Nanosensor quantitative monitoring of ROS/RNS homeostasis in single phagolysosomes of macrophages during bactericidal processes†
Bing-Yi
Guo
a,
Yu-Ting
Qi
a,
Hui-Qian
Wu
a,
Ru-Yan
Zha
a,
Li-Jun
Wang
a,
Xin-Wei
Zhang
 *a and
Wei-Hua
Huang
*a and
Wei-Hua
Huang
 *ab
*ab
aCollege of Chemistry and Molecular Sciences, Wuhan University, Wuhan 430072, China. E-mail: xinweizhang@whu.edu.cn; whhuang@whu.edu.cn
bDepartment of Hepatobiliary and Pancreatic Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, China
First published on 25th November 2024
Abstract
Reactive oxygen and nitrogen species (ROS/RNS) in macrophages have a potent killing effect on pathogens that infect the host. Here, we achieved in situ, quantitative detection of the homeostasis of four primary ROS/RNS (ONOO−, H2O2, NO, and NO2−) and their precursors (O2˙−, NO) in phagolysosomes of single RAW 264.7 macrophages after phagocytosis of Escherichia coli with platinum-black nanoelectrodes. Enhanced bactericidal activity of the macrophages was observed by an increase in the total amount of ROS/RNS as well as the level and proportion of ONOO−, a potent bactericidal species of RNS. Moreover, both the bactericidal process and the steady-state replenishment process were dominated by the production of RNS (NO-based), revealing differences in the enzyme kinetics of the bactericidal process.
Bacteria are the main pathogens of microbial infections, including Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), etc., and cause a wide range of diseases affecting human and animal life and health, such as pyelonephritis, septicemia, meningitis and so on.1 The innate immune response is the host's first line of defense against bacterial infection.2 Macrophages, the primary type of innate immune cell, are capable of recognizing pathogens through the pattern recognition receptor present on their surface, following capture and subsequently destroying pathogens through the formation of phagolysosomes.3 In this process, NADPH oxidase (NOX) and inducible nitric oxide synthase (iNOS) on the phagolysosomes are activated, and a variety of reactive oxygen and nitrogen species (ROS/RNS) are further synthesized. The ROS/RNS can oxidize a range of biological molecules including DNA, lipids, and functional proteins, resulting in the destruction of the pathogens.4 Thus, the composition and levels of ROS/RNS have an important influence on their antimicrobial activity. Quantitatively distinguishing the ROS/RNS during the bactericidal process of macrophages could contribute to the comprehension of cellular immune mechanisms, thereby enhancing the efficacy of anti-infective therapies and mitigating inflammation during infection. In recent decades, ROS/RNS during bacterial infection have been studied using fluorescent imaging and microfluidic chips.5 However, these assays generally perform a semi-quantitative analysis or endpoint detection of single or total levels of various ROS/RNS in the cytoplasm, while the composition, contents, and homeostatic processes of ROS/RNS within phagolysosomes remain unclear.
Nanoelectrodes possess the advantages of small size, high sensitivity, and fast response speed, which can be inserted into cells to realize electrochemical dynamic monitoring of electroactive substances at the level of single cells and organelles with high spatio-temporal resolution, but without severely affecting cell activity.6 The determination of ROS/RNS in individual activated phagolysosomes of lipopolysaccharide (LPS)-stimulated macrophages has been reported previously, and it was revealed that the production of RNS was predominant during continuous inflammatory situations.7 In particular, the platinum-black nanowire electrodes (SiC@Pt NWEs)7,8 were employed in conjunction with intracellular vesicle impact electrochemical cytometry (IVIEC)9 to achieve in situ, quantitative and sub-millisecond dynamic discrimination of four primary ROS/RNS (ONOO−, H2O2, NO, and NO2−) in individual phagolysosomes during RAW 264.7 macrophage sterilization processes. Through comparative analysis of the content and proportion of the produced and initial ROS/RNS and their precursors (O2˙−, NO), the kinetic difference of NOX and iNOS in homeostasis supplementation during host defense was investigated.
