Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect of supramolecular peptide hydrogel scaffold charge on HepG2 viability and spheroid formation†
Yu
Xin
 a,
Cosimo
Ligorio‡
a,
Cosimo
Ligorio‡
 b,
Marie
O’brien
a,
Richard
Collins
b,
Marie
O’brien
a,
Richard
Collins
 c,
Siyuan
Dong
c,
Siyuan
Dong
 d,
Aline F.
Miller
d,
Aline F.
Miller
 d,
Alberto
Saiani
d,
Alberto
Saiani
 e and
Julie E.
Gough
e and
Julie E.
Gough
 *a
*a
aDepartment of Materials & Henry Royce Institute, School of Natural Sciences, Faculty of Science and Engineering, The University of Manchester, UK. E-mail: j.gough@manchester.ac.uk; Tel: +44 0161 3068958
bDepartment of Materials & Manchester Institute of Biotechnology, School of Natural Sciences, Faculty of Science and Engineering, The University of Manchester, UK
cElectron Microscopy Core Facility, Faculty of Biology, Medicine and Health, The University of Manchester, UK
dDepartment of Chemical Engineering & Manchester Institute of Biotechnology, School of Engineering, Faculty of Science and Engineering, The University of Manchester, UK
eDivision of Pharmacy and Optometry & Manchester Institute of Biotechnology, School of Health Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, UK
First published on 24th October 2024
Abstract
Supramolecular bioinspired self-assembling peptide hydrogel (SAPH) scaffolds represent a class of fully defined synthetic materials whose chemical and mechanical properties can be finely engineered. In this study, the relationship between SAPHs physicochemical properties and HepG2 cells viability, spheroid formation and function are discussed. We first report that negatively charged SAPHs promote hepatocyte proliferation and spheroids formation in vitro 3D culture while positively charged SAPHs lead to hepatocyte death irrespective of the hydrogel mechanical properties. More specifically HepG2 cultured in 3D in E(FKFE)2 negatively charged SAPH maintained a differentiated phenotype and assembled into well-defined spheroids with strong cell–cell interactions. Furthermore, HepG2 spheroids responded to acetaminophen exposure with upregulation of key CYP450 enzymes expression clearly showing their potential for drug toxicity testing. These findings demonstrate how fine-tuned functional SAPH scaffolds can be used to identify key scaffolds parameters affecting cells. In this case we demonstrated the potential of negatively charged SAPHs for the 3D culture of HepG2 with potential applications in drug screening.
Introduction
Nature has evolved a variety of creative approaches to many aspects of materials synthesis and microstructural control. One such approach is supramolecular self-assembly, which represents a simple and efficient route to the construction of large, complex structures. This phenomenon is a key process in all living organisms, where many of the building blocks exhibit a hierarchy of structures which are critical to their functions.1 As yet, we are nowhere near achieving the same ability to control and manipulate comparable structures with such skill or variety of end products.A variety of bioinspired molecular building blocks have been devised over the years ranging from protein and polysaccharides to DNA that allow to build a range of nanostructures including sheets, fibres, spheres and tubes.2–5 One such building block that has attracted considerable attention is de-novo-designed peptides. Although at first self-assembling peptides were mainly studied for the role they play in diseases such as Alzheimer's or Parkinson's, in the past two decades scientists have investigated the possibility to actually exploit the self-assembly properties of these short natural molecules to design novel biomaterials.6,7 Peptides offer a number of advantages to the material scientist. Peptide synthesis has become a routine procedure making them easily accessible. The library of 20 natural amino acids offers the ability to play with the intrinsic properties of peptides such as structure, hydrophobicity, charge and functionality allowing the design of materials with a wide range of properties. Synthetic peptides are chemically fully defined and easy to scale through standard processes, which is not always the case for natural polymers. Being built from natural amino acids they can be designed with low toxicity and low immunogenicity for use in vivo and can be degraded and metabolised by the body, which is not always the case for synthetic polymers.
One particular application that has attracted significant interest is the use of self-assembling peptides to design fully defined 3D scaffolds for cell culture applications, i.e. cell niches, with the aim to replace poorly defined and non-translatable animal-derived scaffolds such as mouse Engelbreth–Holm–Swarm (EHS) sarcoma-based matrices that are currently been used across the biomedical field.8,9 Although significant progress has been made over the years, fundamental understanding of, and control over, cell–material interactions remain key challenges.
In this context our group has developed over the past decade a technological platform for the design of biocompatible self-assembling peptide hydrogels (SAPHs) scaffolds with tunable physiochemical properties.10–12 There materials have potential for use are across a range of biomedical fields from cell culture13,14 and tissue engineering15,16 to drug17,18 and cell delivery.15,19 This technology exploits the self-assembly properties of a family of short synthetic peptides originally devised by Zhang and co-workers.20 This design is based on the propensity of peptide with alternating hydrophobic and hydrophilic residues to self-assembled into β-sheet-rich fibers that above a critical gelation concentration (CGC) entangle and associate to form water-entrapping 3D fibrillar networks i.e.: hydrogels (Fig. 1(A)).21,22
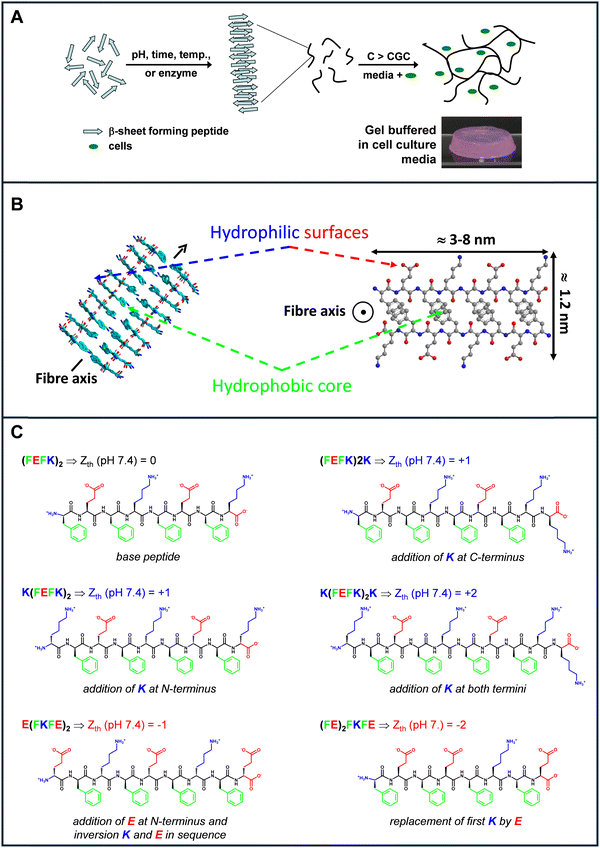 | ||
| Fig. 1 Schematic representation of (A) the self-assembly and gelation process of β-sheet forming self-assembling peptide and (B) the fibres formed (side and top views). The peptide illustrated here is (FEFK)2 (F: phenylalanine, E: glutamic acid, K: lysine); (C) names and chemical structures of the peptides used in the present study (Zth: theoretical charge carried by the peptide at physiological pH 7.4) see Fig. S1 (ESI†); statement describes design changes made in relation to the base peptide (FEFK)2. | ||
The majority of self-assembling peptide hydrogels based on Zhang's design used by us and others in the field will have peptide fibers that carry overall neutral or positive charges. In this current work we were particularly interested in the effect of fiber charge on cell fate and function. We chose the human hepatoma cell lines HepG2 for this study. HepG2 are attractive as they are readily available and exhibit many hepatic functions, such as synthesis and secretion of plasma proteins, cholesterol and triglyceride metabolism, bile and glycogen synthesis.23 It has also been shown that key drug-metabolism gene types in HepG2 cells are similar to primary hepatocytes.24 Thus, HepG2 cells are frequently selected for drug toxicity investigations25 and their reliability has been confirmed for unknown toxins identification.26 Hepatocytes have proven challenging to culture in synthetic hydrogels due to their high sensitivity to scaffolds physicochemical properties27 and currently animal-based matrices remain the scaffold of choice across the industry to culture them, despite the scientific and ethical issues raised by these types of materials.28
We used for this study a series of 6 β-sheet forming self-assembling peptides that carry different charges at physiological pH (Fig. 1(B)) to formulate 8 SAPHs with varying physicochemical properties. (FEFK)2 (F: phenylalanine; K: lysine, E: glutamic acid) was chosen as our base peptide as it has been extensively used in the literature to culture a variety of cells including chondrocytes,13 osteoblasts29 nucleus pulposus16,30 cells and mesenchyme stem cells.31 One particular feature of this peptide design is that when self-assembled into an antiparallel β-sheet, all hydrophobic residue side groups are located on one face of the β-sheet while all the hydrophilic residue side groups are located on the opposite face. As a result, it is thought that two β-sheets come together to bury their hydrophobic faces and form the “elemental” fibers of the network. The fibers formed have a rectangular cross-section with a width ranging from 3 to 10 nm, depending on the length of the peptide used and a thickness of ∼1.5 nm (Fig. 1(B)).
