Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Developing large language models for quantum chemistry simulation input generation†
Pieter Floris
Jacobs
 and
Robert
Pollice
and
Robert
Pollice
 *
*
Stratingh Institute for Chemistry, Nijenborgh 3, 9747 AG Groningen, The Netherlands. E-mail: r.pollice@rug.nl
First published on 5th February 2025
Abstract
Scientists across domains are often challenged to master domain-specific languages (DSLs) for their research, which are merely a means to an end but are pervasive in fields like computational chemistry. Automated code generation promises to overcome this barrier, allowing researchers to focus on their core expertise. While large language models (LLMs) have shown impressive capabilities in synthesizing code from natural language prompts, they often struggle with DSLs, likely due to their limited exposure during training. In this work, we investigate the potential of foundational LLMs for generating input files for the quantum chemistry package ORCA by establishing a general framework that can be adapted to other DSLs. To improve upon 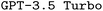 as our base model, we explore the impact of prompt engineering, retrieval-augmented generation, and finetuning via synthetically generated datasets. We find that finetuning, even with synthetic datasets as small as 500 samples, significantly improves performance. Additionally, we observe that finetuning shows synergism with advanced prompt engineering such as chain-of-thought prompting. Consequently, our best finetuned models outperform the formally much more powerful
as our base model, we explore the impact of prompt engineering, retrieval-augmented generation, and finetuning via synthetically generated datasets. We find that finetuning, even with synthetic datasets as small as 500 samples, significantly improves performance. Additionally, we observe that finetuning shows synergism with advanced prompt engineering such as chain-of-thought prompting. Consequently, our best finetuned models outperform the formally much more powerful 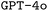 model. In turn, finetuning GPT-4o with the same small synthetic dataset leads to a further substantial performance improvement, suggesting our approach to be more general rather than limited to LLMs with poor base proficiency. All tools and datasets are made openly available for future research. We believe that this research lays the groundwork for a wider adoption of LLMs for DSLs in chemistry and beyond.
model. In turn, finetuning GPT-4o with the same small synthetic dataset leads to a further substantial performance improvement, suggesting our approach to be more general rather than limited to LLMs with poor base proficiency. All tools and datasets are made openly available for future research. We believe that this research lays the groundwork for a wider adoption of LLMs for DSLs in chemistry and beyond.
Introduction
Many researchers are required to master so-called Domain Specific Languages (DSLs) for their work.1 DSLs are specialized programming languages for one specific application domain, for instance simulation instructions for molecular mechanics or quantum chemistry computations. They offer tailored features and field-specific abstractions to streamline research. However, DSLs are hardly ever the primary subject of interest for scientists but rather a means to an end to perform simulations or analyses. This poses a challenge, as researchers have to spend substantial time to learn DSLs, which limits the actual research of interest.Code synthesis offers an appealing solution by automatically generating code from textual instructions. This is achieved via dedicated machine learning models that process the instructions and generate the corresponding code as output. These models are trained on vast amounts of code and corresponding text data.
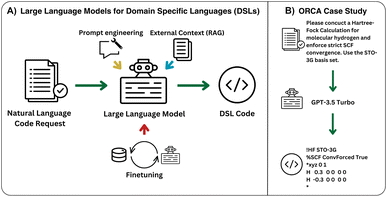
In this work, we implement a general framework (cf.Fig. 1) and develop an open-source package to utilize large language models (LLMs) for DSLs. We apply this framework to the generation of input files for the quantum chemistry software package ORCA.2 Key to this framework is the development of three synthetic data generation schemes. Additionally, we demonstrate the effects of finetuning, prompt engineering, and retrieval-augmented generation (RAG) on the performance of both 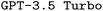 and
and 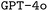 . We find that finetuning, even on small synthetic datasets, significantly boosts performance, especially when combined with chain-of-thought prompting. Our best models significantly outperform both the pristine
. We find that finetuning, even on small synthetic datasets, significantly boosts performance, especially when combined with chain-of-thought prompting. Our best models significantly outperform both the pristine 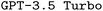 and
and 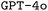 baselines. We believe that our work builds a solid foundation for future improvements in the generation of ORCA input files specifically, and the area of DSL-synthesis for chemistry in general. Accordingly, our framework, which is transferable to any DSL, even outside chemistry, will help to improve the efficiency of researchers in the near future. Expanding our framework could empower researchers to generate specialized code that advances scientific objectives, even in challenging scenarios with limited data access. In contrast to recent studies that create tailored LLMs for dealing with textual data in chemistry, our research offers a foundational approach for teaching LLMs domain-specific code, which the original model could not comprehend, and our approach can be adapted to any DSL across scientific disciplines.
