Open Access Article
Open Access ArticleSpontaneously blinking spiroamide rhodamines for live SMLM imaging of the plasma membrane†
Sonia
Pfister
 a,
Sophie
Walter
a,
Sophie
Walter
 a,
Aurélie
Perrier
a,
Aurélie
Perrier
 b and
Mayeul
Collot
b and
Mayeul
Collot
 *a
*a
aChemistry of Photoresponsive Systems, Laboratoire de Chémo-Biologie Synthétique et Thérapeutique (CBST) UMR 7199, CNRS, Université de Strasbourg, F-67400 Illkirch, France. E-mail: mayeul.collot@unistra.fr
bLaboratoire Interdisciplinaire des Energies de Demain (LIED), UMR 8236, CNRS, Université Paris Cité, F-75013, Paris, France
First published on 21st March 2025
Abstract
We have developed spontaneously blinking fluorescent probes based on the reversible spirolactamization of rhodamine, to efficiently image the plasma membrane (PM) of live cells with enhanced resolution using SMLM. This study demonstrates that the blinking efficiency of spiroamide PM probes is not solely governed by their pKa; the presence of a charged polar group on the amide should also be taken into account.
The advent of super-resolution imaging techniques based on single-molecule localization microscopy (SMLM) has revolutionized optical microscopy as it enables the visualization of biological structures with unprecedented resolution.1,2 While several organelles and lipid-based cellular structures have been successfully imaged in both fixed and live samples,3,4 SMLM imaging of the plasma membrane (PM) remains highly constrained.5,6 Specifically, live-cell SMLM imaging of the PM poses significant challenges due to the free diffusion of fluorescent probes within the lipid bilayer and the inherent movement of the PM during image acquisition. As a result, examples of live SMLM imaging of the PM are exceedingly rare and largely restricted to PAINT-based approaches that utilize transient binding of moderately lipophilic dyes.7–9 This underscores the pressing need for efficient spontaneously blinking probes to advance live SMLM imaging of the PM. In response to this challenge, we have directed our recent research efforts toward developing spontaneously blinking probes tailored for this purpose. Notably, we achieved 3D live SMLM of the PM using directed photooxidation of photoconvertible PM probes.10 Recently, we also introduced a novel concept involving a self-triggered photooxidation cascade of leuco-rhodamine derivatives.11 Indeed, live SMLM imaging demands probes with a spontaneous blinking mechanism characterized by a bright “on” state, a dark “off” state, and a low density of emissive molecules at any given time.12 In this context, rhodamines featuring an intramolecular nucleophilic moiety are particularly well-suited due to their dynamic equilibrium between an emissive open form and a non-emissive spirocyclic form.13 The Urano group has made significant contributions to this field, leveraging hydroxyl groups as intramolecular nucleophiles in various rhodamine derivatives, including Si-,14 C-,15 and O-rhodamines,16 offering a wide range of spectral properties and finely tuned spirocyclization equilibria. In particular, spiroamide O-rhodamines have been extensively studied and shown good efficacies in nanoscopy applications.17–19 Importantly, several studies report that special attention must be given to the design of these probes to reach the most appropriate blinking properties depending on the cellular target.20 Notably, since spiroamide opening depends on the pH, the pKa of the probes is the main factor that governs this fluorogenic equilibrium.21,22
In this work, we investigated the performance of three potential spontaneously blinking spiroamide rhodamine (SR) probes with increasing pKa values and evaluated their efficacy for rapid imaging of live cell contours using SMLM (Fig. 1A). To this end, we first synthesized a clickable rhodamine derivative, R-COOH (see ESI†), which was coupled to three different amines to produce the spiroamide rhodamine SR probes (Fig. 1B). The choice of amines was informed by previous studies reporting SR probes based on rhodamine 6G with pKa values of <423 and 2.8,24 when coupled to ethylenediamine and ethanolamine, respectively. In contrast, 1-adamantylamine, owing to its bulkiness, yielded a pKa of 6.5.24 Once synthesized, the spiroamide rhodamines were converted into PM probes (SR-PM-NH2, SR-PM-OH, and SR-PM-Ad) by coupling them with CAZ, a clickable amphiphilic zwitterion. CAZ, when used in pairs, mimics phospholipids and intercalates into the lipid bilayer, acting as an efficient plasma membrane targeting moiety.6,25,26
