Imaging, Adv. Photonics Res., 2024, 5, 2300245 CrossRef.
- P. Lesani, A. H. Mohamad Hadi, Z. Lu, S. Palomba, E. J. New and H. Zreiqat, Design principles and biological applications of red-emissive two-photon carbon dots, Commun. Mater., 2021, 2, 108 CrossRef.
- K. J. Mintz, Y. Zhou and R. M. Leblanc, Recent development of carbon quantum dots regarding their optical properties, photoluminescence mechanism, and core structure, Nanoscale, 2019, 11, 4634–4652 RSC.
- P. Lesani, G. Singh, C. M. Viray, Y. Ramaswamy, D. M. Zhu, P. Kingshott, Z. Lu and H. Zreiqat, Two-photon dual-emissive carbon dot-based probe: deep-tissue imaging and ultrasensitive sensing of intracellular ferric ions, ACS Appl. Mater. Interfaces, 2020, 12, 18395–18406 CrossRef.
- P. Lesani, S. M. Ardekani, A. Dehghani, M. Hassan and V. G. Gomes, Excitation-independent carbon dot probes for exogenous and endogenous Fe3+ sensing in living cells: Fluorescence lifetime and sensing mechanism, Sens. Actuators, B, 2019, 285, 145–155 CrossRef.
- P. Yang, Z. Zhu, T. Zhang, W. Zhang, W. Chen, Y. Cao, M. Chen and X. Zhou, Orange-emissive carbon quantum dots: toward application in wound pH monitoring based on colorimetric and fluorescent changing, Small, 2019, 15, 1902823 CrossRef.
- F. Zu, F. Yan, Z. Bai, J. Xu, Y. Wang, Y. Huang and X. Zhou, The quenching of the fluorescence of carbon dots: A review on mechanisms and applications, Microchim. Acta, 2017, 184, 1899–1914 CrossRef.
- M. Alafeef, I. Srivastava, T. Aditya and D. Pan, Carbon Dots: From Synthesis to Unraveling the Fluorescence Mechanism, Small, 2024, 20, e2303937 CrossRef PubMed.
- M. L. Liu, B. B. Chen, C. M. Li and C. Z. Huang, Carbon dots: synthesis, formation mechanism, fluorescence origin and sensing applications, Green Chem., 2019, 21, 449–471 RSC.
- P. Lesani, Z. Lu, G. Singh, M. Mursi, M. Mirkhalaf, E. J. New and H. Zreiqat, Influence of carbon dot synthetic parameters on photophysical and biological properties, Nanoscale, 2021, 13, 11138–11149 RSC.
- F. Yan, Y. Jiang, X. Sun, J. Wei, L. Chen and Y. Zhang, Multicolor carbon dots with concentration-tunable fluorescence and solvent-affected aggregation states for white light-emitting diodes, Nano Res., 2020, 13, 52–60 CrossRef.
- T. Gao, S. Guo, J. Zhang, J. Chen, S. Yin, N. Peng, Q. Cai, H. Xu and Y. Liu, Red, yellow, green, and blue light-emitting highly crystallized graphene quantum dots derived from lignin: controllable syntheses and light-emitting diode applications, Green Chem., 2023, 25, 8869–8884 RSC.
- D. López-Díaz, A. Solana, J. L. García-Fierro, M. D. Merchán and M. M. Velázquez, The role of the chemical composition on the photoluminescence properties of N-doped carbon nanoparticles, J. Lumin., 2020, 219, 116954 CrossRef.
- T. A. Duarte, R. F. Pereira, B. Medronho, E. S. Maltseva, E. F. Krivoshapkina, A. Varela-Dopico, P. Taboada, L. Fu, R. A. Ferreira and V. N. de Zea Bermudez, A Glance at Novel Ionanofluids Incorporating Silk-Derived Carbon Dots, Chem. Mater., 2024, 36, 1136–1152 CrossRef.
