Open Access Article
Open Access ArticleSite-specific photo-crosslinking in a double crossover DNA tile facilitated by squaraine dye aggregates: advancing thermally stable and uniform DNA nanostructures†
Shibani
Basu
 a,
Simon K.
Roy
a,
Simon K.
Roy
 a,
Mandeep
Sharma
a,
Mandeep
Sharma
 a,
German
Barcenas
a,
German
Barcenas
 a,
Bernard
Yurke
a,
Bernard
Yurke
 ab,
William B.
Knowlton
ab,
William B.
Knowlton
 ab and
Jeunghoon
Lee
ab and
Jeunghoon
Lee
 *ac
*ac
aMicron School of Materials Science & Engineering, Boise State University, Boise, Idaho 83725, USA. E-mail: jeunghoonlee@boisestate.edu
bDepartment of Electrical & Computer Engineering, Boise State University, Boise, Idaho 83725, USA
cDepartment of Chemistry and Biochemistry, Boise State University, Boise, Idaho 83725, USA
First published on 21st February 2025
Abstract
We investigated the role of dichloro-squaraine (SQ) dye aggregates in facilitating thymine–thymine interstrand photo-crosslinking within double crossover (DX) tiles, to develop thermally stable and structurally uniform two-dimensional (2D) DNA-based nanostructures. By strategically incorporating SQ modified thymine pairs, we enabled site-selective [2 + 2] photocycloaddition under 310 nm UV light. Strong dye–dye interactions, particularly through the formation of aggregates, facilitated covalent bond formation between proximal thymines. To evaluate the impact of dye aggregation on crosslinking efficiency, ten DX tile variants with varying SQ-modified thymine positions were tested. Our results demonstrated that SQ dye aggregates significantly enhanced crosslinking, driven by precise SQ-modified thymine dimer placement within the DNA tiles. Analytical techniques, including denaturing PAGE and UV-visible spectroscopy, validated successful crosslinking in DNA tiles with multiple SQ-modified thymine pairs. This non-phototoxic method offers a potential route for creating thermally stable, homogeneous higher-order DNA–dye assemblies with potential applications in photoactive and exciton-based fields such as optoelectronics, nanoscale computing, and quantum computing. The insights from this study establish a foundation for further exploration of advanced DNA–dye systems, enabling the design of next-generation DNA nanostructures with enhanced functional properties.
1. Introduction
DNA, a versatile and programmable nanomaterial, offers precise control for the construction of two- and three-dimensional structures through Watson–Crick base pairing.1 This capability positions DNA as a promising tool in nanotechnology, molecular biology, and materials science, where it serves as a scaffold for nanostructures, a template for biochemical reactions, and a functional material for smart, responsive systems.2–4 Moreover, the assembly of DNA-based nanostructures, leveraging self-assembly principles with programmable nucleobase sequences and chemical modifications, has become a powerful method for organizing nanoscale materials.5,6Recent advances in programmable self-assembly have introduced strategies such as covalently attaching dye molecules to DNA, resulting in excitonically delocalized molecular dye aggregates.7–9 The close arrangement of dyes that contain chromophores facilitates delocalization of molecular excitons (Frenkel excitons), enabling coherent energy transfer across the dye molecules.10–13 DNA tiles, as stable, and modular building blocks,14–17 are a promising platform for integrating those dye aggregates, enabling precise control in programmable DNA–dye systems. Their robust structures can be utilized for building devices for applications across nanoscale computing, quantum computing, organic optoelectronics, photonics, and light-harvesting technologies by providing a predictable framework for dye placement, which is essential for coherent energy transfer and excitonic coupling in advanced nanoscale devices.18–25
The ability of DNA to form well-defined higher order structures such as DNA tiles, bricks, origamis, and lattices is central to its use in nanotechnology.26–29 This ability relies on the precise hybridization of complementary base pairs, specifically adenine–thymine (A–T) and guanine–cytosine (G–C) pairs, connected through hydrogen bonds.30,31 However, the hydrogen bonds can be unstable under various conditions, such as temperature fluctuations, ionic strength, and sequence composition, leading to dynamic behavior known as “DNA breathing”.32–35 DNA breathing involves the transient dissociation and reassociation of the DNA strands, leading to structural instability and heterogeneity with distinct sub-populations of DNA-based assemblies.36,37 These sub-populations include fully paired, partially denatured, and fully denatured states and intermediary forms that fluctuate between relative orientations of dye aggregate configurations. This structural variability impacts stability, binding sites, and interactions with other molecules, especially when used as templates for assembling functional molecules like dyes.38–41 This dynamic behavior is particularly detrimental to higher-order DNA–dye systems, where precise control over the structure and stability is crucial for applications involving energy transfer processes like exciton-based devices.42–45 The properties of dye aggregates are highly sensitive to the spatial arrangement and their local environment, making a comprehensive understanding of dye–dye and dye–DNA interactions essential for advancing the design and functionality of complex DNA–dye architectures.
To address these challenges, various strategies have been explored to enhance the stability of DNA nanostructures.46–52 One promising approach involves using dye aggregates to facilitate covalent crosslinking between specific nucleotides within the DNA structure, thereby “locking” the DNA in a desired conformation.53,54 This stabilization preserves its structure for use in nanotechnology or molecular applications requiring precise manipulation of DNA structures where maintaining a specific shape is crucial. Traditional crosslinking methods often introduce phototoxic effects or molecular damage to DNA or surrounding cellular components, caused by light exposure, particularly during high-intensity or light-activated processes.55–57 However, recent advances in photochemistry, particularly the use of photoactive dye aggregates, offer opportunities for more controlled and site-specific crosslinking approaches.53,54,58,59 Squaraine dyes,60–64 known for their strong absorption in the visible spectrum and ability to form stable dimers and aggregates with exciton hopping parameters exceeding 100 meV,65–71 have shown to facilitate thymine–thymine crosslinking via [2 + 2] photocycloaddition. By precisely positioning near thymine pair, SQ dimers trigger covalent bond formation between interstrand thymine bases upon light activation by bringing the bases in proximity.53,54
In this context, using site-selective thymine–thymine interstrand photo-crosslinking is an effective strategy for enhancing the thermal stability and structural uniformity of higher order DNA nanostructures.72,73 The [2 + 2] photocycloaddition reaction between thymine bases, resulting in the formation of cyclobutene pyrimidine dimer (CPD), is a well-known photoreaction that can be precisely controlled through UV irradiation.74–80 The formation of dye aggregates plays a critical role in ensuring effective and controlled thymine–thymine photo-crosslinking. However, the efficiency and specificity of this reaction can be significantly influenced by the proximity of the SQ-modified thymine bases.53 Aggregates enhance stability and uniformity by promoting strong dye–dye interactions, which bring thyme pairs into close proximity, facilitating efficient [2 + 2] photocycloaddition reaction.54 Moreover, these dye aggregates can be used as a sensitive probe to monitor crosslinking efficiency and structural homogeneity of the DNA–dye constructs.
Additionally, the SQ-mediated crosslinking offers a non-phototoxic alternative to traditional UV-mediated methods.53,54 Phototoxicity in DNA occurs when light exposure, especially UV light, generates reactive species that cause breakage of DNA strands, and oxidative damage, compromising DNA integrity and potentially reducing crosslinking efficiency.81–86 Specifically, DNA Holliday Junction (HJ)-templated dimers of SQ-attached thymines undergo efficient photo-crosslinking under 310 nm UV light without inducing phototoxicity, thereby mitigating the adverse effects typically associated with conventional UV-mediated crosslinking.53,54 The resulting covalent bonds can enhance the thermal stability and structural homogeneity of the DNA–dye constructs, making them more suitable for various applications, including advanced photoactive materials and exciton-based devices.
In our study, we addressed the pivotal challenge of developing a reproducible technique to enhance thermal stability and reduce structural heterogeneity of DNA–dye constructs via interstrand thymine–thymine photo-crosslinking. We incorporated SQ dye aggregates into 2D double crossover (DX) DNA tiles to achieve site-specific interstrand thymine–thymine photo-crosslinking, enhancing both thermal stability and structural homogeneity of higher order DNA–dye assemblies. Furthermore, this photo-crosslinking method allowed us to obtain a novel five-dimer tile configuration using denaturing PAGE, an approach that would have been challenging to achieve without the enhanced structural stability provided by photo-crosslinking. A range of analytical methods, including denaturing PAGE and UV-visible spectroscopy, were used to evaluate the crosslinking efficiency and homogeneity of the DNA–dye assemblies. Our findings indicate that the controlled placement of SQ aggregates significantly boosts crosslinking yield and stability, establishing a reproducible approach for creating thermally stable, homogenous DNA–dye constructs. This method presents an efficient, scalable approach to constructing thermally stable DNA nanostructures, setting the foundation for their application in photoactive materials, nanoscale computing, and quantum information systems.