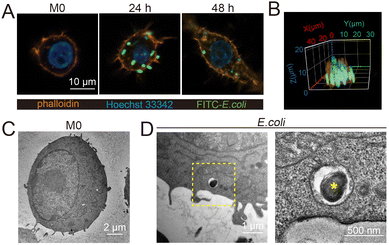
To study the content and proportion of ROS/RNS during bacterial infection, fluorescein isothiocyanate (FITC)-labeled inactivated E. coli (Fig. S1, ESI†) was used to co-culture with RAW 264.7 macrophages. Confocal and TEM images showed that macrophages were able to internalize E. coli after 12 h, and the number of internalized bacteria was positively correlated with time. After 48 h of phagocytosis, the fluorescent spots of E. coli in macrophages showed a reduction in size and intensity, indicating bacterial decomposition (Fig. 1A–D and Fig. S2, S3, ESI†). Accordingly, macrophages after 24 h phagocytosis of E. coli were selected to determine the levels of ROS/RNS generated during the sterilization process, and then DAF-FM DA and DCFH-DA probes were used to characterize ROS and NO after phagocytosis of the pathogens by macrophages. The results showed that the levels of ROS and NO were significantly increased in the macrophages after phagocytosis of E. coli as compared to the M0 type (Fig. S4, ESI†). In addition, calcein-AM and PI staining showed that macrophage activity still remained high under the determined conditions (Fig. S5, ESI†).
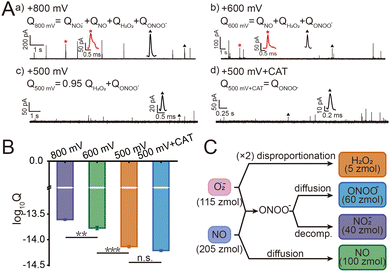
Macrophages phagocytize pathogens to form phagolysosomes and generate ROS/RNS precursors (O2˙− and NO) through NOX and iNOS, respectively. O2˙− is converted to H2O2 by superoxide dismutase (SOD), while O2˙− and NO will further rapidly react to form ONOO− under diffusion control. ONOO− is a more oxidizing and cytotoxic species with a short lifetime, which could further convert to more stable NO2−.10 And the relationship between the production of substances follows the equations (ESI,† eqn (1)–(3)). This series of ROS/RNS has a crucial impact on the defense and killing of pathogens in the phagolysosomes. The SiC@Pt NWEs exhibit distinct oxidation potentials for the four primary ROS/RNS (ONOO−, H2O2, NO, and NO2−) (Fig. S6, ESI†).7 To differentiate individual ROS/RNS while simultaneously reducing interference from other species, the optimal detection potentials for the four primary ROS/RNS were +100 mV, +500 mV, +600 mV, and +800 mV (Fig. S7, ESI†). During the IVIEC events, each phagolysosome in a macrophage ruptured on the electrode surface by electroporation and released its entire contents resulting in the generation of a spike signal.9 The charge value (Q) of a single spike signal was obtained by integrating the signal. The series of spike signals was observed after inserting NWE into the macrophages and applying different potentials (+800 mV, +600 mV, and +500 mV) (Fig. 2A). However, the minimum potential of IVIEC events is +500 mV,9 and thus the detection of ONOO− and H2O2 would be realized simultaneously at this potential and cannot be quantified separately. In order to distinguish ONOO− and H2O2, the specific scavenger of H2O2 (catalase, CAT) was preincubated with macrophages to monitor ONOO− alone (Fig. S8, ESI†). Besides, no spike signal was generated after inserting SiC@Pt NWEs into E. coli solution or inserting carbon-coated SiC nanowire electrodes (SiC@C NWEs)11 into macrophages after 24 h phagocytosis (Fig. S9 and S10, ESI†), suggesting that interference from other electroactive substances as well as bacteria could be ruled out in the assay.