- E. V. Kundelev, N. V. Tepliakov, M. Y. Leonov, V. G. Maslov, A. V. Baranov, A. V. Fedorov, I. D. Rukhlenko and A. L. Rogach, Amino functionalization of carbon dots leads to red emission enhancement, J. Phys. Chem. Lett., 2019, 10, 5111–5116 CrossRef PubMed.
- C. Wang, Y. Fang, M. Zhang, H. Zhuo, Q. Song and H. Zhu, Surface-state controlled synthesis of hydrophobic and hydrophilic carbon dots, Nano Res., 2024, 17, 4391–4399 CrossRef.
- X. Zhang, Y. Liu, C.-H. Kuan, L. Tang, T. D. Krueger, S. Yeasmin, A. Ullah, C. Fang and L.-J. Cheng, Highly fluorescent nitrogen-doped carbon dots with large Stokes shifts, J. Mater. Chem. C, 2023, 11, 11476–11485 RSC.
- F. Yan, Z. Bai, F. Zu, Y. Zhang, X. Sun, T. Ma and L. Chen, Yellow-emissive carbon dots with a large Stokes shift are viable fluorescent probes for detection and cellular imaging of silver ions and glutathione, Microchim. Acta, 2019, 186, 1–11 CrossRef.
- Y. Ma, A. Chen, X. Xie, X. Wang, D. Wang, P. Wang, H. Li, J. Yang and Y. Li, Doping effect and fluorescence quenching mechanism of N-doped graphene quantum dots in the detection of dopamine, Talanta, 2019, 196, 563–571 CrossRef PubMed.
- F. Zu, F. Yan, Z. Bai, J. Xu, Y. Wang, Y. Huang and X. Zhou, The quenching of the fluorescence of carbon dots: a review on mechanisms and applications, Microchim. Acta, 2017, 184, 1899–1914 CrossRef.
- B. B. Chen, M. L. Liu, C. M. Li and C. Z. Huang, Fluorescent carbon dots functionalization, Adv. Colloid Interface Sci., 2019, 270, 165–190 CrossRef PubMed.
- D. G. Smith, N. Sajid, S. Rehn, R. Chandramohan, I. J. Carney, M. A. Khan and E. J. New, A library-screening approach for developing a fluorescence sensing array for the detection of metal ions, Analyst, 2016, 141, 4608–4613 RSC.
- N. Trinh, K. R. Bhuskute, N. R. Varghese, J. A. Buchanan, Y. Xu, F. M. McCutcheon, R. L. Medcalf, K. A. Jolliffe, M. Sunde and E. J. New, A Coumarin-Based Array for the Discrimination of Amyloids, ACS Sens., 2024, 9, 615–621 CrossRef PubMed.
- T. Zhao, C. Song, S. Lu and L. Xu, Prediction of Uniaxial Compressive Strength Using Fully Bayesian Gaussian Process Regression (fB-GPR) with Model Class Selection, Rock Mech. Rock Eng., 2022, 55, 6301–6319 CrossRef.
- H. Harkat, A. E. Ruano, M. G. Ruano and S. D. Bennani, GPR target detection using a neural network classifier designed by a multi-objective genetic algorithm, Appl. Soft Comput., 2019, 79, 310–325 CrossRef.
- D. Sachin, Dimensionality reduction and classification through PCA and LDA, Int. J. Comput. Appl., 2015, 122, 4–8 Search PubMed.
- E. K. Tang, P. N. Suganthan, X. Yao and A. K. Qin, Linear dimensionality reduction using relevance weighted LDA, Pattern Recognit., 2005, 38, 485–493 CrossRef.
- G. T. Reddy, M. P. K. Reddy, K. Lakshmanna, R. Kaluri, D. S. Rajput, G. Srivastava and T. Baker, Analysis of dimensionality reduction techniques on big data, IEEE Access, 2020, 8, 54776–54788 Search PubMed.
- I. Rodriguez-Lujan, G. Bailador, C. Sanchez-Avila, A. Herrero and G. Vidal-de-Miguel, Analysis of pattern recognition and dimensionality reduction techniques for odor biometrics, Knowl.-Based Syst., 2013, 52, 279–289 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00275j |
| This journal is © The Royal Society of Chemistry 2024 |