At physiological pH (7.4) all the hydrophilic residues side groups and the uncapped termini of the peptides will be charged. As a result, the overall theoretical charge, Zth, carried by the peptides/fibers is linked to the difference between the number of positive, in our case K (nK), and negatively, in our case E (nE), residues present along the peptide sequence: Zth (pH 7.4) = nK − nE. Therefore, in order to generate peptide fibers with overall positive charge, K residues were added at the termini of the base peptide to generate the following three sequences: (FEFK)2K, K(FEFK)2 and K(FEFK)2K. These peptides will carry at physiological pH overall charges of +1, +1 and +2 respectively. Finally, to generate peptide fibers with an overall negative charge two new peptide sequences were generated. For the first one a glutamic acid residue was added at the N-terminus and the K and E order was inverted in the base sequence to keep the alternation of positive and negative charges along the peptide sequence to generate E(FKFE)2. This peptide will carry an overall charge of −1 at physiological pH. For the final peptide the first lysine in the base sequence was replaced by a glutamic acid to create a peptide with highly clustered negative charges and carrying an overall negative charge of −2 at physiological pH. All peptide chemical structures and charges are shown in Fig. 1(C).
First, live/dead assay was used to investigate HepG2 viability in the 8 SAPHs formulations over 7 days. The SAPH that promoted the highest viability was subsequently used to further culture HepG2 cells in 3D over 14 days and observe spheroids formation. Hepatic function was then investigated by quantitatively determining the production of albumin and urea while histochemical analysis was used to investigate liver extra cellular matrix (ECM) protein production and presence of cell–cell junctions within the spheroids. Finally, drug metabolism and functionality of the spheroids were assessed by exposure to 2 mM of acetaminophen (i.e.: paracetamol) and investigation of CYP450 enzymes’ gene expression levels as key hepatocyte metabolism markers.
Materials and methods
Materials
All the peptides used were purchased as HCl salts with a purity of >95% (TFA content <0.1%) from Biomatik Corporation, Kitchener, Canada. The peptides purities were confirmed by high performance liquid chromatography (HPLC) and mass spectrometry (MS). All other chemicals used to formulate the SAPHs were purchased from Sigma-Adrich or MERK and used as received.SAPHs formulation
Peptide hydrogels were prepared in 5 mL batches by dissolving the required amount of peptide in 3.5 mL of HPLC grade water and mixing vigorously for 60 s. Once fully dissolved gelation was induced by adjusting the pH to the formulation pH (Table 1) through stepwise addition of 5 to 10 μL of a concentrated NaOH (1 M) solution. After each addition the samples were vigorously agitated to ensure homogeneity. Once the formulation pH was reached the remainder of the HPLC grade water was added to reach the targeted sample volume and the sample mixed once more. Finally, the hydrogels were placed under UV light for 15 min to ensure sterility and then stored at 4 °C for at least 24 hours before use.| Sample | (FEFK)acid2 | (FEFK)basic2 | (FEFK)2K | 1/2(FEFK)2 1/2(FEFK)2K | K(FEFK)2K | (FE)2FKFE | K(FEFK)2 | E(FKFE)2 |
|---|---|---|---|---|---|---|---|---|
| a Formulation pH is the samples’ pH as prepared before the addition of cells and media. b Mechanical spectra for all samples are presented in Fig. S2 (ESI). G′ values were taken at 1 Hz and 0.2% strain. c Theoretical net charge at physiological pH were taken from the charge vs. pH curves (Fig. S1, ESI). | ||||||||
| Peptide concentration (mg mL−1) | 20 | 20 | 20 | 20 | 20 | 16 | 20 | 30 |
| Formulation pHa | 4.0 | 8.9 | 5.3 | 4.5 | 5.9 | 6.3 | 6.7 | 6.9 |
| Initial G′ (kPa)b | 4.9 ± 0.1 | 4.2 ± 0.4 | 2.0 ± 0.2 | 5.2 ± 0.4 | 1.0 ± 0. 1 | 0.8 ± 0.1 | 0.10 ± 0.1 | 0.2 ± 0.1 |
| Theoretical net charge at physiological pH (7.4)c | 0 | 0 | 1.0 | 0.5 | 2.0 | −2.0 | 1.0 | −1.0 |
| G′ after media conditioning (kPa)b | 6.4 ± 0.8 | 4.8 ± 0.5 | 12.0 ± 3.2 | 13.1 ± 4.7 | 0.4 ± 0.1 | 1.0 ± 0.2 | 9.1 ± 1.0 | 1.3 ± 0.1 |
HepG2 cell culture
HepG2 cells (European Collection of Authenticated Cell Cultures, ECACC) were passaged in maintenance medium (Eagle's minimum essential medium) (M2279) with 10% (v/v) fetal bovine serum (FBS, F9665), 1% (v/v) non-essential amino acids solution (NEAA, 11140035 and 1% (v/v) L-glutamine). For sample preparation, cell pellets were resuspended in pre-warmed growth medium (maintenance medium with 1% (v/v) penicillin–streptomycin (P0781)). A cell seeding density of 1 × 106 cells mL−1 was used for hepatocyte encapsulation. The required number of cells was resuspended in cell culture media in a volume equal to 20% (10% for E(FKFE)2) of the hydrogel volume. A piston displacement pipette was used to mix cell and gel homogeneously. 100 μL of the hydrogel–cell mixture was then pipetted into 24-well plate ThinCert inserts. After 5 min, pre-warmed cell culture medium was added on top of the hydrogels and in the well. Samples were then placed in an incubator at 37 °C. Cell culture medium was changed twice within the first hour of incubation and every two days subsequently. Only ¾ of the media on top of the SAPHs was changed each time to avoid damaging the hydrogels surfaces and ensure scaffold stability. Collagen I gel (ECM675, Sigma-Aldrich, UK) scaffold was selected as point of comparison. Collagen I gel preparation and cell encapsulation were performed following the manufacturer protocol. The same cell seeding density as above was used.Oscillatory shear rheology
Viscoelastic properties were measured using a Discovery Hybrid 2 rheometer (DHR-2, TA Instruments). For initial hydrogel stiffness evaluation, 180 cL of sample was directly loaded in the center of the fixed base plate of the rheometer at room temperature. For cell culture medium buffered hydrogel, 180 μL of sample was first pipetted into a 12-well ThinCert insert and buffered in media for 24 hours at 37 °C. The sample was then loaded on the fixed bottom base plate of the rheometer by removing the bottom membrane of the inster and pushing the gel out on the the plate. All buffered hydrogel measurements were performed at 37 °C. The frequency sweep measurements were performed from 0.01 to 10 Hz at 0.2% constant strain. A 20 mm diameter plate geometry was used with a gap of 500 μm. The storage modulus G′ shown were recorded at a frequency of 1 Hz. All measurements were repeated three times on three distinct samples to ensure reproducibility.Transmission electron microscopy (TEM)
SAPH hydrogels were diluted in ddH2O to ∼0.5 mg mL−1. 400 mesh carbon-coated copper grids (Electron Microscopy science, UK) were glow discharged at 25 mA for 30 s and then placed on top of a 10 μL diluted peptide solution droplet for 30 s. Excess solution was removed from the grid edge using Whatman 1 filter paper and then the grids were negatively stained by placing them on top of a 10 μL droplet of 2% uranyl acetate solution for 1 min. Excess reagent was then removed using filter paper and the coated grids were left to air-dry before imaging. The TEM data was recorded using a Talos L120C transmission electron microscope with a ceta 2 camera at 120 kV.Cell viability