2. Materials and methods
2.1. Sample preparation
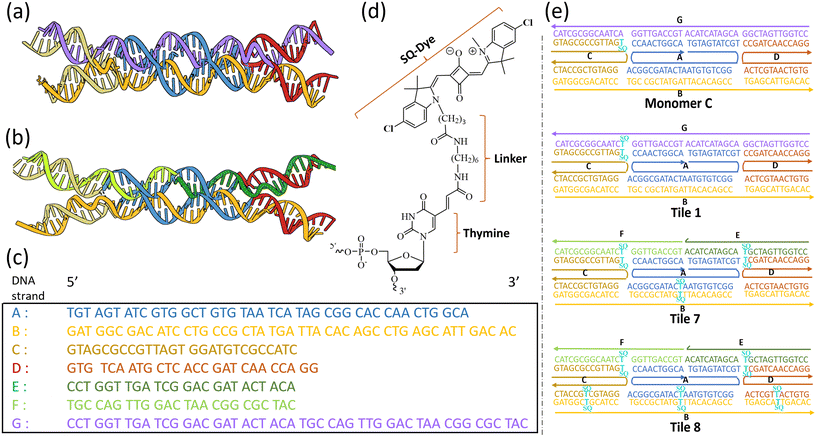
The DNA oligomers, functionalized with indolenine dyes using a single flexible linker, were purified through dual high-performance liquid chromatography and acquired from integrated DNA technology, Inc. (Coralville). Dichloro-squaraine (SQ) dyes were synthesized and supplied by SETA Biomedicals (Urbana, IL).45,69 Unfunctionalized DNA oligomers, purified through desalting, were obtained from Integrated DNA Technology, Inc. All DNA oligomers were rehydrated to prepare approximately 100 μM stock solution in ultrapure water (Barnstead Nanopure, Thermo Scientific). The DNA sample concentrations were determined spectroscopically using calculated extinction coefficients on a NanoDrop One Microvolume UV-Vis system (Thermo Scientific). DNA tiles were hybridized to a final DNA concentration of 1.5 μM by adding equimolar amounts of complementary oligomers in 1× Tris-borate-EDTA (TBE), pH of 10× TBE purchased from Thermo Scientific: 8.3, 15 mM MgCl2 buffer solution. The DNA samples were annealed for 4 minutes at 94 °C, followed by cooling ramps of 1.0 °C min−1 from 94 °C to 64 °C and 10 °C min−1 from 64 °C to room temperature (approximately 22 °C) in a Mastercycler Nexus PCR cycler (Eppendorf).Ten DNA tile variants were synthesized for the study, each differing in the relative position of modified thymine labeled with squaraine dye. We synthesized monomers, in which only one SQ-modified thymine was attached to the DNA (Fig. S2†). Among all the monomers, C (SQ dye was attached to the C strand) and D (SQ dye was attached to the C strand) were used as control samples, as they are the most suitable and available for the study. One set of dimer samples was designed with a continuous G strand, such as Tile 1 and Tile 2 (each with a single pair of SQ-modified thymines), Tile 3 (three pairs of SQ-modified thymines), and Tile 4 (five pairs of SQ-modified thymines). Additionally, another set of dimer samples was designed where the strand G is split into two strands (E, and F): Tile 5 and Tile 6 (each with a single pair of SQ-modified thymines), Tile 7 (three pairs of SQ-modified thymines), and Tile 8 (five pairs of SQ-modified thymines). Five non-tile variants were synthesized and used as controls for the study. Unmodified and SQ-modified single stranded DNA (ssDNA): comprising a single chain of nucleotides, SQ modified double stranded DNA (dsDNA), three stranded DNA, and four stranded DNA that forms incomplete tiles.
2.2. UV-crosslinking
DNA tile samples (2 μM, 100 μL) in 1× TBE, 15 mM MgCl2 were exposed to UV-LED lamp (310 nm; spot size, 1 mm × 1 mm) for 60 minutes at room temperature to monitor photoreaction progress. This was conducted using a T-Cube LED driver (LEDD1B; THORLABS) equipped with a high-power LED light. The UV-LED light's power was measured with a standard photodiode power sensor (S120VC; THORLABS), covering a range of 200–1100 nm and up to 50 mW, and monitored by a compact power and energy meter console (PM100D; THORLABS). The output current was constantly set at 0.2 A, with a maximum forward voltage of 12 V, producing an irradiation power of 0.7 ± 0.04 mW.2.3. Polyacrylamide gel electrophoresis (PAGE)
Denaturing PAGE (12%) was used to measure the progress of photo-crosslink reactions in DNA tile constructs. The gel was prepared in 1× TBE with 20% urea, pre-ran at 600 V and 57 °C for 30 minutes. DNA samples, mixed with a formamide: 100× TBE (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) loading buffer and denatured at 95 °C for 4 minutes, were loaded onto the gel and run at 350 V for 25 minutes at 57 °C. Non-denaturing PAGE was conducted to analyze and compare DNA samples before and after crosslinking. Non-denaturing 12% polyacrylamide gels, 1.5 mm thick, were cast in 1× TBE, 15 mM MgCl2 buffer. DNA constructs were mixed with a loading buffer (20% v/v Ficoll and 20% v/v bromophenol blue, from Sigma-Aldrich). Electrophoresis was performed at a constant voltage of 150 V and a temperature of 17 °C with 1× TBE, 15 mM MgCl2 running buffer. Gels were then washed with ultrapure water, placed on a phosphor plate, and imaged using UV illumination in the Cy5 channel (excitation, 632 nm; emission, 691 nm). Non-SQ samples were stained with SYBR gold and imaged in the Cy2 channel (excitation, 475 nm; emission, 506 nm). Band intensity was quantified using ImageJ 1.53e,87 with background adjustments made by tuning brightness and contrast.
1 v/v) loading buffer and denatured at 95 °C for 4 minutes, were loaded onto the gel and run at 350 V for 25 minutes at 57 °C. Non-denaturing PAGE was conducted to analyze and compare DNA samples before and after crosslinking. Non-denaturing 12% polyacrylamide gels, 1.5 mm thick, were cast in 1× TBE, 15 mM MgCl2 buffer. DNA constructs were mixed with a loading buffer (20% v/v Ficoll and 20% v/v bromophenol blue, from Sigma-Aldrich). Electrophoresis was performed at a constant voltage of 150 V and a temperature of 17 °C with 1× TBE, 15 mM MgCl2 running buffer. Gels were then washed with ultrapure water, placed on a phosphor plate, and imaged using UV illumination in the Cy5 channel (excitation, 632 nm; emission, 691 nm). Non-SQ samples were stained with SYBR gold and imaged in the Cy2 channel (excitation, 475 nm; emission, 506 nm). Band intensity was quantified using ImageJ 1.53e,87 with background adjustments made by tuning brightness and contrast.
2.4. Optical characterization
The steady-state absorption spectra of DNA aggregates were monitored using a Cary 5000 UV-Vis-NIR spectrophotometer (Agilent Technologies) with a dual-beam setup. DNA-templated SQ samples were placed in a quartz cuvette with a 1 cm path length (Starna), and the spectra were recorded over a wavelength range of 230–800 nm. Circular dichroism (CD) measurements were conducted using a JASCO J-1500 spectrometer. For these measurements, DNA-templated SQ samples (100 μL) were placed in a 1 cm path length microcuvette (Jasco), and the CD spectra were recorded in the 200–800 nm wavelength range at a scanning speed of 200 nm min−1.The melting profiles of DNA constructs were recorded using the Cary 5000 UV-Vis-NIR spectrophotometer (Agilent Technologies) equipped with a thermal probe (Agilent Technologies Cary temperature controller). Samples were degassed for 2 minutes at room temperature (22 °C) and then equilibrated at 25 °C for 2 minutes before initiating a temperature ramp of 1 °C from 25 °C to 95 °C.
2.5. Modeling of absorption and circular dichroism spectra
We utilized our in- house KRM model simulation tool, which is based on the theoretical framework developed by Kühn, Renger, and May, to model experimental spectra.88 This tool incorporates dye properties derived from experimental data and employs an extended dipole model within a Holstein like Frenkel Hamiltonian to calculate theoretical absorption and circular dichroism spectra corresponding to specific dye geometries. A stochastic search method was used to iteratively perturb to dye geometry, with each iteration generating spectra that were compared to experimental results. The optimal geometry was selected based on the best fit to the experimental data. A detailed description of the KRM model simulation tool has previously been published.443. Results and discussion
3.1. Molecular design and construct
We designed and synthesized two types of unmodified DNA tiles: one featuring a continuous strand G, and another where strand G is divided into two strands, labeled E and F. The unmodified tiles, composed of single-stranded DNAs (ssDNAs) labeled A, B, C, D, E, F, and G, were designed with carefully selected sequences to ensure precise hybridization and tile formation (Fig. 1a–c) (Fig. S1†). For thymine–thymine (T–T) crosslinking experiments, we designed a series of tile constructs functionalized with dichloro-squaraine dyes (SQ) (Fig. S2 and S3†). The SQ-labeled thymine modifier was attached to the DNA via a C6 dT linker (Fig. 1d). Ten SQ-modified DNA tile variants were designed, each with a unique arrangement and number of SQ-labeled thymines, including monomer C and monomer D. We designed three sets of DX tile samples for crosslinking study: tiles with a single pair of SQ-modified thymines (e.g., Tile 1, Tile 2, Tile 5, and Tile 6), tiles with three pairs (e.g., Tile 3 and Tile 7) of SQ-modified thymines, and tiles with five pairs of SQ-modified thymines (Fig. 1e and S2, S3†).3.2. Formation and structural integrity of DNA tiles
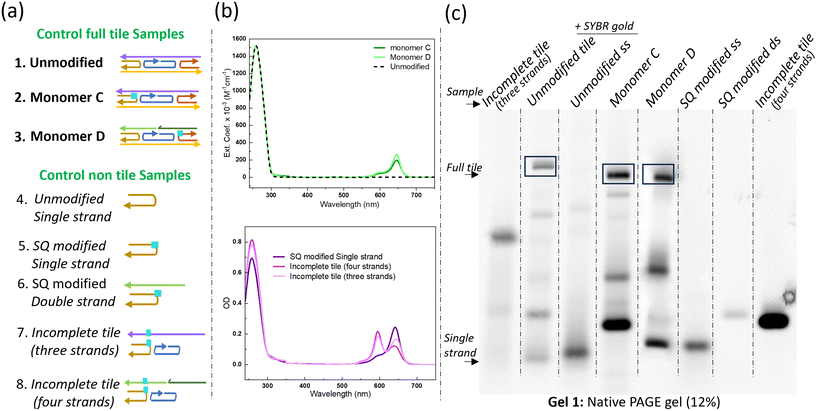
We assessed three types of DNA tiles to serve as controls to investigate successful assembly and the influence of strand continuity on tile formation: the unmodified tile (with a continuous G strand), and SQ-modified monomers C and D. The successful assembly of these tiles was confirmed using absorption spectroscopy and native polyacrylamide gel electrophoresis (PAGE), as illustrated in Fig. 2.Absorption spectroscopy provided an initial validation of tile formation. The absorption peak at 260 nm confirmed the presence of DNA, while the distinct peak profiles observed in the controls (unmodified and modified tiles) suggested successful assembly of properly folded DNA nanostructures, as random or misfolded structures often display broader, less intense peaks due to irregular base stacking or incomplete hybridization. In contrast, the additional peaks observed in the SQ-modified monomers C and D (solid lines) corresponded to absorption from SQ dyes, confirming successful dye incorporation and monomer formation. The monomers exhibited characteristic spectral features of squaraine, including a prominent absorption band with a peak maximum at 645 nm, accompanied by a smaller vibronic shoulder (Fig. 2b and S8†).