baselines. We believe that our work builds a solid foundation for future improvements in the generation of ORCA input files specifically, and the area of DSL-synthesis for chemistry in general. Accordingly, our framework, which is transferable to any DSL, even outside chemistry, will help to improve the efficiency of researchers in the near future. Expanding our framework could empower researchers to generate specialized code that advances scientific objectives, even in challenging scenarios with limited data access. In contrast to recent studies that create tailored LLMs for dealing with textual data in chemistry, our research offers a foundational approach for teaching LLMs domain-specific code, which the original model could not comprehend, and our approach can be adapted to any DSL across scientific disciplines.
Related work
Until recently, specialized code synthesis models were encoder–decoder architectures3,4 using so-called abstract syntax trees (AST).5–8 An AST is a hierarchical, tree-like representation of the syntactic structure of code, where each node represents a component of the source code with specific syntactic or semantic meaning. It captures the underlying structure and connections between different elements. Typically, the encoder compresses this hierarchical structure into a fixed representation, and the decoder is trained to generate the target output, a transformed or synthesized code.Currently, LLMs are increasing in popularity in the context of code synthesis.9 These models have a massive number of parameters and are trained on immense amounts of natural language (NL) data. Most such LLMs are causal decoding-only models10 built on the transformer architecture.11 They do not generate output from a fixed piece of text but instead sequentially, predicting the next character or word based on the context of the preceding text. The transformer blocks enable the model to dynamically focus on different parts of the input, informing the prediction of each subsequent word by relevant portions of the preceding text. Hence, they have to be fed with text, referred to as a prompt, to be continued. In the case of code synthesis, this prompt would generally consist of textual descriptions on how the code should look or function. For instance, these textual descriptions would specify what the purpose of the code to be generated is and what approach is to be adopted to achieve this purpose. Recently released LLMs, such as GPT-4 (ref. 12) and Gemini 1.5,13 have shown remarkable capabilities in code synthesis, translating textual instructions into functional code for popular programming languages like Python and Java.14 These models are trained on vast and diverse datasets and can solve complex tasks given appropriate prompts. Additionally, their chat-like interface provides interactivity to users seeking clarification, troubleshooting, or deeper understanding. Accordingly, they have the potential to lower the barrier of DSLs significantly, thereby boosting overall research productivity.
Additionally, LLMs were shown to possess expert knowledge in many research domains where DSLs are used, including chemistry. Among others, LLMs were found useful for molecular design,15 property prediction,16–18 and chemistry-specific code synthesis. For instance, they can create Python code that can be used to solve chemistry problems19 and perform robotic experiments,20,21 and, in a small case study, were shown to create Python code generating simple inputs for the quantum chemistry program Gaussian.22 There have also been initial efforts to evaluate the capability of LLMs for direct DSL code synthesis. For example, recently, ChatGPT has been qualitatively assessed for generating GPAW code to be used in band structure calculations.23 The authors noted that, despite limited experimentation, the generated DSL code often contained errors or incorrect attributes. A somewhat more formal study evaluated GPT-4 for creating a few input files for LAMMPS, a widely used molecular dynamics software.24 Based on a small number target inputs and a handful of case studies, this study demonstrated that GPT-4 could produce both accurate and functional input files for various molecular dynamics tasks, highlighting the potential of LLMs to assist researchers in navigating DSLs. However, as task complexity increased, GPT-4 struggled to produce fully functional simulation inputs, often requiring significant modifications by the user to achieve desired outcomes. Other issues such as mislabeling of potentials and vague descriptions were also noted, indicating that the model has limited understanding of molecular dynamics simulations.
