Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ring-shaped nanoparticle assembly and cross-linking on lipid vesicle scaffolds†
Gizem
Karabiyik
 a,
Aldo
Jesorka
b and
Irep
Gözen
a,
Aldo
Jesorka
b and
Irep
Gözen
 *a
*a
aCentre for Molecular Medicine Norway, Faculty of Medicine, University of Oslo, 0318 Oslo, Norway. E-mail: irep@uio.no
bDepartment of Chemistry and Chemical Engineering, Chalmers University of Technology, Göteborg, SE-412 96, Sweden
First published on 23rd October 2024
Abstract
We show the assembly of carboxylate-modified polystyrene nanoparticles into flexible circular, ring-shaped structures with micrometer sized diameters around the base of surface-adhered lipid vesicles. The rings remain around the vesicles but disintegrate when the lipid membranes are dissolved in detergent. The aqueous medium allows carbodiimide-based cross-linking chemistry to be applied to the particle assemblies resulting in the preservation of the rings even after the lipid compartments are dissolved.
Research on identifying and characterizing nanoparticle-lipid membrane interactions has sparked considerable interest in recent years.1–5 Lipids are ubiquitous in all domains of life. They can spontaneously self-assemble to lipid bilayers to form planar sheets, spherical vesicles, nanotubes, or other morphologies depending on the components and exact physicochemical conditions.6 Well-established, straightforward protocols to generate synthetic lipid bilayers in the laboratory7–10 have made lipid membranes suitable interfaces for the assembly of various nanostructures. Special attention is currently on design and fabrication of hybrid soft nanodevices4,11,12 for applications in nanomedicine,13 biocomputing14 or environmental sensing and monitoring.15 Combinations of the dynamic properties of molecular lipid films and the functional diversity of nanoparticles enable unique possibilities not attainable with top-down produced solid state devices.16–20 Besides the potential applications, nanoparticle-lipid membrane interfaces serve as model systems for studies focusing on the interaction of the membrane with proteins,21 virus particles22 or drug complexes.23
In order to create biocompatible devices, bottom-up fabrication strategies of hybrid biomembrane nanostructures typically exploit the 2D-fluidic properties of lipid films, most notably bending, autonomous wetting and adhesion, to coat particles and nanoscale surface patterns, and adapt to biological scaffolds, among them protein crystals,24 cellulose fibers25 or DNA.26 However, the curvature of common membrane structures such as liposomes and lipid nanoconduits can also effectively serve as scaffold for nanoparticle association, giving rise to new facile fabrication routes.
Here we report a unique model system consisting of surface-bound giant lipid vesicles and carboxylated polystyrene nanoparticles in an aqueous medium. The nanoparticles preferentially assemble around the base of the vesicle along the contact line between the vesicle and the underlying surface. In untreated samples, the particles are held together only weakly and disintegrate as soon as the system is exposed to a detergent. When the nanoparticles are cross-linked via aqueous carbodiimide conjugation chemistry with a diamine cross-linker, the ring structures are maintained even after the lipid vesicle scaffolds are dissolved in detergent. The remaining structures are a population of structurally flexible nanoparticle loops on a surface.
Results and discussion
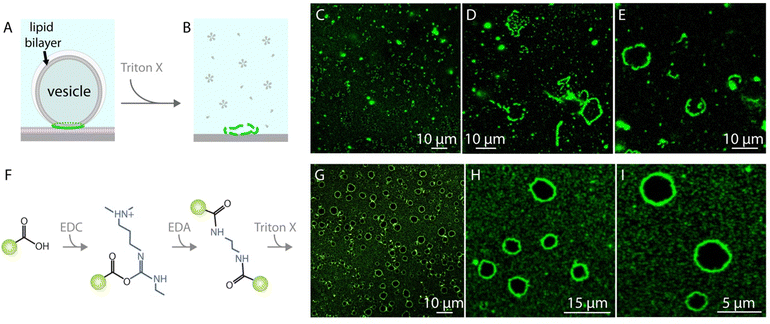