The statistical data of the signals at different potentials showed that the electric quantity at different potentials were log-normally distributed (Fig. S11, ESI†), and their peak values at +800 mV, +600 mV, and +500 mV were significantly different: log10Q800mV = −13.61 ± 0.01, log10Q600mV = −13.77 ± 0.03, log10Q500mV = −14.14 ± 0.02, log10Q800mV+CAT = −14.21 ± 0.02 (Fig. 2B). According to the equations 4–7 and Faraday's law (ESI), Q = nzF where F = 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 C mol−1 is the Faraday constant and z is the electron transfer number (ZH2O2 = ZNO2− = 2, ZNO = ZONOO− = 1), the quantities of four ROS/RNS can be obtained: nH2O2 = 5 zmol, nONOO− = 60 zmol, nNO2 = 40 zmol, nNO = 100 zmol (Fig. 2C). The precursors of four primary ROS/RNS (O2˙− and NO) were calculated by means of the relationship between the substances shown in Fig. 2C:
500 C mol−1 is the Faraday constant and z is the electron transfer number (ZH2O2 = ZNO2− = 2, ZNO = ZONOO− = 1), the quantities of four ROS/RNS can be obtained: nH2O2 = 5 zmol, nONOO− = 60 zmol, nNO2 = 40 zmol, nNO = 100 zmol (Fig. 2C). The precursors of four primary ROS/RNS (O2˙− and NO) were calculated by means of the relationship between the substances shown in Fig. 2C: 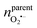 = 115 zmol and nparentNO = 205 zmol, noticed that the amount of O2˙− was ca. half that of NO. The overall quantity of both ROS and RNS was augmented in comparison to LPS-activated phagolysosomes in our previous work,7 suggesting that macrophages exhibit heightened bactericidal efficacy in actual bacterial eradication. Additionally, the content of ONOO−, which plays a significant role in the killing capacity of RNS, was also elevated. These evidences indicated that macrophages initiated a bactericidal process predominantly involving RNS when eliminating pathogens.
= 115 zmol and nparentNO = 205 zmol, noticed that the amount of O2˙− was ca. half that of NO. The overall quantity of both ROS and RNS was augmented in comparison to LPS-activated phagolysosomes in our previous work,7 suggesting that macrophages exhibit heightened bactericidal efficacy in actual bacterial eradication. Additionally, the content of ONOO−, which plays a significant role in the killing capacity of RNS, was also elevated. These evidences indicated that macrophages initiated a bactericidal process predominantly involving RNS when eliminating pathogens.
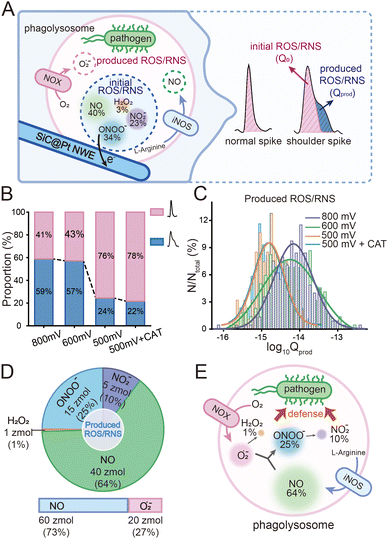
Similar to previous work,7,12 it was noted that a majority of the spikes exhibited a “shoulder”, i.e., current fall-off tails that deviated significantly from the normal exponential expected behavior. Upon the consumption of ROS/RNS by NWEs within their phagolysosomes, a rapid replenishment mechanism of iNOS and NOX was initiated within sub-milliseconds to maintain the killing ability of the phagolysosomes, resulting in the “shoulder” spikes (Fig. 3A). The number of shoulder spikes was further calculated and the percentage of shoulder spikes was found to be ca. 60% at both the +800 mV and +600 mV, while the percentage of shoulder peaks was found to decrease significantly to ca. 20% at +500 mV (Fig. 3B). This indicated that the steady-state replenishment process generated species were predominantly NO-based. These findings are consistent with the kinetics of the two enzymes and previous conclusions from simulated bactericidal models constructed using LPS stimulation.7,13 Subsequently, in order to further investigate the shoulder spikes, the previously constructed kinetic model was employed to split the shoulder peaks.12 Briefly, it deconvolutes mathematically the current time variation, fitted based on the behavior of the spike currents prior to the inflection point of the downward trend (ESI†). The shoulder spikes were then split into two portions with the charge of newly produced (Qprod) and initial (Q0) ROS/RNS.