Controls of non-tile DNA variants including single, double, three, and four-stranded configurations were designed to form incomplete tiles, further demonstrating how variations in strand composition affect the optical properties compared to organized tile-based assemblies. Fully complete tile structures exhibit enhanced absorption due to their organized and densely interconnected arrangements compared to simpler, non-tile DNA structures. Additionally, in the single-stranded configuration, the monomers displayed the characteristic spectral features of SQ, whereas SQ-dimer formation became apparent in the incomplete tile structures comprising three and four strands (Fig. 2a and b). The native PAGE analysis (Fig. 2c, Gel 1) offered more detailed insights into the structural integrity of the tiles. Fully formed tiles, such as unmodified full tile, exhibited a clear and distinct band, indicative of successful assembly. However, the appearance of additional bands in the unmodified tile lane indicates transient structural intermediates or incomplete assemblies, potentially arising from minor deviations in molar ratios of strand. Similarly, monomeric tiles containing SQ dye tethered to C or D strands (Monomer C, MonomerD) also displayed distinct bands, confirming the complete assembly of these modified structures. In contrast, an incomplete tile with three strands showed a band that migrated further in the gel than the fully formed tiles, suggesting lower molecular weight and incomplete assembly. Control non-tile samples including single strands, double strands, and incomplete tiles containing 4 strands, exhibited less distinct bands and smeared patterns, indicating poor formation and assembly compared to the fully formed tiles. Overall, the data obtained from native PAGE gel analysis underscores the formation of fully formed DNA tiles compared to non-tile or incomplete structures. This analysis provides a fundamental understanding of how different strand configurations and modifications influence DNA tile assembly and stability. Specifically, as the complexity and number of strands increase, such as in multi-stranded tile configurations, the structures become more stable and organized due to stronger base-pairing interactions and stacking between nucleotides. These structural reinforcements enhance the cohesion and rigidity of the DNA tile, making it less prone to degradation. In contrast, simpler non-tile configurations, like double strands or incomplete tiles with three or four strands, lack these stabilizing interactions and exhibit lower structural integrity. Such insights are critical for subsequent crosslinking experiments where maintaining the structural integrity of DNA tiles is essential for achieving precise and efficient crosslinking reactions.
3.3. Structural confirmation and purity of SQ-modified tile constructs
To confirm the successful formation and assembly of the SQ-modified dimer constructs used for crosslinking experiments, control samples, monomers C and D were systematically evaluated (Fig. 3a and b). This assessment involved verifying through PAGE gel that the dimers were properly assembled into the intended tile structures. This validation step was critical to confirm that any observed differences during the crosslinking experiments were a result of the crosslinking process itself and not due to pre-existing structural anomalies in the samples.The comparison of SQ-monomer and dimer constructs using native PAGE (Fig. 3c and d) provided clear evidence of the expected formation of these DNA constructs. Monomer C (MmerC) and Monomer D (MmerD) were used as controls to verify the fully formed tile structures and showed distinct bands that confirmed the integrity and correct assembly of the monomers. These bands served as references against which the dimer samples were compared. Gel 2 presents various fully formed tiles with a continuous G strand, such as Tile 1 and Tile 2, Tile 3, and Tile 4. Gel 3 displays the results for dimer samples of the dx tile where strand G is split into two strands (E, and F): Tile 5, Tile 6, Tile 7, and Tile 8. Fully formed tiles exhibited bands at higher molecular weight positions compared to non-tile control ssSQ-C, consistent with their larger size and more complex structure. The increased mass and potential interactions such as base stacking, hydrogen bonding, and hybridization between strands in fully formed tiles were expected to drive this migration pattern in gel electrophoresis. Conversely, incomplete stacking, weak bonding, or poor strand hybridization cause structural inconsistencies, can produce smeared or diffused bands that indicate partial formation, degradation, or heterogeneous structures.
To further assess the structural integrity of the DNA–dye assembly, the fully formed bands were excised from the gel for additional purification. The results of this purification step are shown in the native gels presented in Fig. 3e. These gels reveal clear, distinct bands for each purified sample, with significant differences between purified and non-purified samples. The non-purified controls (Mmer C and ssSQ-C) exhibited smeared bands and less defined structures, indicating the presence of impurities and incomplete assembly. In contrast, the purified samples displayed sharp, well-defined bands, confirming successful purification and the uniformity of the DNA tiles.
3.4. Thymine–thymine photo-crosslinking and its impact on DNA–dye constructs
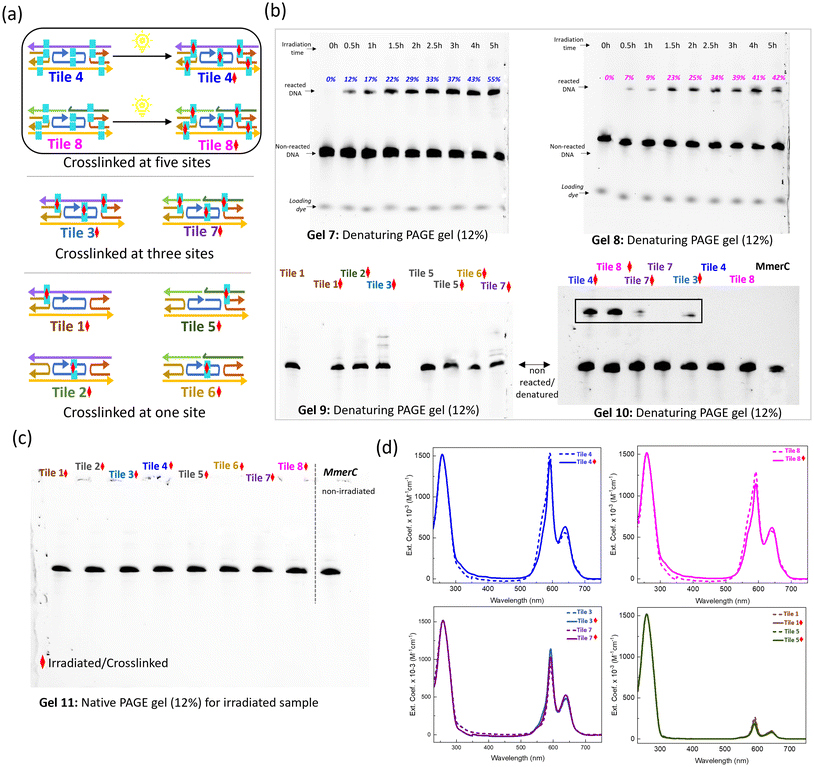
The DNA constructs for the crosslinking study were designed by tethering SQ dyes to the thymine bases, with each construct labeled to indicate specific sites of thymine modification, (cyan rectangles; Fig. 4a). These modifications represented potential sites for thymine–thymine crosslinking upon exposure to UV irradiation. Denaturing PAGE gel electrophoresis was employed to monitor the change in mobilities of DNA constructs after UV irradiation, which induced the thymine–thymine crosslinking process (Fig. 4b). Urea in denaturing PAGE stimulates destabilizing conditions, as urea disrupts hydrogen bonds, challenging DNA stability. Tile 4 and Tile 8, which contained five thymine pairs, exhibited distinct bands with lower mobility post-irradiation, indicating successful crosslinking. The intensity of these bands increased progressively over time, correlating with prolonged UV exposure, indicating an ongoing crosslinking reaction (Gels 7 and 8). The optimal UV irradiation time for achieving maximum crosslinking without side reactions was determined to be 4 hours. During this period, a gradual increase in crosslinking yield was observed at intervals of 0.5h, 1h, 2h, 2.5h, 3h, 4h, and 5h. The crosslinking yields were recorded as 12%, 17%, 22%, 29%, 33%, 37%, 43%, and 55% for Tile 4 (Table S1†) and 7%, 9%, 23%, 25%, 34%, 39%, 41%, and 42% for Tile 8 (Table S2†). Notably, samples irradiated for 5 hours exhibited multiple bands, suggesting the formation of various crosslinked structures.Constructs containing only a single pair of SQ-dimer functionalized thymines, such as Tile 1, Tile 2, Tile 5, and Tile 6, did not show any significant bands corresponding to crosslinked samples, highlighting the need for multiple SQ-dimer functionalized thymines to achieve efficient crosslink formation under UV irradiation (Gel 9 in Fig. 4b and Fig. S6†). A single interstrand crosslink between one SQ-modified thymine pair, connecting only two strands, is insufficient to hold the entire tile together under denaturing conditions. This is because the overall structure requires multiple points of crosslinked attachment or reinforcement to prevent disassembly under environmental conditions due to thermal fluctuations, exposure to urea, or physical agitation. In contrast, incorporating multiple SQ-functionalized thymine pairs enhances the structural stability of the tiles by providing additional crosslinking points, which reinforce the overall assembly. This is evident in the multiple bands displayed by Tile 3 and Tile 7, which contained three SQ-functionalized thymine pairs (Gel 9), indicating the formation of various crosslinked species. Constructs with five SQ-functionalized thymine pairs, Tile 4 and Tile 8, showed a distinct band corresponding to crosslinked structures, with an additional band indicative of higher order structures, suggesting the formation of complex crosslinked assemblies (Gel 10). The absence of multiple bands and smearing indicates that the crosslinked tile is fully formed and structurally homogeneous. The comparison of Tile 3, Tile 7, Tile 4, and Tile 8 demonstrates that the number of SQ-dimers is a critical factor in crosslinking efficiency and determining the structural outcomes of the constructs. While three thymine pairs result in limited and heterogeneous crosslinking structures, five thymine pairs lead to distinct, stable, and higher-order crosslinked assemblies.
