 Open Access Article
Open Access ArticleEnzyme-mediated proximity labeling reveals the co-translational targeting of DLGAP5 mRNA to the centrosome during mitosis†
Gang
Wang
 a,
Mo
Li
*b and
Peng
Zou
a,
Mo
Li
*b and
Peng
Zou
 *acd
*acd
aAcademy for Advanced Interdisciplinary Studies, PKU-Tsinghua Center for Life Science, Peking University, Beijing 100871, China. E-mail: zoupeng@pku.edu.cn
bCenter for Reproductive Medicine, Department of Obstetrics and Gynecology, Peking University Third Hospital, Beijing 100191, China. E-mail: limo@hsc.pku.edu.cn
cCollege of Chemistry and Molecular Engineering, Synthetic and Functional Biomolecules Center, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, PKU-IDG/McGovern Institute for Brain Research, Peking University, Beijing, 100871, China
dChinese Institute for Brain Research (CIBR), Beijing 102206, China
First published on 26th March 2025
Abstract
Subcellular RNA localization is a conserved mechanism in eukaryotic cells and plays critical roles in diverse physiological processes including cell proliferation, differentiation, and embryo development. Nevertheless, the characterization of centrosome-localized mRNAs remains underexplored due to technical difficulties. In this study, we utilize APEX2-mediated proximity labeling to map the centrosome-proximal transcriptome, identifying DLGAP5 mRNA as a novel centrosome-localized transcript during mitosis. Using a combination of drug perturbation, truncation, deletion, and mutagenesis, we demonstrate that microtubule binding of nascent MBD1 polypeptides is required for centrosomal transport of DLGAP5 mRNA. Our data also reveal that mRNA targeting efficiency is tightly linked to the coding sequence (CDS) length. Thus, our study provides a transcriptomic resource for future investigation of centrosome-localized RNAs and sheds light on mechanisms underlying mRNA centrosomal localization.
Introduction
Centrosomes are evolutionarily conserved organelles that play critical roles in diverse biological processes including cell proliferation, adhesion, motility, and polarity maintenance.1 As the main microtubule organizing center in metazoan cells, each centrosome contains two orthogonally positioned centrioles surrounded by an electron-dense proteinaceous matrix known as the pericentriolar material (PCM).2 In proliferating cells, the number, structure, and composition of centrosomes are tightly linked to the cell cycle.3 Structural or functional centrosome abnormalities have been proposed to contribute to tumorigenesis, microcephaly, and ciliopathies.3,4 Our understanding of the structure, function, and dynamics of the centrosome has benefited greatly from a combination of biochemical, proteomic, and bioinformatic approaches, revealing functional roles of centrosomal proteins in microtubule nucleation and cell cycle regulation.5,6 However, the characterization of RNAs localized to the centrosome has remained largely underexplored.Centrosomal mRNA localization might provide an efficient and rapid approach for transporting large centrosomal scaffold proteins to the centrosome and preventing ectopic PCM assembly.7–10 Disruption of centrosomal mRNA accumulation often results in dysfunctional centrosome and deficient ciliogenesis.7–9 For example, mistargeting Drosophila centrosomal mRNA Cen to the anterior cortex of the embryo impaired Cen protein localization to distal centrosomes, thus generating phenotypes similar to Cen loss. Ectopic Cen mRNA enrichment also interfered with local microtubule organization and spindle morphogenesis.8 These observations indicate that the local abundance of centrosomal mRNA is critical to maintain centrosome function. Therefore, identifying centrosome-localized RNAs and clarifying their targeting mechanisms could promote our understanding of their physiological roles.
Fluorescence microscopy and RNA sequencing are two major methods for assaying the subcellular localization of RNAs. The centrosomal localization of specific transcripts has been resolved in various biological contexts via in situ hybridization (ISH), including Drosophila,11,12Xenopus,13Ilyanassa,14Spisula,15 zebrafish,7 and HeLa cell lines.7,10 Genetically encoded RNA tags, such as MS2/MCP, enabled the visualization of ASPM or NUMA1 mRNA to the centrosome in live cells.10 Besides imaging-based methods, transcriptome-wide analysis of purified mitotic spindles further expanded the list of spindle-associated RNAs.16,17 However, due to challenges associated with purifying the centrosome, biochemical fractionation often requires pre-stabilizing the mitotic spindle with taxol to facilitate its precipitation, which limits its applications.16,17 Thus, high-throughput sequencing techniques currently remain under-utilized in discovering centrosome-localized transcripts.
Enzyme-mediated proximity labeling (PL) methods have emerged as a powerful tool for deciphering molecular interactions.18,19 These methods utilize promiscuous labeling enzymes, such as APEX2, to generate reactive intermediates to label neighboring biomolecules including proteins and RNAs.20–22 While the high spatial specificity of APEX2 has enabled the proteomic mapping of centrosomal components,23 PL methods have not been applied to profile the centrosomal transcriptome. We have recently developed the MERR APEX-seq method with improved RNA detection sensitivity through metabolic incorporation of electron-rich nucleosides (s6G or s4U).24 Herein, we applied MERR APEX-seq to decipher the centrosome-proximal transcriptome and identified DLGAP5 as a centrosome-localized mRNA in the mitotic phase. We further demonstrated that the targeting of DLGAP5 depends on the microtubule cytoskeleton and the ribosomal translation of microtubule binding domain 1 (MBD1) polypeptides.
Results
Profiling centrosome-proximal transcriptome with MERR APEX-seq
To target the APEX2 protein to the centrosome, we fused the enzyme with the pericentrin-AKAP450 centrosomal targeting (PACT) domain, a conserved centrosomal localization motif at the C-terminus of the PCNT protein.25 We created a human embryonic kidney 293T (HEK293T) cell line stably expressing the APEX2-PCNT-EGFP fusion protein via lentiviral infection. Immunofluorescence imaging confirmed the centrosomal-specific targeting of the fusion protein, which co-localized with three representative endogenous centrosomal markers, i.e. CEP152, CDK5RAP2, and γ-tubulin (Fig. 1A). Considering that biotin–phenoxyl free radicals might diffuse from the centrosome to the cytosol, we generated a HEK293T cell line stably expressing cytosol-localized APEX2 via fusion with a nuclear export sequence from HIV-1 Rev protein 30 (APEX2-NES), as the non-targeted control.Next, we evaluated the spatial specificity of MERR APEX-seq labeling in APEX2-PCNT-EGFP and APEX2-NES cell lines via immunofluorescence imaging. Both cell lines were pretreated with s6G for 5 hours before incubation with 0.5 mM biotin–phenol (BP) for 30 min, and the biotinylation reaction was triggered by the addition of hydrogen peroxide (H2O2). After 1 min H2O2 treatment, the reaction was quenched by a cocktail of free radical scavengers and peroxidase inhibitors, including sodium ascorbate, sodium azide, and Trolox (Fig. 1B). Thereafter, the cells were either fixed for imaging or lysed for RNA-seq analysis. Immunofluorescence imaging revealed that the BP labeling signal co-localized with APEX2 expression in APEX2-PCNT-EGFP cell lines (Fig. 1C). In the negative control sample omitting the BP probe, only the signal from endogenous biotinylated proteins was observed. In the APEX2-NES cell line, the biotinylation signal mainly distributed throughout the cytosol, with a negligible background in the control (Fig. S1, ESI†).
To obtain the transcriptomes specifically labeled by APEX2-PCNT-EGFP or APEX2-NES fusion proteins, the total RNAs were extracted from labeled cells, digested by DNase I to remove residual DNA, and purified by streptavidin-coated magnetic beads. Following isolation of poly(A)+ RNA, biotinylated RNAs were subjected to fragmentation for cDNA library construction and high-throughput sequencing (Fig. 1B). We performed three independent biological replicate experiments with both cell lines. Enrichment of biotinylated RNAs from the APEX2-PCNT-EGFP and APEX2-NES cell lines was reproducible as revealed by high Pearson's correlation coefficients (>0.94) (Fig. S2A and B, ESI†).