All samples were first washed once in PBS and then incubated with live/dead (L3224, Invitrogen, UK) working solution for 1 h at 37 °C in 5% CO2 in the dark. Triplicate samples were assayed twice. Samples were collected at days 1, 3 and 7. Images were recorded using a Leica SP5 & SP8 confocal microscope. Live cell signal was obtained at excitation/emission wavelength 494/517 nm and dead cell signal was obtained at excitation/emission wavelength 528/617 nm.Cell proliferation
Cell number was determined using PicoGreen™ dsDNA assay kit (P11496, Invitrogen, UK). Samples were transferred to Eppendorfs and pronase solution (10 mg mL−1) was added. Samples were incubated at 37 °C in a water bath for 5 min with agitation every minute. Samples were lysed using 2× TE buffer with 1% Triton-X and were frozen at −20 °C until later use. For quantification, 100 μL of sample was added to 100 μL of PicoGreen reagent (diluted 200× in 1× TE buffer) in wells of a black 96 well plate. Fluorescence was measured using a wavelength of 480 nm using an Optima FluoStar plate reader. Acellular hydrogel was used as a blank and removed from all measurements. Standard curves, fluorescence vs. HepG2 cell number, were established in each SAPHs by seeding a known number of HepG2 cells in the different peptide hydrogels. Triplicate samples were collected at days 1, 3, 7 and 14 and triplicate readings for every sample were performed.Albumin/urea quantification
Triplicate supernatant aliquots were collected from each sample at each time point and frozen at −80 °C. Albumin production was measured using the Human Albumin SimpleStep ELISA® Kit (ab179887, Abcam) following the manufacturer's instructions. Acellular hydrogel controls were cultured in the same conditions as and cell laden hydrogel samples. Cell culture media was changed 48 h before the day of collection. All data was corrected by subtracting the average absorbance of the control (no cells) samples. For urea production evaluation, a urea assay kit (ab83362, Abcam) was used. The samples were collected at the same time points and using the same process as described above for albumin and urea production estimated according to the manufacturer's instructions.Immunohistochemistry (IHC)
Immunohistochemical staining was used to determine the presence of ECM proteins (laminin, fibronectin and collagen I), albumin and E-cadherin up to 14 days post cell seeding. Detail protocol and list of antibodies used (Table S1) are given in ESI.†Immunofluorescent and immunocytochemical (ICC) staining
Phalloidin, DAPI and E-cadherin ICC staining were used to confirm actin filament structure, locate cell nuclei and determine cell–cell adhesion interactions respectively. Detailed protocols are given in ESI.†Response to acetaminophen exposure
HepG2 cells were cultured in 3D for 7 days to allow spheroid formation. On day 7 acetaminophen (A3035) was freshly dissolved in the MEM complete growth medium at 2 mM concentration. Samples were then exposed to 2 mM acetaminophen from day 7 to 9 (48 h). Untreated controls were used as reference.Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
CYP450's genes expression was assessed by RT-qPCR by measuring mRNA expressions level at days 1, 3, 7 and 14. β-Actin gene expression was used as reference and albumin gene expression was chosen as control. Detailed protocol including information on genes and primers used (Table S2) are given in ESI.†Statistical analysis
Origin and GraphPad software were selected to analyze and summarize data by comparing sample results with controls. Graphs were plotted using the average value and mean standard deviation for raw data. The significance of results (ns: P > 0.5; *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001) was evaluated using one-way or two-way ANOVA methods, depending on data set.Results and discussion
When formulating the SAPHs the initial pH needs to be adjusted depending on the charge carried by the peptide. Indeed, we have shown that for this family of self-assembling peptides if the net charge carried by the peptide falls below ± 1 cloudy gels form and in some cases phase separation and precipitation are observed.12,32 As a result, the formulation pH of each SAPH is unique and related to the peptide theoretical net charge vs. pH profile (Fig. S1, ESI†). The formulations’ pH used in this study for each of the 8 SAPHs are given in Table 1. For (FEFK)2, the hydrogel can actually be formulated at both acidic, (FEFK)acid2, and basic, (FEFK)basic2, pHs where the net charge carried by the peptide is positive and negative respectively.14 In addition in order to formulate a hydrogel with a +0.5 theoretical net charge at physiological pH a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of (FEFK)2 and (FEFK)2K was also used to formulate an additional SAPH. Finally, the peptide concentrations were adjusted to target a broad range of mechanical properties and ensure hydrogel stability over the 7 days of cell culture. All SAPH formulations were stable and transparent following media conditioning at day 1. The net charge, pH and storage shear moduli before and after media conditioning are summarized in Table 1. It should also be noted that most of these formulations have been used in previous studies by our group and have been shown to be suitable for the culture of a variety of cells including monocytes,33 fibroblasts,34 osteoblasts,31 cardiac progenitor cells,15 nucleus pulposus cells14,16,19 and mesenchymal stem cells.29,32
1 ratio of (FEFK)2 and (FEFK)2K was also used to formulate an additional SAPH. Finally, the peptide concentrations were adjusted to target a broad range of mechanical properties and ensure hydrogel stability over the 7 days of cell culture. All SAPH formulations were stable and transparent following media conditioning at day 1. The net charge, pH and storage shear moduli before and after media conditioning are summarized in Table 1. It should also be noted that most of these formulations have been used in previous studies by our group and have been shown to be suitable for the culture of a variety of cells including monocytes,33 fibroblasts,34 osteoblasts,31 cardiac progenitor cells,15 nucleus pulposus cells14,16,19 and mesenchymal stem cells.29,32
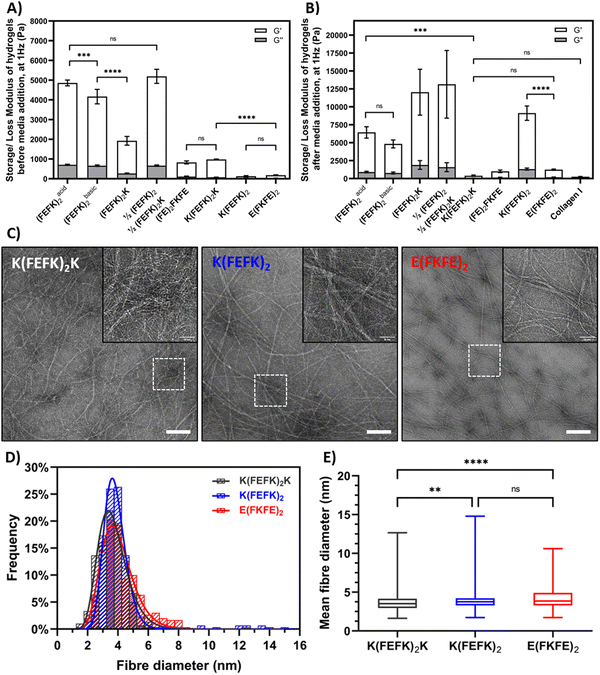
As can be seen from Fig. 2(A) the SAPH formualtions selected had a range of storage shear moduli, G′, before media conditioning ranging from to 0.1 to 5.2 kPa. Following media conditioning the G′ of all the SAPHs were found to increase, to varying degree depending on the fomulation, and resulted in a panel of SAPHs with a broad range of post media G′ ranging from 0.4 to 12 kPa (Fig. 2(B)). The collagen hydrogel used for this study had a G′ of 0.26 ± 0.34 kPa following media conditioning which was similar to the three SAPHs with the lowest G′, (FE)2FKFE, K(FEFK)2K and E(FKFE)2.