The native PAGE gel analysis of all irradiated samples revealed a single band (Fig. 4c, Gel 11), indicating that the overall DNA structure remained intact after irradiation. This finding implies that the crosslinking process occurs without significant disruption to the DNA's native conformation, thereby preserving its structural integrity. Spectroscopic analysis (Fig. 4d and S9, S10†) provided a complementary view of the structural modifications associated with crosslinking. The steady state absorption spectra of non-crosslinked samples serve as controls, offering a baseline for the comparison. All dimer samples exhibited similar optical properties with evidence of strongly coupled H-aggregates between SQ dyes, as indicated by the blue-shift in absorption maxima relative to the monomer absorption maximum. No significant spectral changes were observed in the samples with either single or three pairs of SQ-modified thymine after 4 hours of irradiation at 310 nm. However, crosslinked samples (solid lines) displayed distinct spectral features compared to their non-crosslinked counterparts (dashed lines) in Tile 4 and 8, both containing five pairs of SQ-modified thymine. In the crosslinked Tile 4 and Tile 8 constructs, peak widths in the dye absorption region were slightly reduced, suggesting structural changes possibly due to the formation of covalent bonds between the thymine bases. These covalent bonds likely induced local conformational changes in the DNA or its surrounding environment, such as altering the spacing or orientation of the dye aggregates associated with the thymine pairs. Additionally, crosslinking likely reduced structural flexibility, leading to a more ordered arrangement of the dye aggregates, reflected in the narrower absorption peak compared to the non-crosslinked samples. The broader absorption peaks observed in the non-crosslinked samples suggest structural heterogeneity, where the DNA and dye molecules could adopt a range of configurations. In contrast, crosslinking covalently links thymine bases, effectively locking certain molecular orientations in place and reducing the inhomogeneous broadening typically associated with structural variability. Notably, no significant changes were observed in the DNA absorption region at 260 nm for any of the crosslinked samples, further indicating that the overall DNA structure remained intact post-irradiation. Additional spectral analysis was conducted to confirm the proper formation of the dye aggregates in the crosslinked constructs. Linear combinations of individual dimer spectra were compared with multi-dimer tile absorption spectra, verifying that tiles 3 and 8 contain three and five dye pairs, respectively (Fig. S11†). The analysis of absorption line shapes confirmed that the multi-dimer tiles can support up to five strongly coupled dimers without any unwanted inter-dimer aggregation.
Circular dichroism (CD) spectra of the squaraine labeled constructs showed no significant changes in either DNA region or the dye region after 4 hours of irradiation. This suggests that the crosslinking process did not substantially alter the overall chiral environment of the dimers or the DNA secondary structure. The absence of significant change in the CD spectra implies that crosslinking primarily affected localized regions, such as the thymine pairs, without disrupting the associated dime's chiral structure.
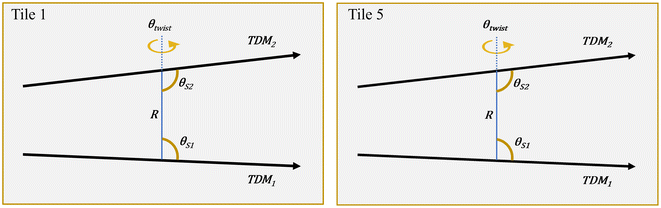
To assess the impact of crosslinking on the optical properties of templated dye aggregates, we employed KRM model simulation tool to model absorption and CD spectra of SQ dimers in Tile 1 and Tile 5 (Table 1, Fig. 5 and Fig. S12†). In all cases, experimental absorption maxima were significantly blue-shifted compared to the monomer absorption maximum, with an induced cotton effect in the CD spectra at the absorption maximum and an optically forbidden lower exciton state. These results are consistent with the expected characteristics of close-packed, face-to-face (H-like) dye arrangements. KRM modeling confirmed the H-like packing geometry, with modeled dimers adopting a nearly parallel, nearly planar, face-to-face packing arrangement. The modeled transition dipole moments (TDMs) showed a total deviation from parallel (α) of less than 11° in all cases, and an out-of-plane twist (θtwist) of less than 10°. Notably, θtwist is reported as a negative value by convention, indicating “right-handed” chirality. The slip angles, measuring the angle between each TDM and the line connecting their centers, were all below 10°, and the center-to-center distance (R) was less than 0.5 nm, collectively describing the close-packed H-like dimer geometry.
| Parameter | J m,n (meV) | θ Slip1 (°) | θ Slip2 (°) | θ twist (°) | α (°) | R (nm) |
|---|---|---|---|---|---|---|
| a See section ESI5† for complete modeling results. ◊ Crosslinked. | ||||||
| Tile 1 | 126.7 | 85.9 | 87.9 | −9.0 | 10.9 | 0.40 |
| Tile 1 ◊ | 127.1 | 81.1 | 86.8 | −6.8 | 8.8 | 0.40 |
| Tile 5 | 126.4 | 81.7 | 87.7 | −9.1 | 10.9 | 0.40 |
| Tile 5 ◊ | 124.1 | 82.8 | 89.5 | −8.1 | 10.5 | 0.41 |
An important dimer parameter derived from KRM modeling is the exciton hopping parameter,45Jm,n, which quantifies the strength of TDM–TDM coupling and exciton delocalization with in the dimer. The modeled dimers exhibited Jm,n values of approximately 125 meV, both before and after irradiation, indicating that irradiation did not impact excitonic coupling within the templated dimers. These values are among the highest reported Jm,n values for DNA-templated dye aggregates and align with previous reports for dichloro-squaraine dimers.67
The consistency of modeling results across all four dimer models suggests that irradiation had no discernable effect on dimer geometry or aggregate coupling behavior. The formation of SQ dimers takes place in the absence of crosslinking. This result supports our explanation that the SQ aggregates attached to thymines promote proto-crosslinking by bringing the thymine bases in proximity. Furthermore, the comparable behavior of Tile 1 and Tile 5 dimers implies favorable dimers interaction with the DNA tile, regardless of their position. We hypothesize that the dimers interact with the nearby crossover junction to minimize solvent exposure, as observed in a previous study. The feasibility of the crossover junction, compared to the canonical double helix, likely facilitates dimer accommodation, while the length of the linkers and the position of attachment points provide sufficient freedom for the dyes to reach the junction. These findings indicate that crosslinking had minimal impact on the overall dimer assembly structure and the templated dimers maintain stable H-aggregate geometry and exciton coupling post-irradiation.
Results from denaturing PAGE gel electrophoresis and spectroscopic studies provide a detailed understanding of thymine–thymine crosslinking in DNA constructs. Distinct bands on denaturing PAGE gels indicate successful crosslink formation in constructs with multiple thymine pairs underscoring the necessity of having multiple thymine pairs for efficient crosslinking. Increasing band intensity over time in the time course study further supports the progressive nature of the crosslinking reaction. The absence of crosslinking bands in constructs with a single SQ-functionalized thymine pair highlights the requirement for multiple thymine sites to achieve significant crosslinking. Constructs with five thymine pairs displayed a distinct higher-order band, suggesting the formation of more complex crosslinked structures. Native PAGE gel analysis, showing a single band in all irradiated samples, confirmed that the overall DNA structure remained intact after irradiation, suggesting that thymine–thymine crosslinking occurs without major disruptions to the DNA's native conformation. The spectroscopic data suggest that crosslinking induces localized modifications by introducing covalent bond formation between thymines without compromising the overall structure of the DNA–dye construct. KRM analysis further corroborates these findings, confirming the preservation of structural integrity post-crosslinking.