We generated unilamellar lipid vesicle networks on solid surfaces as characterized in detail in our previous work27–31 (cf. ESI† for materials and methods). The vesicle networks autonomously form as a result of a series of events after multilamellar lipid reservoirs (MLVs) come in contact with solid supports submerged in aqueous solutions. MLVs initially spread on solid surfaces as double lipid bilayers, followed by the transformation of the distal bilayer to lipid nanotube networks. The nanotubes then partially swell into unilamellar vesicular compartments. Depending on the availability of the excess lipid reservoir in the system, the lipid nanotubes can be completely consumed during swelling,27 or remain connected to the surface-adhered vesicles (arrows in Fig. 1E and in ESI,† Fig. S3A).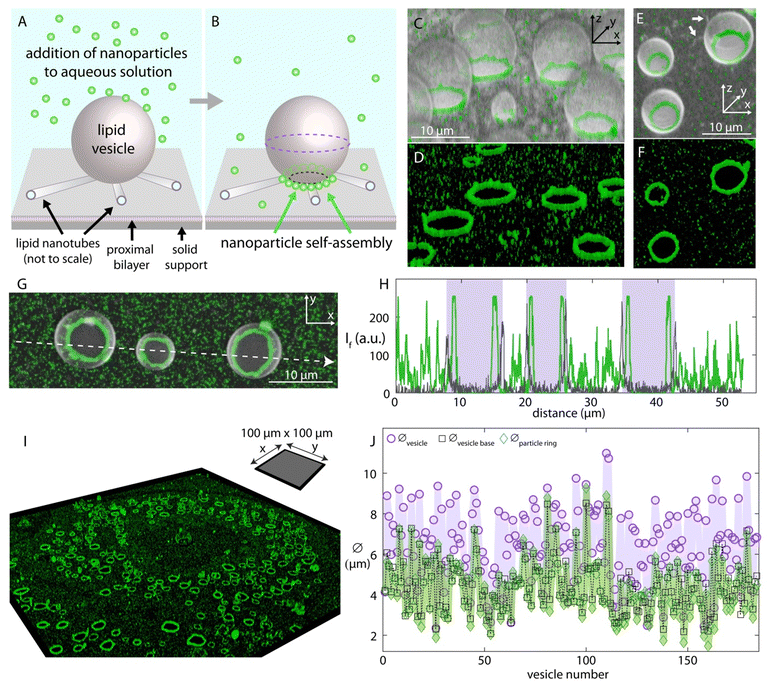 | ||
| Fig. 1 Nanoparticle self-assembly on lipid vesicle networks. Schematic drawing showing (A) the addition- and (B) the self-assembly of the nanoparticles on a surface-adhered unilamellar vesicle. The proximal bilayer and nanotube ends are shown open-ended to reveal the detailed membrane structure where in reality they are continuous, intact bilayer structures. The nanoparticles (Ø:100 nm) are added to the ambient solution of the vesicles via an automatic pipette, and over time (∼hours), assemble around the base of the surface-adhered vesicle. (C)–(F) 3D confocal micrographs showing the ring-like nanoparticle assemblies around the lipid vesicles. (C) and (E) show the fluorescence emission of both the lipid membrane and the particles overlaid, where (D) and (F) show the emission only for the particles in (C) and (E), respectively. (G) Confocal micrograph showing three vesicles with nanoparticle assemblies from the top view. (H) shows the fluorescence intensity in arbitrary units along the white arrow in (G). The plot in gray color shows the fluorescence intensity of the lipid membrane and the plot in green color, the nanoparticles. The three rectangular regions colored in purple mark the interior region of the vesicles between the two bilayer borders of each vesicle across the white arrow, represented by the two distinct spikes. (I) Confocal micrograph of a sample region (100 μm × 100 μm) showing the ring-like particle assemblies. The panel shows the fluorescence emission of only the nanoparticle channel; the full set of micrographs is presented in the ESI.† (J) Size analyses corresponding to (I) and the extended figure in the ESI† (Fig. S1). The graph shows the diameter of a total of 184 vesicles, and of the corresponding vesicle bases and nanoparticle assemblies. The diameter of the particle rings is shown with green diamond-shaped data markers, the diameter across the equator of the vesicles in the network (indicated by the purple dashed lines in panel (B)) is shown with circle data markers, and the diameter of the base of the vesicles in the network is shown with black square data markers (indicated by the black dashed lines in panel (B)). The lines connecting the data points in each data set are to facilitate the comparison between the data sets. | ||
Once the lipid compartments formed and matured, we added fluorescently labeled carboxylated polystyrene nanoparticles (Ø:100 nm) to the aqueous environment containing the vesicles, using an automatic pipette (Fig. 1A). We observed that, over several hours, the nanoparticles assembled around the base of the lipid compartments where the fluorescence intensity analyses confirm the structure and location of the particle arrangements (Fig. 1B–H). The surface attachment provides persistent regions of high curvature that attract particles. The surface-adhered vesicles mature slowly and can remain intact as long as the aqueous solution is continuously supplied to the open top sample chamber compensating for evaporation. This time duration allows, in addition to the assembly and saturation of nanoparticles around the vesicle necks, the application of step-wise cross-linking chemistry to the particles in order to fix this arrangement.