Statistical analysis was performed on the amount of newly produced ROS/RNS during homeostatic replenishment in individual phagolysosomes. The Qprod under different potentials were consistent with log-normal distribution (Fig. 3C), while no significant difference between the value obtained at +800 mV (log10Qprod,800mV = −14.17 ± 0.02) and +600 mV (log10Qprod,600mV = −14.26 ± 0.04), and those obtained at +500 mV (log10Qprod,500mV = −14.80 ± 0.04) and on pre-incubation with CAT at +500 mV (log10Qprod,500mV+CAT = −14.82 ± 0.04) (Fig. S12, ESI†). The amount of the four ROS/RNS species was calculated: 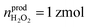 (ca. 1%),
(ca. 1%), 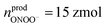 (ca. 25%),
(ca. 25%), 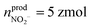 (ca. 10%), nprodNO = 40 zmol (ca. 64%) (Fig. 3D), and the precursors of ROS/RNS were calculated according to the conversion relationship between ROS/RNS:
(ca. 10%), nprodNO = 40 zmol (ca. 64%) (Fig. 3D), and the precursors of ROS/RNS were calculated according to the conversion relationship between ROS/RNS: 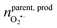 = 20 zmol, nparent,prodNO = 60 zmol (Fig. 3D). It is shown that the replenishment process is dominated by the production of NO, with a less amount of O2˙− being produced. Moreover, only a very small amount of O2˙− would be converted to H2O2 by SOD, and most of the O2˙− would be rapidly generated with NO to ONOO− under diffusion control (Fig. 3E).
= 20 zmol, nparent,prodNO = 60 zmol (Fig. 3D). It is shown that the replenishment process is dominated by the production of NO, with a less amount of O2˙− being produced. Moreover, only a very small amount of O2˙− would be converted to H2O2 by SOD, and most of the O2˙− would be rapidly generated with NO to ONOO− under diffusion control (Fig. 3E).
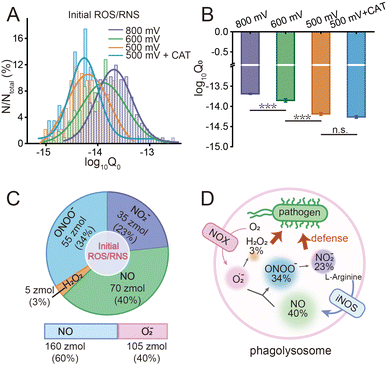
Moreover, the statistical analysis of initial content revealed that the Q0 under different potentials also conformed to the log-normal distribution (Fig. 4A), and the peak values at +800 mV, +600 mV and +500 mV potential were significantly different: log10Q0,800mV = −13.69 ± 0.01, log10Q0,600mV = −13.89 ± 0.04, log10Q0,500mV = −14.19 ± 0.04, log10Q0,500mV+CAT = −14.26 ± 0.03 (Fig. 4B). The amounts of four primary ROS/RNS were calculated by using the equations: 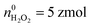 (ca. 3%),
(ca. 3%), 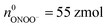 (ca. 34%),
(ca. 34%), 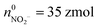 (ca. 23%), nNO = 70 zmol (ca. 40%) (Fig. 4C), and their precursors were further calculated according to the conversion relationship between ROS/RNS:
(ca. 23%), nNO = 70 zmol (ca. 40%) (Fig. 4C), and their precursors were further calculated according to the conversion relationship between ROS/RNS: 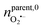 = 105 zmol and nparent,0NO = 160 zmol (Fig. 4C). These evidences implied that the ROS/RNS in phagolysosomes accumulated over time, with the amount of ROS increasing but remaining significantly less than that of RNS. The proportion of ONOO− increased, while the conversion ratio of ONOO− to NO2− (ca. 41%) also showed an increasing trend compared with the production process (ca. 30%) (Fig. 4D). This phenomenon may be attributed to the continuous reaction of O2˙− and NO to ONOO− during the bactericidal process to maintain the bactericidal capacity of the phagolysosomes. At the same time, the ONOO− in the phagolysosomes was partially consumed under the bactericidal effect for a period of time.10a,14 On the other hand, due to its strong oxidative activity and cytotoxicity of ONOO−, it will bring some damage to the host itself while achieving host defense.10c Therefore, in order to prevent excessive levels of ONOO− from causing severe injury to the host, part of the ONOO− was converted to the more stable NO2−. Thus, the combination of antimicrobial activity and host cell protection mechanism led to an increase in the conversion of ONOO− to NO2− after a period of storage.