First, the photophysical properties of the SR precursors were determined and are presented in Fig. 2 (table). As expected, under acidic conditions (pH 2), the probes exhibited absorption with a λAbs max around 560 nm and emission with a λEm max around 582 nm, along with similar fluorescence quantum yields. This indicates that the coupled amine does not interfere with the fluorescence process. At a pH close to physiological extracellular conditions (pH 7), their absorbance and emission intensities in the visible region decreased significantly due to spirolactamization. The fluorescence spectra of the three SR probes were recorded at various pH values (Fig. 2A), providing their respective pKa values (Fig. 2B). While SR-Ad showed a lower pKa compared to previously reported SR probes based on rhodamine 6G (5.09 versus 6.4),24 SR-NH2 and SR-OH displayed significantly lower pKa values (2.06 and 2.82, respectively), consistent with their rhodamine 6G analogues.23,24 Before proceeding with live cellular imaging, a viability test was performed on HeLa cells, revealing that the three SR-PM probes did not induce any observable cytotoxicity at 200 nM (Fig. S1, ESI†).
The probes were first evaluated using laser scanning confocal microscopy (Fig. S2, ESI†). As expected, owing to their low pKa values favoring the off-spiroform, the resulting images displayed weak signal-to-noise ratios along with low Pearson's colocalization coefficients compared with a reference PM probe (MemBright Cy5.56). Interestingly, while cells in the presence of SR-PM-Ad and SR-PM-OH exhibited a dim fluorescence signal mainly due to autofluorescence, those stained with SR-PM-NH2 showed a weak additional signal at the cell surface and the highest Pearson's colocalization coefficient, despite its lowest pKa value among the three probes. This observation suggests that factors beyond pKa influence the ring-opening equilibrium at the PM.
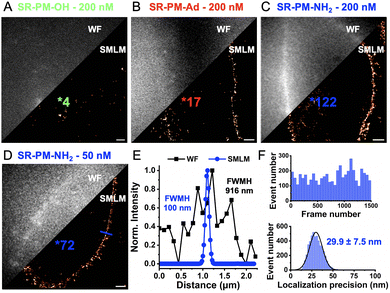
Single-molecule localization microscopy (SMLM) imaging was then performed on live HeLa cells in the presence of 200 nM SR-PM probes (Fig. 3 and Supplementary Movies, ESI†) to rapidly image their contours with enhanced resolution. The results revealed significant differences. As expected, SR-PM-OH, due to its low pKa, produced virtually no blinking events and failed to reconstruct the cell surface in SMLM (Fig. 3A). Surprisingly, SR-PM-Ad, despite being ≈180-fold more basic, provided only a slightly higher number of blinking events per cell surface (17, Fig. 3B). Once again, SR-PM-NH2 distinguished itself with unique behavior, generating significantly more blinking events in the single-molecule regime and successfully reconstructing the cell surface of live HeLa cells within only 14 seconds of acquisition (Fig. 3C).
Although the bulk concentration of the probe might seem low (200 nM), the local concentration, due to accumulation in the membrane, can become relatively high. Consequently, the concentration of SR-PM-NH2 was reduced to 50 nM. At this reduced concentration, the number of events per cell surface decreased; however, the resulting SMLM images improved, likely due to reduced phototoxicity and lower blinking density, and a clear resolution enhancement was achieved compared to widefield microscopy (Fig. 3E). This improvement was observed in only 21 seconds of acquisition (1500 frames, 13.9 ms per frame) (Fig. 3F). At a 532 nm laser irradiance of 500 W cm−2, SR-PM-NH2 provided a stable number of blinks per frame with a localization precision of 29.9 ± 7.5 nm (Fig. 3F).