As mentioned above most of the SAPH formulations used here have been the subject of previous studies and have already been characterized in detail.10–12,14,15,17,19,29,31 Hence here we focused on characterizing two novel symmetric SAPHs: K(FEFK)2 and E(FKFE)2, which fibers will carry opposite charges at physiological pH,32 in parallel with K(FEFK)2K, which has been characterized in previous studies,12 as point of comparison. The TEM images revealed the presence in all three samples of long semi-flexible fibers that entangle and associate in larger bundles to form crosslinked networks (Fig. 2(C)). Size analysis of the thinnest fibers observed showed that all three samples had similar elemental fibers width distributions (Fig. 2(D)) with means around ∼4 nm (Fig. 2(E)) in agreement with adoption by these peptides of β-sheet conformations and formation of β-sheet rich fibers, and in the case of K(FEFK)2K in agreement with our previous results.12 The fiber morphologies and sizes observed for K(FEFK)2 and E(FKFE)2 were in good agreement to the ones observed in a previous study32 and similar to elemental fibers sizes observed all the other SAPHs used here.10–12,14,15,17,19,29,31 The differences in mechanical properties between the different SAPHs is therefore not linked to the fiber morphology but to the network topologies formed. We have done significant work investigating the relationship between network topologies and hydrogel mechanical properties and have shown that increased mechanical properties in these systems is linked to the formation of large fiber bundles that create strong crosslinks leading to higher G′.10–12 The detailed investigation of the correlation between the SAPHs used in this study and their network topologies is beyond the scope of this work which focusses on their use in the culture of HepG2.
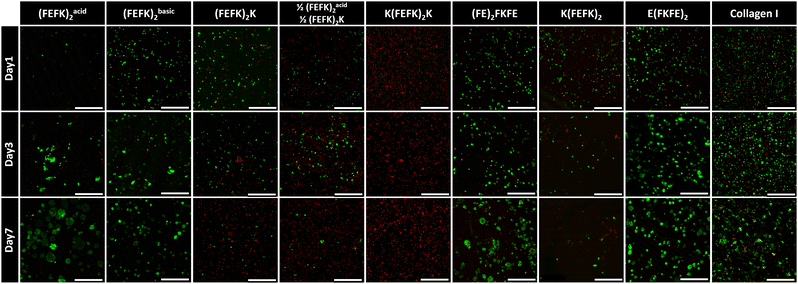
Viability and morphology of HepG2 cells cultured in SAPHs and collagen were observed at days 1, 3, and 7 using live/dead staining (Fig. 3). In collagen hydrogels HepG2 cells were viable and aggregated to form irregularly shaped spheroids, which increased in size over time. When HepG2 were encapsulated in (FEFK)acid2, (FEFK)basic2, E(FKFE)2 and (FE)2FKFE, cells were viable and formed more distinct regular spheroids. Particularly, in E(FKFE)2, HepG2 spheroids showed rounded shape, uniform distribution throughout the hydrogel and similar sizes which increased with time: average spheroid sizes in E(FKFE)2 were ∼163 μm2, ∼640 μm2 and ∼1067 μm2 at day 1, 3 and 7 respectively. When encapsulated in (FEFK)2K, 1/2(FEFK)2: 1/2(FEFK)2K, K(FEFK)2K and K(FEFK)2 cell viability was found to be poor.
There are 3 key parameters that are varied when using this panel of SAPHs: storage modulus, charge carried by the peptide fibers and the encapsulation pH, i.e.: pH before media conditioning. From previous work we know that encapsulation pH does not affect cell viability as long as the plating and conditioning with cell culture media is performed within a few minutes.13,14,29 The viability results obtained here show no obvious correlation with hydrogels’ storage moduli. Indeed, good viability was observed in E(FKFE)2 while poor viability was observed in K(FEFK)2K which both have similar G′. Overall, our results suggest that hepatocytes could maintain good viability within SAPHs with stiffnesses ranging from ∼1 kPa to ∼6.4 kPa. On the other hand, a clear direct correlation between the charge carried by the peptide fibers and HepG2 viability was found. Indeed, good cell viability was observed in negatively charged SAPHs: E(FKFE)2 and (FE)2FKFE, irrespective of their G′ while poor cell viability was observed in positively charged SAPHs: 1/2(FEFK)2: 1/2(FEFK)2K, (FEFK)2K, K(FEFK)2K and K(FEFK)2, once again irrespective of their G′. In addition, cell viability was found to decrease with increasing positive charge, with K(FEFK)2K which peptide carries the highest positive net charge (+2) showing the lowest cell viability (Fig. 3).
This sensitivity of HepG2 to fiber charge is clearly demonstrated by (FEFK)acid2 and (FEFK)basic2. These two SAPHs are formulated using the same peptide (same sequence) but at acidic pH 4, (FEFK)acid2, where fibers carry a positive charge and basic pH 9, (FEFK)basic2, where fibers carry a negative charge respectively. As can be seen good viability was found on day 1 for (FEFK)basic2 while poor viability was observed for (FEFK)acid2. Once conditioned with media the fibers in these two SAPHs carry an overall neutral charge (Fig. S1, ESI†). As can be seen from Fig. 3 good viability is observed at day 3 and 7 with spheroids forming in both systems showing that HepG2 cells recovered after media addition in (FEFK)acid2. These results also show that neutrally charged SAPHs are also suitable for HepG2 culture.
Collagen hydrogels were used as point of comparison as commonly used in 3D cell culture.35 The net charge carried by collagen fibers has been reported by various researchers. It is commonly agreed that the net charge decreases with increasing pH, turning from positive to negative around physiological pH.36 A recent study found that the isoelectric point of collagen is ∼7.5 suggesting that under cell culture condition the overall net charge carried by collagen fibers is neutral.37 As can be seen in Fig. 3, a significant number of dead cells were present at day 1 in collagen compared to (FE)2FKFE and E(FKFE)2. HepG2 cells then recovered and formed loose cell spheroids at later time points. The cell behavior observed in collagen is reminiscent of the cell behavior observed in (FEFK)acid2 suggesting that indeed collagen gel had an isoelectric point between pH 7 and 8. Collagen fibers probably carried a weak positive charge when the cells were encapsulated resulting in low HepG2 viability at day 1. After adding cell culture media, their charge was likely 0 or slightly negative allowing HepG2 cells to recover from the encapsulation stress and proliferate.
The net charge of SAPHs appears to play the most pivotal role in HepG2 3D culture. Previous studies on a range of scaffolds have shown that the charge carried by fibers in 3D scaffolds can affects the behavior of a range cell types but usually these effects are linked to improved cell adhesion and as a result improved viability38,39 Here the effect observed on HepG2 is significantly more marked leading to cell death in the case of negatively charges scaffolds underlying the high sensitive of these cells to scaffolds physicochemical properties beyond mechanical properties.
(FEFK)basic2 and (FE)2FKFE showed low mechanical stability after day 7 with significant hydrogel degradation being observed at day 14. No swelling was observed in any of the hydrogels used over time. Degradation occurred through physical erosion of the hydrogel surface resulting from the media changes leading to the thinning of the hydrogel layer. (FEFK)acid2 and E(FKFE)2 showed good stability and resistance to media changes and therefore, good potential for the culture of HepG2 over longer time periods. PicoGreen assay (Fig. 4(A)) was used to quantify cell number in these two SAPHs. The results indicate that a significantly higher number of cells were present and increased proliferation was observed in E(FKFE)2 compared to (FEFK)acid2. E(FKFE)2 was therefore deemed to be the optimal SAPH scaffold from our panel to culture HepG2 in terms of hydrogel stability, cell viability and proliferation and was selected for further in vitro assessments of the spheroids formed.