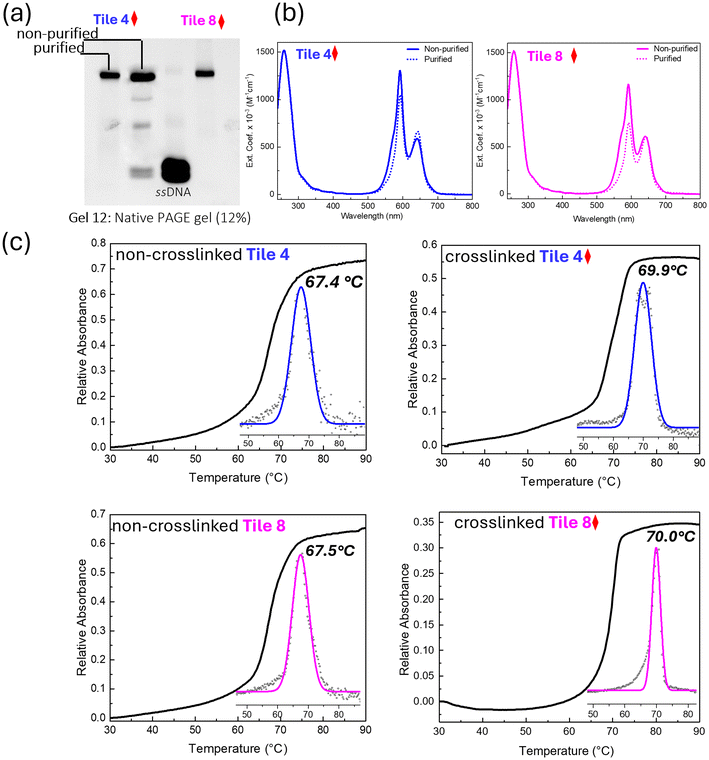
3.5. Analysis of purified crosslinked tile
The crosslinked DNA samples with five crosslinking points, Tile 4 and Tile 8, were subjected to purification to obtain structurally uniform crosslinked constructs. Denaturing PAGE gel analysis (Fig. 6a) provided clear evidence of the successful purification (Fig. S7†). The presence of a distinct sharp band in both purified Tile 4 and Tile 8 samples, comparable to the ssDNA and non-purified Tile 4 controls (Gel 12), indicates high purity and homogeneity in the purified samples, with no detectable contamination or degradation. In contrast, the multiple bands and smearing observed in the non-purified Tile 4 control suggest the presence of impurities, incomplete products, or degraded DNA fragments. This contrast between purified and non-purified samples underscores the effectiveness of the purification process in isolating high-quality, structurally uniform crosslinked DNA. The enhanced stability from crosslinking enabled the successful isolation and purification of the novel five dimer configuration, a process that would have been otherwise challenging due to potential dissociation or partial assembly in non-crosslinked constructs.Spectroscopic analysis further highlighted the differences between purified and non-purified samples. The spectroscopic profiles of the purified Tile 4 and Tile 8 samples (dotted line; Fig. 6b) exhibited distinct and sharp peaks and absorption features reflecting strongly coupled H aggregates, indicative of a high degree of structural integrity and minimal degradation or modification. Crosslinking likely enhances the excitonic interaction by locking the chromophores into a fixed, favorable orientation, thereby reducing the number of possible electronic transitions and contributing to the narrower peak. Conversely, the broader peaks observed in the non-purified samples (solid line), particularly in the 500–700 nm range where dye absorption is observed, indicate the presence of aggregated or partially assembled structures. A heterogeneous mixture of H-like aggregate subpopulations and residual impurities such as single strands, or other intermediates in the non-purified samples likely contributed to increased peak widths. These impurities likely stem from incomplete crosslinking or the presence of unreacted compounds, compromising structural uniformity, or from strong dye–dye interaction that promotes the multiple dimer tiles to capture additional dye-labeled strands from the solution.
The thermal stability of the crosslinked and non-crosslinked DNA samples was evaluated using DNA melting experiments, monitored by absorption at 260 nm (Fig. 6c). The crosslinked samples, Tile 4 and Tile 8, exhibited higher melting temperatures (Tm) compared to their non-crosslinked counterparts (69.9 °C and 70.0 °C vs. 67.4 °C and 67.5 °C respectively), indicating that the introduction of crosslinks enhanced the thermal stability of the DNA tile. This enhancement likely arises from the crosslinks reinforcing the structural integrity of the DNA, thereby reducing the flexibility and inhibiting the unwinding of the double helix at elevated temperatures. The first derivative of the melting curves (insets; Fig. 6c), and Gaussian-fitted curves further confirmed the increased thermal stability in the crosslinked samples. The consistent Tm values across different crosslinked samples suggest a reproducible enhancement of thermal stability due to crosslinking.
3.6. Stability of crosslinked DNA constructs
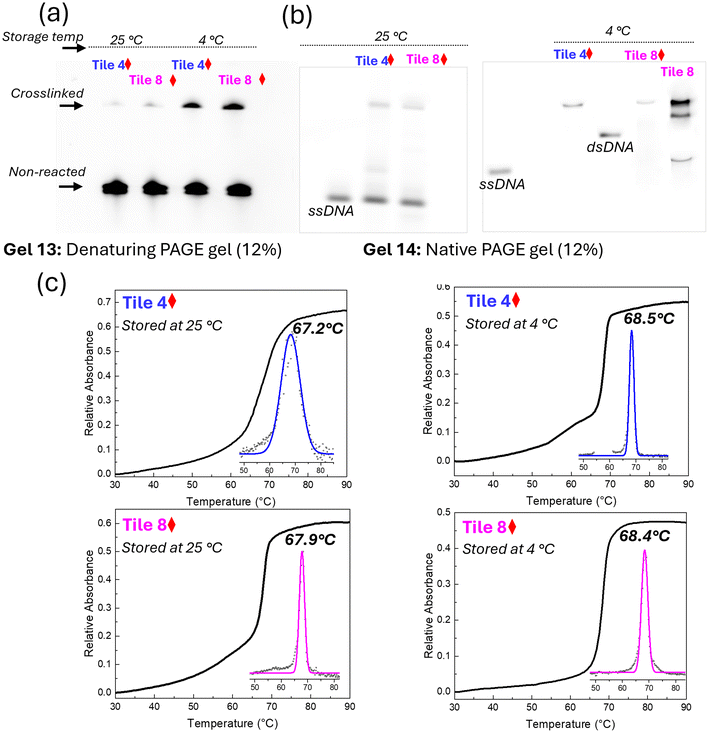
The structural stability of crosslinked DNA samples was evaluated using denaturing PAGE, native PAGE, and thermal denaturation profiles under different storage temperatures (4 °C and 25 °C). Denaturing PAGE with urea assesses the structural stability by disrupting hydrogen bonds, allowing covalently bonded crosslinked structures to resist dissociation under denaturing conditions. Native PAGE, which preserves DNA's native conformation, allows stable complexes to migrate as expected without dissociation, indicating intact structures. Thermal denaturation profiles reveal the temperature at which DNA strands separate, with higher melting points signifying greater stability, thereby providing insight into the thermal resilience of the crosslinked samples. Together, these methods offer a comprehensive evaluation of structural stability in crosslinked DNA constructs.Denaturing PAGE analysis revealed that crosslinked samples stored at both 4 °C and 25 °C retained consistent band patterns after 5 days, indicating that the crosslinked structures remained largely intact under these conditions (Fig. 7a). However, a decrease in band intensity was observed due to elevated storage temperatures. In contrast, samples stored at 4 °C maintained consistent band intensity, affirming the preservation of structural integrity at lower storage temperatures. These results indicate that while crosslinking significantly enhances stability, higher storage temperatures may still induce some degree of structural degradation.
Further insights into the structural stability were gained through native PAGE analysis. The crosslinked samples exhibited consistent banding patterns at 4 °C, indicating the preservation of their native 2D structure (Fig. 7b). In contrast, samples stored at 25 °C displayed a higher mobility band, similar to that observed for the ssDNA control, suggesting partial denaturation of the DNA structure. The non-crosslinked Tile 8 control exhibited multiple bands, indicative of structural deformation over time, further highlighting the stabilizing effect of crosslinking. The clear, distinct bands observed for crosslinked samples at 4 °C suggest that these structures are highly stable under cold storage, making them suitable for long-term storage applications.
The thermal stability of samples stored at 4 °C and 25 °C was further assessed through melting profile analysis. Samples stored at 4 °C showed sharp transitions and consistent Tm values, with Tile 4 and Tile 8 displaying Tm values of 68.5 °C and 68.4 °C, respectively (Fig. 7c). This stability indicates that the crosslinked structures are not only preserved at lower temperatures but also maintain their thermal robustness. In contrast, samples stored at 25 °C exhibited slightly lower Tm values (67.2 °C for Tile 4 and 67.9 °C for Tile 8), suggesting that prolonged exposure to higher temperatures can slightly reduce thermal stability. However, the decrease in Tm was modest, indicating that crosslinking still provides substantial protection against thermal degradation, even at elevated storage temperatures.