Importantly, this supports the notion that, despite their proficiency in general-purpose code synthesis and expertise in fields like chemistry, LLMs tend to struggle with DSLs. We hypothesise that this is due to the specialized nature of DSLs and their limited coverage in the training data of the LLM, as the corresponding model parameters are optimized during training. While LLMs are trained on vast data, it consists predominantly of widely used languages and general concepts. In contrast, DSLs typically have limited documentation, little public code, and a small user base. Consequently, this information scarcity could translate to poor performance of LLMs on DSLs. Furthermore, the syntax and semantics of DSLs can differ significantly from popular languages like Python, making it challenging for LLMs to generalise their knowledge to DSLs.
Model architecture
An overview of our model architecture is provided in Fig. 2. We selected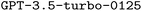 as our base LLM due to its near state-of-the-art performance when we initiated this work and its support for finetuning, which allows updating the model parameters based on small training datasets. During the reviewing process of this work, finetuning more advanced models such as
as our base LLM due to its near state-of-the-art performance when we initiated this work and its support for finetuning, which allows updating the model parameters based on small training datasets. During the reviewing process of this work, finetuning more advanced models such as 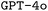 became available. Hence, we also performed finetuning with
became available. Hence, we also performed finetuning with 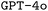 for a limited number of model configurations. A thorough description of
for a limited number of model configurations. A thorough description of 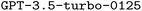 and our parameter choices for both
and our parameter choices for both 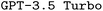 and
and 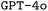 is provided in the ESI Material.†
is provided in the ESI Material.†
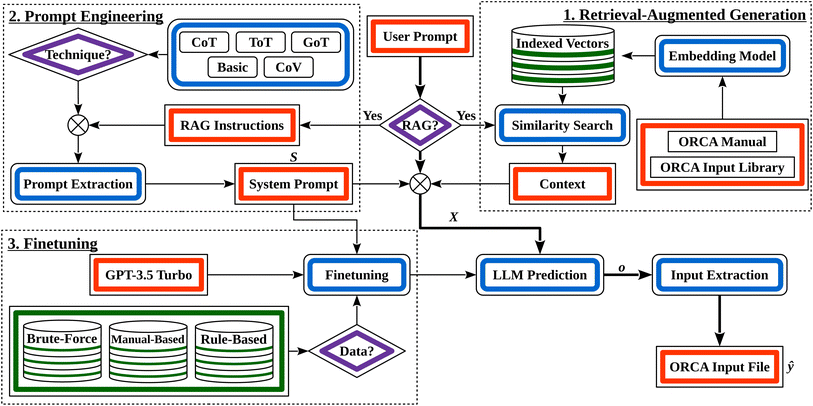
Given a prompt, a textual instruction, describing the requested chemistry simulation (X), our model aims to create the corresponding ORCA input file (ŷ). We rely on three techniques to achieve this: prompt engineering to best extract the existing knowledge of the LLM, Naive Retrieval-Augmented Generation (RAG)25 to provide the model with external documentation about ORCA inputs, and LLM finetuning on three synthetically generated datasets to teach it the syntax and semantics of an ORCA input (Section 1 in Fig. 2).