Serum albumin is the major plasma protein in the human body and is produced by hepatocytes in the liver.40 There was a consistent increase in albumin production levels in both E(FKFE)2 and collagen gels, with E(FKFE)2 showing significantly higher production levels at days 7 and 14 compared to collagen (Fig. 4(B)). Overall, albumin protein production from 3D cultured HepG2 cells within E(FKFE)2 increased 4.4-folds from day 3 to day 14. Our albumin secretion results after normalizing to per million cells closely matched in order of magnitude results of previous studies.41,42 However, in our case a more sustained and continuous increase in albumin production over 14 days was observed compared to studies that used other scaffolds types in which albumin production levels were found usually to decrease after day 7.43,44 Urea production, another important marker of hepatocyte function, was also evaluated. As shown in Fig. 4(C), the urea concentration in E(FKFE)2 was found to increase with time and was higher than for collagen hydrogels with the difference becoming statistically significantly at days 7 and 14. As for albumin, urea results suggest that E(FKFE)2 lead to higher cellular function compared to collagen.
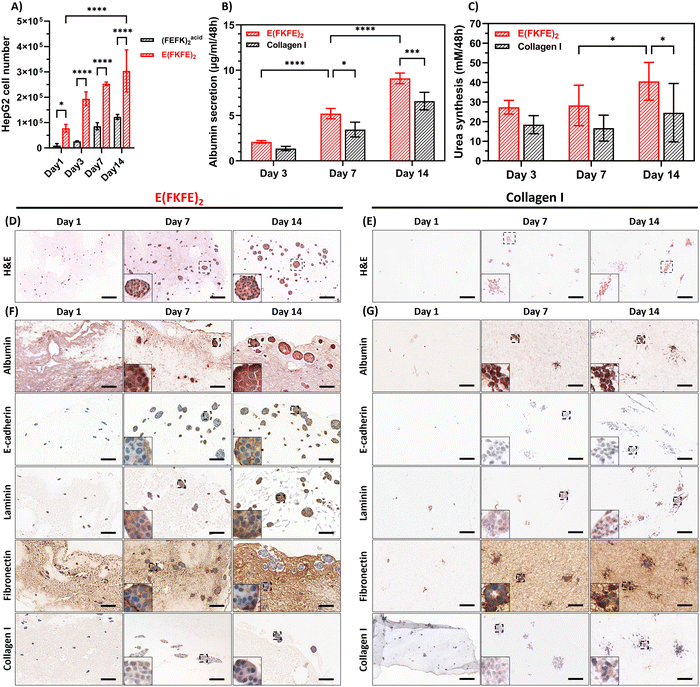
H&E staining (Fig. 4(D)) showed that HepG2 cells arranged themselves into well-defined spheroids in E(FKFE)2 over 14 days culture. For comparison, the H&E staining images of cell-free E(FKFE)2 are presented in Fig. S3A (ESI†). The HepG2 spheroids were regular and compact and had a clear and smooth border. The spheroid morphology observed here agreed well with previous description of HepG2 spheroid morphology,45 and also resembled primary human hepatocytes spheroid morphology.46 H&E staining of collagen hydrogels (Fig. 4(E)) on the other hand showed a different cell organization with weak staining and no regular rounded cell clusters being observed at day 1. At day 7 HepG2 showed some partial cell organization, with some cell stretching at the edges of irregular and sparse clusters. By day 14, this partial organization had loosened with a more spread-out clusters morphology being observed. Our collagen H&E staining results were in agreement with a previous study by Luckert et al. that also reported the formation by HepG2 of irregular cell clusters when cultures in collagen hydrogels.44
Immunohistochemical staining of E(FKFE)2 over 14 days are shown in Fig. 4(F), with negative controls shown in Fig. S3B–S5 (ESI†). Albumin staining confimed that in E(FKFE)2 HepG2 cells produced albumin over the 14 days of culture, with increased positive staining being observed over time. Albumin staining was found to be present intracellulary and extracellularly. E-cadherin staining was also found to increase over time. In this case at day 1, no obvious E-cadherin staining was seen, however from day 7 onwards stronger positive staining was observed. At day 14, the strong E-cadherin staining revealed clear cell outlines within the spheroids suggesting strong cell–cell interactions. Positive laminin staining in E(FKFE)2 samples was observed at all timepoints within the spheroids mainly, with day 14 spheroids showing the strongest positive staining. For fibronectin positive staining was found at every timepoint too, in this case inside and outside of the spheroids. While at day 7 uniform fibronectin staining was observed across the spheroids, at day 14, weaker positive staining was observed in the center of the spheroids, but strong positive staining was retained at the spheroid edges. In normal liver tissue, weak fibronectin staining is observed within area of arranged hepatocyte strips, but strong positive staining is observed in sinusoidal area.47 Our results suggest that as in normal liver tissue, HepG2 cells with time increasingly expressed fibronectin extracellularly. Finally, collagen I positive staining was also evident in E(FKFE)2 and seen to become stronger with time both intracellularly and extracellularly.
As far as collagen gels were concerned significant difference in staining levels compared to E(FKFE)2 were observed. The most striking is the lack of positive E-cadherin staining which point towards the absence in collagen of strong cell–cell interactions and confirming the results above which pointed towards the formation of loose cell clusters. Finally, weaker positive staining was observed in collagen for laminin and collagen I compared to E(FKFE)2 while for fibronectin similar level of staining were seen (Fig. 4(G)).
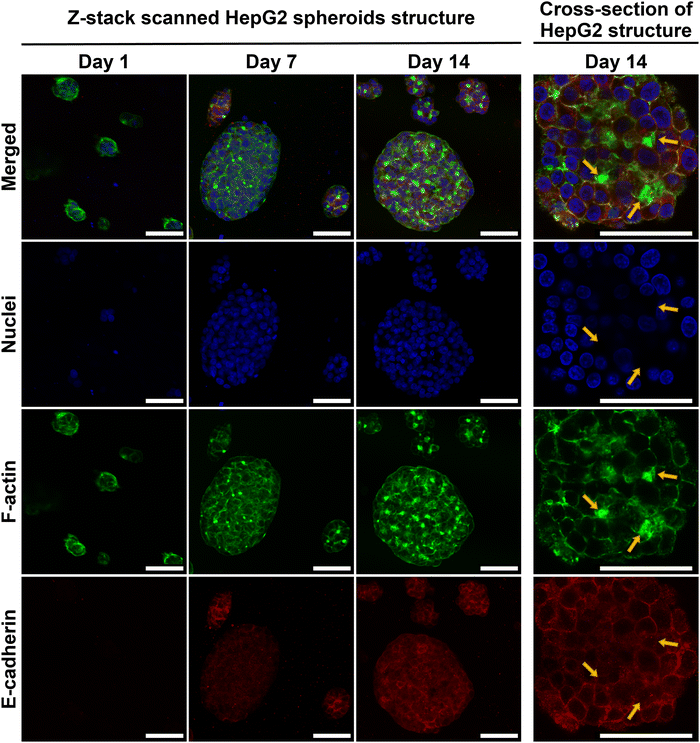
HepG2 spheroid morphology, cross-section structure and cell–cell adhesions within E(FKFE)2 SAPH were further investigated via ICC (Fig. 5). Negative controls for E-cadherin staining are presented in Fig. S6 (ESI†). The fluorescent images confirmed that HepG2 cells aggregated into rounded spheroids over 14 days culture. F-actin was mainly seen at the cell–cell boundaries. In addition, by comparing F-actin deposition for similar sized spheroids across different time points, it was observed that F-actin also accumulated at some specific cell junctions, and that with time these junctional regions enriched as shown by the increase in green-fluorescence intensity from day 7 to day 14. F-actin staining showed sharper green-fluorescent outlines around bigger spheroids than small spheroids. Strong positive E-cadherin staining was evident in all spheroids at day 7 and 14 with E-cadherin being clearly present at the cells’ borders pointing once again to the formation over time of strong cell–cell interactions in these spheroids. Interestingly when observing the co-localization of F-actin (green) and E-cadherin (red) at day 14 (Fig. 5, yellow arrows) it was seen that E-cadherin was concentrated on the cell borders and overlapped with F-actin, however, E-cadherin was also found to surround and concentrate around the F-actin junctional accumulation regions (bright green regions discussed above). The immunofluorescence staining results obtained showed a similar spheroid morphology as reported by Ramaiahgari et al.45 and Hiemstra et al.48 Our results also agreed with 3D HepaRG spheroid structure noted by Malinen et al.49 and primary human hepatocyte spheroid structure determined by Tostoes et al.50 These previous studies confirmed formation of functional bile-canaliculi-like structures through observation of F-actin enrichment regions. Therefore, our results suggest that bile-canaliculi-like structures also likely formed in our SAPH scaffolds. Confirmation of the presence of these structures will require further investigations.