Since the vesicles are spherical, the diameter of the cross section of their base at the surface contact is smaller than the diameter at the equator of the vesicle. The particle assemblies around the base of the vesicles, corresponding to the spikes in the green-colored plot inside the purple-colored regions, are therefore located inside the bilayer borders of each vesicle. Note that the interior volume of the lipid vesicles shows no signal of nanoparticles, indicating that the lipid membrane is not permeable to the nanoparticles and the particles assemble at the outer membrane leaflet.
The ring formation is consistently observed over the entire surface of the substrate (Fig. 1I, cf. ESI,† Fig. S1 for all micrographs corresponding to this section including the vesicles). The intensity analyses confirms that the rings form not on the equator of the vesicle but at the neck region at the interface to the substrate (Fig. 1J and ESI,† Fig. S1B and C). Similar results were obtained with nanoparticles of 20 nm diameter (ESI,† Fig. S2).
There may be charge interactions between the carboxyl groups on the particles and the membrane. This is facilitated by the Ca2+ ions in the solution, which typically act as a bridging agent32,33 (cf. ESI† for materials and methods). However, we do not think that this attraction is the dominating factor for the assembly, as in that case the nanoparticles would not only concentrate at the neck region. The observation that the spherical nanoparticles preferentially assemble at the negatively curved interface of the vesicle base indicates that the assembly of nanoparticles is dominantly curvature-driven.34 The surface free energy of the vesicle-nanoparticle system is lowered if the membrane curvature has the same sign as the deformation induced by the particle.34 The circumference of the base of the vesicular membrane in our experimental system intrinsically has a negative curvature where the spherical nanoparticles we utilized would impose a concave deformation on the lipid membrane. The localization of the nanoparticles on the lipid vesicles in our system is therefore energetically favorable. We note that particle self-assembly on lipid vesicles may also be due to membrane-mediated interaction of partially wrapped particles. Theoretical and experimental work reports multiple pathways of that phenomenon.35–38
The nanoparticles assemble into rings also vertically with respect to the surface plane, along the contact line of two or more adjacent vesicles, (ESI,† Fig. S4). The nanoparticle localization at the bilayer–bilayer boundary of two vesicles is also curvature-driven, similar to the ring-like alignment of nanoparticles on the neck of dumbbell-shaped vesicles previously reported by Koltover et al.34
When the vesicles with the nanoparticle assemblies are exposed to a detergent solution, i.e., Triton X, the lipids dissolve and the ring-like nanoparticle structures disintegrate (Fig. 2A–E). In order to prevent the disintegration of the nanoparticle assemblies and maintain the ring-like structures as a whole we cross-linked the nanoparticles with carbodiimide chemistry39–41 (Fig. 2F, cf. ESI† for details of materials and methods). Once the nanoparticles assembled around lipid vesicles, we first added an N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) solution to the ambient buffer. EDC reacts with the carboxyl groups on the nanoparticles and forms an amine reactive intermediate. One hour following the EDC addition, we added ethylenediamine (EDA) to the ambient solution which reacted with the activated esters on the particles and cross-linked them. The cross-linked nanoparticles remain intact even after exposure to Triton X (Fig. 2G–I and ESI,† Fig. S3).
Conclusions
We showed a unique synthetic model system where spherical nanoparticles preferentially localize along the negative curvature of the contact line of lipid vesicles on a planar bilayer surface. In biology, there are multiple examples of proteins that are sensing- and assembling on- the negatively curved cellular membranes.42,43 Our experimental setup can support the detailed understanding of organization, assembly and activity of proteins at negative membrane curvatures between a vesicular and a planar membrane. If extremely dense vesicle suspensions such as vesicle gels44 would be used in a bulk environment, the inter-vesicular interfaces could dramatically increase the yield of ring production.Furthermore, we retrieved the nanoparticle assemblies by chemically cross-linking the particles followed by the removal of lipids by a detergent. Our technique can be further developed using different lipid membrane compositions or nanoparticle types, and employed as a practical method to generate micro-ring structures without the need of microfabrication facilities and training, for example for surface-enhanced Raman spectroscopy.45
The phenomenon we report also has implications for the origin of life. The lipid compartments we use are commonly employed as primitive cell models among coacervates and emulsion droplets.46 How exactly metabolic and genetic material interacted with the boundary of a protocell compartment in the prebiotic world before they were encapsulated is still a pending question. Previous studies discussed that such compartment boundaries could have served as interfaces for the molecules to adsorb and react47 while stabilizing the compartment.48 The results we show provide an additional argument of membrane curvature, which would facilitate the adsorption and concentration of biomolecules that are of comparable size to the curvature on surface-adhered prebiotic compartments.29
Data availability
The data supporting this article have been included in the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. J. acknowledges funding by the European Union Marie Sklodowska-Curie Actions program, grant ID 812868, as well as the European Union Excellent Science—Future and Emerging Technologies (FET) program, grant ID 899205. I. G. acknowledges funding by the Research Council of Norway, project number: 324630. We acknowledge Dr Elif Koksal for technical assistance at the initial stage of the project.References
- C. S. Martin, M. D. Maximino, J. F. V. A. Martins, W. M. Pazin and C. J. L. Constantino, J. Mol. Liq., 2024, 406, 125081 CrossRef CAS.