An example of an ORCA input is provided in Fig. 3. It typically contains: keyword lines, which start with an exclamation mark 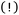 and define global options; input blocks, which are enclosed between a percent sign
and define global options; input blocks, which are enclosed between a percent sign 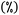 and the phrase
and the phrase  , and offer fine control over settings; and a coordinate block, which is enclosed within asterisks
, and offer fine control over settings; and a coordinate block, which is enclosed within asterisks 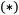 and specifies the molecular geometry, charge, and multiplicity. Comments start with
and specifies the molecular geometry, charge, and multiplicity. Comments start with 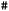 and are ignored by ORCA. A more extensive explanation of ORCA input is given in the ESI Material.†
and are ignored by ORCA. A more extensive explanation of ORCA input is given in the ESI Material.†
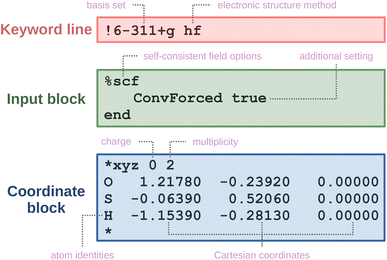 | ||
| Fig. 3 An ORCA input file for a restricted Hartree–Fock calculation with the corresponding sections highlighted in different colors. | ||
Prompt engineering
Prompt engineering has become essential to make optimal use of LLMs,26–28 also in code synthesis.19,29–32 An extensive overview of related work in prompt engineering is provided in the ESI Materials.† Previous work does not offer a clear best method, which is why we explore various approaches. To optimally make use of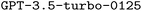 , we implemented the six prompt engineering methodologies detailed below. Each follows a different approach to structure the prompt. To make the results as comparable as possible, the corresponding system prompts all have a similar structure and utilize comparable instructions.
, we implemented the six prompt engineering methodologies detailed below. Each follows a different approach to structure the prompt. To make the results as comparable as possible, the corresponding system prompts all have a similar structure and utilize comparable instructions.
The full prompts, together with an explanation of their structure, are provided in the ESI Material.† Note that we employ these techniques in the so-called system prompt of the LLM (Section 2 in Fig. 2). It cannot be overridden by the user and is utilized for every response of the LLM.
Finetuning
Finetuning (Section 3 in Fig. 2) is a form of transfer learning,40 where knowledge gained from initial training on a large general dataset is adapted for a specific task. The model is trained in a supervised manner on a curated dataset containing examples of desired inputs and the corresponding outputs. When applied to teaching a model generating ORCA inputs, this dataset should consist of ORCA input files (y, the desired outputs) and their corresponding textual descriptions (X, the inputs).Unfortunately, for our targeted problem, these data pairs are generally unavailable. Much like the reason LLMs struggle with code synthesis for DSLs in the first place, this derives from the niche nature of DSLs. Therefore, we synthetically created our training data for finetuning, starting with input files. We devised three methods:
(1). Brute-force: this method does not consider any interactions between keywords and input blocks, and instead combines documentation randomly. Specifically, it randomly combines keywords, options, and settings to produce valid input files. To facilitate this, we scraped all options and keywords from version 5.0.4 of the ORCA manual. Additionally, we extracted all input blocks from the manual, capturing both the setting option and setting value as a full line, and documented which setting lines belonged to which option block.
(2) Manual-based: this method relies on the code blocks provided in the ORCA manual. We extracted all unique individual keyword lines and input blocks from these examples. The method randomly combines these entire input sections to create valid inputs. By extracting entire sections, this assumes that meaningful combinations of keywords, options, and settings are utilized.
(3) Rule-based: this method generates input files for specific ORCA calculation types, using predefined rules to create coherent and accurate inputs, and uses information from the ORCA manual to formulate these rules. As this is the most labor-intensive approach, we decided to only implement frequently used calculation types. We refer the reader to the ESI Materials† for a full overview of the implemented calculation types and their functionality.
