Open Access Article
Open Access ArticleCytotoxic and molecular effects of soil extracts from the Agbogbloshie electronic-waste site on fish and human cell lines†
Krittika
Mittal
a,
Ke
Xu
 a,
Jingyun
Zheng
b,
Stephane
Bayen
a,
Jingyun
Zheng
b,
Stephane
Bayen
 b,
Julius
Fobil
c and
Niladri
Basu
b,
Julius
Fobil
c and
Niladri
Basu
 *a
*a
aDepartment of Natural Resource Sciences, McGill University, 21111 Lakeshore Road, Sainte-Anne-de-Bellevue, Quebec, H9X3V9, Canada. E-mail: niladri.basu@mcgill.ca
bDepartment of Food Science and Agricultural Chemistry, McGill University, 21111 Lakeshore Road, Sainte-Anne-de-Bellevue, Quebec, H9X3V9, Canada
cDepartment of Biological, Environmental and Occupational Health Science, University of Ghana School of Public Health, Accra, Ghana
First published on 29th October 2024
Abstract
Effect-based methods (EBM) are of growing interest in environmental monitoring programs. Few EBM have incorporated transcriptomics even though these provide a wealth of biological information and can be modeled to yield transcriptomic points of departure (tPODs). The study objectives were to: (A) characterize cytotoxic effects of soil extracts on the rainbow trout RTgill-W1 and the human Caco-2 cell lines; (B) measure gene expression changes and calculate tPODs; and (C) compare in vitro responses to available measures of plastic-related compounds and metals. Extracts were prepared from 35 soil samples collected at the Agbogbloshie E-waste site (Accra, Ghana). Cells were exposed to six soil concentrations (0.3 to 9.4 mg dry weight of extract (eQsed) per mL). Many samples caused cytotoxicity with RTgill cells being more sensitive than Caco-2 cells. Eleven samples were analyzed for transcriptomics in both cell lines, with responses measured in all samples (52 to 5925 differentially expressed genes) even in the absence of cytotoxicity. In RTgill cells there was concordance between cytotoxic measures in tPOD values (spearman = 0.85). Though trends between in vitro measures and contaminant data were observed, more work is needed in this area before definitive conclusions are drawn. Nonetheless, this study helps support ongoing efforts in establishing alternative testing strategies (e.g., alternative to animal methods; toxicogenomics) for the assessment of complex environmental samples.
Environmental significanceEffect-based methods (EBM) are of growing interest to evaluate the hazard potential of real-world environmental samples. Some EBM studies have included a small number of gene targets but these do not take advantage of advances in the field in which next generation sequencing technologies and bioinformatics can reveal rich and untargeted insights into potential mechanisms of action. Further, transcriptomic data can be modeled using benchmark dose statistical techniques to yield a discrete concentration at which a concerted molecular change occurs (= transcriptomic point of departure, or tPOD). Accordingly, the objective of this study was to assess the implementation of the tPOD concept into an EBM study situated at the Agbogbloshie electronic waste site involving human and fish cell lines. |
Introduction
Contaminants in environmental matrices often exist as complex mixtures, and to assess the hazard potential of such matrices, researchers tend to focus on select priority chemicals.1 In doing so, this approach underestimates risk as it neither accounts for the universe of chemicals that may be present in the sample, nor does it consider mixture interactions well. Given these deficiencies, there is increased scientific and regulatory interest to improve the hazard and risk characterization of real-world environmental samples that tend to exist as complex mixtures.2,3In contrast to approaches strictly reliant on chemical measures, effect-based methods (EBM) interrogate responses in biological systems following exposure to environmental samples.4 To date there has been a focus on bioanalytical tests that characterize apical measures in exposed fish, invertebrate, and algae, as well as in in vitro assays designed to interrogate specific mode of action such as endocrine disruption.4,5 Some of these latter approaches may rely on genomic techniques though to date have tended to focus on a small number of gene targets. Next generation sequencing and associated bioinformatic methods continue to advance and are now penetrating into regulatory toxicology. Transcriptome-wide data can provide rich information on a chemical's mechanism of action by returning data on thousands of individual genes, as well as on focused gene sets or biological pathways.6 In addition, transcriptomic data can be modeled using benchmark dose (BMD) statistical techniques to yield a discrete concentration at which a concerted molecular change occurs.7 The resulting value, referred to as a transcriptomic point of departure (tPOD), tends to be lower in concentration (i.e., protective) than values that would be associated with causing adverse outcomes in long-term studies. For example, the US EPA's Transcriptomic Assessment Product (ETAP) is based on the concept that tPOD values from short term (5 day) rodent tests are comparable to apical point of departure values from long-term (e.g., 2 year) studies. Despite the promise of transcriptomic approaches to be used as EBMs, its penetration for scientific, monitoring, and regulatory purposes remains limited.5
The overall objective of this study was to assess the implementation of the tPOD concept into an EBM study situated at a contaminated site involving human and fish cell lines. Specifically, this work was positioned at the Agbogbloshie electronic waste site (e-waste) in Ghana which is widely viewed as one of the most contaminated sites worldwide8 and one in which we have conducted extensive research.9,10 Rainbow trout (Oncorhynchus mykiss) gill (RTgill-W1) and human intestinal epithelial cells (Caco-2) were exposed to chemical extracts of soil samples from this site in a concentration-dependent manner, following which cytotoxic and transcriptomic responses were measured and compared. The resulting data were also compared to available measures of select priority chemicals of concern in these same samples, namely a panel of plastic-related organic compounds and inorganic metals.
Methods
As a brief introduction to the methods section, fish and human cells were exposed to extracts prepared from soil samples collected at various locations within the Agbogbloshie e-waste site (i.e., sites for dumping waste, trading materials, and burning items as well as various control sites) to examine effects on cell viability and gene expression. We focused on rainbow trout gill cells under OECD Test Guideline 249 (ref. 11) as the method has been validated to study risks to fish following waterborne exposures, and also human Caco-2 cells which are commonly used as an intestinal cell model in toxicity studies involving ingestion of substances due to their similar microvilli structures to the intestinal epithelial cells and biological functions in absorption.12 Soil pollution at the Agbogbloshie e-waste site is well-established from which the adjacent Korle Lagoon gets contaminated.13 In addition, e-waste workers and community members may inhale or ingest chemicals from contaminated soil or food.8Dimethyl sulfoxide (DMSO; Sigma-Aldrich) was used as the solvent since the dried extracts were reconstituted in DMSO. 3,4-Dichloroaniline (DCA) was used as the positive control for the RTgill-W1 cytotoxicity assays (as recommended by OECD 249) while copper sulphate (CuSO4) was used as the positive control for the Caco-2 cytotoxicity assays. Untreated cells were used as negative controls. All materials were obtained from Thermo Fisher Scientific unless otherwise specified.
Cell culture
Experiments on both cell lines followed past publications14,15 from our group as well as guidance16,17 from ATCC. The cytotoxicity experiments for RTgill cells were performed in 24-well plates following the OECD 249 guidelines.11 Subsequently, to increase throughput and improve efficiency of this assay we adapted the protocol to be conducted in 96-well plates. Thus, the transcriptomics assay for RTgill cells, and all exposures for the Caco-2 cells were conducted in 96-well plates.E-Waste soil and contaminants
The verification of data quality involves internal validation, focusing on both precision and accuracy. Precision is assessed by calculating coefficient of variance (CV) from three replicates of each selected sample (eqn (1)), namely samples 14, 37, 65, and 88. The coefficient of variance provides a measure of the variability among these replicates, helping ensure the reproducibility of the results.
 | (1) |
Accuracy is another critical aspect of data quality and is verified by calculating the percentage difference relative to a reference value. In this study, soil reference materials (QC 98-04 and 98-05) were measured in triplicates and compared to the respective certified values. The percentage error calculated by eqn (2) provides an insight between the level of accuracy achieved in the experimental measurements, indicating how closely the results align with the expected reference.