4. Conclusion
This study collectively examines the formation, stability, and crosslinking efficiency of DNA double (DX) tiles, comparing continuous and split strand designs and evaluating the role of SQ dye aggregation in site-specific interstrand thymine–thymine photo-crosslinking. Successful assembly of DNA tiles was confirmed using PAGE analysis, with distinct bands indicating fully formed tiles. Thymine–thymine crosslinking efficiency under UV irradiation was analyzed by denaturing PAGE, revealing that multiple thymine pairs are required for effective interstrand thymine–thymine photo-crosslinking through [2 + 2] photocycloaddition reaction. Notably, urea in denaturing PAGE was used to stimulate destabilizing conditions by disrupting hydrogen bonds, and the resilience of crosslinked structures in urea demonstrated the effectiveness of SQ dye-mediated crosslinking.PAGE purification improved structural uniformity, and crosslinked samples exhibited thermal stability and resilience under challenging conditions, such as urea and light exposure. By facilitating site-specific crosslinking through strategically placed SQ dye aggregates, the study addresses a key challenge in DNA nanotechnology: achieving stable, homogeneous structures that can withstand varying environmental conditions.
Moreover, SQ dye aggregates function as structural stabilizers and functional probes, allowing monitoring of crosslinking efficiency and structural uniformity via PAGE and spectroscopic analysis. Results indicate that increasing the number of SQ-modified thymine pairs improves crosslinking efficiency and structural integrity, providing insights for optimizing DNA–dye constructs for specific nanotechnological applications. This scalable, non-phototoxic crosslinking approach offers new possibilities for designing robust, stable DNA nanostructures suitable for advanced applications in optoelectronics, photonics, and quantum computing.
Future research will focus on exploring the adaptability of SQ dimer-mediated crosslinking across diverse DNA structures and further elucidating the mechanisms governing dye aggregate interactions within DNA scaffolds, aiming to optimize these constructs for a wide range of nanotechnological and molecular computing applications.
Author contributions
SB, SKR, JL, BY, and WBK contributed to the original conceptualization and design of this study. SKR performed and analyzed KRM modeling. GB modeled the unmodified tile structure. SB designed and prepared all samples and conducted characterizations and analyses. SB prepared the original draft, with JL and MS providing feedback on the draft. All authors contributed to reviewing and editing the final manuscript.Data availability
The data supporting this article has been included as part of ESI.† The ESI† includes comprehensive supporting materials such as DNA sequences, PAGE gel images, additional absorption data, and KRM modeling methodologies.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We thank the U.S. Department of Energy (DOE), Office of Basic Energy Sciences, Division of Materials Science and Engineering through the Established Program to Stimulate Competitive Research (EPSCoR) via Award No. DE-SC0020089 for fully supporting this research.References
- N. C. Seeman, Nanomaterials Based on DNA, Annu. Rev. Biochem., 2010, 65–87, DOI:10.1146/annurev-biochem-060308-102244.
- N. C. Seeman, Nucleic Acid Junctions and Lattices, J. Theor. Biol., 1982, 99(2), 237–247, DOI:10.1016/0022-5193(82)90002-9.
- B. Yurke and A. P. Mills, Using DNA to Power Nanostructures; 2003, vol. 4 Search PubMed.
- M. R. Wasielewski, Self-Assembly Strategies for Integrating Light Harvesting and Charge Separation in Artificial Photosynthetic Systems, Acc. Chem. Res., 2009, 42(12), 1910–1921, DOI:10.1021/ar9001735.
- A. V. Pinheiro, D. Han, W. M. Shih and H. Yan, Challenges and Opportunities for Structural DNA Nanotechnology, Nat. Nanotechnol., 2011, 763–772, DOI:10.1038/nnano.2011.187.
- N. R. Kallenbach, R. Ma and N. C. Seeman, An immobile nucleic acid junction constructed from oligonucleotides, Nature, 1983, 305, 829–831 CrossRef CAS https://doi.org/10.1038/305829a0 .
- S. M. Hart, W. J. Chen, J. L. Banal, W. P. Bricker, A. Dodin, L. Markova, Y. Vyborna, A. P. Willard, R. Häner, M. Bathe and G. S. Schlau-Cohen, Engineering Couplings for Exciton Transport Using Synthetic DNA Scaffolds, Chem, 2021, 7(3), 752–773, DOI:10.1016/j.chempr.2020.12.020.
- S. M. Hart, X. Wang, J. Guo, M. Bathe and G. S. Schlau-Cohen, Tuning Optical Absorption and Emission Using Strongly Coupled Dimers in Programmable DNA Scaffolds, J. Phys. Chem. Lett., 2022, 13(7), 1863–1871, DOI:10.1021/acs.jpclett.1c03848.
- X. Wang, R. Sha, W. B. Knowlton, N. C. Seeman, J. W. Canary and B. Yurke, Exciton Delocalization in a DNA-Templated Organic Semiconductor Dimer Assembly, ACS Nano, 2022, 16(1), 1301–1307, DOI:10.1021/acsnano.1c09143.
- M. Kasha, Energy Transfer Mechanisms and the Molecular Exciton Model for Molecular Aggregates, 1963, vol. 20. https://www.jstor.org/stable/3571331 Search PubMed.
- L. Blancafort and A. A. Voityuk, Exciton Delocalization, Charge Transfer, and, Electronic Coupling for Singlet Excitation Energy Transfer between Stacked Nucleobases in DNA: An MS-CASPT2 Study, J. Chem. Phys., 2014, 140(9), 095102, DOI:10.1063/1.4867118.
- M. Kasha, H. R. Rawls and M. Ashraf El-Bayoumi, The Exciton Model in Molecular Spectroscopy, Pure Appl. Chem., 1965, 11, 371–392, DOI:10.1351/pac196511030371.
- B. Yurke, R. Elliott and A. Sup, Implementation of a Frenkel Exciton-Based Controlled Phase Shifter, Phys. Rev. A, 2023, 107(1), 012603, DOI:10.1103/PhysRevA.107.012603.
- T.-J. Fu and N. C. Seeman, DNA Double-Crossover Molecules1, 1993, vol. 32. https://pubs.acs.org/sharingguidelines Search PubMed.
- C. Jiang, B. Lu, W. Zhang, Y. P. Ohayon, F. Feng, S. Li, N. C. Seeman and S. J. Xiao, Regulation of 2D DNA Nanostructures by the Coupling of Intrinsic Tile Curvature and Arm Twist, J. Am. Chem. Soc., 2022, 144(15), 6759–6769, DOI:10.1021/jacs.1c13601.
- C. Ashworth, Building Big with DNA Bricks, Nat. Rev. Mater., 2018, 3, 17092, DOI:10.1038/natrevmats.2017.92.
- Q. Yang, X. Chang, J. Y. Lee, M. Saji and F. Zhang, DNA T-Shaped Crossover Tiles for 2D Tessellation and Nanoring Reconfiguration, Nat. Commun., 2023, 14(1), 7675, DOI:10.1038/s41467-023-43558-8.
- B. L. Cannon, D. L. Kellis, L. K. Patten, P. H. Davis, J. Lee, E. Graugnard, B. Yurke and W. B. Knowlton, Coherent Exciton Delocalization in a Two-State DNA-Templated Dye Aggregate System, J. Phys. Chem. A, 2017, 121(37), 6905–6916, DOI:10.1021/acs.jpca.7b04344.
- N. P. D. Sawaya, D. Rappoport, D. P. Tabor and A. Aspuru-Guzik, Excitonics: A Set of Gates for Molecular Exciton Processing and Signaling, ACS Nano, 2018, 12(7), 6410–6420, DOI:10.1021/acsnano.8b00584.
- F. Garo and R. Häner, A DNA-Based Light-Harvesting Antenna, Angew. Chem., Int. Ed., 2012, 51(4), 916–919, DOI:10.1002/anie.201103295.
- G. D. Scholes and C. Smyth, Perspective: Detecting and Measuring Exciton Delocalization in Photosynthetic Light Harvesting, J. Chem. Phys., 2014, 140, 110901, DOI:10.1063/1.4869329.
- R. Monshouwer, M. Abrahamsson, F. Van Mourik and R. Van Grondelle, Superradiance and Exciton Delocalization in Bacterial Photosynthetic Light-Harvesting Systems, 1997. https://pubs.acs.org/sharingguidelines Search PubMed.
- K. L. Lau and H. F. Sleiman, Minimalist Approach to Complexity: Templating the Assembly of DNA Tile Structures with Sequentially Grown Input Strands, ACS Nano, 2016, 10(7), 6542–6551, DOI:10.1021/acsnano.6b00134.
- B. Yurke, DNA Assembly of Dye Aggregates—A Possible Path to Quantum Computing, 2023; 125–169. DOI:10.1007/978-981-19-9891-1_9.
- X. Zhou, S. Lin and H. Yan, Interfacing DNA Nanotechnology and Biomimetic Photonic Complexes: Advances and Prospects in Energy and Biomedicine, J. Nanobiotechnol., 2022, 20, 257, DOI:10.1186/s12951-022-01449-y.
- A. Jaekel, P. Lill, S. Whitelam and B. Saccà, Insights into the Structure and Energy of Dna Nanoassemblies, Molecules, 2020, 25(23), 5466, DOI:10.3390/molecules25235466.
- D. Mathur, S. A. Díaz, N. Hildebrandt, R. D. Pensack, B. Yurke, A. Biaggne, L. Li, J. S. Melinger, M. G. Ancona, W. B. Knowlton and I. L. Medintz, Pursuing Excitonic Energy Transfer with Programmable DNA-Based Optical Breadboards, Chem. Soc. Rev., 2023, 7848–7948, 10.1039/d0cs00936a.
- Y. Ke, L. L. Ong, W. Sun, J. Song, M. Dong, W. M. Shih and P. Yin, DNA Brick Crystals with Prescribed Depths, Nat. Chem., 2014, 6(11), 994–1002, DOI:10.1038/nchem.2083.