To identify centrosome-localized transcripts, we performed DESeq226 analysis comparing the abundance of RNAs enriched from the centrosome-localized APEX2 sample (enrich) versus two separate negative controls: (1) RNAs from unlabeled samples omitting BP (control); or (2) RNAs labeled by cytosol-localized APEX2 to remove the background signals arising from free radical diffusion. Furthermore, to normalize gene expression levels, RNAs with significantly higher abundance (log2Fold change >1 and an FDR-adjusted p-value < 0.05) in the APEX2-PCNT-EGFP cell line were removed from the second dataset (see Methods and Data S1, ESI†). The above analysis yielded 112 transcripts, among which 15 mRNAs encode known centrosome- or mitosis-associated proteins (Fig. 1D and E). The percentage (13%) of transcripts with centrosome-associated GOCC annotations in our dataset was about two-fold of that (7%) in mRNAs expressed in the APEX2-PCNT-EGFP cell line (Fig. S3, ESI†). NIN, a previously reported centrosome-localized transcript,9,10 was included in our dataset (Fig. 1E and Table S3, ESI†). We confirmed its centrosomal enrichment during interphase via smFISH, which was consistent with previous studies.9,10 (Fig. S4, ESI†). Other known centrosome-targeted mRNAs (ASPM, PCNT, NUMA1, etc.) were not identified as enriched transcripts in our proximity labeling experiments (Table S3, ESI†).
DLGAP5 mRNA is localized at the centrosome during mitosis
To further validate our centrosome-proximal RNA dataset, we selected 13 transcripts with the highest fold change values (CLGN, KRT8, BNIP3L) or centrosome-associated GOCC annotations (DLGAP5, KAT2B, TOP2A, etc.) for further smFISH imaging analysis. Surprisingly, a majority of these transcripts did not display centrosomal localization throughout the cell cycle (Fig. S5, ESI†). Nevertheless, DLGAP5 mRNA, scoring the highest enrichment fold among the 15 transcripts, was identified as a novel centrosome-localized transcript in smFISH validation (Fig. 2A). Thus, we focus on the localization of DLGAP5 mRNA in the rest of this study.The DLGAP5/HURP protein encoded by DLGAP5 mRNA co-localizes with the mitotic spindle when cells enter into mitosis. It is a Ran-Importin β-regulated protein that stabilizes the kinetochore microtubules in the vicinity of chromosomes.27–31 Consistent with the previous study,31 smFISH imaging revealed that the level of DLGAP5 expression was more variable across cells during interphase than during the mitotic phase (Fig. S6, ESI†). smFISH imaging also confirmed the centrosomal targeting of DLGAP5 mRNA, and revealed that its subcellular localization was tightly linked to the cell cycle. During interphase and early mitosis, DLGAP5 mRNA was randomly distributed in the cytoplasm. When cells enter prometaphase and metaphase, prominent centrosomal localization of DLGAP5 mRNA was observed. As the cell cycle progresses into the late mitotic stages, DLGAP5 mRNA became gradually diffusive throughout the cell (Fig. 2A and B).
We next examined whether the centrosomal localization of DLGAP5 mRNA during mitosis is conserved among other cell types and species. To this end, we investigated the localization of DLGAP5 mRNA in five human cancer cell lines: HeLa, MCF-7, MDA-MB-231, U-2 OS, and SH-SY5Y. Remarkably, except for SH-SY5Y cells, we observed centrosomal or spindle pole localization of DLGAP5 mRNA in all other cell lines during mitosis (Fig. 2C). Additionally, we explored the subcellular localization of Dlgap5 mRNA, the mouse orthologs of the human DLGAP5 mRNA, in the mouse Neuro-2a cell line and observed the centrosomal targeting of Dlgap5 mRNA in metaphase (Fig. 2C). Together, these observations demonstrate that the centrosomal localization of DLGAP5 mRNA is evolutionarily conserved, suggesting a common mechanism of centrosomal mRNA targeting in both human and mouse cells.
Translation of DLGAP5 mRNA is both necessary and sufficient for its centrosomal targeting
We sought to explore the centrosomal targeting mechanism of DLGAP5 mRNA. Recent studies have revealed that the centrosomal targeting of mRNAs generally requires translation in mammalian cells.7,9,10,32 We therefore examined whether centrosomal localization depends on DLGAP5 translation. We treated HEK293T cells for 20 min with either cycloheximide, which blocks translation elongation but preserves the ribosome-nascent chain complex (RNC), or puromycin, which generates premature translation termination and disrupts the RNC.33,34 Following drug perturbation, we visualized endogenous DLGAP5 mRNA via smFISH imaging and the centrosome using the antibody against the endogenous PCNT protein. Whereas puromycin nearly abolished the centrosomal localization of DLGAP5 mRNA, cycloheximide slightly promoted centrosomal localization (Fig. 3A and B). Since the nascent peptides remain associated with ribosomes after treatment with cycloheximide but not with puromycin, we suspected that centrosomal targeting of DLGAP5 mRNA requires the nascent peptides. Additionally, our data suggested that as long as the RNCs are kept intact, the maintenance of DLGAP5 mRNA localization does not require active translational elongation.Active transport of mRNPs usually involves cytoskeleton networks.35,36 Centrosomes localize at the minus-end of microtubules and function as the main microtubule-organizing centers.1 Thus, we next examined whether the localization of DLGAP5 mRNA relies on the microtubules. When HEK293T cells were treated for 20 min with nocodazole, an inhibitor of microtubule polymerization, the microtubule cytoskeleton was disintegrated while the centrosome remained intact (Fig. S7, ESI†). Consistent with the role of the cytoskeleton in mRNA localization, nocodazole significantly disrupted the centrosomal enrichment of DLGAP5 mRNA (Fig. 3A and B).
Taken together, our data suggested that the DLGAP5 mRNA localization depends on the intact RNC and microtubule cytoskeleton in HEK293T cells. We next asked whether the localization mechanism is conserved in other cell types. For this purpose, we repeated puromycin, cycloheximide, and nocodazole perturbation in HeLa cells. Consistent with our observation in HEK293T cells, puromycin and nocodazole, but not cycloheximide, significantly affected the centrosomal accumulation of DLGAP5 mRNA during mitosis (Fig. S8, ESI†). These data indicated that DLGAP5 mRNA is co-translationally targeted to the centrosome via cytoskeletons in HEK293T and HeLa cells.
To further elucidate how DLGAP5 mRNA was transported to the centrosomes, we designed the following EGFP reporter mRNAs to examine which region of DLGAP5 mRNA was sufficient and necessary for its localization. We started by testing whether the untranslated regions (UTRs) of DLGAP5 mRNA is capable of targeting EGFP to centrosomes. We fused the EGFP ORF to either the 5′UTR of DLGAP5 (DLGAP5 5′UTR-EGFP) or the 3′UTR of DLGAP5 (EGFP-DLGAP5 3′UTR). When these reporters were transiently expressed in HeLa cells and detected using smFISH probes against the EGFP ORF sequence, we did not observe enrichment of EGFP mRNAs around the centrosomes in mitotic cells (Fig. 3C and D). When fusing EGFP ORF to the DLGAP5 CDS (EGFP-DLGAP5 CDS), smFISH imaging of the reporter showed substantial localization of the fusion mRNA to the centrosomes in metaphase, which was similar to endogenous DLGAP5 mRNA. Moreover, consistent with the previous study, the EGFP-DLGAP5 fusion proteins co-localized with the mitotic spindle (Fig. 3C and D).29,37 As a negative control, the mRNAs of EGFP ORF alone were randomly distributed throughout the cytoplasm in HeLa cells (Fig. 3C and D). Together, the above data demonstrated that the UTRs of DLGAP5 mRNA are dispensable for its centrosomal localization.