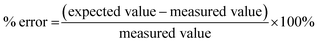 | (2) |
Integrated assessment
Holistic examination of the measured contaminants in soil extracts involved considering both plastic-related and metal contaminants. To aid in this, we categorized each contaminant in a given sample into quintiles based on the detected concentrations of the respective contaminants (i.e., assigned scores ranged from 1 to 5, corresponding to the first to the last quintile). Subsequently, the scores for all contaminants within a single sample were equally weighed and summed to yield a composite score (CS). The composite scoring system used in this study was adapted from composite indicators commonly employed in public communications of academic reports to summarize complex or multidimensional variables.24Hazard Quotient (HQ) is a common tool used in human health risk assessment to evaluate the potential adverse effects of exposure to a chemical or substance.25 The general equation for calculating HQ is listed below as eqn (3),
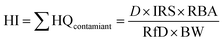 | (3) |
For the RTgill cell data, we used the risk index (RI), the sum the risk quotient (RQ), approach by the US EPA for environmental risk assessment of a chemical. It is calculated by dividing an exposure concentration (D) by an effect threshold at which adverse effects are expected to occur (eqn (4)).
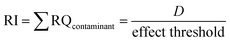 | (4) |
Cytotoxicity exposures
In vitro exposures were conducted according to guidelines prescribed in the OECD Test Guideline # 249 for chemical exposures using the RTgill-W1 cell line11 with modifications. A working stock of the extract was prepared in DMSO at a concentration equivalent to 1.875 g dry weight of soil (eQsed) per mL. The stock was diluted two-fold in the respective exposure medium (L-15/ex for RTgill-W1 and serum-free DMEM for Caco-2) to obtain the final test concentrations equivalent to 9.38, 4.69, 2.34, 1.17, 0.59, and 0.29 mg dry weight of extract (eQsed) per mL. For RTgill cells, a working stock of DCA prepared in DMSO and diluted in L-15/ex to obtain concentrations of 6.25 mg L−1, 12.5 mg L−1, 25 mg L−1, 50 mg L−1, and 100 mg L−1 was used as the positive control. For Caco-2 cells, CuSO4 was diluted in serum-free DMEM medium to 200 mg L−1, 100 mg L−1, 50 mg L−1, 25 mg L−1, 12.5 mg L−1, 6.3 mg L−1, 3.1 mg L−1, and 1.6 mg L−1 as the positive control. DMSO was used as the solvent control and added to the exposure medium at a final concentration that did not exceed 0.5% v/v in all exposure solutions.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL complete medium per well in 22 wells of 24-well plates and incubated for 24 hours (h). Two wells per plate were designated as no-cell controls and contained only the culture medium. On day 2, the old complete medium was discarded and replaced with 1 mL of L-15/ex for the negative control (one well), or L-15/ex containing either the extract, solvent or positive control at appropriate concentrations (in triplicate). One of the two no-cell control wells received 1 mL of the highest concentration of the e-waste extract exposure solution being tested and the other well received 1 mL L-15/ex. The cells were then incubated for 24 h. On day 3, the exposure medium was discarded, and Alamar Blue (AB) cytotoxicity assay was conducted immediately. Briefly, the cells were rinsed with ice-cold phosphate buffered saline (PBS) and AB dye (5% v/v in PBS) was added to the wells and incubated for 30 min following which fluorescence was read at 530 nm/595 nm using the Synergy HT Gen5 microplate reader (BioTek, Winooski, USA).
000 cells per mL complete medium per well in 22 wells of 24-well plates and incubated for 24 hours (h). Two wells per plate were designated as no-cell controls and contained only the culture medium. On day 2, the old complete medium was discarded and replaced with 1 mL of L-15/ex for the negative control (one well), or L-15/ex containing either the extract, solvent or positive control at appropriate concentrations (in triplicate). One of the two no-cell control wells received 1 mL of the highest concentration of the e-waste extract exposure solution being tested and the other well received 1 mL L-15/ex. The cells were then incubated for 24 h. On day 3, the exposure medium was discarded, and Alamar Blue (AB) cytotoxicity assay was conducted immediately. Briefly, the cells were rinsed with ice-cold phosphate buffered saline (PBS) and AB dye (5% v/v in PBS) was added to the wells and incubated for 30 min following which fluorescence was read at 530 nm/595 nm using the Synergy HT Gen5 microplate reader (BioTek, Winooski, USA).
Caco-2 cells
On day 1, Caco-2 cells were seeded at a density of 350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL complete medium in 85 wells of a 96-well plates (100 μL volume) and incubated for 24 h. The remaining 11 wells contained complete culture medium designated as no-cell controls for each tested extract, solvent control, and positive control. On day 2, the growth medium was replaced by 100 μL of serum-free medium containing either the extract, solvent, or positive control at appropriate concentrations (in triplicates). The 11 no-cell control wells received 100 μL of the highest exposure concentration of the e-waste extract, solvent, or positive control. Cells were then incubated for 24 h. On day 3, the exposure medium was discarded, rinsed with ice-cold PBS, and replaced by 100 μL dye mixtures of AB (5% v/v). Plates were incubated for 60 min following which fluorescence was read at 530 nm/595 using the Synergy HT Gen5 microplate reader (BioTek, Winooski, USA).
000 cells per mL complete medium in 85 wells of a 96-well plates (100 μL volume) and incubated for 24 h. The remaining 11 wells contained complete culture medium designated as no-cell controls for each tested extract, solvent control, and positive control. On day 2, the growth medium was replaced by 100 μL of serum-free medium containing either the extract, solvent, or positive control at appropriate concentrations (in triplicates). The 11 no-cell control wells received 100 μL of the highest exposure concentration of the e-waste extract, solvent, or positive control. Cells were then incubated for 24 h. On day 3, the exposure medium was discarded, rinsed with ice-cold PBS, and replaced by 100 μL dye mixtures of AB (5% v/v). Plates were incubated for 60 min following which fluorescence was read at 530 nm/595 using the Synergy HT Gen5 microplate reader (BioTek, Winooski, USA).
Exposures for transcriptomics
Based on the cell viability results, 11 sites were chosen to represent a mix of site types (e-waste work activities and geographic proximity), contaminant levels, and cytotoxicity assay results (Table 1). In vitro exposures for transcriptomics in both cell lines were conducted in 96-well plates following the same timeline as the cytotoxicity exposures. Cells were seeded at a density of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and no-cell controls were excluded. For each site, the concentration equivalent to the EC20 (effect concentration of a chemical that induces 20% cell death) or (the highest concentration tested in cases where extracts caused no cytotoxicity) was the highest concentration chosen from which an 8-point, 2-fold dilution series was prepared. This concentration range ensures the detection of meaningful biological responses while minimizing the risk of data quality issues that arise from cell death.31 Of the 96 wells, two wells were left empty for the Xpressref internal control RNA (see library preparation section). After the 24 h exposure, exposure solutions were removed and 40 μL lysis premix (UPX 3′ lysis buffer, RNase inhibitor and nuclease-free water; QIAGEN) was added to each well, pipetted up and down 10 times and plates were placed on a shaker at room temperature for 10 min to facilitate lysis. Cells were checked visually under a microscope to make sure they were detached, following which the pipetting and shaking steps were repeated if needed. The cell lysate plates were stored at −80 °C until library preparation.