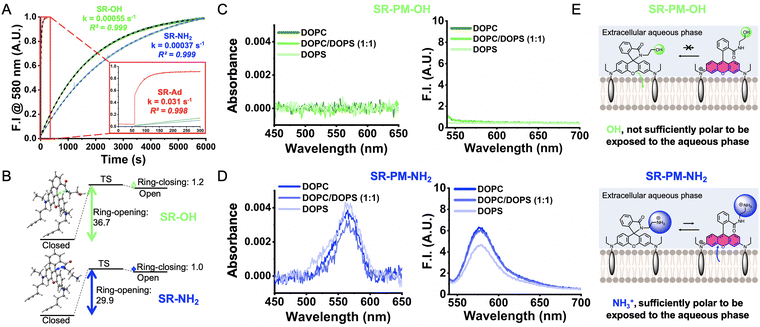
To understand the enhanced performance of SR-PM-NH2 compared to the other SR-PM probes, additional experiments were conducted. Initially, we aimed to assess their opening and closing kinetics. While the ring-closure kinetics could not be determined due to the rapidity of the process (Fig. S3, ESI†), the ring-opening kinetics could be measured (Fig. 4A). As expected, based on its pKa value, SR-Ad exhibited the highest ring-opening rate from pH 7.4 to 2 of 0.031 s−1. Conversely, SR-OH and SR-NH2, which have comparable low pKa values, displayed much lower ring-opening kinetics of ≈3–4 × 10−4 s−1. These results were expected and did not provide further insights into the differences observed in SMLM performance between SR-PM-OH and SR-PM-NH2. To gain valuable insights into the differences in probe performance, we attempted to investigate the influence of environmental polarity on the spirocyclization equilibrium using water/dioxane mixtures, as previously reported.27 However, due to their pronounced lipophilic nature, the SR probes exhibited poor solubility under these conditions, preventing such an analysis. To address these limitations, the reaction described in Fig. 1A can be decomposed into a three-step process based on (1) a ring-opening process, (2) a protonation reaction and (3) a rotation of the amide group (see Scheme S1, ESI†). The ring-opening process being expected to be the rate-determining step,19 the Gibbs free energies of the former reaction (Fig. 4B) were calculated with DFT using an implicit solvent computational model to qualitatively compare the activation energies of the two compounds.28 For this reaction step, it is worth noting that the relative pKa values of the two probes have no impact on these results. The calculated barriers revealed significant differences, indicating that SR-NH2 requires considerably less energy to open compared to SR-OH. This is likely due to the presence of the ammonium group in the protonated form of SR-NH2 (Fig. 4E), which stabilizes the highly polar open-ring zwitterionic structure and lowers its relative free energy.
Finally, we hypothesized that the ring-opening efficiency might be influenced by the membrane environment. Indeed, in this interfacial environment, factors other than pH could govern the ring opening of SR. To explore this, 80 nm large unilamellar vesicles (LUVs) composed of neutral zwitterionic DOPC and anionic DOPS were prepared at various ratios: 1/0, 1/1, and 0/1 (Fig. S4, ESI†). The probes were incubated with the LUVs at a fixed pH (PBS, pH 7.4), and their absorption and emission spectra were recorded (Fig. 4C and D). The results revealed that within these membrane models, the pKa was not the primary determinant of ring opening, as the intensity of the absorption signal at 560 nm—indicative of ring opening—did not correlate with the pKa values. For instance, SR-PM-Ad, which has the highest pKa (5.09), predominantly existed in its closed form (Fig. S5, ESI†). Interestingly, when embedded in the negatively charged DOPS bilayer, a signal appeared, suggesting that the negative charges of the lipid polar heads can stabilize the cationic rhodamine after ring opening (Fig. S5, ESI†). Notably, SR-PM-NH2, despite having the lowest pKa, exhibited the highest absorbance (Fig. 4C) and fluorescence intensity (Fig. 4D) in LUVs, irrespective of their composition. In contrast, SR-PM-OH, which has a higher pKa, showed neither absorbance nor fluorescence signals in any type of LUV (Fig. 4C). These results suggest that at the lipid bilayer interface, SR probes with non-polar amides, even those with high pKa values (SR-PM-Ad), tend to orient within the lipid phase, favoring their hydrophobic