Two of these approaches strongly rely on the ORCA manual. Software documentation usually focuses on common use cases and simple input examples to demonstrate the core functionality. Hence, this introduces a bias towards frequent input files and likely neglects edge cases for both the manual-based and the rule-based input file generation. However, the brute-force approach does not suffer significantly from this bias. A more in-depth comparison of the generated input files can be found in the ESI.† Across all three approaches, we employed the same method to generate a coordinate block. We created the  dataset with coordinate sections for random small molecules and selected one of them stochastically. This dataset consists of valid molecules with up to three atoms and was created from random SELFIES,41 followed by conversion to SMILES.42 The Cartesian coordinates were then generated with
dataset with coordinate sections for random small molecules and selected one of them stochastically. This dataset consists of valid molecules with up to three atoms and was created from random SELFIES,41 followed by conversion to SMILES.42 The Cartesian coordinates were then generated with  .43 We opted for small molecules with up to three atoms because this dramatically reduces simulation times while allowing for the full range of ORCA simulations (except for dihedral angle constraints). Notably, we also generated radicals to support open-shell methods such as unrestricted Hartree–Fock calculations. However, we excluded ions and heavy atoms like bromine and iodine.
.43 We opted for small molecules with up to three atoms because this dramatically reduces simulation times while allowing for the full range of ORCA simulations (except for dihedral angle constraints). Notably, we also generated radicals to support open-shell methods such as unrestricted Hartree–Fock calculations. However, we excluded ions and heavy atoms like bromine and iodine.
After we appended the coordinate block, we ran the generated ORCA input to see whether it is executable and ORCA finishes the corresponding simulation without errors, ensuring they represent valid data. Importantly, for executable files, we replaced the coordinate block with a comment containing the SMILES of the molecule. We did this because generating meaningful Cartesian coordinates with LLMs is challenging as they consist solely of floating-point numbers. Since tools like 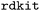 readily generate valid coordinates, training an LLM for this would be inefficient. Additionally, we believe that LLMs would not be well-suited for that. By including the SMILES in our modified inputs, we focused instead on teaching the model to associate certain basis sets with specific elements.
readily generate valid coordinates, training an LLM for this would be inefficient. Additionally, we believe that LLMs would not be well-suited for that. By including the SMILES in our modified inputs, we focused instead on teaching the model to associate certain basis sets with specific elements.
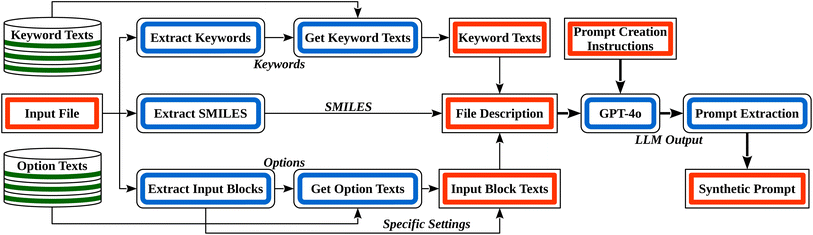
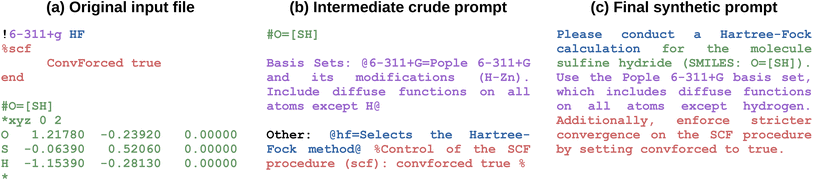
Overall, for each method, we generated 500 input files: 500 for the brute-force approach, 500 for the manual-based method, and 500 for the rule-based technique. We kept these inputs separate to compare the performance of each input generation method. With the synthetic input files in hand, we still needed to generate the corresponding NL prompts. Given an augmented input (i.e., the coordinate block is replaced with a SMILES comment), we first extract all individual parts: the keywords, the options of the different input blocks, the corresponding setting lines, and the SMILES. We map these to textual descriptions via documentation from the ORCA manual using the following hardcoded steps:
(1) Write down the SMILES of the molecule after a ‘#’ character.
(2) Write down all keywords with their description in the format ‘@keyword = description@’.