- B. L. Cannon, D. L. Kellis, P. H. Davis, J. Lee, W. Kuang, W. L. Hughes, E. Graugnard, B. Yurke and W. B. Knowlton, Excitonic and Logic Gates on DNA Brick Nanobreadboards, ACS Photonics, 2015, 2(3), 398–404, DOI:10.1021/ph500444d.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Programmable Materials and the Nature of the DNA Bond, Science, 2015, 347, 1744–1752, DOI:10.1126/science.1260901.
- K. E. Bujold, A. Lacroix and H. F. Sleiman, DNA Nanostructures at the Interface with Biology, Chem, 2018, 495–521, DOI:10.1016/j.chempr.2018.02.005.
- F. Albalawi, M. Z. Hussein, S. Fakurazi and M. J. Masarudin, Engineered Nanomaterials: The Challenges and Opportunities for Nanomedicines, Int. J. Nanomed., 2021, 161–184, DOI:10.2147/IJN.S288236.
- V. Hippel, P. H. Johnson, N. P. Marcus and A. H, Fifty Years of DNA “Breathing”: Reflections on Old and New Approaches, Biopolymers, 2013, 923–954, DOI:10.1002/bip.22347.
- R. Metzler and T. Ambjörnsson, Dynamic Approach to DNA Breathing, J. Biol. Phys., 2005, 31, 339–350, DOI:10.1007/s10867-005-2410-y.
- A. Kabir, M. Bhattarai, K.Ø Rasmussen, A. Shehu, A. Usheva, A. R. Bishop and B. S. Alexandrov, Examining DNA Breathing with PyDNA-EPBD, Bioinformatics, 2023, 39(11) DOI:10.1101/2023.09.09.557010.
- T. Ambjörnsson, S. K. Banik, O. Krichevsky and R. Metzler, Breathing Dynamics in Heteropolymer DNA, Biophys. J., 2007, 92(8), 2674–2684, DOI:10.1529/biophysj.106.095935.
- C. R. Simmons, T. MacCulloch, M. Krepl, M. Matthies, A. Buchberger, I. Crawford, J. Šponer, P. Šulc, N. Stephanopoulos and H. Yan, The Influence of Holliday Junction Sequence and Dynamics on DNA Crystal Self-Assembly, Nat. Commun., 2022, 13(1), 3112, DOI:10.1038/s41467-022-30779-6.
- H. Asanuma, K. Shirasuka, T. Takarada, H. Kashida and M. Komiyama, DNA-Dye Conjugates for Controllable H* Aggregation, J. Am. Chem. Soc., 2003, 125(8), 2217–2223, DOI:10.1021/ja021153k.
- H. Asanuma, T. Fujii, T. Kato and H. Kashida, Coherent Interactions of Dyes Assembled on DNA, J. Photochem. Photobiol., C, 2012, 124–135, DOI:10.1016/j.jphotochemrev.2012.04.002.
- J. S. Huff, D. B. Turner, O. A. Mass, L.K. Patten, C. K. Wilson, S. K. Roy, M. S. Barclay, B. Yurke, W. B. Knowlton, P.H. Davis and R. D. Pensack, Excited-State Lifetimes of DNA-Templated Cyanine Dimer, Trimer, and Tetramer Aggregates: The Role of Exciton Delocalization, Dye Separation, and DNA Heterogeneity, J. Phys. Chem. B, 2021, 125(36), 10240–10259, DOI:10.1021/acs.jpcb.1c04517.
- E. Graugnard, D. L. Kellis, H. Bui, S. Barnes, W. Kuang, J. Lee, W. L. Hughes, W. B. Knowlton and B. Yurke, DNA-Controlled Excitonic Switches, Nano Lett., 2012, 12(4), 2117–2122, DOI:10.1021/nl3004336.
- B. S. Alexandrov, V. Gelev, A. R. Bishop, A. Usheva and K. Rasmussen, DNA Breathing Dynamics in the Presence of a Terahertz Field, Phys. Lett. A, 2010, 374(10), 1214–1217, DOI:10.1016/j.physleta.2009.12.077.
- B. L. Cannon, L. K. Patten, D. L. Kellis, P. H. Davis, J. Lee, E. Graugnard, B. Yurke and W. B. Knowlton, Large Davydov Splitting and Strong Fluorescence Suppression: An Investigation of Exciton Delocalization in DNA-Templated Holliday Junction Dye Aggregates, J. Phys. Chem. A, 2018, 122(8), 2086–2095, DOI:10.1021/acs.jpca.7b12668.
- S. K. Roy, O. A. Mass, D. L. Kellis, C. K. Wilson, J. A. Hall, B. Yurke and W. B. Knowlton, Exciton Delocalization and Scaffold Stability in Bridged Nucleotide-Substituted, DNA Duplex-Templated Cyanine Aggregates, J. Phys. Chem. B, 2021, 125(50), 13670–13684, DOI:10.1021/acs.jpcb.1c07602.
- O. A. Mass, C. K. Wilson, S. K. Roy, M. S. Barclay, L. K. Patten, E. A. Terpetschnig, J. Lee, R. D. Pensack, B. Yurke and W. B. Knowlton, Exciton Delocalization in Indolenine Squaraine Aggregates Templated by DNA Holliday Junction Scaffolds, J. Phys. Chem. B, 2020, 124(43), 9636–9647, DOI:10.1021/acs.jpcb.0c06480.
- S. Manuguri, M. K. Nguyen, J. Loo, A. K. Natarajan and A. Kuzyk, Advancing the Utility of DNA Origami Technique through Enhanced Stability of DNA-Origami-Based Assemblies, Bioconjugate Chem., 2023, 6–17, DOI:10.1021/acs.bioconjchem.2c00311.
- V. Linko and A. Keller, Stability of DNA Origami Nanostructures in Physiological Media: The Role of Molecular Interactions, Small, 2023, 19, 6–17, DOI:10.1002/smll.202301935.
- D. A. Rusling, I. S. Nandhakumar, T. Brown and K. R. Fox, Triplex-Directed Covalent Cross-Linking of a DNA Nanostructure, Chem. Commun., 2012, 48(77), 9592–9594, 10.1039/c2cc35407a.
- T. Sakamoto, Y. Tanaka and K. Fujimoto, DNA Photo-Cross-Linking Using 3-Cyanovinylcarbazole Modified Oligonucleotide with Threoninol Linker, Org. Lett., 2015, 17(4), 936–939, DOI:10.1021/acs.orglett.5b00035.
- A. Tavakoli and J. H. Min, Photochemical Modifications for DNA/RNA Oligonucleotides, RSC Adv., 2022, 6484–6507, 10.1039/d1ra05951c.
- Y. Watanabe and K. Fujimoto, Complete Photochemical Regulation of 8–17 DNAzyme Activity by Using Reversible DNA Photo-Crosslinking, ChemBioChem, 2020, 21(22), 3244–3248, DOI:10.1002/cbic.202000227.
- M. Ye, J. Guillaume, Y. Liu, R. Sha, R. Wang, N. C. Seeman and J. W. Canary, Site-Specific Inter-Strand Cross-Links of DNA Duplexes, Chem. Sci., 2013, 4(3), 1319–1329, 10.1039/c2sc21775a.
- S. Basu, K. Cervantes-Salguero, B. Yurke, W. B. Knowlton, J. Lee and O. A. Mass, Photocrosslinking Probes Proximity of Thymine Modifiers Tethering Excitonically Coupled Dye Aggregates to DNA Holliday Junction, Molecules, 2022, 27(13), 4006, DOI:10.3390/molecules27134006.
- S. Basu, S. K. Roy, G. Barcenas, L. Li, B. Yurke, W. B. Knowlton and J. Lee, Enhanced Photo-Cross-Linking of Thymines in DNA Holliday Junction-Templated Squaraine Dimers, Biochemistry, 2023, 62(22), 3234–3244, DOI:10.1021/acs.biochem.3c00471.
- T. M. Brown, H. H. Fakih, D. Saliba, J. Asohan and H. F. Sleiman, Stabilization of Functional DNA Structures with Mild Photochemical Methods, J. Am. Chem. Soc., 2023, 145(4), 2142–2151, DOI:10.1021/jacs.2c08808.
- J. Hu and S. Adar, The Cartography of UV-Induced DNA Damage Formation and DNA Repair, Photochem. Photobiol., 2017, 199–206, DOI:10.1111/php.12668.
- H. Ding and M. M. Greenberg, DNA Damage and Interstrand Cross-Link Formation upon Irradiation of Aryl Iodide C-Nucleotide Analogues, J. Org. Chem., 2010, 75(3), 535–544, DOI:10.1021/jo902071y.
- J. Sun and X. Tang, Photouncaged Sequence-Specific Interstrand DNA Cross-Linking with Photolabile 4-Oxo-Enal-Modified Oligonucleotides, Sci. Rep., 2015, 5, 10473, DOI:10.1038/srep10473.
- M. M. Haque, H. Sun, S. Liu, Y. Wang and X. Peng, Photoswitchable Formation of a DNA Interstrand Cross-Link by a Coumarin-Modified Nucleotide, Angew. Chem., Int. Ed., 2014, 53(27), 7001–7005, DOI:10.1002/anie.201310609.
- L. I. Markova, V. L. Malinovskii, L. D. Patsenker and R. Häner, Synthesis and Properties of Squaraine-Modified DNA, Org. Biomol. Chem., 2012, 10(45), 8944–8947, 10.1039/c2ob26787j.