Given our previous observation that active protein translation is required for efficient DLGAP5 mRNA localization to the centrosomes, we next asked whether the DLGAP5 CDS sequence alone or its translation product mediates the centrosomal localization of its mRNA. By inserting a stop codon (UAG) between the EGFP ORF and DLGAP5 CDS, we created a reporter mRNA (EGFP-stop-DLGAP5 CDS) that differs from the previous EGFP-DLGAP5 CDS reporter by only the translation of the DLGAP5 CDS. Immuno-smFISH imaging revealed that the introduction of the stop codon abolished both the spindle localization of the EGFP protein and the centrosomal targeting of reporter mRNA (Fig. 3C and D). Taken together, we concluded that the translation of DLGAP5 CDS is both sufficient and necessary for trafficking its transcript to the centrosome during mitosis.
Nascent MBD1 polypeptide is required but not sufficient for the centrosomal localization of DLGAP5 mRNA
We then sought to identify the amino acid motifs/domains responsible for targeting DLGAP5 mRNA to the centrosome. As a member of the DLGAP family, DLGAP5 protein contains a conserved guanylate kinase associated protein (GKAP) domain (254–601 aa) with an unknown function.38 The N-terminus of DLGAP5 consists of two microtubule binding domains (MBDs), MBD1 (65–174 aa) and MBD2 (1–69 aa),37,39 while its C-terminus is intensively post-translationally phosphorylated by cell cycle-dependent kinases (Fig. 4A).40,41Based on the DLGAP5 protein structure, we designed two panels of EGFP reporters to identify the domain involved in its mRNA localization. The first panel contains C-terminally truncated DLGAP5 CDS with varying lengths, which aims for identifying the minimal CDS fragment that is sufficient to localize the reporter mRNA. (Fig. 4A). smFISH imaging analysis reveals that fusions with DLGAP5 CDS encoding the first 624, 425, or 308 aa are sufficient to localize the reporter mRNA to the centrosome. In contrast, fusion with only the first 231 aa of the protein failed to achieve mRNA targeting to the centrosome (Fig. 4A and B). These observations strongly suggest that the first 308 aa in the DLGAP5 polypeptide, which include both MBD1/2 domains, might mediate DLGAP5 mRNA localization, and the segment between 232 and 308 aa likely plays a key role in this process.
In the second panel, several CDS fragments are individually deleted from the full-length DLGAP5 CDS, allowing the examination of peptide segments that are necessary for the centrosomal mRNA targeting (Fig. 4A). Consistent with our results from the above truncation analysis, deletion of amino acids after the site 308 (Δ309–425 or Δ426–624) from the full-length CDS had minimal effects on the centrosomal enrichment of the reporter mRNA (Fig. 4A and C). It was expected that deletion of the peptide segment between amino acid 232 and 308 (Δ232–308) should abolish the DLGAP5 mRNA localization to the centrosomes. However, no significant effect was observed in this deletion on DLGAP5 mRNA targeting (Fig. 4A and C). As the deletion sites moved further to the N-terminus, our data showed that both the segments of 2–69 aa (i.e. MBD2 domain) and 175–231 aa are dispensable for the mRNA targeting (Fig. 4A and C). However, removal of the MBD1 domain (Δ65–174) led to almost complete abolishment of the centrosomal mRNA targeting (Fig. 4A and D). Taken together, the above data demonstrated that MBD1 domain is necessary but not sufficient to drive DLGAP5 mRNA targeting to the centrosome.
We next sought to determine whether the interaction between MBD1 and microtubules is involved in DLGAP5 mRNA localization to the centrosome. The binding of the DLGAP5 protein to the microtubules has been shown to regulate its localization, degradation, and function. While MBD1 constitutively binds to microtubules with high affinity, the interaction between MBD2 and microtubules is weaker and inhibited by Importin β.30,39,42 We replaced six critical positively charged residues in MBD1 (K105, K107, R110, K112, K114, and R115) with alanine to ablate its microtubule binding activity.39 Introducing these mutations into full-length CDS (MBD1*) significantly disrupts the localization of the reporter mRNA and EGFP fusion protein (Fig. 4A and D). Consistent with the above observation, removal of the peptide segment that harbors the above critical residues (Δ90–120) also abolishes centrosomal mRNA targeting (Fig. 4A and D). Together, these results confirm that the microtubule binding activity of MBD1 is critical for the centrosomal localization of DLGAP5 mRNA.
The centrosomal-targeting efficiency of DLGAP5 mRNA is tightly linked with its CDS length
The observation that the microtubule-binding of MBD1 is necessary but insufficient to drive DLGAP5 mRNA localization prompted us to speculate the involvement of factors other than amino acid sequences. We hypothesized that the nascent peptide length might influence the centrosomal targeting efficiency of reporter mRNAs. This hypothesis is inspired by the fact that most of the reported centrosome-localized mRNAs in HeLa cells contain long CDS (Fig. S9, ESI†). In the targeting deficient reporter containing CDS encoding the first 231 aa, when we replaced the C-terminal EGFP (475 aa) with a tandem dimer GFP (tdGFP, 723 aa), the centrosomal localization is modestly but significantly improved (Fig. S10, ESI†), thus offering preliminary experimental support to the length hypothesis.We then created three reporter mRNAs containing the CDS of MBD1 fused to the N-terminus of EGFP (MBD1-EGFP, 355 aa), tdGFP (MBD1-tdGFP, 603 aa), or tdGFP-Halotag (MBD1-tdGFP-HT, 907 aa) to carefully verify our hypothesis. Among these reporters, the longest fusion MBD1-tdGFP-HT exhibited the strongest centrosomal localization of the EGFP mRNA reporter (Fig. 5A and B). Replacing the MBD1 domain with the microtubule-binding-deficient MBD1* abolished centrosomal enrichment of reporter mRNA, further confirming the vital role of the microtubule binding activity of MBD1 (Fig. 5A and B). When the CDS sequence is shuffled with the MBD1 domain moved further to the C-terminus (tdGFP-MBD1-HT and tdGFP-HT-MBD1), the centrosomal localization of reporter EGFP mRNA is also significantly reduced (Fig. 5A and B). Taken together, we concluded that the centrosomal targeting of DLGAP mRNA requires the translation of its CDS encoding MBD1 along with a long peptide (approximately 600 aa) at the C-terminus.
Finally, we sought to understand why the CDS length is linked to mRNA localization efficiency. Generally, longer CDS means that the ribosomes stay associated with mRNA for a longer time. Nevertheless, in our reporter assay, cells transiently expressing EGFP reporters were treated with cycloheximide prior to fixation, resulting in the freezing of ribosomes on the mRNAs. Thus, the longer engagement of ribosomes with mRNAs might not be a main contributor. Alternatively, we supposed that the nascent MBD1 peptide dwell time and MBD1 peptide concentration might play vital roles in mRNA localization. For instance, when MBD1 is positioned at the N-terminus of the construct, increasing CDS length simultaneously prolongs the dwell time of nascent MBD1 peptides on the mRNA and elevates the number of translating ribosomes. Both factors increase the local concentration of nascent MBD1 peptides in the polysome, synergistically enhancing centrosomally targeting of reporter mRNA.
To verify our hypothesis, we fused tandem MBD1 to the N-terminus of EGFP, thus producing tandem nascent MBD1 polypeptides. The reporter mRNA transcribed from this construct localized to the centrosome. However, when replacing the second MBD1 with MBD1*, the centrosomal accumulation of reporter mRNA was disrupted without affecting the spindle localization of EGFP fusion proteins (Fig. 5B and C). The distinct localization of the above two reporters with the same CDS length further implied that the residence time of ribosomes on the mRNAs may not play key roles. Taken together, these observations supported the role of the local concentration of nascent MBD1 polypeptides in promoting the centrosomal targeting of DLGAP5.
Discussion
In summary, we have applied MERR APEX-seq to profile the centrosome-proximal transcriptome in cultured mammalian cells. From the transcripts significantly enriched by MERR APEX-seq, we confirmed the centrosomal localization of DLGAP5 mRNA by smFISH imaging across multiple mammalian cell types, suggesting a conserved targeting mechanism. The sensitivity of DLGAP5 mRNA localization towards puromycin treatment indicated the involvement of the intact ribosome-nascent chain complex, which is consistent with the previously reported co-translational transport mechanism of centrosome-localized mRNAs.10,32 Our model suggested that DLGAP5 mRNAs undergo local translation in pericentrosomal regions. Indeed, ribosomes are consistently observed near centrosomes across species. Electron microscopy reveals their proximity to centrioles and basal bodies,43 while the biochemical isolation of mitotic spindles from sea urchins confirms their evolutionary conservation in this compartment.44 Immunofluorescence further corroborates localization of ribosomal components to the centrosome.7,16We further identified microtubule interactions with nascent MBD1 polypeptides as the mediator for the centrosomal localization of DLGAP5 mRNA (Fig. 6). Interestingly, several centrosome-localized mRNAs have been reported to encode proteins that can directly bind microtubules (e.g. ASPM, NUMA1, HMMR, and CEP350) or the motor protein dynein (e.g. NIN, BICD2, CCDC88C, and NUMA1).45–49 It is likely that the microtubule- or dynein-interacting nascent polypeptides might participate in the transport of their mRNA. This hypothesis could be tested using reporter mRNA assays in future studies.
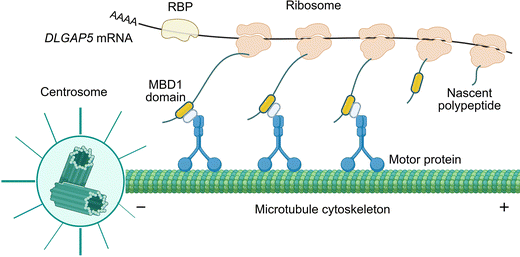 | ||
| Fig. 6 Proposed localization mechanism for DLGAP5 mRNA. Created in BioRender. https://BioRender.com/omwo3kq. | ||
An intriguing finding in this study is the dependence of DLGAP5 mRNA centrosomal targeting on its CDS length, the dwell time and copy number of the MBD1 domain, all of which likely affect the local concentration of nascent MBD1 polypeptides on the polysomes. Increasing the CDS length or shifting MBD1 to the N-terminus drives the centrosomal targeting of the reporter mRNA. Indeed, emerging evidence links the transcript length to subcellular RNA localization. For instance, shorter transcripts are enriched in nuclear speckles, while longer transcripts associate with the nuclear lamina or G1-phase processing bodies.50,51 However, whether the CDS length influences co-translational mRNA targeting via nascent polypeptide quantities remains unexplored in other subcellular compartments. Furthermore, MBD1 contains a conserved coiled-coil motif (90–120 aa), which might undergo liquid–liquid phase separation (LLPS) in a concentration manner.52 A recent study revealed that PCNT undergoes phase separation through its coiled-coils and low-complexity regions during co-translational transport of its mRNA to the centrosome.53 Whether similar LLPS occurs during the centrosomal targeting of DLGAP5 mRNA warrants further investigation.
While we have identified the nascent MBD1 domain as a necessary element for DLGAP5 mRNA localization, several questions remain regarding the localization mechanism. On the one hand, the transport machinery responsible for centrosomal targeting and interacting partners of MBD1 polypeptides remain unknown. Dynein, which transports cargoes to the microtubule minus-end, is implicated in the centrosomal localization of PCNT and NIN mRNA.7,9 However, our preliminary experiments showed that the inhibition of dynein had little effect on DLGAP5 mRNA localization (data not shown). Thus, whether dynein is involved in DLGAP5 mRNA transport is still an open question. On the other hand, centrosomal targeting of DLGAP5 mRNA is tightly linked to the cell cycle, but regulators related to its cell cycle dynamics remain elusive. Phosphorylation of the C-terminus of the DLGAP5 protein by Aurora A was found to control its microtubule binding activity of the N-terminus through the autoinhibition mechanism.37 Thus, cell-cycle-regulated post-translational modifications in the nascent DLGAP5 polypeptide might play a role in the mRNA localization. Moreover, mitosis-specific translation regulators might be involved in the localization process.
Subcellular RNA localization generally enables precise spatio-temporal control of gene expression.35,36 The biological significance of DLGAP5 mRNA localized to the centrosome is unresolved in this study. Considering that the local abundance of the DLGAP5 protein on the mitotic spindle is related to spindle stability and mitosis progression, its mRNA localization might be an ideal approach to finely tune the local protein dosage.28,30,42 Approaches including transcript-specific translation inhibition and the ectopic targeting of aptamer-tagged mRNAs should help us unearth the physiological role of DLGAP5 mRNA localization.
Materials and methods
Reagents and resources
A full description of the reagents and resources utilized in this study is listed in Table S1 (ESI†).Mammalian cell culture
HEK293T, HeLa, MCF-7, SH-SY5Y, Neuro-2a, and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, C11995500BT) supplemented with 10% fetal bovine serum (FBS, Gibco, 10099141). U-2 OS cells were cultured in McCoy's 5A Medium (BI, 01-075-1ACS) supplemented with 10% FBS. The HEK293T, HeLa S3, SH-SY5Y, U-2 OS, and Neuro-2a cell lines were obtained from the National Infrastructure of Cell Line Resource, Beijing, China. The MCF-7 and MDA-MB-231 cell lines were generous gifts from Prof. Mo Li, Peking University. The source of the cell lines utilized in the study is available in Table S1 (ESI†). All the cell lines were maintained at 37 °C under 5% CO2.Generation of HEK293T cell lines stably expressing APEX2 fusion proteins
To construct cells stably expressing APEX2-PCNT-EGFP or APEX2-NES, HEK293T cells cultured in 6-well plates at 60–80% confluency were transfected with a lentiviral vector (1 μg) and two packaging plasmids pVSVG (0.7 μg) and dR8.91 (1 μg) with the assistance of the Lipofectamine 3000 Transfection reagent (5.4 μL) (Invitrogen, L3000015) in a total of 200 μL Opti-MEM I Reduced Serum Medium (Gibco, 31985062). 6 hours after transfection, the medium was replaced with a fresh complete medium. 48 hours after transfection, the lentivirus-containing medium was collected and filtered through a 0.45 μm syringe filter. 1 mL of the lentivirus-containing medium was added to a wild-type HEK293T cell culture at 60–70% confluency in a 6-well plate. About 48 hours after lentiviral infection, the medium was changed to a complete medium supplemented with 5 μg mL−1 blasticidin (Selleck, S7419). The infected cells were maintained in a blasticidin-containing complete medium for about two weeks. Immunofluorescence imaging was utilized to validate the expression of APEX2-NES or APEX2-PCNT-EGFP fusion proteins.Genetic plasmid construction
For construction of EGFP reporters, DLGAP5 CDS was amplified from cDNA reverse transcribed from the total RNAs of the HEK293T cell line using a ProtoScript II First Strand cDNA Synthesis Kit (NEB, E6560). DLGAP5 5′UTR and 3′UTR were cloned from genomic DNA extracted from HEK293T. The tdGFP fragment was amplified from pUbC-stdMCP-stdGFP (a generous gift from Prof. Wulan Deng, Peking University). All the PCR reactions were performed with a Phanta Max Super-Fidelity DNA Polymerase (Vazyme, P505-d1). The fragments were assembled into the scaffold of pUbC-stdMCP-stdGFP using a Lightning Cloning Kit (Biodragon, BDIT0014). The information of EGFP reporter plasmids used in this study are listed in Table S2 (ESI†). The sequences of reporter plasmids were confirmed by Sanger sequencing at Ruibiotech (Beijing).MERR APEX-seq labeling in living cells
HEK293T cells stably expressing APEX2-NES or APEX2-PCNT-EGFP at 80–90% confluency were pre-treated with 100 μM s6G in a complete medium for 5 hours before incubation with a 0.5 mM BP probe for 30 min at 37 °C under 5% CO2. Biotinylation was initiated by addition of 1 mM H2O2 for 1 min. To halt the APEX2 labeling, the cells were treated with a quencher solution containing 10 mM sodium ascorbate, 5 mM Trolox, and 10 mM sodium azide for 2 min. After quenching the reaction three times, the cells were either fixed for imaging or lysed for RNA extraction.Immunofluorescence microscopy
For general immunostaining, the cells were fixed with 4% formaldehyde in PBS at room temperature for 20 min and washed three times with PBS. Next, the cells were permeabilized by PBS containing 0.2% Triton X-100 (Aladdin, T109026) for 15 min. After 3 times’ washing, the cells were incubated with the blocking buffer (0.5% Tween-20 (Sigma, P1379) and 3% bovine serum albumin (BSA, Sangon Biotech, A500023) in PBS) at room temperature for 1 hour. Then, the cells were incubated with primary antibodies (see below) diluted in a blocking buffer at 4 °C overnight. After being washed with PBST (0.5% Tween-20 in PBS) three times, the cells were further incubated with secondary antibodies (see below) diluted in PBS for 1 hour at room temperature. Following washing with PBST three times, the cells were stained with DAPI (ThermoFisher, D1306) diluted in PBS for 15 min, washed twice with PBS, and maintained in PBS at 4 °C before imaging. For immunostaining of APEX2-mediated biotinylation, the cells were fixed and permeabilized with pre-chilled methanol (−20 °C) for 10 min at room temperature. Then, the cells were blocked, incubated with primary and secondary antibodies, and stained with DAPI as mentioned above.Primary antibodies used in this study are listed as below: mouse V5-tag monoclonal antibody (3C8) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, Biodragon, B1005), rabbit anti-CEP152 antibody (1
800, Biodragon, B1005), rabbit anti-CEP152 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Sigma-Aldrich, HPA039408), rabbit anti-CDK5RAP2 antibody (1
200, Sigma-Aldrich, HPA039408), rabbit anti-CDK5RAP2 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Sigma-Aldrich, HPA046529), mouse monoclonal anti-γ-tubulin antibody (1
200, Sigma-Aldrich, HPA046529), mouse monoclonal anti-γ-tubulin antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Sigma-Aldrich, T5326), rabbit anti-PCNT antibody (1
1000, Sigma-Aldrich, T5326), rabbit anti-PCNT antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, abcam, ab4448), and the rat anti-α-tubulin monoclonal antibody (YL1/2) (1
500, abcam, ab4448), and the rat anti-α-tubulin monoclonal antibody (YL1/2) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Invitrogen, MA180017). Secondary antibodies against the above primary antibodies were: Alexa Fluor 647-conjugated anti-mouse goat IgG (H + L) (1
500, Invitrogen, MA180017). Secondary antibodies against the above primary antibodies were: Alexa Fluor 647-conjugated anti-mouse goat IgG (H + L) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Invitrogen, A-21236), Alexa Fluor 568-conjugated anti-rabbit goat IgG (H + L) (1
1000, Invitrogen, A-21236), Alexa Fluor 568-conjugated anti-rabbit goat IgG (H + L) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Invitrogen, A-11036), and Alexa Fluor 488-conjugated anti-rat goat IgG (H + L) (1
1000, Invitrogen, A-11036), and Alexa Fluor 488-conjugated anti-rat goat IgG (H + L) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Invitrogen, A-11006). Alexa Fluor 568-conjugated Streptavidin (1
1000, Invitrogen, A-11006). Alexa Fluor 568-conjugated Streptavidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Invitrogen, S11226) was utilized to counterstain biotinylation.
1000, Invitrogen, S11226) was utilized to counterstain biotinylation.
Affinity purification of biotinylated RNAs
Following APEX2 labeling, the cells were lysed by the addition of the TRIzol reagent (Invitrogen, 15596018). After homogenizing via pipetting, total RNAs were extracted via addition of chloroform, isopropanol precipitation, ethanol washing, and air drying according to the manufacturer's protocol. The extracted RNAs were further digested with DNase I (NEB, M0303) at 37 °C for 30 min. The reaction mix was purified using a RNA Clean & Concentrator kit (Zymo, R1018). A Fragment Analyzer (Agilent) was used to analyze the integrity of purified RNAs. Only samples of RQN > 9.0 were subjected to downstream affinity purification. 10 μL purified RNA was set as the input sample.The enrichment of biotinylated RNAs was performed as previously reported.54 Briefly, Dynabeads MyOne Streptavidin C1 beads (Invitrogen, 65002), using 10 μL beads per 50 μg of RNA, were washed three times with a bead washing buffer (5 mM Tris, pH7.5, 1 M NaCl, 0.5 mM EDTA, 0.1% v/v Tween-20), followed by twice in Solution A (0.1 M NaOH and 0.05 M NaCl) and once in Solution B (0.1 M NaCl). The beads were then blocked with a bead washing buffer supplemented with 1 mg mL−1 BSA and 1 mg mL−1 yeast tRNA (Invitrogen, 15401011) on the vortex at room temperature for two hours. Then, the beads were washed three times with a bead washing buffer, followed by incubation with purified RNAs in a bead washing buffer supplemented with 1 U μL−1 RiboLock RNase Inhibitor at room temperature for 40–50 min with thorough mixing. The biotinylated RNA-bound beads were next washed three times with a bead washing buffer, twice with urea buffer (4 M urea and 0.1% SDS in PBS), and twice with PBS at room temperature to remove non-specific absorption. Finally, biotinylated RNAs were eluted with RNA elution buffer (95% formamide, 10 mM EDTA, and 1.5 mM D-biotin) at 50 °C for 5 min, followed by 90 °C for 5 min on a shaker. The eluted RNAs were purified with TRIzol reagent as per manufacturer's instructions. 20 μg glycogen was added to the aqueous phase to assist precipitation before performing isopropanol precipitation. The purified biotinylated RNAs were dissolved in 20 μL RNase-free water and termed as the enrich sample for labeling samples and control sample for the negative control omitting the BP probe. Before the library construction of input, enrich, and control samples, we generally performed reverse transcription and qRT-PCR to evaluate the enrichment efficiency of biotinylated RNAs.
Drug perturbation assay
HEK293T or HeLa cells cultured on Matrigel-coated coverslips were treated with drugs at the following final concentrations: 0.2% for DMSO, 100 μg mL−1 for puromycin (J&K, 168086), 200 μg mL−1 for cycloheximide (Fluorochem, 375034), and 5 μg mL−1 for nocodazole (TargetMol, T14965). Treatment of cells with all drugs lasted for 20 min. After drug treatment, the cells were washed with PBS once, followed by fixation for smFISH imaging.EGFP reporter mRNA assay
HeLa cells were transfected with the corresponding constructs with the Lipofectamine 3000 transfection reagent according to the manufacturer's protocol. Briefly, ∼100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded into a 24-well plate. ∼24 hours later, the cells at 80–90% confluence were transfected with 0.5 μg plasmids using the 1 μL Lipofectamine 3000 transfection reagent in a total of 50 μL OPTI-MEM. 4 hours after transfection, the medium was replaced with a fresh complete medium. 18–24 hours after medium change, the transfected cells were trypsinized and replated on coverslips coated with Matrigel. 18–24 hours after cell adhesion, the cells were treated with a complete medium supplemented with 200 μg mL−1 cycloheximide for 20 min, followed by washing with PBS and fixation for smFISH imaging.
000 cells were seeded into a 24-well plate. ∼24 hours later, the cells at 80–90% confluence were transfected with 0.5 μg plasmids using the 1 μL Lipofectamine 3000 transfection reagent in a total of 50 μL OPTI-MEM. 4 hours after transfection, the medium was replaced with a fresh complete medium. 18–24 hours after medium change, the transfected cells were trypsinized and replated on coverslips coated with Matrigel. 18–24 hours after cell adhesion, the cells were treated with a complete medium supplemented with 200 μg mL−1 cycloheximide for 20 min, followed by washing with PBS and fixation for smFISH imaging.
Single-molecule FISH imaging
smFISH probes against endogenous mRNAs or EGFP ORF were designed using Oligostan.55 Each primary probe has a gene-specific region that is complementary to RNAs of interest, flanked by a common overhang called Flap X or Flap Y. Primary probes were synthesized by Genewiz (Tianjin). The lyophilized primary probes were dissolved in Tris-EDTA (TE) buffer (pH 8.0), at a final concentration of 100 μM. Then, an equimolar mixture of all primary probes against the same RNA were prepared, and diluted five times with TE buffer. The Alexa Fluor 568- or Alexa Fluor 647-conjugated Flap probes were synthesized by Invitrogen (Shanghai). The lyophilized fluorescent probes were resuspended with TE buffer and diluted to 10 μM for following experiments. Sequences of primary probes and Flap probes used in this study are listed in Data S2 (ESI†).The smFISH samples were prepared according to a previous report.56 Briefly, cells expressing EGFP reporters or treated with drugs were fixed with 3.2% paraformaldehyde (PFA) dissolved in PBSM (1 mM MgCl2 in PBS) at room temperature for 10 min. The fixed cells were then washed with pre-chilled PBSM supplemented with 10 mM glycine three times, followed by permeabilization with PBSM containing the 0.1% Triton X-100 and 2 mM vanadyl ribonucleoside complex (VRC) on ice for 20 min. After washing with PBSM twice, cells were incubated with prehyb-30 buffer (30% formamide, 2× SSC) at room temperature for 10 min with gentle shaking. Next, the cells were stained with the primary probe hybridization buffer (10% dextran sulfate, 30% formamide, 2× SSC, 2 mM VRC, 10 μg mL−1 salmon sperm DNA, 10 μg mL−1E. coli tRNA, 10 μg mL−1 BSA, and a 200 ng primary probe mix) at 37 °C overnight. After primary probe hybridization, cells were washed with prehyb-30 buffer at 37 °C three times, followed by washing with 2× SSC buffer once at room temperature. Then, cells were fixed with 1% PFA dissolved in PBSM at room temperature for 5 min. After two times’ washing, cells were incubated with prehyb-10 buffer (10% formamide, 2× SSC) at 37 °C for 10 min. Cells were hybridized with fluorescent Flap X or Flap Y probes in a secondary probe hybridization buffer (10% dextran sulfate, 10% formamide, 2× SSC, 2 mM VRC, 10 μg mL−1 salmon sperm DNA, 10 μg mL−1E. coli tRNA, 10 μg mL−1 BSA, and 10 ng FLAP probes) at 37 °C for at least three hours. Stained cells were then washed twice with prehyb-10 buffer at 37 °C, followed by a final wash in 2× SSC buffer.
For smFISH imaging in APEX2-PCNT-EGFP cells, the cells were stained with DAPI diluted in PBSM at room temperature for 10 min and mounted in a Fluoromount-G anti-fade mounting medium (SouthernBiotech, 0100-35) for following imaging. For smFISH imaging in wild-type HEK293T or HeLa cell lines, the endogenous PCNT protein was immunostained to mark the centrosome. Following washing in 2× SSC buffer, the cells were washed twice with PBSM, and incubated with the anti-PCNT antibody diluted in PBSM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) at 4 °C overnight. After three times washing with PBSM, the cells were incubated with the Alexa Fluor 568-conjugated goat anti-rabbit secondary antibody at room temperature for one hour. After washing with PBSM and staining with DAPI, cells were mounted for the following imaging.
500) at 4 °C overnight. After three times washing with PBSM, the cells were incubated with the Alexa Fluor 568-conjugated goat anti-rabbit secondary antibody at room temperature for one hour. After washing with PBSM and staining with DAPI, cells were mounted for the following imaging.
Image acquiring and analysis
Immunofluorescence images and smFISH images were collected via an inverted fluorescence microscope (Nikon-TiE) equipped with a 40× 1.3 NA oil immersion objective lens, a 60× 1.4 NA oil immersion objective lens, a spinning disk confocal unit (Yokogawa CSU-X1), and a scientific CMOS camera (Hamamatsu ORCA-Flash 4.0 v2). The imaging platform was controlled using customized software written in LabVIEW v15.0 (National Instruments). The power of lasers used to excite the fluorescent dyes or proteins was 1–6 W cm−2. In this study, the smFISH images were acquired under a 60× oil objective lens with an intermediate magnification switch (1.5×) on to increase spatial resolution. Additionally, images were taken as z-stacks with one plane every 0.5 μm.All the fluorescence images were analyzed using Fiji software.57 Mitotic cells were identified based on high DNA compaction and the existence of two pairs of centrioles manually. Quantitative analysis of smFISH images were performed using FISH-quant according to the manufacturer's manual.55,58 Briefly, the intensity of the DAPI channel was used to identify the nucleus and membrane boundary manually. For drug perturbation assays and EGFP reporter assays, only prometaphase, metaphase, and anaphase (estimated via DAPI staining) cells were chosen for following FISH-quant analysis. mRNA spots were detected from the smFISH channel and centrosomes from the anti-PCNT channel. mRNA spots were detected and counted using FISH-quant. For EGFP reporter assays, only cells with less than 500 spots per cell were subjected to co-localization analysis (Fig. S10B, S11, S12, and S13, ESI†). The colocalization of mRNA spots and centrosomes were analyzed using an FQ_DualColor module. The maximum allowed distance between the two spots was set as 2 μm according to a previous study.10 In other words, RNAs, less than 2 μm away from the nearest centrosome, were identified as centrosome-proximal RNAs. Significance of the proportion of centrosome-proximal mRNAs was evaluated with a two-sided Mann–Whitney test in this study.