IHC and ICC staining consistently confirmed increase presence of E-cadherin at the HepG2 cells border suggesting that stronger junctions formed between cells within the spheroids over time and that cells became more tightly connected in agreement with work by Bell et al.46 The presence of E-cadherin on hepatocyte membranes usually points towards maintenance of the differentiated phenotype as well as functions relevant to protein transportation across membranes.51 Our results overall point towards HepG2 cells maintaining their polarity and differentiated state within the spheroids when cultured within E(FKFE)2 SAPH over 14 days.
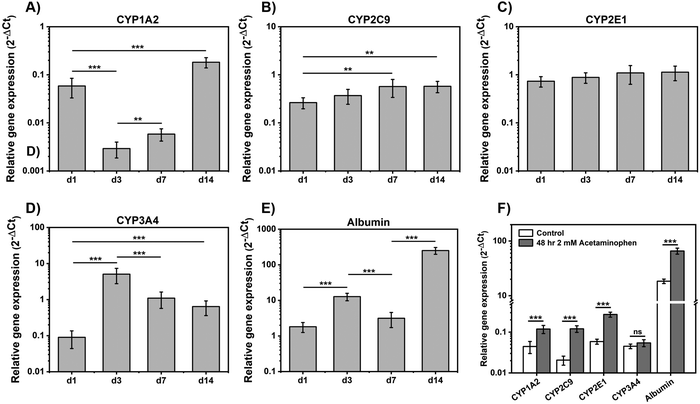
RT-qPCR was used to investigate the relative gene expression levels of cytochrome P450 (CYP450) enzymes and evaluate spheroids functionality (Fig. 6). CYP3A4, CYP2C9, CYP1A2 and CYP2E1 are the main CYP450 enzymes used to identify toxins response.52 As can be seen there was a significant fold-increase in mRNA expressions of CYP1A2 (P < 0.001) over 14 days, following an initial drop from day 1 to day 3 (Fig. 6(A)) and CYP2C9 (P < 0.01) mRNA level increased steadily up to day 7 and then stabilized (Fig. 6(B)). CYP1A2 and CYP2C9 gene expression levels in our study were in agreement with those found by Ramaiahgari et al.,45 where CYP2C9 expression increased significantly after 21 days culture. CYP2E1 gene expression showed a small increased over 14 days although results were not statistically significant (Fig. 6(C)). High level of CYP2E1 expression was not expected as over expressed CYP2E1 is related to HepG2 cell apoptosis which can be triggered by oxidative stress or toxicity.53 CYP3A4 expression increased from day 1 to day 3 and then decreased from day 3 to day 14 (Fig. 6(D)), in agreement with findings from previous studies that also observed an increase followed by a decrease in this gene expression over 14 days culture.45,49 Finally, albumin gene expression levels were found to increase with cell culture time in agreement with our results above that showed increasing level of albumin production by HepG2 when cultured in E(FKFE)2 (Fig. 6(E)).
CYP450 enzymes response following hepatocytes exposure to drugs was investigated following 48 h exposure to 2 mM acetaminophen. As can be seen from Fig. 6(F) following acetaminophen treatment, enhanced gene expression of CYP1A2 (P < 0.001), CYP2C9 (P < 0.001) and CYP2E1 (P < 0.001) was observed. There was no difference detected for CYP3A4 gene expression levels between control and acetaminophen-treated samples. Albumin gene expression level on the other hand was also found to increase (P < 0.001).
CYP1A2 and CYP2E1 are the main enzymes involved in acetaminophen clearance.46 Exposure to low-concentration of acetaminophen (<2 mM) was shown in previous studies to result in enhanced enzyme activity of CYP1A2 and CYP2E1.46,54,55 The absence of increase in CYP3A4 activity was also in agreement with the common understanding of acetaminophen metabolism.54 Finally the increased level of albumin gene expression upon exposure to 2 mM of acetaminophen suggests that the drug did not cause any visible hepatocyte damage or apoptosis and that albumin may contribute to the clearance of acetaminophen. Overall, these preliminary studies suggest that HepG2 spheroids formed in E(FKFE)2 SAPH displayed drug metabolism functions after 7 days of culture, and therefore that E(FKFE)2 SAPH could be a good fully synthetic scaffold candidate for the design of HepG2 spheroids for drug toxicity testing.
Conclusions
Hepatocyte behavior was investigated within a variety of SAPHs which properties were tuned by design. HepG2 were found to be sensitive to the charge carried by the fibers with negatively charged networks leading to good cell viability while positively charged networks lead to cell death independently from encapsulation pH and hydrogel storage shear-moduli. The negatively charged E(FKFE)2 was found to be the optimal SAPH in our panel to develop an HepG2 based in vitro spheroid model to mimic normal hepatic function. Albumin secretion and urea synthesis functions of hepatocytes were improved and higher than in collagen hydrogels. Production of hepatic ECM proteins: laminin, collagen I and fibronectin were observed. In addition, cell–cell junction marker E-cadherin was clearly visible from day 7 onwards showing strong cellular interactions within the observed spheroids. Significantly, hepatocytes continuously expressed specific CYP450 genes and presented a drug metabolism response to acetaminophen.Although previous work by us and others has clearly demonstrated the suitability of neutrally and positively charged SAPHs for the culture of a range of cells, the results presented here clearly show that only neutrally and negatively charged SAPHs are suitable for the culture of HepG2. The biological reason behind the sensitivity of cells to SAPHs fiber charge is still unclear and to our knowledge has not been investigated in the literature until now. Further biological studies are currently being undertaken to elucidate the relationship between cell fate and fiber charge in these SAPHs.
The use of bioinspired fully defined SAPHs as 3D scaffolds for cell culture offers the potential to investigate cell–material interactions in a controlled and defined way. In particular they allow careful design of scaffold's physicochemical properties, beyond simple mechanical stiffness changes, to understand their effects on cell viability and function and therefore improve our understand of material–cell interactions.
Data availability
All data generated or analyzed during this study are included in this published article and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the support from the Henry Royce Institute for Advanced Materials (EPSRC grants no.: EP/R00661X/1, EP/P025021/1 and EP/P025498/1). The authors also acknowledge the support from Prof. Judith Hoyland, School of Biological Sciences for access to and support with qPCR as well as from the Biological Electron Microscopy Core Facility (RRID: SCR_021147), Faculty of Biology, Medicine and Health.References
- G. M. Whitesides and B. Grzybowski, Self-Assembly at All Scales, Science, 2002, 295(5564), 2418–2421 CrossRef CAS PubMed.
- W. Liu, H. Duan, D. Zhang, X. Zhang, Q. Luo, T. Xie, H. Yan, L. Peng, Y. Hu, L. Liang, G. Zhao, Z. Xie and J. Hu, Concepts and Application of DNA Origami and DNA Self-Assembly: A Systematic Review, Appl. Bionics Biomech., 2021, 2021, 9112407 Search PubMed.
- A. Solomonov, A. Kozell and U. Shimanovich, Designing Multifunctional Biomaterials via Protein Self-Assembly, Angew. Chem., Int. Ed., 2024, 63(14), e202318365 CrossRef CAS PubMed.