(3) Write down all options in question and provide their definition in the format used for keywords. For each option, write down the corresponding settings and enclose the description of the full block between two ‘%’ characters.
Through these steps, we obtain a crude prompt, which we use as starting point to create a user-like prompt with GPT-4o. The full process of user prompt creation is depicted in Fig. 4, with a simple example illustrated in Fig. 5. Further details are provided in the ESI Material.† Manual checking of a small subset of the generated user prompts showed them to incorporate the simulation instructions of the input files with high fidelity. However, the total number of generated prompts is too high for efficient manual checking.
These datasets were the foundation for refining the base LLM. When finetuning on any of our three synthetic datasets, we used the same system prompt across all examples for the model to learn generating consistent responses. With the  prompt engineering technique, the user prompt is the input of the model and the desired ORCA input file is the corresponding output (i.e., the label). In contrast, for systematic prompt engineering, it is necessary to create additional synthetic responses as the model is taught to reason about the desired ORCA input in its output. Hence, we altered y to start with the steps defined in all our prompts and used hard-coded rules to create answers based on the target input. This process is detailed in the ESI Material.†
prompt engineering technique, the user prompt is the input of the model and the desired ORCA input file is the corresponding output (i.e., the label). In contrast, for systematic prompt engineering, it is necessary to create additional synthetic responses as the model is taught to reason about the desired ORCA input in its output. Hence, we altered y to start with the steps defined in all our prompts and used hard-coded rules to create answers based on the target input. This process is detailed in the ESI Material.†
RAG
RAG is a general means to provide LLMs with additional context by adding information retrieved from external data sources to the system prompt.44 Our version of RAG, illustrated in Section 1 in Fig. 2, uses a database of two documents that we considered useful context: the ORCA manual (version 5.0.4) and the ORCA Input Library website.45 Such a documentation-based version of RAG has previously been shown to be effective for code synthesis.46 The manual is the main documentation for ORCA. It provides extensive and authoritative information on the available functionalities, commands, and parameters. We used all pages except for the parts where keyword and option definitions were provided. We excluded these because they were already used to construct our user prompts. Hence, including them could inflate model performance artificially if the model learned to rely on direct overlaps between the prompts and documentation. By omitting this data, we ensure a more realistic performance evaluation. In addition, the ORCA Input Library offers a comprehensive overview of the possible calculations in ORCA. It contains many practical examples and input file templates as guidance for users. However, a significant part of its content was created and tested for ORCA version 4.2.1. This means that some input file changes between ORCA versions 5.0.4 and 4.2.1 have not been adapted. This can cause misleading RAG context in some cases when inconsistent information is retrieved from the two data source. Nevertheless, the corresponding differences are sufficiently small to warrant the inclusion of the ORCA Input Library because of its otherwise highly valuable information. To be able to use the ORCA Input Library as a document, we saved all its pages as PDF.Both these documents were split by page, each of which was embedded using the 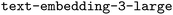 model, the state-of-the-art embedding model by OpenAI at the time of writing.47 The resulting embeddings were indexed using FAISS,48 a library designed to readily cluster dense vectors. These clustered vectors permit a FAISS similarity search between the user prompt and the indexed embedding vectors to quickly retrieve relevant documentation. The K most similar pages (one of our hyperparameters) are retrieved and added to the user prompt for the model to make use of during the ORCA input generation.
model, the state-of-the-art embedding model by OpenAI at the time of writing.47 The resulting embeddings were indexed using FAISS,48 a library designed to readily cluster dense vectors. These clustered vectors permit a FAISS similarity search between the user prompt and the indexed embedding vectors to quickly retrieve relevant documentation. The K most similar pages (one of our hyperparameters) are retrieved and added to the user prompt for the model to make use of during the ORCA input generation.