Next generation sequencing
1 μg input, 14 μL enrich, and 14 μL control from each biological replicate experiment were subjected to library construction by the NEBNext Ultra II RNA Library Prep Kit for Illumina (NEB, E7770) with a poly(A) mRNA Magnetic Isolation Module (NEB, E7490) according to manufacturer's instructions. Briefly, after isolation of poly(A)+ RNA using magnetic oligo(dT) beads, the RNAs were fragmented at 94 °C for 10 min. The fragmented RNAs were subjected to first strand cDNA synthesis, followed by second strand cDNA synthesis. Double-stranded cDNA fragments were purified with 1.8× VAHTS DNA Clean Beads (Vazyme, N411). The purified cDNA was used for end repair, followed by immediate adaptor ligation. The reaction mix was further purified with 0.9× DNA Clean Beads to remove excess adaptors. The purified adaptor-ligated cDNA was proceeded to PCR amplification. The PCR cycles for input, enrich, and control samples are 10, 16–18, and 20–22, respectively. The PCR reaction was purified with 0.9× DNA Clean Beads. Two rounds of size fractionation were performed to obtain cDNA segments distributed between 300 and 400 bp with 0.7× and 0.2× DNA Clean Beads. Size distribution was further analyzed on the Fragment Analyzer. Samples with good quality were deep-sequenced for 150 bp paired reads on the Illumina HiSeq X Ten platform.NGS data analysis
The sequencing reads for each library were aligned against the human genome assembly GRCh38 (hg38) that was downloaded from Ensembl project using HISAT2 (v2.1.0).59 Then, the results of alignments were sorted by read names. The read counts of each gene were measured with the matching gene annotation (v.87) from Ensembl by HTSeq v0.6.1, using the option “-stranded no”.60 FPKM of each gene was calculated based on raw counts to estimate the reproducibility between biological replicates.To identify centrosome-localized RNAs, differential expression analysis between the labeled samples and control samples was carried out using DESeq2.26 The arbitrary cutoff for DESeq2 analysis was log2Fold change > 1, FDR-adjusted p-value < 0.05, and baseMean > 100. When applying the above cutoff to PCNT_enrich vs. PCNT_control analysis, 1903 RNAs were significantly enriched. For PCNT_enrich vs. NES_enrich analysis, 1595 RNAs were significantly enriched. Among these 1595 RNAs, 773 RNAs have significantly higher expression levels (log2Fold change >1 and FDR-adjusted p-value < 0.05) in the APEX2-PCNT-EGFP cell line. Thus, 822 RNAs were more preferentially labeled by centrosome-localized APEX2 compared to cytosol-localized APEX2. Overlapping the above 1903 and 822 RNAs generated the final dataset containing 112 RNAs. The list of these 112 RNAs is provided in Data S1 (ESI†). Centrosome-associated proteins were defined as proteins containing centrosome- or mitosis-related GOCC terms, i.e. “centrosome”, “centriole”, “pericentriolar material”, “microtubule”, “equatorial cell cortex”, “midbody”, “spindle”, “mitotic spindle”, “cell division site part”, “kinetochore”, “condensed chromosome”, “centromere”, and “telomere”.
Data availability
All data other than RNA sequencing data are incorporated into the article and its ESI.† The sequencing data underlying this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession code: GSE260463).Conflicts of interest
The authors declare no competing interests.Acknowledgements
We thank Dr B. Xie for assistance with cell line construction, Dr L. Peng for assistance with image acquiring and analysis, and Drs Y. Zhou and R. Li for helpful discussions. This work was supported by the Ministry of Science and Technology (2022YFA1304700) and the National Natural Science Foundation of China (32088101). PZ is sponsored by the Bayer Investigator Award. The authors thank the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with the Fragment Analyzer 12.References
- P. T. Conduit, A. Wainman and J. W. Raff, Nat. Rev. Mol. Cell Biol., 2015, 16, 611–624 CrossRef CAS PubMed.
- M. Bornens, Science, 2012, 335, 422–426 CrossRef CAS PubMed.
- E. A. Nigg and J. W. Raff, Cell, 2009, 139, 663–678 CrossRef CAS PubMed.
- E. A. Nigg and A. J. Holland, Nat. Rev. Mol. Cell Biol., 2018, 19, 297–312 CrossRef CAS PubMed.
- C. Arquint, A. M. Gabryjonczyk and E. A. Nigg, Philos. Trans. R. Soc. London, Ser. B, 2014, 369, 20130464 CrossRef PubMed.
- J. B. Woodruff, O. Wueseke and A. A. Hyman, Philos. Trans. R. Soc. London, Ser. B, 2014, 369, 20130459 CrossRef PubMed.
- G. Sepulveda, M. Antkowiak, I. Brust-Mascher, K. Mahe, T. Y. Ou, N. M. Castro, L. N. Christensen, L. Cheung, X. E. Jiang, D. Yoon, B. Huang and L. E. Jao, eLife, 2018, 7, e34959 CrossRef PubMed.
- P. V. Ryder, J. Fang and D. A. Lerit, J. Cell Biol., 2020, 219, e202004101 CrossRef CAS PubMed.
- O. S. Kwon, R. Mishra, A. Safieddine, E. Coleno, Q. Alasseur, M. Faucourt, I. Barbosa, E. Bertrand, N. Spassky and H. Le Hir, Nat. Commun., 2021, 12, 1351 CrossRef CAS PubMed.
- A. Safieddine, E. Coleno, S. Salloum, A. Imbert, A. M. Traboulsi, O. S. Kwon, F. Lionneton, V. Georget, M. C. Robert, T. Gostan, C. H. Lecellier, R. Chouaib, X. Pichon, H. Le Hir, K. Zibara, F. Mueller, T. Walter, M. Peter and E. Bertrand, Nat. Commun., 2021, 12, 1352 CrossRef CAS PubMed.
- J. W. Raff, W. G. Whitfield and D. M. Glover, Development, 1990, 110, 1249–1261 CrossRef CAS PubMed.
- E. Lécuyer, H. Yoshida, N. Parthasarathy, C. Alm, T. Babak, T. Cerovina, T. R. Hughes, P. Tomancak and H. M. Krause, Cell, 2007, 131, 174–187 CrossRef PubMed.
- I. Groisman, Y. S. Huang, R. Mendez, Q. P. Cao, W. Theurkauf and J. D. Richter, Cell, 2000, 103, 435–447 CrossRef CAS PubMed.
- J. D. Lambert and L. M. Nagy, Nature, 2002, 420, 682–686 CrossRef CAS PubMed.
- M. C. Alliegro, M. A. Alliegro and R. E. Palazzo, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9034–9038 CrossRef CAS PubMed.
- M. D. Blower, E. Feric, K. Weis and R. Heald, J. Cell Biol., 2007, 179, 1365–1373 CrossRef CAS PubMed.
- S. Hassine, F. Bonnet-Magnaval, L. P. B. Bouvrette, B. Doran, M. Ghram, M. Bouthillette, E. Lecuyer and L. DesGroseillers, J. Cell Sci., 2020, 133, jcs247155 CrossRef CAS PubMed.
- W. Qin, K. F. Cho, P. E. Cavanagh and A. Y. Ting, Nat. Methods, 2021, 18, 133–143 CrossRef CAS PubMed.
- Y. Zhou and P. Zou, Curr. Opin. Chem. Biol., 2021, 60, 30–38 CrossRef CAS PubMed.
- H. W. Rhee, P. Zou, N. D. Udeshi, J. D. Martell, V. K. Mootha, S. A. Carr and A. Y. Ting, Science, 2013, 339, 1328–1331 CrossRef CAS PubMed.
- S. S. Lam, J. D. Martell, K. J. Kamer, T. J. Deerinck, M. H. Ellisman, V. K. Mootha and A. Y. Ting, Nat. Methods, 2015, 12, 51–54 CrossRef CAS PubMed.
- F. M. Fazal, S. Han, K. R. Parker, P. Kaewsapsak, J. Xu, A. N. Boettiger, H. Y. Chang and A. Y. Ting, Cell, 2019, 178, 473–490 CrossRef CAS PubMed.
- B. Xie, Y. Pu, F. Yang, W. Chen, W. Yue, J. Ma, N. Zhang, Y. Jiang, J. Wu, Y. Lin, X. Liang, C. Wang, P. Zou and M. Li, Cancer Res., 2022, 82, 2576–2592 CrossRef CAS PubMed.
- R. Li, Z. Zou, W. Wang and P. Zou, Cell Chem. Biol., 2022, 29, 1218–1231 CrossRef CAS PubMed.
- A. K. Gillingham and S. Munro, EMBO Rep., 2000, 1, 524–529 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- M. D. Koffa, C. M. Casanova, R. Santarella, T. Köcher, M. Wilm and I. W. Mattaj, Curr. Biol., 2006, 16, 743–754 CrossRef CAS PubMed.
- H. H. Silljé, S. Nagel, R. Körner and E. A. Nigg, Curr. Biol., 2006, 16, 731–742 CrossRef PubMed.
- J. Wong and G. W. Fang, J. Cell Biol., 2006, 173, 879–891 CrossRef CAS PubMed.
- K. Tsuchiya, H. Hayashi, M. Nishina, M. Okumura, Y. Sato, M. T. Kanemaki, G. Goshima and T. Kiyomitsu, Curr. Biol., 2021, 31, 115–127 CrossRef CAS PubMed.
- A. P. Tsou, C. W. Yang, C. Y. F. Huang, R. C. T. Yu, Y. C. G. Lee, C. W. Chang, B. R. Chen, Y. F. Chung, M. J. Fann, C. W. Chi, J. H. Chiu and C. K. Chou, Oncogene, 2003, 22, 298–307 CrossRef CAS PubMed.
- R. Chouaib, A. Safieddine, X. Pichon, A. Imbert, O. S. Kwon, A. Samacoits, A. M. Traboulsi, M. C. Robert, N. Tsanov, E. Coleno, I. Poser, C. Zimmer, A. Hyman, H. Le Hir, K. Zibara, M. Peter, F. Mueller, T. Walter and E. Bertrand, Dev. Cell, 2020, 54, 773–791 CrossRef CAS PubMed.
- M. E. Azzam and I. D. Algranati, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 3866–3869 CrossRef CAS PubMed.
- T. Schneider-Poetsch, J. Ju, D. E. Eyler, Y. Dang, S. Bhat, W. C. Merrick, R. Green, B. Shen and J. O. Liu, Nat. Chem. Biol., 2010, 6, 209–217 CrossRef CAS PubMed.
- S. Das, M. Vera, V. Gandin, R. H. Singer and E. Tutucci, Nat. Rev. Mol. Cell Biol., 2021, 22, 483–504 CrossRef CAS PubMed.
- A. R. Buxbaum, G. Haimovich and R. H. Singer, Nat. Rev. Mol. Cell Biol., 2015, 16, 95–109 CrossRef CAS PubMed.
- J. Wong, R. Lerrigo, C. Y. Jang and G. Fang, Mol. Biol. Cell, 2008, 19, 2083–2091 CrossRef CAS PubMed.
- A. H. Rasmussen, H. B. Rasmussen and A. Silahtaroglu, Mol. Brain, 2017, 10, 43 CrossRef PubMed.
- L. Song, A. Craney and M. Rape, Mol. Cell, 2014, 53, 179–192 CrossRef CAS PubMed.
- J. M. Hsu, Y. C. Lee, C. T. Yu and C. Y. Huang, J. Biol. Chem., 2004, 279, 32592–32602 CrossRef CAS PubMed.
- C. T. Yu, J. M. Hsu, Y. C. Lee, A. P. Tsou, C. K. Chou and C. Y. Huang, Mol. Cell. Biol., 2005, 25, 5789–5800 CrossRef CAS PubMed.
- L. Song and M. Rape, Mol. Cell, 2010, 38, 369–382 CrossRef CAS PubMed.
- S. Sorokin, J. Cell Biol., 1962, 15, 363–377 CrossRef CAS PubMed.
- R. D. Goldman and L. I. Rebhun, J. Cell Sci., 1969, 4, 179–209 CrossRef CAS PubMed.
- V. Assmann, D. Jenkinson, J. F. Marshall and I. R. Hart, J. Cell Sci., 1999, 112, 3943–3954 CrossRef CAS PubMed.
- Q. Du, L. Taylor, D. A. Compton and I. G. Macara, Curr. Biol., 2002, 12, 1928–1933 CrossRef CAS PubMed.
- A. Hoppeler-Lebel, C. Celati, G. Bellett, M. M. Mogensen, L. Klein-Hitpass, M. Bornens and A. M. Tassin, J. Cell Sci., 2007, 120, 3299–3308 CrossRef CAS PubMed.
- K. Jiang, L. Rezabkova, S. Hua, Q. Liu, G. Capitani, A. F. M. Altelaar, A. J. R. Heck, R. A. Kammerer, M. O. Steinmetz and A. Akhmanova, Nat. Cell Biol., 2017, 19, 480–492 CrossRef CAS PubMed.
- S. L. Reck-Peterson, W. B. Redwine, R. D. Vale and A. P. Carter, Nat. Rev. Mol. Cell Biol., 2018, 19, 382–398 CrossRef CAS PubMed.
- A. R. Barutcu, M. Wu, U. Braunschweig, B. J. A. Dyakov, Z. Luo, K. M. Turner, T. Durbic, Z.-Y. Lin, R. J. Weatheritt, P. G. Maass, A.-C. Gingras and B. J. Blencowe, Mol. Cell, 2022, 82, 1035–1052 CrossRef CAS PubMed.
- A. Safieddine, M. N. Benassy, T. Bonte, F. Slimani, O. Pourcelot, M. Kress, M. Ernoult-Lange, M. Courel, E. Coleno, A. Imbert, A. Laine, A. M. Godebert, A. Vinit, C. Blugeon, G. Chevreux, D. Gautheret, T. Walter, E. Bertrand, M. Bénard and D. Weil, Mol. Cell, 2024, 84, 4191–4208.e4197 CrossRef CAS PubMed.
- D. A. Ramirez, L. E. Hough and M. R. Shirts, Biophys. J., 2024, 123, 703–717 CrossRef CAS PubMed.
- X. Jiang, D. B. T. Ho, K. Mahe, J. Mia, G. Sepulveda, M. Antkowiak, L. Jiang, S. Yamada and L. E. Jao, J. Cell Sci., 2021, 134, 258897 CrossRef PubMed.
- Y. Zhou, G. Wang, P. Wang, Z. Li, T. Yue, J. Wang and P. Zou, Angew. Chem., Int. Ed., 2019, 58, 11763–11767 CrossRef CAS PubMed.
- N. Tsanov, A. Samacoits, R. Chouaib, A. M. Traboulsi, T. Gostan, C. Weber, C. Zimmer, K. Zibara, T. Walter, M. Peter, E. Bertrand and F. Mueller, Nucleic Acids Res., 2016, 44, e165 CrossRef PubMed.
- J. C. Wheat, Y. Sella, M. Willcockson, A. I. Skoultchi, A. Bergman, R. H. Singer and U. Steidl, Nature, 2020, 583, 431–436 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- F. Mueller, A. Senecal, K. Tantale, H. Marie-Nelly, N. Ly, O. Collin, E. Basyuk, E. Bertrand, X. Darzacq and C. Zimmer, Nat. Methods, 2013, 10, 277–278 CrossRef CAS PubMed.
- D. Kim, B. Langmead and S. L. Salzberg, Nat. Methods, 2015, 12, 357–360 CrossRef CAS PubMed.
- S. Anders, P. T. Pyl and W. Huber, Bioinformatics, 2015, 31, 166–169 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00155a |
| This journal is © The Royal Society of Chemistry 2025 |