- J. Y. Li, C. Y. Chen, W. C. Ho, S. H. Chen and C. G. Wu, Unsymmetrical Squaraines Incorporating Quinoline for near Infrared Responsive Dye-Sensitized Solar Cells, Org. Lett., 2012, 14(21), 5420–5423, DOI:10.1021/ol302481k.
- Y. D. Lee, C. K. Lim, S. Kim, I. C. Kwon and J. Kim, Squaraine-Doped Functional Nanoprobes: Lipophilically Protected near-Infrared Fluorescence for Bioimaging, Adv. Funct. Mater., 2010, 20(17), 2786–2793, DOI:10.1002/adfm.201000650.
- K. Ilina, W. M. MacCuaig, M. Laramie, J. N. Jeouty, L. R. McNally and M. Henary, Squaraine Dyes: Molecular Design for Different Applications and Remaining Challenges, Bioconjugate Chem., 2020, 31(2), 194–213, DOI:10.1021/acs.bioconjchem.9b00482.
- G. Wei, X. Xiao, S. Wang, J. D. Zimmerman, K. Sun, V. V. Diev, M. E. Thompson and S. R. Forrest, Arylamine-Based Squaraine Donors for Use in Organic Solar Cells, Nano Lett., 2011, 11(10), 4261–4264, DOI:10.1021/nl2022515.
- G. Xia and H. Wang, Squaraine Dyes: The Hierarchical Synthesis and Its Application in Optical Detection, J. Photochem. Photobiol., C, 2017, 84–113, DOI:10.1016/j.jphotochemrev.2017.03.001.
- M. S. Barclay, C. K. Wilson, S. K. Roy, O. A. Mass, O. M. Obukhova, R. P. Svoiakov, A. L. Tatarets, A. U. Chowdhury, J. S. Huff, D. B. Turner, P. H. Davis, E. A. Terpetschnig, B. Yurke, W. B. Knowlton, J. Lee and R. D. Pensack, Oblique Packing and Tunable Excitonic Coupling in DNA-Templated Squaraine Rotaxane Dimer Aggregates, ChemPhotoChem, 2022, 6(7), e202200039, DOI:10.1002/cptc.202200039.
- O. A. Mass, C. K. Wilson, G. Barcenas, E. A. Terpetschnig, O. M. Obukhova, O. S. Kolosova, A. L. Tatarets, L. Li, B. Yurke, W. B. Knowlton, R. D. Pensack and J. Lee, Influence of Hydrophobicity on Excitonic Coupling in DNA-Templated Indolenine Squaraine Dye Aggregates, J. Phys. Chem. C, 2022, 126(7), 3475–3488, DOI:10.1021/acs.jpcc.1c08981.
- G. Barcenas, A. Biaggne, O. A. Mass, W. B. Knowlton, B. Yurke and L. Li, Molecular Dynamic Studies of Dye–Dye and Dye–DNA Interactions Governing Excitonic Coupling in Squaraine Aggregates Templated by DNA Holliday Junctions, Int. J. Mol. Sci., 2023, 24(4), 4059, DOI:10.3390/ijms24044059.
- O. A. Mass, S. Basu, L. K. Patten, E. A. Terpetschnig, A. I. Krivoshey, A. L. Tatarets, R. D. Pensack, B. Yurke, W. B. Knowlton and J. Lee, Exciton Chirality Inversion in Dye Dimers Templated by DNA Holliday Junction, J. Phys. Chem. Lett., 2022, 10688–10696, DOI:10.1021/acs.jpclett.2c02721.
- A. K. Kędziera, M. Ziegler-Borowska, D. Chełminiak, P. Kuchnicki and H. Kaczmarek, Effect of UV-Irradiation on Spectral Properties of Squaraine Dye in Diluted Solutions, J. Photochem. Photobiol., A, 2016, 318, 77–89, DOI:10.1016/j.jphotochem.2015.11.011.
- G. Barcenas, A. Biaggne, O. A. Mass, C. K. Wilson, O. M. Obukhova, O. S. Kolosova, A. L. Tatarets, E. Terpetschnig, R. D. Pensack, J. Lee, W. B. Knowlton, B. Yurke and L. Li, First-Principles Studies of Substituent Effects on Squaraine Dyes, RSC Adv., 2021, 11(31), 19029–19040, 10.1039/d1ra01377g.
- A. Rajendran, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, Photo-Cross-Linking-Assisted Thermal Stability of DNA Origami Structures and Its Application for Higher-Temperature Self-Assembly, J. Am. Chem. Soc., 2011, 133(37), 14488–14491, DOI:10.1021/ja204546h.
- T. Gerling, M. Kube, B. Kick and H. Dietz, Sequence-Programmable Covalent Bonding of Designed DNA Assemblies, Sci. Adv., 2018, 4(8), eaau1157, DOI:10.1126/sciadv.aau1157.
- S. Poplata, A. Tröster, Y. Q. Zou and T. Bach, Recent Advances in the Synthesis of Cyclobutanes by Olefin [2 + 2] Photocycloaddition Reactions, Chem. Rev., 2016, 14, 9748–9815, DOI:10.1021/acs.chemrev.5b00723.
- S. Ha, Y. Lee, Y. Kwak, A. Mishra, E. Yu, B. Ryou and C. M. Park, Alkyne–Alkene [2 + 2] Cycloaddition Based on Visible Light Photocatalysis, Nat. Commun., 2020, 11(1), 2509 (2020), DOI:10.1038/s41467-020-16283-9.
- D. M. Noll, A. M. Noronha, C. J. Wilds and P. S. Miller, Preparation of Interstrand Cross-Linked DNA Oligonucleotide Duplexes, 2004, DOI:10.2741/1246.
- Z. Liu, C. Tan, X. Guo, Y.-T. Kao, J. Li, L. Wang, A. Sancar and D. Zhong, Dynamics and Mechanism of Cyclobutane Pyrimidine Dimer Repair by DNA Photolyase, Biophys. Comput. Biol., 2011, 108(36), 14831–14836, DOI:10.1073/pnas.1110927108/-/DCSupplemental.
- H. O. Abdallah, Y. P. Ohayon, A. R. Chandrasekaran, R. Sha, K. R. Fox, T. Brown, D. A. Rusling, C. Mao and N. C. Seeman, Stabilisation of Self-Assembled DNA Crystals by Triplex-Directed-Cross-Linking, Chem. Commun., 2016, 52, 8014–8017, 10.1039/C6CC03695C.
- J. Yamamoto, K. Nishiguchi, K. Manabe, C. Masutani, F. Hanaoka and S. Iwai, Photosensitized [2 + 2] Cycloaddition of N-Acetylated Cytosine Affords Stereoselective Formation of Cyclobutane Pyrimidine Dimer, Nucleic Acids Res., 2011, 39(3), 1165–1175, DOI:10.1093/nar/gkq855.
- D. G. T. Su, J. L. F. Kao, M. L. Gross and J. S. A. Taylor, Structure Determination of an Interstrand-Type Cis-Anti Cyclobutane Thymine Dimer Produced in High Yield by UVB Light in an Oligodeoxynucleotide at Acidic PH, J. Am. Chem. Soc., 2008, 130(34), 11328–11337, DOI:10.1021/ja8010836.
- E. Markkanen, Not Breathing Is Not an Option: How to Deal with Oxidative DNA Damage, DNA Repair, 2017, 82–105, DOI:10.1016/j.dnarep.2017.09.007.
- G. Pizzino, N. Irrera, M. Cucinotta, G. Pallio, F. Mannino, V. Arcoraci, F. Squadrito, D. Altavilla and A. Bitto, Oxidative Stress: Harms and Benefits for Human Health, Oxid. Med. Cell. Longevity, 2017, 8416763, DOI:10.1155/2017/8416763.
- A. J. Fornace, K. W. Kohnt and H. E. Kann, DNA Single-Strand Breaks during Repair of UV Damage in Human Fibroblasts and Abnormalities of Repair in Xeroderma Pigmentosum (Alkaline Elution of DNA/DNA Crosslinking/x-Ray Sensitivity/Excision Repair), 1976; vol. 73. https://www.pnas.org Search PubMed.
- R. P. Rastogi, Richa, A. Kumar, M. B. Tyagi and R. P. Sinha, Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair, J. Nucleic Acids, 2010, 2010, 592980, DOI:10.4061/2010/592980.
- H. J. Shields, A. Traa and J. M. Van Raamsdonk, Beneficial and, Detrimental Effects of Reactive Oxygen Species on Lifespan: A Comprehensive Review of Comparative and Experimental Studies, Front. Cell Dev. Biol., 2021, 9, 628157, DOI:10.3389/fcell.2021.628157.
- H. L. Lo, S. Nakajima, L. Ma, B. Walter, A. Yasui, D. Ethell and L. B. Owen, Differential Biologic Effects of CPD and 6–4PP UV-Induced DNA Damage on the Induction of Apoptosis and Cell-Cycle Arrest, BMC Cancer, 2005, 5, 135, DOI:10.1186/1471-2407-5-135.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, NIH Image to ImageJ: 25 Years of Image Analysis HHS, Public Access, 2012, vol. 9 Search PubMed.
- O. Kühn, T. Renger and V. May, Theory of Exciton-Vibrational Dynamics in Molecular Dimers, Chem. Phys., 1996, 204, 99–114 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01695e |
| This journal is © The Royal Society of Chemistry 2025 |