Finally, we opted to add extra instructions to the system prompt whenever RAG was used. Aside from introducing what type of data we retrieve the external context from, we instruct the LLM to indicate what part of the context is used for what part of the input with explanation. We also instructed the model only to use the context when relevant. The exact instructions we used are found in the ESI Material.†
Results and discussion
To evaluate the performance of both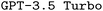 and
and 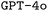 in generating ORCA input files, we explored the effect of finetuning with different synthetic datasets, different prompt engineering methods, and RAG on model performance. The full experimental setup, including hyperparameters, data overviews, and explanations of all our metrics is provided in the ESI Materials.†
in generating ORCA input files, we explored the effect of finetuning with different synthetic datasets, different prompt engineering methods, and RAG on model performance. The full experimental setup, including hyperparameters, data overviews, and explanations of all our metrics is provided in the ESI Materials.†
Experiments
Whereas finetuning was performed by training with our synthetic data, we wanted to have independent data to compare model performance during hyperparameter optimization (validation) and evaluate final model performance (testing). Thus, validation and testing were performed using the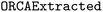 dataset, which comprises 588 input files gathered from both ioChem49 and internal sources. Therefore, all input files used in validation and testing were real-world data. These real-world input files were more complex than the synthetic input files, and generally tend to contain more keywords, more input blocks, and more setting lines (details in the ESI Materials†) These input files were preprocessed for model compatibility and the corresponding prompts were created using our prompt generation scheme. Finally,
dataset, which comprises 588 input files gathered from both ioChem49 and internal sources. Therefore, all input files used in validation and testing were real-world data. These real-world input files were more complex than the synthetic input files, and generally tend to contain more keywords, more input blocks, and more setting lines (details in the ESI Materials†) These input files were preprocessed for model compatibility and the corresponding prompts were created using our prompt generation scheme. Finally, 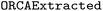 was split 50/50 to create both a validation and a test set.
was split 50/50 to create both a validation and a test set.
Using the validation set, we conducted a grid search by evaluating all systematic combinations of hyperparameters on a pre-defined grid of allowed values to find the best hyperparameters. Afterwards, we finetuned 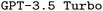 , our base LLM, independently using the three training datasets (brute-force, manual-based, rule-based), and probed the impact of both RAG and prompt engineering on model performance in all possible configurations. At the time of writing, GPT-4o could not be finetuned yet, however, it became available for finetuning during the review process. Accordingly, we used it both in its standard configuration as a strong baseline and finetuned in a limited number of model configurations.
, our base LLM, independently using the three training datasets (brute-force, manual-based, rule-based), and probed the impact of both RAG and prompt engineering on model performance in all possible configurations. At the time of writing, GPT-4o could not be finetuned yet, however, it became available for finetuning during the review process. Accordingly, we used it both in its standard configuration as a strong baseline and finetuned in a limited number of model configurations.
Importantly, ORCA inputs lack the hierarchical complexity and linguistic properties that typical code and NL translation metrics assess (e.g., word order), making these metrics unsuitable here. Instead, the presence of specific terms necessary for correct execution is key for ORCA inputs. Hence, we framed this task as a classification and evaluated accuracy solely based on the presence or absence of predicted words via the F1 score, which accounts for both errors resulting from missing words and errors resulting from superfluous words. Accordingly, we evaluated all models via the F1total and F1avg scores on the 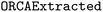 test set, and determined the fraction of executable input files. In contrast to F1total, F1avg weighs all three ORCA sections equally rather than assigning more weight to the section containing more terms.
test set, and determined the fraction of executable input files. In contrast to F1total, F1avg weighs all three ORCA sections equally rather than assigning more weight to the section containing more terms.