- A. Sood, A. Gupta and G. Agrawal, Recent advances in polysaccharides based biomaterials for drug delivery and tissue engineering applications, Carbohydr. Polym. Technol. Appl., 2021, 2, 100067 CAS.
- A. C. Mendes, E. T. Baran, R. L. Reis and H. S. Azevedo, Self-assembly in nature: using the principles of nature to create complex nanobiomaterials, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5(6), 582–612 CAS.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Biomimetic peptide self-assembly for functional materials, Nat. Rev. Mater., 2020, 4(11), 615–634 CAS.
- G. Fichman and E. Gazit, Self-assembly of short peptides to form hydrogels: Design of building blocks, physical properties and technological applications, Acta Biomater., 2014, 10(4), 1671–1682 CrossRef CAS PubMed.
- E. A. Aisenbrey and W. L. Murphy, Synthetic alternatives to Matrigel, Nat. Rev. Mater., 2020, 5(7), 539–551 CrossRef CAS PubMed.
- M. T. Kozlowski, C. J. Crook and H. T. Ku, Towards organoid culture without Matrigel, Commun. Biol., 2021, 4(1), 1387 CrossRef PubMed.
- M. A. Elsawy, A. M. Smith, N. Hodson, A. Squires, A. F. Miller and A. Saiani, Modification of beta-Sheet Forming Peptide Hydrophobic Face: Effect on Self-Assembly and Gelation, Langmuir, 2016, 32(19), 4917–4923 CrossRef CAS PubMed.
- J. Gao, C. Tang, M. A. Elsawy, A. M. Smith, A. F. Miller and A. Saiani, Controlling Self-Assembling Peptide Hydrogel Properties through Network Topology, Biomacromolecules, 2017, 18(3), 826–834 CrossRef CAS PubMed.
- J. K. Wychowaniec, A. M. Smith, C. Ligorio, O. O. Mykhaylyk, A. F. Miller and A. Saiani, Role of Sheet-Edge Interactions in beta-sheet Self-Assembling Peptide Hydrogels, Biomacromolecules, 2020, 21(6), 2285–2297 CrossRef CAS PubMed.
- A. Mujeeb, A. F. Miller, A. Saiani and J. E. Gough, Self-assembled octapeptide scaffolds for in vitro chondrocyte culture, Acta Biomater., 2013, 9(1), 4609–4617 CrossRef CAS PubMed.
- C. Ligorio, A. Vijayaraghavan, J. A. Hoyland and A. Saiani, Acidic and basic self-assembling peptide and peptide-graphene oxide hydrogels: characterisation and effect on encapsulated nucleus pulposus cells, Acta Biomater., 2022, 143, 145–158 CrossRef CAS PubMed.
- K. A. Burgess, C. Frati, K. Meade, J. Gao, L. Castillo Diaz, D. Madeddu, G. Graiani, S. Cavalli, A. F. Miller, D. Oceandy, F. Quaini and A. Saiani, Functionalised peptide hydrogel for the delivery of cardiac progenitor cells, Mater. Sci. Eng., C, 2021, 119, 111539 CrossRef CAS PubMed.
- C. Ligorio, M. O’Brien, N. W. Hodson, A. Mironov, M. Iliut, A. F. Miller, A. Vijayaraghavan, J. A. Hoyland and A. Saiani, TGF-β3-loaded graphene oxide - self-assembling peptide hybrid hydrogels as functional 3D scaffolds for the regeneration of the nucleus pulposus, Acta Biomater., 2021, 127, 116–130 CrossRef CAS PubMed.
- M. A. Elsawy, J. K. Wychowaniec, L. A. Castillo Diaz, A. M. Smith, A. F. Miller and A. Saiani, Controlling Doxorubicin Release from a Peptide Hydrogel through Fine-Tuning of Drug-Peptide Fiber Interactions, Biomacromolecules, 2022, 23(6), 2624–2634 CrossRef CAS PubMed.
- C. Tang, A. F. Miller and A. Saiani, Peptide hydrogels as mucoadhesives for local drug delivery, Int. J. Pharm., 2014, 465(1–2), 427–435 CrossRef CAS PubMed.
- C. Ligorio, M. Zhou, J. K. Wychowaniec, X. Zhu, C. Bartlam, A. F. Miller, A. Vijayaraghavan, J. A. Hoyland and A. Saiani, Graphene oxide containing self-assembling peptide hybrid hydrogels as a potential 3D injectable cell delivery platform for intervertebral disc repair applications, Acta Biomater., 2019, 92, 92–103 CrossRef CAS PubMed.
- S. Zhang, Self-assembling peptides: From a discovery in a yeast protein to diverse uses and beyond, Protein Sci., 2020, 29(11), 2281–2303 CrossRef CAS PubMed.
- M. R. Caplan, E. M. Schwartzfarb, S. Zhang, R. D. Kamm and D. A. Lauffenburger, Control of self-assembling oligopeptide matrix formation through systematic variation of amino acid sequence, Biomaterials, 2002, 23(1), 219–227 CrossRef CAS PubMed.
- S. Zhang, T. Holmes, C. Lockshin and A. Rich, Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane, Proc. Natl. Acad. Sci. U. S. A., 1993, 90(8), 3334–3338 CrossRef CAS PubMed.
- M. T. Donato; L. Tolosa and M. J. Gómez-Lechón, Culture and Functional Characterization of Human Hepatoma HepG2 Cells, in Protocols in In Vitro Hepatocyte Research, ed. M. Vinken and V. Rogiers, Springer, New York, NY, 2015, pp. 77–93 Search PubMed.
- S. Wilkening, F. Stahl and A. Bader, Comparison of Primary Human Hepatocytes And Hepatoma Cell Line HEPG2 With Regard to Their Biotransformation Properties, Drug Metab. Dispos., 2003, 31(8), 1035–1042 CrossRef CAS PubMed.
- M. Štampar, B. Breznik, M. Filipič and B. Žegura, Characterization of In Vitro 3D Cell Model Developed from Human Hepatocellular Carcinoma (HepG2) Cell Line, Cells, 2020, 9(12), 2557 CrossRef PubMed.
- J. Saito, A. Okamura, K. Takeuchi, K. Hanioka, A. Okada and T. Ohata, High content analysis assay for prediction of human hepatotoxicity in HepaRG and HepG2 cells, Toxicol. In Vitro, 2016, 33, 63–70 CrossRef CAS PubMed.
- J. A. Heslop, C. Rowe, J. Walsh, R. Sison-Young, R. Jenkins, L. Kamalian, R. Kia, D. Hay, R. P. Jones, H. Z. Malik, S. Fenwick, A. E. Chadwick, J. Mills, N. R. Kitteringham, C. E. Goldring and B. Kevin Park, Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile, Arch. Toxicol., 2017, 91(1), 439–452 CrossRef CAS PubMed.
- A. C. Duarte, E. C. Costa, H. A. L. Filipe, S. M. Saraiva, T. Jacinto, S. P. Miguel, M. P. Ribeiro and P. Coutinho, Animal-derived products in science and current alternatives, Biomater. Adv., 2023, 151, 213428 CrossRef CAS PubMed.
- L. A. C. Diaz, J. Gough, A. Saiani and A. Miller, Human osteoblasts within soft peptide hydrogels promote mineralisation in vitro, J. Tissue Eng. Regener. Med., 2014, 8, 159–160 Search PubMed.
- S. Wan, S. Borland, S. M. Richardson, C. L. R. Merry, A. Saiani and J. E. Gough, Self-assembling peptide hydrogel for intervertebral disc tissue engineering, Acta Biomater., 2016, 46, 29–40 CrossRef CAS PubMed.
- L. A. Castillo Diaz, M. Elsawy, A. Saiani, J. E. Gough and A. F. Miller, Osteogenic differentiation of human mesenchymal stem cells promotes mineralization within a biodegradable peptide hydrogel, J. Tissue Eng., 2016, 7, 2041731416649789 CrossRef PubMed.