closed form (Fig. 4E). SR-PM-NH2, on the other hand, contains a polar ammonium (NH3+) at physiological pH, which promotes the orientation of the rhodamine toward the aqueous phase (Fig. 4E). This hypothesis was confirmed by monitoring the fluorescence intensity at 580 nm upon the addition of SR-PM to a solution of DOPC and DOPS LUVs (Fig. S6, ESI†). While SR-PM-OH showed no signal over time, SR-PM-NH2 produced an immediate and constant low signal. Interestingly, SR-PM-Ad rapidly reached its maximum intensity before decreasing with a kinetics depending on the LUV composition, indicating that its open form is stabilized by the negatively charged phosphatidylserine before ring closure (Fig. S6, ESI†). Overall, at the lipid–water interface, SR probes with non-charged and non-polar amides (SR-OH and SR-Ad) experience a shift in their apparent pKa to much lower values. In contrast, SR-PM-NH2, which primarily orients toward the extracellular aqueous phase, retains its ability to protonate, thereby enabling a higher number of transient and reversible ring openings at the cell surface (Fig. 4E). In conclusion, we have demonstrated that SR-PM-NH2 is an efficient spontaneously blinking fluorescent probe for imaging the plasma membrane and contours of live cells with short acquisition times (<30 s) and enhanced resolution using SMLM. Its superior performance can be attributed not only to its low pKa, which ensures the low density of emissive dye required for SMLM, but also to the presence of a charged ammonium group, which: (1) lowers the free energy barrier for ring opening, and (2) promotes the orientation of the rhodamine moiety toward the aqueous phase, where ring opening is favored compared to being embedded in lipids. This study underscores the importance of considering structural and physical properties beyond pKa when designing spontaneously blinking fluorescent probes based on spirolactamization for SMLM, tailored to their intended cellular localization.
Conceptualization SP, MC; formal analysis SP, SW, AP; funding acquisition MC; investigation SP, SW; project administration MC; supervision MC; validation MC; visualization SP MC; writing SP, AP, MC.
This work was funded by the Agence Nationale de la Recherche: ANR 5D-SURE (21-CE42-0015) and was granted access to the HPC resources of IDRIS under the allocations 2024-AD010814608 made by Grand Equipement National de Calcul Intensif. The synthesis protocol, characterizations of the products and supplementary figures and movies can be found in the ESI.†
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- M. Lelek, M. T. Gyparaki, G. Beliu, F. Schueder, J. Griffié, S. Manley, R. Jungmann, M. Sauer, M. Lakadamyali and C. Zimmer, Nat. Rev. Methods Primers, 2021, 1, 39 CrossRef CAS PubMed.
- T. Klein, S. Proppert and M. Sauer, Histochem. Cell Biol., 2014, 141, 561–575 CrossRef CAS PubMed.
- X. Chen, X. Wang, F. Huang and D. Ma, PhotoniX, 2024, 5, 29 CrossRef.
- A. Jimenez, K. Friedl and C. Leterrier, Methods, 2020, 174, 100–114 CAS.
- M. Collot, S. Pfister and A. S. Klymchenko, Curr. Opin. Chem. Biol., 2022, 69, 102161 CAS.
- M. Collot, P. Ashokkumar, H. Anton, E. Boutant, O. Faklaris, T. Galli, Y. Mély, L. Danglot and A. S. Klymchenko, Cell Chem. Biol., 2019, 26, 600–614.e7 CAS.
- D. I. Danylchuk, S. Moon, K. Xu and A. S. Klymchenko, Angew. Chem., Int. Ed., 2019, 58, 14920–14924 CAS.
- I. O. Aparin, R. Yan, R. Pelletier, A. A. Choi, D. I. Danylchuk, K. Xu and A. S. Klymchenko, J. Am. Chem. Soc., 2022, 144, 18043–18053 CAS.
- R. Yan, K. Chen and K. Xu, J. Am. Chem. Soc., 2020, 142, 18866–18873 CAS.
- L. Saladin, V. Breton, V. Le Berruyer, P. Nazac, T. Lequeu, P. Didier, L. Danglot and M. Collot, J. Am. Chem. Soc., 2024, 146, 17456–17473 CAS.