- F. Cecchet, Colloids Surf., B, 2024, 241, 114013 CrossRef CAS PubMed.
- A. Sharma, Y. Zhu, E. J. Spangler, T. B. Hoang and M. Laradji, ACS Nano, 2024, 18, 12957–12969 CrossRef CAS.
- M. Mendozza, L. Caselli, A. Salvatore, C. Montis and D. Berti, Soft Matter, 2019, 15, 8951–8970 RSC.
- M. Schulz, A. Olubummo and W. H. Binder, Soft Matter, 2012, 8, 4849–4864 RSC.
- T. Kunitake, Angew. Chem., Int. Ed. Engl., 1992, 31, 709–726 CrossRef.
- W. Yuan, J. Piao and Y. Dong, Mater. Chem. Front., 2021, 5, 5233–5246 RSC.
- J. Kurniawan, J. F. Ventrici de Souza, A. T. Dang, G.-Y. Liu and T. L. Kuhl, Langmuir, 2018, 34, 15622–15639 CrossRef CAS PubMed.
- K. Yasuhara and K. Morigaki, Biophys. Physicobiol., 2020, 17, 125–129 CrossRef CAS PubMed.
- C. G. Siontorou, G. P. Nikoleli, D. P. Nikolelis and S. K. Karapetis, Membranes, 2017, 7, 38 CrossRef PubMed.
- Y. Li, Y. Gan, C. Li, Y. Y. Yang, P. Yuan and X. Ding, J. Mater. Chem. B, 2020, 8, 5578–5596 RSC.
- S. Kim, J. Seo, H. H. Park, N. Kim, J.-W. Oh and J.-M. Nam, Acc. Chem. Res., 2019, 52, 2793–2805 CrossRef CAS.
- A. Noureddine, A. Maestas-Olguin, L. Tang, J. I. Corman-Hijar, M. Olewine, J. A. Krawchuck, J. Tsala Ebode, C. Edeh, C. Dang, O. A. Negrete, J. Watt, T. Howard, E. N. Coker, J. Guo and C. J. Brinker, ACS Nano, 2023, 17, 16308–16325 CrossRef CAS.
- J. Seo, S. Kim, H. H. Park, D. Y. Choi and J.-M. Nam, Sci. Adv., 2019, 5, eaau2124 CrossRef CAS PubMed.
- G. P. Nikoleli, D. Nikolelis, C. G. Siontorou and S. Karapetis, Sensors, 2018, 18, 284 CrossRef PubMed.
- C. H. Lee, A. J. Crosby, R. C. Hayward and T. Emrick, ACS Appl. Mater. Interfaces, 2014, 6, 4850–4855 CrossRef CAS PubMed.
- F. Nan, F. Han, N. F. Scherer and Z. Yan, Adv. Mater., 2018, 30, 1803238 CrossRef PubMed.
- G. Lin, X. Zhu, U. Anand, Q. Liu, J. Lu, Z. Aabdin, H. Su and U. Mirsaidov, Nano Lett., 2016, 16, 1092–1096 CrossRef CAS PubMed.
- Y. Bao, T. A. Witten and N. F. Scherer, ACS Nano, 2016, 10, 8947–8955 CrossRef CAS.
- Y.-X. Chang, C.-F. Wang, C.-J. Chang, C.-H. Lu and J.-K. Chen, Sens. Actuators, B, 2023, 375, 132875 CrossRef CAS.
- Y. Jiang, B. Thienpont, V. Sapuru, R. K. Hite, J. S. Dittman, J. N. Sturgis and S. Scheuring, Nat. Commun., 2022, 13, 7373 CrossRef CAS PubMed.
- Y. Zhang, S. Anbir, J. McTiernan, S. Li, M. Worcester, P. Mishra, M. E. Colvin, A. Gopinathan, U. Mohideen, R. Zandi and T. E. Kuhlman, Sci. Adv., 2024, 10, eadm7030 CrossRef CAS PubMed.