- S. Dong, S. L. Chapman, A. Pluen, S. M. Richardson, A. F. Miller and A. Saiani, Effect of Peptide–Polymer Host–Guest Electrostatic Interactions on Self-Assembling Peptide Hydrogels Structural and Mechanical Properties and Polymer Diffusivity, Biomacromolecules, 2024, 25(6), 3628–3641 CrossRef CAS PubMed.
- A. Markey, V. L. Workman, I. A. Bruce, T. J. Woolford, B. Derby, A. F. Miller, S. H. Cartmell and A. Saiani, Peptide hydrogel in vitro non-inflammatory potential, J. Pept. Sci., 2017, 23(2), 148–154 CrossRef CAS PubMed.
- L. Szkolar, J. B. Guilbaud, A. F. Miller, J. E. Gough and A. Saiani, Enzymatically triggered peptide hydrogels for 3D cell encapsulation and culture, J. Pept. Sci., 2014, 20(7), 578–584 CrossRef CAS PubMed.
- D. Yip and C. H. Cho, A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing, Biochem. Biophys. Res. Commun., 2013, 433(3), 327–332 CrossRef CAS PubMed.
- Y. Li, A. Asadi, M. R. Monroe and E. P. Douglas, pH effects on collagen fibrillogenesis in vitro: Electrostatic interactions and phosphate binding, Mater. Sci. Eng., C, 2009, 29(5), 1643–1649 CrossRef CAS.
- K. Guo, H. Wang, S. Li, H. Zhang, S. Li, H. Zhu, Z. Yang, L. Zhang, P. Chang and X. Zheng, Collagen-Based Thiol-Norbornene Photoclick Bio-Ink with Excellent Bioactivity and Printability, ACS Appl. Mater. Interfaces, 2021, 13(6), 7037–7050 CrossRef CAS PubMed.
- G. B. Schneider, A. English, M. Abraham, R. Zaharias, C. Stanford and J. Keller, The effect of hydrogel charge density on cell attachment, Biomaterials, 2004, 25(15), 3023–3028 CrossRef CAS PubMed.
- C. Sinthuvanich, L. A. Haines-Butterick, K. J. Nagy and J. P. Schneider, Iterative design of peptide-based hydrogels and the effect of network electrostatics on primary chondrocyte behavior, Biomaterials, 2012, 33(30), 7478–7488 CrossRef CAS PubMed.
- R. K. Jagdish, J. S. Maras and S. K. Sarin, Albumin in Advanced Liver Diseases: The Good and Bad of a Drug!, Hepatology, 2021, 74(5), 2848–2862 CrossRef CAS PubMed.
- J. Christoffersson, C. Aronsson, M. Jury, R. Selegard, D. Aili and C. F. Mandenius, Fabrication of modular hyaluronan-PEG hydrogels to support 3D cultures of hepatocytes in a perfused liver-on-a-chip device, Biofabrication, 2018, 11(1), 015013 CrossRef PubMed.
- S. M. Coward, C. Selden, A. Mantalaris and H. J. F. Hodgson, Proliferation Rates of HepG2 Cells Encapsulated in Alginate Are Increased in a Microgravity Environment Compared With Static Cultures, Artif. Organs, 2005, 29(2), 152–158 CrossRef CAS PubMed.
- P. Gunness, D. Mueller, V. Shevchenko, E. Heinzle, M. Ingelman-Sundberg and F. Noor, 3D organotypic cultures of human HepaRG cells: a tool for in vitro toxicity studies, Toxicol. Sci, 2013, 133(1), 67–78 CrossRef CAS PubMed.
- C. Luckert, C. Schulz, N. Lehmann, M. Thomas, U. Hofmann, S. Hammad, J. G. Hengstler, A. Braeuning, A. Lampen and S. Hessel, Comparative analysis of 3D culture methods on human HepG2 cells, Arch. Toxicol., 2017, 91(1), 393–406 CrossRef CAS PubMed.
- S. C. Ramaiahgari, M. W. den Braver, B. Herpers, V. Terpstra, J. N. Commandeur, B. van de Water and L. S. Price, A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies, Arch. Toxicol., 2014, 88(5), 1083–1095 CAS.
- C. C. Bell, D. F. Hendriks, S. M. Moro, E. Ellis, J. Walsh, A. Renblom, L. Fredriksson Puigvert, A. C. Dankers, F. Jacobs, J. Snoeys, R. L. Sison-Young, R. E. Jenkins, A. Nordling, S. Mkrtchian, B. K. Park, N. R. Kitteringham, C. E. Goldring, V. M. Lauschke and M. Ingelman-Sundberg, Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease, Sci. Rep., 2016, 6, 25187 CrossRef CAS PubMed.
- M. Torbenson, J. Wang, M. Choti, R. Ashfaq, A. Maitra, R. E. Wilentz and J. Boitnott, Hepatocellular carcinomas show abnormal expression of fibronectin protein, Mod. Pathol., 2002, 15(8), 826–830 CrossRef PubMed.
- S. Hiemstra, S. C. Ramaiahgari, S. Wink, G. Callegaro, M. Coonen, J. Meerman, D. Jennen, K. van den Nieuwendijk, A. Dankers, J. Snoeys, H. de Bont, L. Price and B. van de Water, High-throughput confocal imaging of differentiated 3D liver-like spheroid cellular stress response reporters for identification of drug-induced liver injury liability, Arch. Toxicol., 2019, 93(10), 2895–2911 CrossRef CAS PubMed.
- M. M. Malinen, L. K. Kanninen, A. Corlu, H. M. Isoniemi, Y. R. Lou, M. L. Yliperttula and A. O. Urtti, Differentiation of liver progenitor cell line to functional organotypic cultures in 3D nanofibrillar cellulose and hyaluronan-gelatin hydrogels, Biomaterials, 2014, 35(19), 5110–5121 CrossRef CAS PubMed.
- R. M. Tostoes, S. B. Leite, M. Serra, J. Jensen, P. Bjorquist, M. J. Carrondo, C. Brito and P. M. Alves, Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing, Hepatology, 2012, 55(4), 1227–1236 CrossRef CAS PubMed.
- D. Theard, M. Steiner, D. Kalicharan, D. Hoekstra and S. C. van Ijzendoorn, Cell polarity development and protein trafficking in hepatocytes lacking E-cadherin/beta-catenin-based adherens junctions, Mol. Biol. Cell, 2007, 18(6), 2313–2321 CrossRef CAS PubMed.
- U. M. Zanger and M. Schwab, Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation, Pharmacol. Ther., 2013, 138(1), 103–141 CrossRef CAS PubMed.
- K. Chandrasekaran, K. Swaminathan, S. M. Kumar, D. L. Clemens and A. Dey, Increased oxidative stress and toxicity in ADH and CYP2E1 overexpressing human hepatoma VL-17A cells exposed to high glucose, Integr. Biol., 2012, 4(5), 550–563 CrossRef CAS PubMed.
- H. Raza, A. John and J. Shafarin, Potentiation of LPS-Induced Apoptotic Cell Death in Human Hepatoma HepG2 Cells by Aspirin via ROS and Mitochondrial Dysfunction: Protection by N-Acetyl Cysteine, PLoS One, 2016, 11(7), e0159750 CrossRef PubMed.
- L. Schyschka, J. J. Sanchez, Z. Wang, B. Burkhardt, U. Muller-Vieira, K. Zeilinger, A. Bachmann, S. Nadalin, G. Damm and A. K. Nussler, Hepatic 3D cultures but not 2D cultures preserve specific transporter activity for acetaminophen-induced hepatotoxicity, Arch. Toxicol., 2013, 87(8), 1581–1593 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01701c |
| ‡ Current address: Biodiscovery Institute and Department of Chemical and Environmental Engineering, The University of Nottingham, UK. |
| This journal is © The Royal Society of Chemistry 2024 |