- S. Pfister, V. Le Berruyer, K. Fam and M. Collot, bioRxiv, 2024, 2024.05.28.596159 Search PubMed.
- H. Li and J. C. Vaughan, Chem. Rev., 2018, 118, 9412–9454 CrossRef CAS PubMed.
- D. Si, Q. Li, Y. Bao, J. Zhang and L. Wang, Angew. Chem., Int. Ed., 2023, 62, e202307641 CAS.
- S. Uno, M. Kamiya, T. Yoshihara, K. Sugawara, K. Okabe, M. C. Tarhan, H. Fujita, T. Funatsu, Y. Okada, S. Tobita and Y. Urano, Nat. Chem., 2014, 6, 681–689 CAS.
- R. Tachibana, M. Kamiya, A. Morozumi, Y. Miyazaki, H. Fujioka, A. Nanjo, R. Kojima, T. Komatsu, T. Ueno, K. Hanaoka, T. Yoshihara, S. Tobita and Y. Urano, Chem. Commun., 2020, 56, 13173–13176 CAS.
- S. Uno, M. Kamiya, A. Morozumi and Y. Urano, Chem. Commun., 2018, 54, 102–105 CAS.
- V. N. Belov, M. L. Bossi, J. Fölling, V. P. Boyarskiy and S. W. Hell, Chem. – Eur. J., 2009, 15, 10762–10776 CrossRef CAS PubMed.
- P. J. Macdonald, S. Gayda, R. A. Haack, Q. Ruan, R. J. Himmelsbach and S. Y. Tetin, Anal. Chem., 2018, 90, 9165–9173 CrossRef CAS PubMed.
- Q. Qi, W. Chi, Y. Li, Q. Qiao, J. Chen, L. Miao, Y. Zhang, J. Li, W. Ji, T. Xu, X. Liu, J. Yoon and Z. Xu, Chem. Sci., 2019, 10, 4914–4922 RSC.
- Z. Ye, H. Yu, W. Yang, Y. Zheng, N. Li, H. Bian, Z. Wang, Q. Liu, Y. Song, M. Zhang and Y. Xiao, J. Am. Chem. Soc., 2019, 141, 6527–6536 CrossRef CAS PubMed.
- Q. Qiao, W. Liu, J. Chen, X. Wu, F. Deng, X. Fang, N. Xu, W. Zhou, S. Wu, W. Yin, X. Liu and Z. Xu, Angew. Chem., Int. Ed., 2022, 61, e202202961 CrossRef CAS PubMed.
- Z. Ye, Y. Zheng, X. Peng and Y. Xiao, Anal. Chem., 2022, 94, 7990–7995 CrossRef CAS PubMed.
- Z. Li, S. Wu, J. Han and S. Han, Analyst, 2011, 136, 3698–3706 CAS.
- L. Yuan, W. Lin and Y. Feng, Org. Biomol. Chem., 2011, 9, 1723–1726 CAS.
- M. Collot, E. Boutant, M. Lehmann and A. S. Klymchenko, Bioconjugate Chem., 2019, 30, 192–199 CAS.
- S. Pfister, J. Lesieur, P. Bourdoncle, M. Elhassan, P. Didier, N. Anton, H. Anton and M. Collot, Anal. Chem., 2024, 96, 12784–12793 CrossRef CAS PubMed.
- N. Lardon, L. Wang, A. Tschanz, P. Hoess, M. Tran, E. D’Este, J. Ries and K. Johnsson, J. Am. Chem. Soc., 2021, 143, 14592–14600 CAS.
- R. Tachibana, M. Kamiya, A. Morozumi, Y. Miyazaki, H. Fujioka, A. Nanjo, R. Kojima, T. Komatsu, T. Ueno, K. Hanaoka, T. Yoshihara, S. Tobita and Y. Urano, Chem. Commun., 2020, 56, 13173–13176 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary figures, synthesis protocols, characterization & spectra. See DOI: https://doi.org/10.1039/d5cc00151j |
| This journal is © The Royal Society of Chemistry 2025 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: