Open Access Article
Open Access ArticleAutomating stochastic antibody–drug conjugation: a self-driving lab approach for enhanced therapeutic development†
Liam
Roberts
a,
Matthew E.
Reish
a,
Jerrica
Yang
a,
Wenyu
Zhang
 a,
Joshua S.
Derasp
*a and
Jason E.
Hein
a,
Joshua S.
Derasp
*a and
Jason E.
Hein
 *abcd
*abcd
aDepartment of Chemistry, The University of British Columbia, Vancouver, BC V6T 1Z1, Canada. E-mail: jhein@chem.ubc.ca; jderasp@chem.ubc.ca
bDepartment of Chemistry, University of Bergen, Norway
cAcceleration Consortium, University of Toronto, Toronto, ON, Canada
dTelescope Innovations Corp., Vancouver, BC, Canada
First published on 24th February 2025
Abstract
Antibody–drug conjugates (ADCs) have become a promising cancer treatment over the past two decades due to their on-target drug-release capabilities. However, labor-intensive manual conjugations currently limit the throughput of ADC synthesis. Herein, we introduce a Self-Driving Lab (SDL) for automated stochastic antibody–drug conjugation and characterization. The robotic platform performs conjugations and determines drug to antibody ratios from chromatography data, enabling the production of target ADCs iteratively in a closed loop. Our SDL establishes a robust foundation for increasing ADC production throughput and accelerating the development of cancer therapeutics.
Introduction
Cancer remains a significant global health issue, resulting in 10 million deaths in 2020 and ranking as the second most common cause of mortality worldwide.1 Among various treatment strategies, antibody–drug conjugates (ADCs) have emerged as a highly promising category of cancer therapies.2,3 By directly delivering drugs to cancer cells, the antibody minimizes toxicity to healthy tissues.2,3Several methods facilitate the attachment of linker-bound drugs (drug-linkers) to the antibody's amino acid residues.4 Site-specific conjugation often uses artificially inserted amino acid residues to achieve uniform attachment of drug-linkers.4 In contrast, stochastic conjugation uses the antibody's native lysine or cysteine residues, resulting in a non-uniform binding of drug-linkers.4 Stochastic cysteine conjugation has proven effective for five of the eleven FDA-approved ADCs and many ADCs that are currently in clinical trials.5,6
IgG1 antibodies contain 8 key cysteine residues which are present as interchain disulfide bridges.4 The stochastic reduction of these bonds exposes the nucleophilic thiols which can then be reacted with a maleimide drug-linker containing a chemotherapeutic payload.4 The average drug-to-antibody ratio (DAR) is a key parameter that influences efficacy and toxicity of the resulting ADC.5–7 However, the laborious nature of manual antibody conjugation is a bottleneck to ADC screening and development.8 This bottleneck is particularly evident because ADC precursors can be produced at high throughput through modern drug and antibody screening methods.9–11
Previous studies have focused on optimizing isolated aspects of the conjugation and characterization protocols for stochastic ADC production.12–14 For example, Endo et al. employed flow reactors to accelerate reduction and conjugation reactions,12 while Yang et al. introduced an automated buffer exchange system to streamline ADC preparation for mass spectrometry analysis.13,14 Goyon et al. developed a rapid characterization approach using online 2D and 4D liquid chromatography.14 Despite these advancements, the development of a fully autonomous platform capable of synthesizing ADCs with precise drug-to-antibody ratios (DAR), a key developmental parameter, remains a significant challenge. A platform with integrated feedback control, enabling on-the-fly testing and optimization of experimental conditions, would represent a major advancement for the field. Further, reliable synergy between data extraction algorithms and hardware would drastically improve the ability to design ADCs with precise drug-to-antibody ratio (DAR) outputs, ultimately enhancing the production of targeted therapeutics.
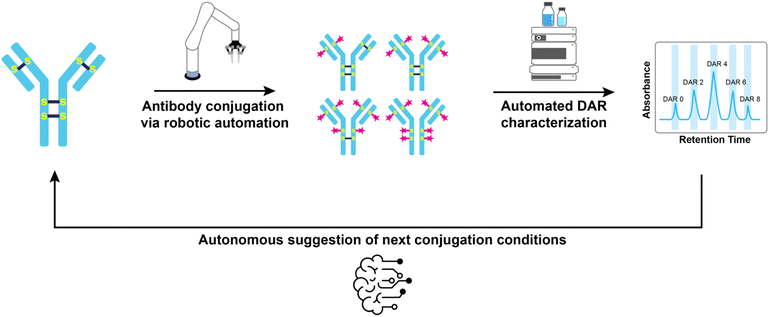
In this work, we introduce an automated robotic platform for the synthesis of stochastic antibody–drug conjugates (Fig. 1). This platform integrates a custom liquid handling system for performing the reactions with hydrophobic interaction chromatography-high-performance liquid chromatography (HIC) for drug-to-antibody ratio (DAR) characterization. We developed an algorithm capable of extracting the average DAR from chromatographic data, streamlining the analysis process. Additionally, our platform optimizes conjugation conditions providing access to ADCs with precise DARs. Together, these advancements demonstrate a proof-of-concept for significantly increasing the throughput of ADC production, with potential implications for accelerating cancer therapeutic development.
Results and discussion
Conjugation procedure
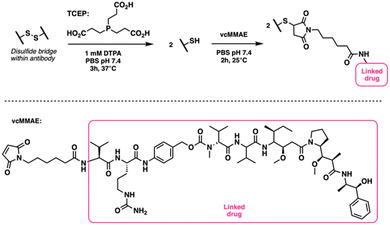
We began by testing a fully manual workflow to verify that both the chemistry and analytical processes were functioning as expected. This manual execution allowed us to establish a baseline for the conjugation protocol in terms of error rates, analytical precision, and the operational limits of each unit operation. This baseline assessment was critical for identifying potential bottlenecks, ensuring reliable performance of each step—reduction, conjugation, and purification—and confirming that the HIC analysis for drug-to-antibody ratio (DAR) determination was accurate. By understanding the inherent variability and limitations of the manual process, we could subsequently refine the automation, making targeted adjustments to ensure the system operated consistently. The manual workflow also provided a reference point for comparing the performance of the automated platform and guiding optimization efforts during the automation phase.To generalize our approach, we adopted a standard procedure for stochastic cysteine conjugation as the foundation for automating ADC synthesis (Fig. 2). This process starts with the incubation of an antibody solution with a phosphine-based reductant, tris(2-carboxyethyl)phosphine (TCEP), at 37 °C for 3 hours. The reduction cleaves the disulfide bonds within the antibody, freeing the cysteine thiol groups. After incubation, the reduced antibody solution is passed through a gel-filtration column to remove excess TCEP and other small molecules.
The reduced antibody is then reacted with an excess of maleimide drug-linker, maleimidocaproyl-valine-citrulline-p-amino-benzyloxycarbonyl monomethylauristatin E (vcMMAE), at 25 °C for 2 hours. This allows the maleimide groups to covalently attach to the free thiol groups on the antibody. After the conjugation step, the mixture is filtered through another gel-filtration column to remove any unreacted drug-linker, yielding the purified antibody–drug conjugate (ADC). The purified ADC is then analyzed using HIC to determine the DAR. For this study, we used trastuzumab as the antibody and vcMMAE as the drug-linker due to their common use in ADC development.4–6,15,16
One of the key challenges in ADC conjugation is that the reactions are typically carried out on a low microliter (μL) scale, which demands highly accurate and precise liquid handling. This is particularly important when transitioning to an automated workflow, as small deviations in liquid handling can significantly affect the outcome of the reaction. To address this, we used dilute solutions during manual experimentation. This approach allowed for the use of larger reagent volumes during the automation phase, minimizing costs while maintaining feasibility. Through repeated manual conjugations, we verified that the low antibody concentration did not cause significant variability in the ADC's DAR (see ESI Table 3†). Additionally, a crucial post-conjugation step involves removing excess drug-linker to prevent interference with downstream ADC applications. Consequently, the automated system must have a reliable purification protocol to ensure that unreacted drug-linker is effectively removed without compromising the yield or integrity of the ADC.
Automated conjugation platform
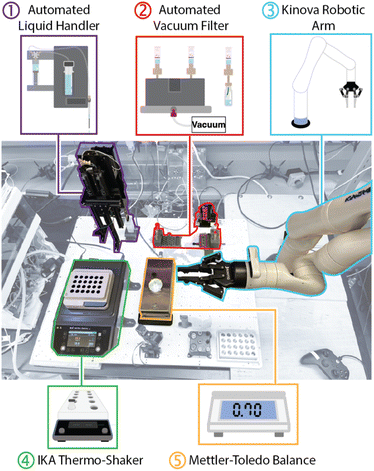
The challenges of precise liquid handling and reliable automated purification were central to our platform development. Managing small reaction volumes with high accuracy and minimizing variability in reagent dispensing were crucial for reproducibility. At the same time, ensuring that our purification system could effectively remove unreacted drug-linker while maintaining high antibody yields was equally important. These technical hurdles shaped our design choices, which aimed to create a robust, modular system capable of performing autonomous ADC synthesis.Our automated conjugation platform, shown in Fig. 3, was designed with modularity in mind, allowing flexibility in execution and workflow design. At the core of the system is a Kinova Gen 3 robot arm, which transfers samples between the liquid handler, filtration blocks, and other modules.
The robot arm, with its seven degrees of freedom, allows for precise manipulation of micro-scale volumes across the various components. While some modules, such as the IKA thermo-shaker, were easily integrated from commercially available units for temperature control during reactions, key operations like liquid handling and antibody purification required custom in-house solutions (ESI Fig. 3 and 7†).
Central to this platform was a custom mobile liquid handler, developed in collaboration with Telescope Innovations. This unit is battery-powered, wirelessly operated, and equipped with 3D-printed handles to interface with the robotic arm. Its mobility across the deck, constrained only by the range of the robotic arm, enabled flexible reagent handling and integration into the workflow without requiring manual repositioning.
While the liquid handler performed accurately for larger volumes, early tests showed significant inaccuracies when handling smaller volumes, particularly in the 20–40 μL range, where error rates were around 30% (see Fig. 4 and ESI Table 1,† average error %).
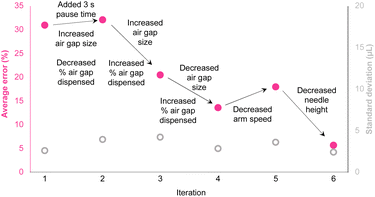 | ||
| Fig. 4 Liquid handling parameter optimization to reduce average error (pink) and maintain low standard deviation (grey). Variables included air gap size and dispense percentage, pause time to stabilize aspiration, speed of the robotic arm, and the height of the needle after priming. Full parameter details are available in ESI Table 1.† | ||
The early performance limitations led us to probe the impact of several parameters including:
(A) Airgap size optimization: by varying the size of the airgap used during aspiration, we were able to prevent mixing between the aspirated reagent and the backing solvent. Testing various airgap sizes also allowed us to fine-tune how much air was dispensed with each reagent, ensuring full ejection while minimizing variability.
(B) Aspiration stabilization: we hypothesized that introducing a three-second pause after aspiration could help stabilize the needle, reducing vibrations that could disturb the reagent in the syringe. No significant impact was observed however this change was carried forward as a precautionary measure.
(C) Robotic arm movement: in the same vein, the robotic arm movement could itself potentially impact reagent mixing/airgap mixing during reagent translocation. Although no significant impact on the error was observed, this change was carried through as a precautionary measure.
(D) Needle height adjustment: finally, we adjusted the height of the needle after priming to avoid residual water buildup on the needle guide. This fine-tuning was essential to ensure that each drop of reagent was fully ejected without leaving trace amounts behind that could skew the volumes in subsequent steps.
(E) Gravimetric cross checking and verification: we incorporated an analytical balance (Mettler-Toledo) into the workflow to measure the mass of transferred reagents and enable real-time corrections to reagent volumes (ESI Fig. 15†). By recording the exact mass of solution transferred at each step, the system could dynamically adjust subsequent volumes, ensuring accurate liquid handling despite any small discrepancies in aspiration or dispensing, dramatically increasing the precision of the liquid transfer steps, particularly in the lower microliter range.
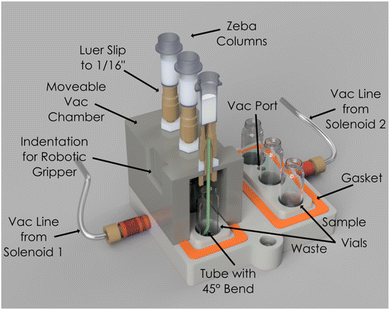
To address these challenges, we developed custom-built filtration modules (Fig. 5 and ESI Fig. 7†) that utilized vacuum-driven gel-filtration columns. A mobile vacuum filtration block was designed to integrate with the robotic arm and liquid handler. This enabled the initial washing of the stabilizing solution from the column into waste vials. The robot arm then moves the filtration block to the sample vials to conduct the filtration and acquisition of the desired sample, all in the absence of manual intervention. However, achieving consistent yields from the small volumes typical in ADC synthesis proved particularly difficult. Early tests revealed substantial variability in filtration efficiency, with yields fluctuating based on factors such as vacuum strength, filtration time, and tubing geometry. Furthermore, the air flow from the tubing line was found to splatter the sample along the walls of the vial and tubing line. This is a particular problem on such small scale as it impedes subsequent sampling as well as introducing contamination issues on the tubing line when moving from the stabilizing solution filtration to the sample filtration. These inconsistencies led us to systematically optimize the process by iteratively testing and refining various configurations. Critical adjustments—including modifying vacuum strength and tubing geometry—were essential in minimizing liquid splatter and maintaining sample integrity.
During the optimization process, we tested several key parameters to improve filtration efficiency, including the inner diameter (ID) of the tubing, vacuum strength, and vacuum duration (Fig. 6, ESI Fig. 8b, c and Table 2†). Initial trials using a 0.03 inch ID tube at vacuum strengths of 400 mbar and 600 mbar for 30 and 60 seconds, respectively, resulted in high antibody yields but caused excessive liquid splatter, compromising sample integrity. In an attempt to resolve this, we tested narrower tubing (0.01 inch and 0.02 inch IDs), which successfully eliminated splatter but reduced antibody recovery.
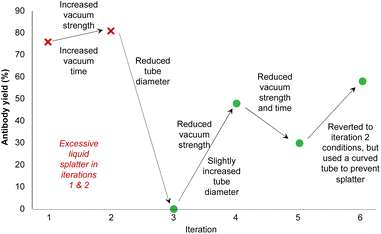 | ||
| Fig. 6 Vacuum filtration parameter optimization to increase antibody yield. Variables include the inner diameter of the tubing, vacuum strength and time, and the geometry of the tube. Full parameter details are available in ESI Table 2.† | ||
Ultimately, we reverted to the 0.03 inch ID tubing and modified its geometry by bending the tube to direct liquid flow toward the side of the waste vial (ESI Fig. 8c†). This adjustment successfully prevented splatter. Using this configuration at 600 mbar for 60 seconds, we achieved optimal antibody recovery while minimizing drug-linker carryover, ensuring the integrity of the final product. These refinements allowed us to address the primary challenges of purification—efficient filtration and consistent antibody recovery from small sample volumes.
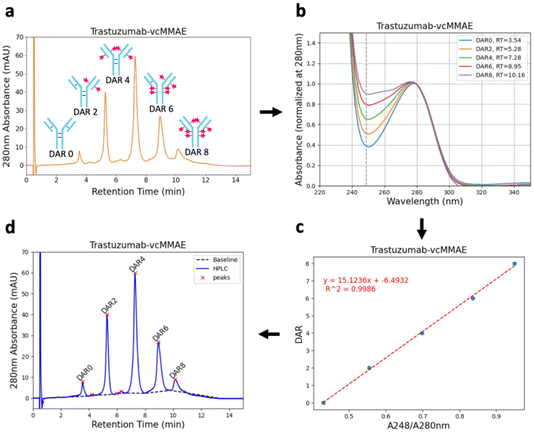
DAR determination algorithm
The average drug-to-antibody ratio (DAR) is a pivotal factor in determining the therapeutic efficacy and safety profile of antibody–drug conjugates (ADCs). Even slight deviations in the DAR can significantly shift the balance between potency and toxicity, impacting both clinical outcomes and patient safety.5–7 Stochastic cysteine conjugation inherently results in a heterogeneous mixture of ADCs, each displaying a distinct level of drug attachment, known as DAR species (typically DAR 0, 2, 4, 6, or 8) (Fig. 7a)4 where variability arises from the reduction of native interchain disulfide bonds to expose two reactive thiols, enabling conjugation with excess drug-linker molecules. The prevalence of even-numbered DAR species reflects this dual-thiol reactivity.8 However, inherent heterogeneity in conjugation underscores the critical need for precise DAR characterization, as even minor discrepancies in DAR can drastically alter the therapeutic window, affecting both the efficacy and tolerability of the resulting ADCs. Achieving consistent, optimized DAR is thus essential for advancing the next generation of targeted cancer therapies.Our automated platform leverages HIC to separate and quantify DAR species based on their hydrophobicity profiles. An increase in the conjugated drug load correlates with increased hydrophobicity, which is readily resolved in chromatographic analysis. The resulting chromatograms are rich in information yet interpreting them accurately has traditionally been a labour-intensive task. Here, we automate both the analytical acquisition and data interpretations process with a robust algorithm that dynamically assigns DAR values, significantly enhancing the throughput and precision of ADC characterization.18,19
The DAR determination algorithm replicates the nuanced decision-making process a skilled researcher would use when interpreting HIC data, translating it into a streamlined digital workflow (see ESI† for algorithm details).18,19 Initially, it assigns DAR species based on the elution order of dominant peaks (Fig. 7a). However, the complexity of the chromatogram—often populated with minor peaks from degradation products or residual unreacted drug-linker—can lead to erroneous DAR assignments if based purely on elution sequence.20–22 To circumvent this challenge, our algorithm integrates ultraviolet (UV) absorbance analysis at 280 nm (tryptophan and tyrosine residues in antibody) and 248 nm (drug-linker vcMMAE) to validate each assignment (Fig. 7b).23 As the number of conjugated vcMMAE molecules increases, the absorbance at 248 nm rises linearly.18 After correlating the ratio of A248/A280 absorbance with the degree of conjugation, the algorithm applies a linear regression model to confirm the accuracy of DAR assignments, ensuring that only peaks with a strong linear relationship (R2 > 0.99) are accepted (Fig. 7c).
Notably, this step relies on the presence of different absorbance maxima between the drug-linker and the antibody. However, similar DAR determination strategies, such as UV-Vis spectroscopy, have been employed manually with differences as small as 10 nm.23,24 Furthermore, a peak deconvolution algorithm is included in our workflow to enable its use in settings with difficult HIC separations, a known problem in ADC developments. This provides a broadly applicable automated workflow for DAR determination as a key feature of our platform.
To further refine accuracy, the algorithm automatically excludes peaks that deviate from the expected linear correlation, which are typically associated with degradation products or unbound drug-linker. This iterative reassignment ensures that only the most reliable peaks contribute to the final weighted DAR calculation. The result not only enhances the precision of DAR determination but also seamlessly integrates into our autonomous conjugation workflow, providing real-time feedback for optimizing reaction conditions.
After peak assignment, the algorithm integrates the peak areas using tangential skims (Fig. 7d and ESI Fig. 17†) and calculates the weighted average DAR of the ADC (using the equation in ESI Fig. 12†).18 A limitation of the algorithm is that it requires at least three DAR peaks to confirm the assignments, though this is typically not an issue given the broad distribution of DAR species produced through stochastic conjugation.8,25 All HIC data in this project were analyzed using this algorithm, which was validated across a wide range of average DAR values, from 1.97 to 6.60 (ESI Fig. 18†). This fully automated approach to DAR analysis exemplifies the power of digital innovation in streamlining ADC development, paving the way for faster and more efficient synthesis of targeted cancer therapeutics.
Autonomous conjugation workflow
Manual synthesis of an ADC with a specific drug-to-antibody ratio (DAR) involves a lengthy, iterative process where researchers adjust reaction conditions based on the outcomes of initial conjugations.26 This process relies on assumed predictable linear correlation between DAR and the concentration of reducing agents, such as tris(2-carboxyethyl)phosphine (TCEP). Our automated system replicates this decision-making process, transforming it into a fully autonomous workflow.In our platform, the process begins with the user specifying the target DAR and reaction conditions, including the required TCEP equivalents (Fig. 8(1)). The system then calculates the appropriate reagent volumes (Fig. 8(2)) and initiates the conjugation sequence. Once the reaction is complete, the resulting ADC is analyzed using HIC, with our algorithm determining the average DAR (Fig. 8(9)). If the measured DAR does not match the target, the system constructs a zero-intercept linear relationship between the DAR and TCEP concentration. This enables the platform to dynamically adjust TCEP equivalents for subsequent iterations (Fig. 8(10)), iteratively refining conditions to converge on the desired DAR.
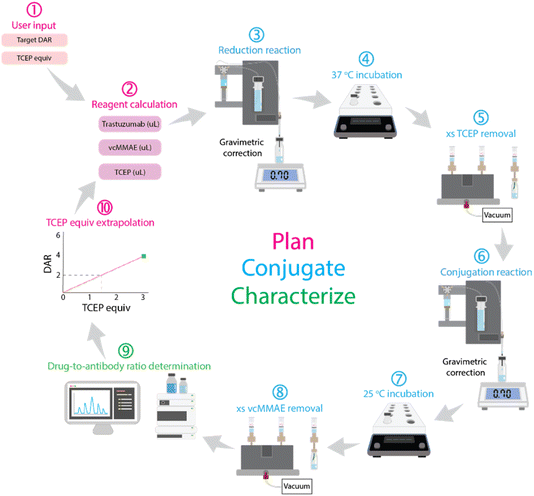 | ||
| Fig. 8 Schematic of the autonomous workflow for antibody–drug conjugation, integrating automated reduction, conjugation, purification, and real-time DAR analysis with iterative optimization. | ||
The key innovation of our workflow lies in its integration of real-time, data-driven decision-making. The system continuously adjusts reaction parameters based on analytical feedback, rather than relying on predefined conditions. This adaptability creates a continuous feedback loop where reaction planning, execution, and characterization inform one another, significantly reducing human intervention and enhancing reproducibility.
Application to trastuzumab and vcMMAE conjugation
To validate the platform, we applied it to the synthesis of ADCs using trastuzumab and the drug-linker vcMMAE. First, we assessed the platform's ability to produce and characterize an ADC without specifying a target DAR. Using an arbitrary amount of TCEP, the system successfully synthesized an ADC with a DAR of 3.85 (ESI Fig. 19a†), confirming the accuracy of the end-to-end process including synthesis, purification, and characterization.Next, we evaluated the closed-loop workflow for producing an ADC with a target DAR of 6. In the initial attempt, the system synthesized an ADC with a DAR of 3.39 (ESI Fig. 19b†). Based on this result, the system established a linear correlation between DAR and TCEP equivalents and adjusted the concentration accordingly for the next iteration. This adjustment produced an ADC with a DAR of 6.6 (ESI Fig. 19c†), which slightly exceeded the target due to a liquid handling error—9.9 TCEP equivalents (43.1 μL) were added instead of the intended 8.1 (35.2 μL). These results highlight the need for further refinement of the liquid handling module to improve precision at low dosing volumes which are on-going in our laboratory (see ESI† for further discussion).
In summary, our autonomous workflow successfully demonstrated its ability to optimize ADC synthesis iteratively. However, improving the precision of the liquid handling system will be essential to achieve consistent target DARs within a narrow margin of error (±0.2 DAR).
Conclusion
In this work, we developed a self-driving laboratory (SDL) platform that autonomously performs stochastic antibody–drug conjugate (ADC) synthesis and precise drug-to-antibody ratio (DAR) characterization. The automated workflow integrates modular hardware components for stirring, temperature control, sample transfer, and purification, along with a custom liquid handling unit and an in-house developed DAR determination algorithm. This highly flexible system was validated using stochastic cysteine-based conjugations—one of the most commonly employed methods for ADC production.The modular design of the platform will allow for straightforward adaptation, such as running optimizations of the conjugation conditions, or broadening to other settings such as lysine-based conjugations, with minimal reconfiguration. By leveraging real-time data feedback and iterative optimization, the system significantly reduces the manual effort and time typically required for ADC synthesis.
Looking ahead, our efforts will focus on enhancing the precision of the liquid handling module to reliably achieve target DARs with tighter tolerances. This advancement will further optimize the platform's capability to produce ADCs with consistent therapeutic profiles. Ultimately, our SDL prototype demonstrates the potential to streamline the production of ADCs, thereby accelerating the development of next-generation cancer therapies. This work lays the foundation for broader applications of autonomous systems in pharmaceutical development, driving both efficiency and innovation in the field.
Data availability
The hardware design files are available at https://gitlab.com/heingroup/adc-automation-hardware. The automation system and DAR determination algorithm are available at https://gitlab.com/heingroup/adc-automation. A combined dataset DOI is available at https://doi.org/10.5281/zenodo.14902831.Author contributions
Liam Roberts: conceptualization, methodology, investigation, analysis, writing – original draft, visualization. Matthew E. Reish: hardware development, data curation, formal analysis, validation, writing – review & editing. Jerrica Yang: software development, methodology, investigation. Wenyu Zhang: software development, data analysis, algorithm development, validation, writing – review & editing. Joshua S. Derasp: project administration, experimental design, methodology, writing – review & editing. Jason E. Hein: conceptualization, supervision, project administration, funding acquisition, writing – review & editing, visualization. All authors have read and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge Paloma Prieto and Dr Ekaterina Trushina for their insightful guidance and conversation during the preparation of this manuscript. Financial support for this work was provided The University of British Columbia, the Canada Foundation for Innovation (CFI-35883, CFI-44843), the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2021-03168, Discovery Accelerator Supplement), and the Canada First Research Excellence Fund (CFREF2022-00042).Notes and references
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- S. C. Alley, N. M. Okeley and P. D. Senter, Curr. Opin. Chem. Biol., 2010, 14, 529–537 CrossRef CAS PubMed.
- J. Z. Drago, S. Modi and S. Chandarlapaty, Nat. Rev. Clin. Oncol., 2021, 18, 327–344 CrossRef PubMed.
- H. Yao, F. Jiang, A. Lu and G. Zhang, Int. J. Mol. Sci., 2016, 17, 194 CrossRef PubMed.
- L. Gauzy-Lazo, I. Sassoon and M.-P. Brun, SLAS Discovery, 2020, 25, 843–868 CrossRef CAS PubMed.
- H. Maecker, V. Jonnalagadda, S. Bhakta, V. Jammalamadaka and J. R. Junutula, mAbs, 2023, 15, 2229101 CrossRef PubMed.
- T. D. Nguyen, B. M. Bordeau and J. P. Balthasar, Cancers, 2023, 15, 713 CrossRef CAS PubMed.
- M. M. C. Sun, K. S. Beam, C. G. Cerveny, K. J. Hamblett, R. S. Blackmore, M. Y. Torgov, F. G. M. Handley, N. C. Ihle, P. D. Senter and S. C. Alley, Bioconjugate Chem., 2005, 16, 1282–1290 CrossRef CAS PubMed.
- P. Szymański, M. Markowicz and E. Mikiciuk-Olasik, Int. J. Mol. Sci., 2012, 13, 427–452 CrossRef PubMed.
- Y. Wang, R. Jin, B. Shen, N. Li, H. Zhou, W. Wang, Y. Zhao, M. Huang, P. Fang, S. Wang, P. Mary, R. Wang, P. Ma, R. Li, Y. Tian, Y. Cao, F. Li, L. Schweizer and H. Zhang, Sci. Adv., 2021, 7, eabe3839 CrossRef CAS PubMed.
- N. J. Martinez, S. A. Titus, A. K. Wagner and A. Simeonov, Expert Opin. Drug Discovery, 2015, 10, 1347–1361 CrossRef CAS PubMed.
- Y. Endo, Y. Nakahara, R. Shiroma, K. Takimoto and Y. Matsuda, Org. Process Res. Dev., 2024, 28, 1806–1813 CrossRef CAS.
- Y. Yang, R. Rao, J. Valliere-Douglass and G. Tremintin, J. Chromatogr. B, 2024, 1235, 124007 CrossRef CAS PubMed.
- A. Goyon, M. Kim, L. Dai, C. Cornell, F. Jacobson, D. Guillarme and C. Stella, Anal. Chem., 2019, 91, 14896–14903 CrossRef CAS PubMed.
- E. Ferraro, J. Z. Drago and S. Modi, Breast Cancer Res., 2021, 23, 84 CrossRef CAS PubMed.
- M. Abdollahpour-Alitappeh, M. Lotfinia, T. Gharibi, J. Mardaneh, B. Farhadihosseinabadi, P. Larki, B. Faghfourian, K. S. Sepehr, K. Abbaszadeh-Goudarzi, G. Abbaszadeh-Goudarzi, B. Johari, M. R. Zali and N. Bagheri, J. Cell. Physiol., 2019, 234, 5628–5642 CrossRef CAS PubMed.
- S. Andris, M. Wendeler, X. Wang and J. Hubbuch, J. Biotechnol., 2018, 278, 48–55 CrossRef CAS PubMed.
- J. Ouyang, in Antibody-Drug Conjugates, ed. L. Ducry, Humana Press, Totowa, NJ, 2013, pp. 275–283 Search PubMed.
- S. Fekete, J.-L. Veuthey, A. Beck and D. Guillarme, J. Pharm. Biomed. Anal., 2016, 130, 3–18 CrossRef CAS PubMed.
- M. Sarrut, A. Corgier, S. Fekete, D. Guillarme, D. Lascoux, M.-C. Janin-Bussat, A. Beck and S. Heinisch, J. Chromatogr. B, 2016, 1032, 103–111 CrossRef CAS PubMed.
- M. Sarrut, S. Fekete, M.-C. Janin-Bussat, O. Colas, D. Guillarme, A. Beck and S. Heinisch, J. Chromatogr. B, 2016, 1032, 91–102 CrossRef CAS PubMed.
- Y. Yan, T. Xing, S. Wang, T. J. Daly and N. Li, J. Pharm. Biomed. Anal., 2020, 186, 113313 CrossRef CAS PubMed.
- A. Wagh, H. Song, M. Zeng, L. Tao and T. K. Das, mAbs, 2018, 10, 222–243 CrossRef CAS PubMed.
- Y. Chen, in Antibody-Drug Conjugates, ed. L. Ducry, Humana Press, Totowa, NJ, 2013, pp. 267–273 Search PubMed.
- V. Metrangolo and L. H. Engelholm, Cancers, 2024, 16, 447 CrossRef CAS PubMed.
- Z. Tawfiq, Y. Matsuda, M. J. Alfonso, C. Clancy, V. Robles, M. Leung and B. A. Mendelsohn, Anal. Sci., 2020, 36, 871–875 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00363b |
| This journal is © The Royal Society of Chemistry 2025 |