000 cells per well and no-cell controls were excluded. For each site, the concentration equivalent to the EC20 (effect concentration of a chemical that induces 20% cell death) or (the highest concentration tested in cases where extracts caused no cytotoxicity) was the highest concentration chosen from which an 8-point, 2-fold dilution series was prepared. This concentration range ensures the detection of meaningful biological responses while minimizing the risk of data quality issues that arise from cell death.31 Of the 96 wells, two wells were left empty for the Xpressref internal control RNA (see library preparation section). After the 24 h exposure, exposure solutions were removed and 40 μL lysis premix (UPX 3′ lysis buffer, RNase inhibitor and nuclease-free water; QIAGEN) was added to each well, pipetted up and down 10 times and plates were placed on a shaker at room temperature for 10 min to facilitate lysis. Cells were checked visually under a microscope to make sure they were detached, following which the pipetting and shaking steps were repeated if needed. The cell lysate plates were stored at −80 °C until library preparation.
| Species (cell line) | Sites | # of DEGs | DEGs w/BMDs | tPOD (eQsed per mL) | EC20 (eQsed per mL) | EC50 (eQsed per mL) | ||
|---|---|---|---|---|---|---|---|---|
| 20th gene | Max 1st peak | 10th percentile | ||||||
| a The FDR adjusted differential expression analysis resulted in 15 or fewer DEGs in seven out of 11 sites, and hence for the RTgill cells, the FDR adjusted filter was turned off. | ||||||||
| Rainbow trouta (RTgill-W1) | 8 | 187 | 75 | 0.052 | 0.059 | 0.027 | 0.27 | 0.50 |
| 16 | 52 | 17 | NA | 0.018 | 0.003 | 0.67 | 2.27 | |
| 2 | 181 | 33 | 0.130 | 0.140 | 0.038 | 1.12 | 1.90 | |
| 5 | 941 | 209 | 0.052 | 0.096 | 0.058 | 1.26 | 2.28 | |
| 48 | 1279 | 565 | 0.029 | 0.220 | 0.140 | 1.27 | 2.28 | |
| 3 | 196 | 87 | 0.019 | 0.510 | 0.065 | 1.40 | 2.21 | |
| 7 | 678 | 111 | 0.260 | 0.640 | 0.072 | 1.93 | 3.02 | |
| 30 | 529 | 197 | 0.17 | 0.890 | 0.200 | 5.90 | 9.79 | |
| 20 | 1999 | 105 | 0.640 | 0.870 | 0.210 | 8.88 | 10.79 | |
| 74 | 496 | 107 | 1.100 | 4.700 | 0.160 | >9.38 | >9.38 | |
| 49 | 1797 | 513 | 1.500 | 4.900 | 2.400 | >9.38 | >9.38 | |
| Human (Caco-2) | 49 | 1835 | 43 | 0.820 | 0.260 | 0.160 | 2.85 | >9.38 |
| 8 | 842 | 217 | 0.025 | 0.240 | 0.056 | 3.10 | 5.00 | |
| 48 | 1505 | 25 | 1.700 | 0.770 | 0.160 | 5.82 | >9.38 | |
| 2 | 175 | 21 | 1.300 | 1.1 | 0.800 | >9.38 | >9.38 | |
| 3 | 1490 | 128 | 0.230 | 0.920 | 0.270 | >9.38 | >9.38 | |
| 5 | 1645 | 52 | 0.170 | 0.410 | 0.072 | >9.38 | >9.38 | |
| 7 | 1828 | 44 | 0.058 | 0.280 | 0.041 | >9.38 | >9.38 | |
| 16 | 18 | NA | NA | NA | NA | >9.38 | >9.38 | |
| 20 | 1639 | 38 | 0.630 | 0.820 | 0.33 | >9.38 | >9.38 | |
| 30 | 104 | 7 | NA | NA | NA | >9.38 | >9.38 | |
| 74 | 1119 | 8 | NA | NA | NA | >9.38 | >9.38 | |
Library preparation
Libraries were prepared from the cell lysates following the QIAGEN UPXome protocol for 3′ partial transcriptome sequencing.32 Cell lysates were reverse transcribed using an Oligo-dT (ODT) primer with the following temperature profile: 1 min at 4 °C, 5 min at 25 °C, 90 min at 42 °C, 10 min at 70 °C, and 1 min at 4 °C. In the 96-well plate, 94 wells contained 94 samples from the e-waste exposure plate, and 2 wells contained an internal RNA control (RNA isolated from RT-gill-W1 cells following RNeasy protocol with on-column DNAse I digestion or human Xpressref RNA control obtained from QIAGEN). The reverse transcription step incorporates a unique sample ID into each individual cDNA sample thus allowing up to 24 cDNA samples to be pooled for library preparation. The pooling scheme for each microplate was as follows: 3 pools of 24 wells each, 1 pool of 22 wells, and 1 pool of 2 Xpressref control wells. This was followed by a two-step magnetic bead clean-up that included two ethanol washes using a magnetic tube rack (Permagen Labware, Amesbury, USA). Subsequently, qPCR amplification was performed on each pool with the following temperature profile: 30 s at 98 °C, 17 cycles of 5 s at 98 °C, 10 s at 55 °C and 20 s at 72 °C, 2 min at 72 °C and 1 min 4 °C, followed by another bead clean-up including one ethanol wash. The amplification step incorporates a RUDI unique library index to each pooled library, thus allowing libraries to be sequenced together on one lane. At this step, the Xpressref control wells were pooled as an internal quality control library but were not sequenced. In total, there were six 96-well plates exposed to e-waste soil extracts (three for each cell line), resulting in 24 libraries (12 for each cell line) for sequencing. The prepared libraries were assessed for quality on a Tapestation using the D1000 screen tape (Agilent). Libraries were then shipped to the Genome Quebec Innovation Center, quantified using the PicoGreen assay prior to pooling and sequenced to produce paired-end 2 × 150 bp reads on a Novaseq 6000 S4 platform (Illumina).Data analysis
Cell viability of the exposed cells was determined as the percentage fluorescence compared to the DMSO treated solvent-control cells. Cell viability of the DMSO treated cells was determined as the percentage fluorescence compared to the negative untreated control cells. Exposure response curves were plotted by the Logit method using the drc package for dose–response analysis in R.33Raw FASTQ sequencing reads from the 24 libraries were submitted to QIAGEN's online RNASeq analysis portal (RAP, Analysis workflow version 1.0) for demultiplexing according to the corresponding sample IDs to obtain FASTQ files for each individual sample. Raw reads from each individual FASTQ file were trimmed and filtered (ESI Table ST2†) and mapped to the rainbow trout genome version Omyk_1.0.105 (RTgill-W1 cells) and the human genome version GRCh38.noalt.106 (Caco-2 cells). Raw counts, obtained from RAP as the total exon reads, were submitted to ExpressAnalyst (https://www.expressanalyst.ca) for differential expression and tPOD analysis. Counts were normalized (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 counts per million) and filtered for abundance (cut-off 4) and variance (cut-off 15). Differential expression was determined using Limma as the statistical method first with a log
2 counts per million) and filtered for abundance (cut-off 4) and variance (cut-off 15). Differential expression was determined using Limma as the statistical method first with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC and false discovery rate (FDR) adjusted p-value threshold of 1 and 0.05, respectively. For RTgill-W1 cells, due to lower number of DEGs in many cases, the FDR adjusted filter was turned off. Whereas for Caco-2 cells, these thresholds resulted in many more than 2000 DEGs. Curve-fitting for dose–response analysis is performed on a maximum of 2000 DEGs which represents the maximum allowable number of genes for optimal curve fitting.34 Hence for Caco-2 cells, a uniform, more-stringent threshold of log
2FC and false discovery rate (FDR) adjusted p-value threshold of 1 and 0.05, respectively. For RTgill-W1 cells, due to lower number of DEGs in many cases, the FDR adjusted filter was turned off. Whereas for Caco-2 cells, these thresholds resulted in many more than 2000 DEGs. Curve-fitting for dose–response analysis is performed on a maximum of 2000 DEGs which represents the maximum allowable number of genes for optimal curve fitting.34 Hence for Caco-2 cells, a uniform, more-stringent threshold of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 1.5 and FDR adjusted p-value of 0.01 was used to obtain at most 2000 DEGs. Curve-fitting was performed on these lists of DEGs with lack-of-fit p-value and BMR factor thresholds of 0.1 and 1.0, respectively. Depending on the best fit, a gene-level BMD relative to the mean expression of the control samples was calculated from which tPODs were derived as the (1) BMD of the 20th most sensitive gene (tPOD20thgene), (2) first mode of the gene-level BMD distribution (tPODmode), and (3) 10th percentile of all gene-level BMDs (tPOD10th
2FC = 1.5 and FDR adjusted p-value of 0.01 was used to obtain at most 2000 DEGs. Curve-fitting was performed on these lists of DEGs with lack-of-fit p-value and BMR factor thresholds of 0.1 and 1.0, respectively. Depending on the best fit, a gene-level BMD relative to the mean expression of the control samples was calculated from which tPODs were derived as the (1) BMD of the 20th most sensitive gene (tPOD20thgene), (2) first mode of the gene-level BMD distribution (tPODmode), and (3) 10th percentile of all gene-level BMDs (tPOD10th![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) percentile). Genes with BMDs were then examined for pathway level BMDs using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (adj p-value <0.05 and minimum number of genes = 3) and for enriched gene ontologies (GO) under the three categories – biological processes (BP), cellular components (CC) and molecular functions (MF) (http://www.geneontology.org). Raw sequencing data files are available through NCBI GEO Accession Number GSE279464.
percentile). Genes with BMDs were then examined for pathway level BMDs using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (adj p-value <0.05 and minimum number of genes = 3) and for enriched gene ontologies (GO) under the three categories – biological processes (BP), cellular components (CC) and molecular functions (MF) (http://www.geneontology.org). Raw sequencing data files are available through NCBI GEO Accession Number GSE279464.
Results and discussion
Contaminant levels
The measured concentrations were cross-referenced with reference values to assess potential harm. While the CCME reference only provides aquatic threshold value for DEP and DBP, and no specific soil guidelines, the US EPA regional soil screening level (EPA-RSL) offers reference concentrations for DEHP, BBzP, DBP, DEP and BPA. The values used for comparison in this study were based on carcinogenic target risk (TR) = 1 × 10−6 and HI = 0.1.22 Notably, all analytes were at least one magnitude below the reference threshold with the exception of DEHP. However, as illustrated in ESI Fig. SF4,† 75% of the DEHP results fell below the reference threshold. The findings indicate that, despite the detection of plastic-related chemicals, their individual concentrations may not pose a concern for human health.
Of the six metals included in our analysis, Cu and Pb were among the most prominent with concentrations ranging from 20.1 to 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672.1 μg g−1 and 16.4 to 4347.0 μg g−1 dry weight, respectively. While there were certain site types with relatively high levels of metals detected including dump sites (2, 5, 18) and upstream sites (30, 46), in general, trade sites were again the ones with highest concentrations of most metals (ESI Table ST5†). The trade sites also influenced the most variation for PC1 (ESI Fig. SF5†). Furthermore, we observed that all selected metals for deeper investigation here were positively correlated to each other and the plastic-related contaminants (ESI Fig. SF3†). Out of all selected metals, Cd and Pb seemed to contribute the most to PC1.
672.1 μg g−1 and 16.4 to 4347.0 μg g−1 dry weight, respectively. While there were certain site types with relatively high levels of metals detected including dump sites (2, 5, 18) and upstream sites (30, 46), in general, trade sites were again the ones with highest concentrations of most metals (ESI Table ST5†). The trade sites also influenced the most variation for PC1 (ESI Fig. SF5†). Furthermore, we observed that all selected metals for deeper investigation here were positively correlated to each other and the plastic-related contaminants (ESI Fig. SF3†). Out of all selected metals, Cd and Pb seemed to contribute the most to PC1.
The metal concentrations measured in this study were compared to respective metal concentrations measured in other studies situated at the Agbogbloshie area (ESI Table ST5†) (ref. 13, 35–37) and regulatory standards from EPA22 and CCME21 (ESI Fig. SF6†). Compared to other studies, the measured levels of Cu, Zn, and Pb showed great variations and were close to the lower concentration range (ESI Fig. SF6†). However, the measured levels of As, Cr, and Cd were much lower than the detected ranges reported in the literature. While the measured values here were relatively low compared to previous studies (with reasons for this difference not entirely known), the majority of soil samples had metal concentrations above both EPA and CCME standards.21,22 For example, out of the 35 sites, all contained Cr levels higher than both EPA and CCME standards. As for specific metals, all sites had As levels higher than the EPA standard, but only 11.4% exceeded the CCME standard. Further, approximately half of the sites detected Cd levels higher than both EPA and CCME standards. For Cu, Zn, and Pb, fewer than half of the sites reported metal concentrations higher than the EPA standard, while over 70% exceeded the CCME standard.
Composite scores, hazard indexes and risk indexes
Composite scores are widely employed to integrate and summarize data with multiple parameters, and has been suggested for the communication of cumulative risk.24,38 In the exposure assessment of this study, composite scores were particularly valuable for synthesizing exposure data related to both metal and plastic-related contaminants. Reducing multiple contaminant data into a single numerical value for each site facilitates the comparison and ranking of exposure levels across different sites. The average composite score across all sites was 38.5 and ranged from 19 to 65. Within this, the average score was highest for trade sites (averaged 57.6, ranged from 51–65). Notably, four out of the five highest composite scores were associated with trade sites (Sites #7, 8, 10, and 11, ESI Fig. SF7†). Following the trade sites, the composite score decreasing rank-order was: upstream sites (average 38.2, between 26–59), dump site (average 33.2, between 18–55), community site (average 30, between 20–37), burn site (average 23, between 18–27), and downstream sites (average 21, between 19–23). This observation may also be examined by a PCA (ESI Fig. SF3†), and also the indexes calculated for human (HI, ESI Table ST6†) and aquatic species (RI, ESI Table ST7†). The PCA showed that the trade sites were distinctly separated from the rest of the sites (ESI Fig. SF2 and SF5†). The loading score of the six selected metals were closely clustered with the plastics and were distinctly separated from half of the detected metals in PC1 and PC2 (ESI Fig. SF3†). Notably, DEHP had the greatest loading among all the plastics-related chemicals. These observations suggested that the selected metals and DEHP were the primary contributors to the variation across identified sites. Furthermore, the aquatic RI (ESI Table ST7†) for the selected metals significantly exceeded 1 (RI ranged from 5.9 to 1004.4), and half of the RI values for plastics were higher the level of concern (LOC > 1). Notably, only DEHP had a RQ that exceeded 1 across all sites, and the majority of the trade sites detected DEHP at levels surpassing the threshold. As a result, from an exposure perspective, the selected six metals and DEHP posed higher level of concern compared to other contaminants. Trade sites, which collectively contained the highest concentrations of all measured contaminants, were consequently ranked as of greatest concern across all sites. The dismantling activities around the trade sites might be one of reasons for the increased contaminant content as this is an activity that produces contaminants that can then settle into the soil.39,40Cell viability
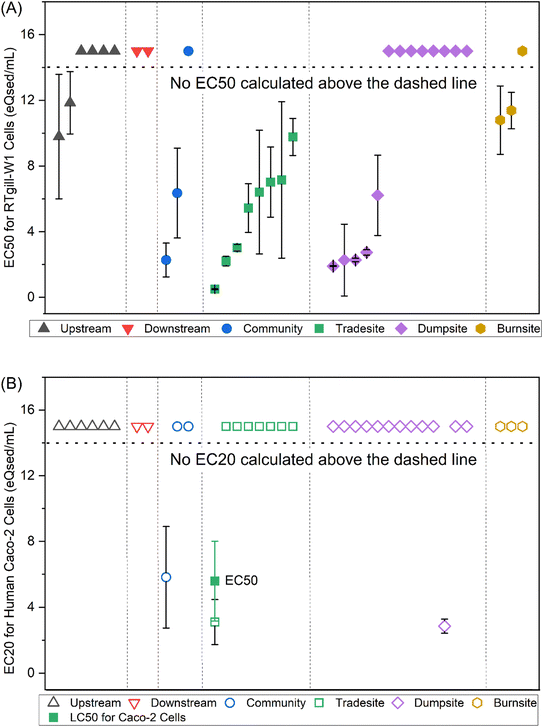
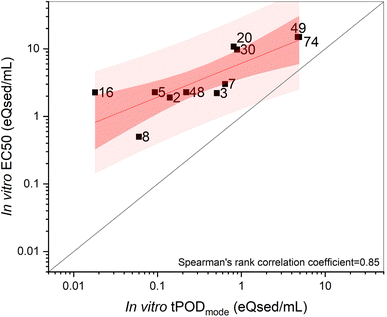
The cell viability results for both cell lines are presented in ESI Table ST8.† For the RTgill-W1 cells, the cell viability in DMSO solvent controls compared to the untreated controls was 99.7% ± 3.4%, and the variability among the no-cell control wells was under 20% for all experiments. The EC50 for the DCA positive control was 47 mg L−1, thus meeting the quality control requirements (28.4–58.9 mg L−1) as specified by the OECD guidelines for Test No. 249 (ref. 11) and similar to our previous study14 (43.6 mg L−1). For the Caco-2 cells, the cell viability in DMSO solvent controls compared to the untreated controls was 96.6% ± 7.7%, the variability among the no-cell control wells was under 20% for all experiments, and the EC50 for the CuSO4 positive control was 201.6 mg L−1 (ESI Tables ST8 and ST9†).RTgill-W1 cells were more sensitive to the e-waste soil extracts than the Caco-2 cells (Fig. 1). For the RTgill-W1 cells, 17 of the 35 e-waste soil extracts were cytotoxic enough to derive EC50s (Table 1). Trade site #8 was the most cytotoxic site with an EC50 of 0.5 eQsed per mL. Trade sites were also the most cytotoxic site type with EC50s derived for all eight sites. Dump sites were the next most cytotoxic with EC50s derived for seven of the 13 sites. Among the burn site, community, and upstream sites, EC50s were derived for isolated sites while neither of the two downstream sites were cytotoxic. Unlike the RTgill-W1 cells, for the Caco-2 cells, only one site (trade site #8) was cytotoxic with an EC50 of 5.59 eQsed per mL (Table 1). There are relatively few studies comparing cytotoxic responses across these two cell lines. One study concerning magnetite and palladium/magnetite nano-catalysts found minor exposure-related decreases in metabolic activity in both cell lines after 1 h of exposure, though following three days of exposure the changes disappeared in the RTgill cells but were sustained in the Caco-2 cells.41 Another study concerning parabens, including methylparaben, ethylparaben and propylparaben, and their chlorinated by-products found that many compounds were cytotoxic to RTgill cells but not to Caco-2 cells in terms of absolute responses (cytotoxic versus non cytotoxic) and relative responses (lower EC50s in RTgill cells).42 While not overly conclusive, these findings are generally similar to what we observed here.
RNA sequencing
The raw reads per sample ranged from 0.5–14 million for RTgill cells and 0.2–26 million for Caco-2 cells. For RTgill cells, 50–78% were mapped in pairs of which 21–49% were mapped to genes for downstream analysis. For Caco-2 cells, 76–99% were mapped in pairs of which 53–98% were mapped to genes. In total, 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 and 61
260 and 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 552 features were annotated for RTgill-W1 and Caco-2 cells, respectively. Further details on the sequencing run are provided in ESI Tables ST2 and ST10.†
552 features were annotated for RTgill-W1 and Caco-2 cells, respectively. Further details on the sequencing run are provided in ESI Tables ST2 and ST10.†
Overall, we noticed some differences in sequencing metrics between the RTgill-W1 and Caco-2 cells. For example, the percentage of reads mapped to the rainbow trout and the human genome were lower for the RTgill cells than the Caco-2 cells, respectively. Of the reads that mapped to the corresponding genomes, the percentage that mapped to exons and protein coding genes were lower for the RTgill compared to the Caco-2 cells. Further, the percentage of reads mapping to ribosomal RNA (rRNA) were much higher in the RTgill (∼90%) than the Caco-2 cells (∼5%). The rainbow trout genome is known to be less annotated than the human genome which might explain the lower percentage of reads mapping for the RTgill cells. Additionally, in traditional RNA sequencing, RNA is isolated from the starting biological material and is depleted of rRNA. In this protocol, the library preparation was conducted directly on lysed cells without RNA isolation or rRNA depletion but using an oligo-dT primer to selectively bind to the poly-A tail. While the oligo-dT binding was successful in eliminating a large percentage of the rRNA in Caco-2 libraries, this was not the case in the RTgill cells. Further, the percentage of reads mapping to introns and intergenic regions were higher in the Caco-2 cells.
Transcriptomic responses
For the Caco-2 cells, although cytotoxic responses were not measured following exposure to e-waste site extracts, there were many transcriptomic effects observed. Differential expression analysis resulted in 226 to 5925 DEGs per sample (ESI Table ST11†). Exposure to extracts from site #s 7 and 16 resulted in the most and least number of DEGs. Site #8, which was the most cytotoxic, resulted in 3458 DEGs. Some of the identified DEGs were related to processes such as immune and stress response, inhibition of cell proliferation, cell fate, xenobiotic metabolism.
For the Caco-2 cells, in many cases we observed more than 2000 DEGs. Thus, for curve fitting we employed more stringent statistical analysis (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC and FDR adjust p-value threshold of 1.5 and 0.01 to obtain at most 2000 DEGs), which resulted in gene BMDs calculated for 2–26% of DEGs (Table 1). The lowest tPODmode calculated was for trade site #8 (0.24 eQsed per mL), which exhibited the strongest dose-dependent effect with 217 gene BMDs. Unlike the RTgill-W1 tPODs, the Caco-2 derived tPODs were within 5-fold of each other except for site #s 16, 30 and 74 for which no tPOD could be calculated due to lack of DEGs or genes with BMDs.
2FC and FDR adjust p-value threshold of 1.5 and 0.01 to obtain at most 2000 DEGs), which resulted in gene BMDs calculated for 2–26% of DEGs (Table 1). The lowest tPODmode calculated was for trade site #8 (0.24 eQsed per mL), which exhibited the strongest dose-dependent effect with 217 gene BMDs. Unlike the RTgill-W1 tPODs, the Caco-2 derived tPODs were within 5-fold of each other except for site #s 16, 30 and 74 for which no tPOD could be calculated due to lack of DEGs or genes with BMDs.
To further explore this dataset, we performed Gene Ontology (GO) annotation analysis based on the genes with BMDs (ESI Table ST12†). Due to the lack of a reference in the gene ontology database for RTgill-W1 cells, we utilized the zebrafish database as an alternative. While there were no significantly enriched GO terms identified for the RTgill cells, we observed several enriched terms for Caco-2 cells exposed to site #s 7 (2 terms), 20 (3 terms), 3 (22 terms), and 8 (158 terms) (ESI Table ST12†). These included regulation of hormone levels and fibrinogen complex in site #7, and system development, anatomical structure development and multicellular organism development in site #20. Exposure to site #3 resulted in 10 enriched biological processes, 1 molecular function and 10 cellular component terms including regulation of hormone levels, response to stimulus, and several extracellular component terms. Exposure to site #8 resulted in the highest number of gene ontology terms – 121 biological processes, 27 cellular components and 10 molecular functions including several related to response to metal ions, response to stress, detoxification and binding. None of the GO terms were commonly enriched across all sites. However, seven biological processes and nine cellular component terms were commonly identified between site #s 3 and 8. These included response to stimulus, and metabolic processes related to organic substances with a specific emphasis on organic acids, and all detected extracellular components.
A few studies have examined the transcriptomic effects of exposure to e-waste related contaminants. One studying effects of key e-waste contaminants including Pb and decabromodiphenyl ether (BDE) in zebrafish larvae (Danio rerio) reported several enriched GO terms that were also found in our study including protein binding, ion binding and intracellular membrane-bound organelle, in addition to many development and metabolic processes that were similar though not identical.44 Another, examining effects of in situ exposure to e-waste contaminants in climbing perch (Anabus testudineus), a native fish species, also reported several enriched terms that were found here including protein folding, regulation of apoptotic process, response to unfolded protein and regulation of transcription from RNA polymerase II promoter.45 More recently, a study examining the potential of a bacteria (Pseudomonas plecoglossicida) to biodegrade BDEs reported several enriched GO terms including response to toxic substance, ion binding, plasma membrane and cytoplasm.46 While these studies differed from ours in many ways (e.g., different species, in vivo exposures), the perturbed biological functions (i.e., main GO terms being hormone metabolic process and regulation, amino acid metabolism such as alanine, aspartate and glutamate metabolism) following exposure to e-waste materials were somewhat consistent.
Application of effect-based methods at contaminated sites
The ultimate goal of this study was to assess the implementation of the tPOD concept into an in vitro EBM study involving human and fish cell lines situated at a contaminated site. Accordingly, we need to consider how the tPODs data compare to the cytotoxicity measures, and also how the tPODs data compare to available measures of select priority chemicals of concern in these same samples.First, the in vitro tPOD data correlated well with corresponding EC50 values of cytotoxicity. We note that this was only possible for the trout cells given the lack of cytotoxicity results in the human cells (i.e., only one case). Across all cases of cytotoxicity, we noted that the corresponding tPOD value was lower in concentration than the EC50 value. In studies of chemical exposures versus exposures to complex mixture extracts, this has also been observed for RTgill-W1 (ref. 14) and Caco-2 cells.15 Such is not surprising given that molecular responses indicate adaptive cellular responses and precede adverse apical effects, and is thus a principle that underlies growing interest in the tPODs approach.7 We also note a few cases in which cytotoxicity was not measured though tPODs were calculated, which is similar to what has been reported in a previous study where exposure to certain silicon dioxide and silver nanoparticles caused no cytotoxicity in Caco-2 cells, however elicited a measurable transcriptomic response.15 This reinforces the notion that quantitative tPODs may be derived in the absence of adverse measures.
In addition to yielding quantitative data that may be used in a risk assessment context, the tPODs approach can also yield helpful qualitative information to elucidate a sample's mechanisms of action. If sufficient number of gene BMDs exist in a biological pathway, then quantitative pathway-based BMDs may be derived (as found here) which provide even deeper insights. For example, our results show that biological pathways such as mineral absorption and GO terms including responses to inorganic metal ions were activated, specifically, cellular responses to copper, zinc, and cadmium ions. These three metals were detected at site #8 at concentrations exceeding standard values for agricultural soil and sediment from CCME21,29 and US EPA.22 Other identified GO terms included those associated with metabolic processes involving (di)carboxylic acids, oxoacids, and organic acids. One potential source of organic acids may be the diverse types of phthalate esters present in the plastic compounds measured. Phthalate esters in soil, via processes such as biodegradation, photolysis, and/or hydrolysis47 may be converted to phthalic acid and subsequently hydrogenated to carboxylic acids.48
Another consideration is whether this approach (in vitro tPOD EBM) yields similar conclusions to the typical chemistry-based method focused on select priority chemicals when assessing environmental samples. The chemistry results show that trade sites, followed by dump sites, and certain isolated sites from the upstream and community sites showed high levels of some contaminants, with some of these above a guideline value. Among the shortlisted sites assessed for transcriptomics, we observed that the most cytotoxic and contaminated sites generally resulted in the most sensitive tPODs. Based on the chemistry data, the only site with a HI above 1 was site #8 for human references, and the highest RI was also site #8 based on aquatic references. The in vitro results also flagged site #8. In Caco-2 cells, the extract from this site was the only one to return an EC50 value and also had the lowest tPOD value. Further, for RTgill cells, the lowest EC50 was observed from site #8. A linear regression and non-parametric correlation were used to capture the potential relationships between the composite scores and the in vitro results (ESI Fig. SF8†). All three regressions showed reasonable negative correlation, suggesting higher composite scores correspond to lower benchmark values. The only statistically significant correlation (Spearman, p < 0.05) was between the composite score and the EC50. The R value was −0.72, suggesting a relative strong correlation.49 While there seems to be some concordance between the in vitro results and the chemistry data, we only studied a few e-waste sites with limited numbers of targeted chemicals measured. Thus, more work is required in this area with larger sample sizes and in-depth chemical analysis.
When comparing the two approaches, there are some advantages of the EBM approach to note. While the two approaches may be similarly priced (∼$1000–2000 USD per sample), the in vitro work covering cytotoxicity and transcriptomics took about 4–6 weeks whereas the chemical analysis took longer. The in vitro method also yielded much more biological data. However, despite the promise of a more resource efficient and informative method, some challenges remain in comparing the methods (which also represent some limitations of our work). First, the chemical analysis focused on a select number of organic and inorganic substances. E-Waste sites are notoriously contaminated with many more chemicals. Therefore, trying to rank and prioritize the study sites using such (incomplete) information poses challenges. Second, reducing multiple chemical data into a single composite score, while necessary, is overly simplistic. Third, reference concentrations were challenged. Typically, reference concentrations in water are used for fish and other aquatic species. However, these values may not be directly applicable to this study which focused on soil extract. Given that relevant information is limited, human reference doses were utilized for estimation purposes. Fourth, the transcriptomic analysis went well for the human cell line but was more challenging for the trout cell given that its genome is less resolved and annotated. In terms of the library preparation, we attempted to skip the rRNA depletion step by selectively binding to the poly-A tail for 3′ sequencing. While we were able to obtain meaningful information, a substantial percentage of the reads were lost to rRNA, introns or intergenic regions, thus reducing overall efficiency of the workflow. Hence, for future studies, the workflow could be optimized for higher efficiency by including the rRNA depletion step.
Beyond the limitations noted in the previous paragraph, there are some additional methodological ones that merit discussion. First, the metals analyses did not account for speciation nor were we able examine the bioavailability of compounds from our extracts. This makes it challenging to understand potential exposures to humans or fish, and so we focus the work on differences across the sites. Further, while we conducted these studies using relevant cell lines to gauge exposures in fish and humans, these represented single modes of exposure. The inclusion of different cell lines such as epidermal, hepatic, and neuronal may help increase understanding of other exposure routes and also potential mechanisms of action. Finally, our studies focused on single cell lines grown in monolayers which may represent an oversimplification of complex biological responses, and so future studies could consider more sophisticated models such as co-cultures or spheroids.
Concluding remarks
In this study, we used an EBM approach to study the cytotoxic and transcriptomic effects of soil extracts from e-waste contaminated sites on the rainbow trout RTgill-W1 and human Caco-2 cell lines. In general, the molecular responses were not only in line with the cytotoxic results observed but also with contaminant levels measured at the selected sites. We demonstrated the potential of using such methods on multiple levels – as screening platforms to prioritize environmental samples, and to gain better understanding of the sub-lethal effects of complex mixtures. However, some challenges still remain (as detailed near the end of the Discussion) and thus more case studies like this are needed to assess the performance of cell- and transcriptomics-based methods with a diverse range of substances to build more confidence on their use.Data availability
Sequencing data for this article are available at NCBI GEO under accession number GSE279464 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE279464). The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by West Africa-Michigan CHARTER in GEO-Health with United States National Institutes of Health/Fogarty International Center (paired grant number: 1U2RTW010110-01/5U01TW010101), Canada's International Development Research Center (grant number: 108121-001), and the NSERC Discovery Grants program award (RGPIN-2019-04842). The authors would like to acknowledge members of the GEOHealth team for sample collection. We also thank Jenny Eng and Sophie Emberley-Korkmaz for technical support, and Drs Markus Brinkmann and Lucy Lee for support with the cell cultures.References
- B. I. Escher, H. M. Stapleton and E. L. Schymanski, Tracking complex mixtures of chemicals in our changing environment, Science, 2020, 367(6476), 388–392 CrossRef CAS PubMed.
- S. K. Bopp, A. Kienzler, A.-N. Richarz, S. C. van der Linden, A. Paini and N. Parissis, et al., Regulatory assessment and risk management of chemical mixtures: challenges and ways forward, Crit. Rev. Toxicol., 2019, 49(2), 174–189 CrossRef PubMed.
- A. Kortenkamp and M. Faust, Regulate to reduce chemical mixture risk, Science, 2018, 361(6399), 224–226 CrossRef CAS PubMed.
- W. Brack, S. A. Aissa, T. Backhaus, V. Dulio, B. I. Escher and M. Faust, et al., Effect-based methods are key. The European Collaborative Project SOLUTIONS recommends integrating effect-based methods for diagnosis and monitoring of water quality, Environ. Sci. Eur., 2019, 31(1), 1–6 CrossRef CAS.
- A.-S. Wernersson, M. Carere, C. Maggi, P. Tusil, P. Soldan and A. James, et al., The European technical report on aquatic effect-based monitoring tools under the water framework directive, Environ. Sci. Eur., 2015, 27, 1–11 CrossRef CAS.
- J. A. Head, J. D. Ewald and N. Basu, The DIKW of Transcriptomics in Ecotoxicology: Extracting Information, Knowledge, and Wisdom From Big Data, Environ. Toxicol. Chem., 2024, 1–7, DOI:10.1002/etc.5954.
- K. J. Johnson, S. S. Auerbach, T. Stevens, T. S. Barton-Maclaren, E. Costa and R. A. Currie, et al., A transformative vision for an omics-based regulatory chemical testing paradigm, Toxicol. Sci., 2022, 190(2), 127–132 CrossRef CAS.
- M. Heacock, B. Trottier, S. Adhikary, K. A. Asante, N. Basu and M.-N. Brune, et al., Prevention-intervention strategies to reduce exposure to e-waste, Rev. Environ. Health, 2018, 33(2), 219–228 Search PubMed.
- L. Kwarteng, A. M. Devasurendra, Z. Laskaris, J. Arko-Mensah, A. A. A. Nti and S. Takyi, et al., Occupational exposures to particulate matter and PM2. 5-associated polycyclic aromatic hydrocarbons at the Agbogbloshie waste recycling site in Ghana, Environ. Int., 2022, 158, 106971 CrossRef CAS.
- S. A. Takyi, N. Basu, J. Arko-Mensah, D. Dwomoh, K. G. Houessionon and J. N. Fobil, Biomonitoring of metals in blood and urine of electronic waste (E-waste) recyclers at Agbogbloshie, Ghana, Chemosphere, 2021, 280, 130677 CrossRef CAS.
- OECD, Test No. 249: Fish Cell Line Acute Toxicity – the RTgill-W1 Cell Line Assay, Organization for Economic Co-operation and Development, 2021 Search PubMed.
- Y. Sambuy, I. De Angelis, G. Ranaldi, M. L. Scarino, A. Stammati and F. Zucco, The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics, Cell Biol. Toxicol., 2005, 21, 1–26 CrossRef CAS.
- C. Moeckel, K. Breivik, T. H. Nøst, A. Sankoh, K. C. Jones and A. Sweetman, Soil pollution at a major West African E-waste recycling site: Contamination pathways and implications for potential mitigation strategies, Environ. Int., 2020, 137, 105563 CrossRef CAS.
- K. Mittal, J. Ewald and N. Basu, Transcriptomic points of departure calculated from rainbow trout gill, liver, and gut cell lines exposed to methylmercury and fluoxetine, Environ. Toxicol. Chem., 2022, 41(8), 1982–1992 CrossRef CAS PubMed.
- K. Xu, K. Mittal, J. Ewald, S. Rulli, J. L. Jakubowski and S. George, et al., Transcriptomic points of departure calculated from human intestinal cells exposed to dietary nanoparticles, Food Chem. Toxicol., 2022, 170, 113501 CrossRef CAS.
- ATCC[Internet], RTgill-W1 handling information, available from: https://www.atcc.org/products/crl-2523 Search PubMed.
- ATCC[Internet], Caco-2 handling information, available from: https://www.atcc.org/products/htb-37 Search PubMed.
- J. Zheng, K. Mittal, J. N. Fobil, N. Basu and S. Bayen, Simultaneous targeted and non-targeted analysis of plastic-related contaminants in e-waste impacted soil in Agbogbloshie, Ghana, Sci. Total Environ., 2024, 917, 170219 CrossRef CAS PubMed.
- L. Liu, N. M. Aljathelah, H. Hassan, B. W. Giraldes, A. Leitão and S. Bayen, Targeted and suspect screening of contaminants in coastal water and sediment samples in Qatar, Sci. Total Environ., 2021, 774, 145043 CrossRef CAS.
- B. Nfor, P. B. A. Fai, S. A. Tamungang, J. N. Fobil and N. Basu, Soil contamination and bioaccumulation of heavy metals by a tropical earthworm species (Alma nilotica) at informal E-waste recycling sites in Douala, Cameroon, Environ. Toxicol. Chem., 2022, 41(2), 356–368 CrossRef CAS PubMed.
- CCME, Canadian Environmental Quality Guidelines (CEQGs), Canadian Council of Ministers of the Environment, 2018 Search PubMed.
- EPA, Regional Screening Levels (RSLs)–Generic Tables, United States Environmental Protection Agency, 2023 Search PubMed.
- ATSDR, The ATSDR 2022 Substance Priority List, Agency for Toxic Substances and Disease Registry, 2022 Search PubMed.
- OECD EUJRC, Handbook on Constructing Composite Indicators: Methodology and User Guide, Organization for Economic Co-operation and Development, 2008 Search PubMed.
- ATSDR, Guidance Manual for the Assessment of Joint Toxic Action of Chemical mixtures. Atlanta: US Department of Health and Human ServicesPublic Health Service, Agency for Toxic Substances and Disease Registry, 2004 Search PubMed.
- EPA, Exposure Factors Handbook 2011, United States Environmental Protection Agency, 2011 Search PubMed.
- N. Healey, H. Jones-Otazo, M. Walker and A. Knafla, Toxicological Review and Recommended Toxicological Reference Values for Lead Exposure in Canada, Report Prepared for Health Canada, Healthy Environments and Consumer Safety Branch, Environmental Health Bureau, Ottawa, 2010 Search PubMed.
- EPA, Dibutyl phthalate, Integrated Risk Information System, United States Environmental Protection Agency, 1987 Search PubMed.
- CCME, Sediment Quality Guidelines for the Protection of Aquatic Life, Canadian Council of Ministers of the Environment, 1998 Search PubMed.
- Washinton State, Sediment Management Standards: Chapter 173-204 WAC, Department of Ecology, 2013 Search PubMed.
- EFSA, Comparison of NOEC values to EC10/EC20 values, including confidence intervals, in aquatic and terrestrial ecotoxicological risk assessment, EFSA Supporting Publications, 2015, vol. 12(12), p. 906E Search PubMed.
- QIAGEN[Internet], QIAseq UPXome RNA library kit handbook, 2024, available from https://www.qiagen.com/us/resources/resourcedetail?id=5e6e5735-1f9b-4ef6-9074-4652ed4d715c%26lang=en Search PubMed.
- C. B. F. Ritz, J. C. Streibig and D. Gerhard, Dose-Response Analysis Using R, PLoS One, 2015, 10, e0146021 CrossRef PubMed.
- J. Ewald, G. Zhou, Y. Lu and J. Xia, Using ExpressAnalyst for comprehensive gene expression analysis in model and non-model organisms, Curr. Protoc., 2023, 3(11), e922 CrossRef CAS PubMed.
- M. Ackah, Soil elemental concentrations, geoaccumulation index, non-carcinogenic and carcinogenic risks in functional areas of an informal e-waste recycling area in Accra, Ghana, Chemosphere, 2019, 235, 908–917 CrossRef CAS PubMed.
- P. Cao, T. Fujimori, A. Juhasz, M. Takaoka and K. Oshita, Bioaccessibility and human health risk assessment of metal (loid) s in soil from an e-waste open burning site in Agbogbloshie, Accra, Ghana, Chemosphere, 2020, 240, 124909 CrossRef CAS.
- M. Dodd, L. O. Amponsah, S. Grundy and G. Darko, Human health risk associated with metal exposure at Agbogbloshie e-waste site and the surrounding neighbourhood in Accra, Ghana, Environ. Geochem. Health, 2023, 45(7), 4515–4531 CrossRef CAS.
- US EPA, Framework for Cumulative Risk Assessment, United States Environmental Protection Agency, 2003 Search PubMed.
- D.-Y. Huang, H.-Q. Zhao, C.-P. Liu and C.-X. Sun, Characteristics, sources, and transport of tetrabromobisphenol A and bisphenol A in soils from a typical e-waste recycling area in South China, Environ. Sci. Pollut. Res., 2014, 21, 5818–5826 CrossRef CAS.
- T. T. Ma, P. Christie, Y. M. Luo and Y. Teng, Phthalate esters contamination in soil and plants on agricultural land near an electronic waste recycling site, Environ. Geochem. Health, 2013, 35(4), 465–476 CrossRef CAS.
- H. Hildebrand, D. Kühnel, A. Potthoff, K. Mackenzie, A. Springer and K. Schirmer, Evaluating the cytotoxicity of palladium/magnetite nano-catalysts intended for wastewater treatment, Environ. Pollut., 2010, 158(1), 65–73 CrossRef CAS.
- A. L. Ball, M. E. Solan, M. E. Franco and R. Lavado, Comparative cytotoxicity induced by parabens and their halogenated byproducts in human and fish cell lines, Drug Chem. Toxicol., 2023, 46(4), 786–794 CrossRef CAS.
- D. L. Villeneuve, M. Le, M. Hazemi, A. Biales, D. C. Bencic and K. Bush, et al., Pilot testing and optimization of a larval fathead minnow high throughput transcriptomics assay, Curr. Res. Toxicol., 2023, 4, 100099 CrossRef CAS.
- L. Chen, B. Zhu, Y. Guo, T. Xu, J.-S. Lee and P.-Y. Qian, et al., High-throughput transcriptome sequencing reveals the combined effects of key e-waste contaminants, decabromodiphenyl ether (BDE-209) and lead, in zebrafish larvae, Environ. Pollut., 2016, 214, 324–333 CrossRef CAS PubMed.
- W. Zhang, H. Q. Xie, X. Zou, J. Li, L. Xu and Y. Li, et al., The toxic effects of in situ exposure of a native fish species (Anabas testudineus) to electronic waste pollution, Sci. Total Environ., 2019, 690, 1170–1177 CrossRef CAS.
- X. Qi, M. Zhu, Y. Yuan, X. Rong, Z. Dang and H. Yin, Integrated toxicology, metabolomics, and transcriptomics analyses reveal the biodegradation and adaptation mechanisms to BDE-47 in Pseudomonas plecoglossicida, Chem. Eng. J., 2023, 454, 140412 CrossRef CAS.
- Y.-C. Chung and C.-Y. Chen, Degradation of Di-(2-ethylhexyl) phthalate (DEHP) by TiO2 photocatalysis, Water, Air, Soil Pollut., 2009, 200, 191–198 CrossRef CAS.
- J. Schwarzbauer, S. Heim, B. Krooss and R. Littke, Analysis of undisturbed layers of a waste deposit landfill–Insights into the transformation and transport of organic contaminants, Org. Geochem., 2006, 37(12), 2026–2045 CrossRef CAS.
- J. Fowler, L. Cohen and P. Jarvis, Practical Statistics for Field Biology, John Wiley & Sons, 2013 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4va00178h |
| This journal is © The Royal Society of Chemistry 2024 |