= 105 zmol and nparent,0NO = 160 zmol (Fig. 4C). These evidences implied that the ROS/RNS in phagolysosomes accumulated over time, with the amount of ROS increasing but remaining significantly less than that of RNS. The proportion of ONOO− increased, while the conversion ratio of ONOO− to NO2− (ca. 41%) also showed an increasing trend compared with the production process (ca. 30%) (Fig. 4D). This phenomenon may be attributed to the continuous reaction of O2˙− and NO to ONOO− during the bactericidal process to maintain the bactericidal capacity of the phagolysosomes. At the same time, the ONOO− in the phagolysosomes was partially consumed under the bactericidal effect for a period of time.10a,14 On the other hand, due to its strong oxidative activity and cytotoxicity of ONOO−, it will bring some damage to the host itself while achieving host defense.10c Therefore, in order to prevent excessive levels of ONOO− from causing severe injury to the host, part of the ONOO− was converted to the more stable NO2−. Thus, the combination of antimicrobial activity and host cell protection mechanism led to an increase in the conversion of ONOO− to NO2− after a period of storage.
In conclusion, the SiC@Pt NWE was employed in combination with the IVIEC to monitor the contents, composition, and homeostatic complement of the four primary ROS/RNS within individual phagolysosomes throughout bactericidal processes in single RAW 264.7 macrophages. The experimental results showed that macrophages initiate an RNS-based bactericidal process after recognizing and phagocytosing pathogens. Additionally, the content of ROS/RNS in phagolysosomes with pathogens was higher than that in activated macrophages stimulated by LPS.7 Meanwhile, the level and proportion of ONOO−, which plays a major bactericidal role in RNS, also showed an increasing trend. Upon the rapid depletion of ROS/RNS within phagolysosomes, NOX and iNOS promptly initiated a homeostatic replenishment mechanism within sub-milliseconds to maintain the killing capacity of phagolysosomes, as evidenced by the production of large quantities of NO. These findings revealed the complex interactions and bactericidal processes of ROS/RNS during bacterial infection and host defense, thereby providing a basis for the regulation of ROS/RNS during macrophage immunity. It is anticipated that this will lead to improvements in the therapeutic efficacy of bacterial infections by targeted regulation of ROS/RNS in macrophages and may provide new ideas for the prevention of bacterial infections and the development of new therapeutic strategies.
We gratefully acknowledge financial support from the National Key Research and Development Program of China (2022YFA1104800) and the National Natural Science Foundation of China (Grants 22090050, 22090051, and 224340002). We also thank the Core Facility of Wuhan University for TEM characterizations.
Data availability
The data supporting the findings of this study are available within the article and its ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
-
(a) M.-A. Croxen and B.-B. Finlay, Nat. Rev. Microbiol., 2010, 8, 26–38 CrossRef CAS PubMed
; (b) L. Thomer, O. Schneewind and D. Missiakas, Annu. Rev. Pathol. Mech. Dis., 2016, 11, 343–364 CrossRef CAS PubMed
; (c) L. Radoshevich and P. Cossart, Nat. Rev. Microbiol., 2018, 16, 32–46 CrossRef CAS PubMed
.
-
(a) A.-J. Wolf and D.-M. Underhill, Nat. Rev. Immunol., 2018, 18, 243–254 CrossRef CAS
; (b) C.-A. Thaiss, N. Zmora, M. Levy and E. Elinav, Nature, 2016, 535, 65–74 CrossRef CAS PubMed
.
- D.-M. Sivaloganathan and M.-P. Brynildsen, Annu. Rev. Chem. Biomol. Eng., 2021, 12, 309–331 CrossRef CAS PubMed
.
-
(a) A. Panday, M.-K. Sahoo, D. Osorio and S. Batra, Cell. Mol. Immunol., 2015, 12, 5–23 CrossRef CAS PubMed
; (b) R.-S. Flannagan, G. Cosio and S. Grinstein, Nat. Rev. Microbiol., 2009, 7, 355–366 CrossRef CAS PubMed
; (c) G. Weiss and U.-E. Schaible, Immunol. Rev., 2015, 264, 182–203 CrossRef CAS PubMed
; (d) P.-J. Murray and T.-A. Wynn, Nat. Rev. Immunol., 2011, 11, 723–737 CrossRef CAS
.
-
(a) J. Heijden, E.-S. Bosman, L.-A. Reynolds and B.-B. Finlay, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 560–565 CrossRef
; (b) X.-Y. Bai, K.-K. Ng, J.-J. Hu, S. Ye and D. Yang, Annu. Rev. Biochem., 2019, 88, 605–633 CrossRef CAS PubMed
; (c) Z.-M. Wang, T.-D. Cong, W.-B. Zhong, J.-W. Lau, G. Kwek, M.-B. Chan-Park and B.-G. Xing, Angew. Chem., Int. Ed., 2021, 60, 16900–16905 CrossRef CAS PubMed
; (d) Z.-Y. Jiang, S.-D. Liu, X. Xiao, G.-M. Jiang, Q. Qu, X.-X. Miao, R.-F. Wu, R. Shi, R.-C. Guo and J. Liu, Lab Chip, 2022, 22, 2944–2953 RSC
.
-
(a) C. Amatore, S. Arbault, M. Guille and F. Lemaitre, Chem. Rev., 2008, 108, 2585–2621 CrossRef CAS
; (b) Y.-X. Wang, J.-M. Noel, J. Velmurugan, W. Nogala, M.-V. Mirkin, C. Lu, M.-G. Collignon, F. Lemaitre and C. Amatore, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11534–11539 CrossRef CAS
; (c) K.-K. Hu, Y.-L. Liu, A. Oleinick, M.-V. Mirkin, W.-H. Huang and C. Amatore, Curr. Opin. Electrochem., 2020, 22, 44–50 CrossRef CAS
.
- Y.-T. Qi, H. Jiang, W.-T. Wu, F.-L. Zhang, S.-Y. Tian, W.-T. Fan, Y.-L. Liu, C. Amatore and W.-H. Huang, J. Am. Chem. Soc., 2022, 144, 9723–9733 CrossRef CAS
.
-
(a) Y.-T. Qi, F.-L. Zhang, S.-Y. Tian, H.-Q. Wu, Y. Zhao, X.-W. Zhang, Y.-L. Liu, P.-Q. Fu, C. Amatore and W.-H. Huang, Nat. Nanotechnol., 2024, 19, 524–533 CrossRef CAS
; (b) H.-Q. Wu, Y.-T. Qi, B.-Y. Guo, Y. Zhao, X.-W. Zhang and W.-H. Huang, Chem. Commun., 2024, 60, 5546–5549 RSC
.
-
(a) X.-C. Li, S. Majdi, J. Dunevall, H. Fathali and A.-G. Ewing, Angew. Chem., Int. Ed., 2015, 54, 11978–11982 CrossRef CAS PubMed
; (b) J. Lovric, N. Najafinobar, J. Dunevall, S. Majdi, I. Svir, A. Oleinick, C. Amatore and A.-G. Ewing, Faraday Discuss., 2016, 193, 65–79 RSC
; (c) N.-T.-N. Phan, X.-C. Li and A.-G. Ewing, Nat. Rev. Chem., 2017, 1, 1–18 CrossRef
.
-
(a) P. Pacher, J.-S. Beckman and L. Liaudet, Phys. Rev., 2007, 87, 315–424 CAS
; (b) C. Nathan and M.-U. Shiloh, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8841–8848 CrossRef CAS
; (c) L. Piacenza, A. Zeida, M. Trujillo and R. Radi, Physiol. Rev., 2022, 102, 1881–1906 CrossRef CAS PubMed
.
- X.-W. Zhang, Q.-F. Qiu, H. Jiang, F.-L. Zhang, Y.-L. Liu, C. Amatore and W.-H. Huang, Angew. Chem., Int. Ed., 2017, 56, 12997–13000 CrossRef CAS
.
- X.-W. Zhang, A. Oleinick, H. Jiang, Q.-L. Liao, Q.-F. Qiu, I. Svir, Y.-L. Liu, C. Amatore and W.-H. Huang, Angew. Chem., Int. Ed., 2019, 58, 7753–7756 CrossRef CAS PubMed
.
-
(a) T.-E. DeCoursey and E. Ligeti, Cell. Mol. Life Sci., 2005, 62, 2173–2193 CrossRef CAS PubMed
; (b) C.-C. Winterbourn, A.-J. Kettle and M.-B. Hampton, Annu. Rev. Biochem., 2016, 85, 765–792 CrossRef CAS PubMed
.
- R.-G. Allen, W.-P. Lafuse, N.-D. Powell, J.-I.-W. Markenton, L.-M. Stiner-Jones, J.-F. Sheridan and M.-T. Bailey, Infect. Immun., 2012, 80, 3429–3437 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc05423g |
| This journal is © The Royal Society of Chemistry 2025 |