- C. Peetla, A. Stine and V. Labhasetwar, Mol. Pharmaceutics, 2009, 6, 1264–1276 CrossRef CAS PubMed.
- M. Künzle, M. Lach and T. Beck, Dalton Trans., 2018, 47, 10382–10387 RSC.
- H. Tian and J. He, Langmuir, 2016, 32, 12269–12282 CrossRef CAS PubMed.
- Y. Tian, T. Wang, W. Liu, H. L. Xin, H. Li, Y. Ke, W. M. Shih and O. Gang, Nat. Nanotechnol., 2015, 10, 637–644 CrossRef CAS PubMed.
- E. S. Köksal, S. Liese, I. Kantarci, R. Olsson, A. Carlson and I. Gözen, ACS Nano, 2019, 13, 6867–6878 CrossRef.
- E. S. Köksal, S. Liese, L. Xue, R. Ryskulov, L. Viitala, A. Carlson and I. Gözen, Small, 2020, 16, 2002529 CrossRef.
- E. S. Köksal, I. Põldsalu, H. Friis, S. J. Mojzsis, M. Bizzarro and I. Gözen, ChemSystemsChem, 2022, 4, e202100040 CrossRef.
- I. Põldsalu, E. S. Köksal and I. Gözen, Phys. Chem. Chem. Phys., 2021, 23, 26948–26954 RSC.
- I. J. Schanke, L. Xue, K. Spustova and I. Gözen, Nanoscale, 2022, 14, 10418–10427 RSC.
- K. Akashi, H. Miyata, H. Itoh and K. Kinosita, Jr., Biophys. J., 1996, 71, 3242–3250 CrossRef CAS PubMed.
- I. Gözen, P. Dommersnes, I. Czolkos, A. Jesorka, T. Lobovkina and O. Orwar, Nat. Mater., 2010, 9, 908–912 CrossRef.
- I. Koltover, J. O. Rädler and C. R. Safinya, Phys. Rev. Lett., 1999, 82, 1991–1994 CrossRef CAS.
- A. Šarić and A. Cacciuto, Phys. Rev. Lett., 2012, 108, 118101 CrossRef.
- A. Azadbakht, B. Meadowcroft, T. Varkevisser, A. Šarić and D. J. Kraft, Nano Lett., 2023, 23, 4267–4273 CrossRef CAS.
- J. Midya, T. Auth and G. Gompper, ACS Nano, 2023, 17, 1935–1945 CrossRef CAS PubMed.
- J. Agudo-Canalejo and R. Lipowsky, Nano Lett., 2015, 15, 7168–7173 CrossRef CAS PubMed.
- S. Y. Yang, D. Lee, R. E. Cohen and M. F. Rubner, Langmuir, 2004, 20, 5978–5981 CrossRef CAS.
- M. Himmelhaus and H. Takei, Phys. Chem. Chem. Phys., 2002, 4, 496–506 RSC.
- C. Gautier, I. López and T. Breton, Mater. Adv., 2021, 2, 2773–2810 RSC.
- D. H. Johnson, O. H. Kou, N. Bouzos and W. F. Zeno, Trends Biochem. Sci., 2024, 49, 401–416 CrossRef CAS PubMed.
- M. Simunovic, G. A. Voth, A. Callan-Jones and P. Bassereau, Trends Cell Biol., 2015, 25, 780–792 CrossRef CAS.
- N. R. Agrawal, M. Omarova, F. Burni, V. T. John and S. R. Raghavan, Langmuir, 2021, 37, 7955–7965 CrossRef CAS PubMed.
- H. Lee, W. Ye, J. Lee, H. Kim and D. Byun, Appl. Sci., 2020, 10, 8018 CrossRef CAS.
- I. Gözen, E. S. Köksal, I. Põldsalu, L. Xue, K. Spustova, E. Pedrueza-Villalmanzo, R. Ryskulov, F. Meng and A. Jesorka, Small, 2022, 18, 2106624 CrossRef PubMed.
- A. Fallah-Araghi, K. Meguellati, J.-C. Baret, A. E. Harrak, T. Mangeat, M. Karplus, S. Ladame, C. M. Marques and A. D. Griffiths, Phys. Rev. Lett., 2014, 112, 028301 CrossRef PubMed.
- C. E. Cornell, R. A. Black, M. Xue, H. E. Litz, A. Ramsay, M. Gordon, A. Mileant, Z. R. Cohen, J. A. Williams, K. K. Lee, G. P. Drobny and S. L. Keller, Proc. Natl. Sci., 2019, 116, 17239–17244 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sm01010h |
| This journal is © The Royal Society of Chemistry 2024 |