Hyperparameter optimization
Due to the large number of possible model configurations with all the prompt engineering techniques considered, to reduce computational expense, we decided to identify the most effective approach for prompt engineering with the base model before finetuning with any particular system prompt (Table 1). Importantly, all prompt engineering techniques improved performance over using no prompt engineering and lead to similar levels of performance. This suggests that providing the model with information about ORCA inputs and guidance on what to do is essential for enabling the model to employ its limited pre-trained knowledge productively. CoT outperformed the other techniques, with a marginal, but likely insignificant, improvement over ToT. As every prompt engineering method requires separate finetuning later on, we decided to only proceed with CoT for all subsequent experiments. Nevertheless, CoT and ToT show essentially equal performance and would be equally good choices going forward.
base model before finetuning with any particular system prompt (Table 1). Importantly, all prompt engineering techniques improved performance over using no prompt engineering and lead to similar levels of performance. This suggests that providing the model with information about ORCA inputs and guidance on what to do is essential for enabling the model to employ its limited pre-trained knowledge productively. CoT outperformed the other techniques, with a marginal, but likely insignificant, improvement over ToT. As every prompt engineering method requires separate finetuning later on, we decided to only proceed with CoT for all subsequent experiments. Nevertheless, CoT and ToT show essentially equal performance and would be equally good choices going forward.
| Prompt engineering | Validation F1avg |
|---|---|
| None | 0.083 |
| Basic | 0.199 |
| CoT | 0.214 |
| CoVe | 0.199 |
| GoT | 0.207 |
| ToT | 0.212 |
Apart from identifying the best prompt engineering technique, we also made two key observations during hyperparameter optimization. First, Fig. 6 shows a scatter plot correlating the validation loss obtained from the OpenAI API during finetuning against the corresponding validation F1avg score. Note that the loss metric used for training  is not made public by OpenAI. We observe significant fluctuations in validation performance over different parameter settings, often resulting in extremely low F1avg scores. This observation aligns with the findings of Yao et al.,50 who demonstrated that finetuning is very meticulous and can increase the tendency towards hallucinations. The plot also illustrates that, counter to our expectations, a lower validation loss does not necessarily correspond to a better F1avg score. Many models achieving the best F1avg scores exhibited high losses, while, conversely, some of the worst models showed low losses. From this, we concluded that the training loss of
is not made public by OpenAI. We observe significant fluctuations in validation performance over different parameter settings, often resulting in extremely low F1avg scores. This observation aligns with the findings of Yao et al.,50 who demonstrated that finetuning is very meticulous and can increase the tendency towards hallucinations. The plot also illustrates that, counter to our expectations, a lower validation loss does not necessarily correspond to a better F1avg score. Many models achieving the best F1avg scores exhibited high losses, while, conversely, some of the worst models showed low losses. From this, we concluded that the training loss of  was not a reliable metric for selecting our best model. Therefore, we decided against the validation loss available through the OpenAI API for determining the best parameters for testing. Instead, we optimized the validation F1avg score directly.
was not a reliable metric for selecting our best model. Therefore, we decided against the validation loss available through the OpenAI API for determining the best parameters for testing. Instead, we optimized the validation F1avg score directly.
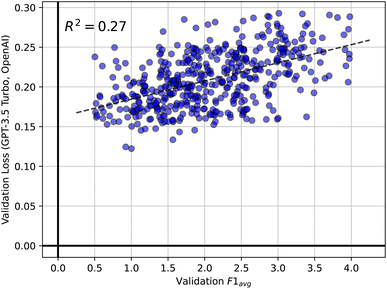 | ||
Fig. 6 Scatter plot of validation loss for  against F1avg score during the hyperparameter search over 600 different configurations. against F1avg score during the hyperparameter search over 600 different configurations. | ||
Second, Fig. 7 shows the validation performance of all our model configurations utilizing RAG as a function of the number of retrieved documentation pages. It shows that non-finetuned models tend to slightly increase in validation performance as K increases. However, for the finetuned models, we generally observe a decline in performance as K grows until the model context window is filled (K =Limit). The only exception is the brute-force model with CoT, where retrieving five documentation pages shows best validation performance. It seems that the finetuned models handle external context poorly. We hypothesize that they either ‘forget’ how to handle the additional context or get ‘confused’ by it. This could be explained by the models not being trained to use RAG. Alternatively, RAG might only be really useful when the model has little knowledge about ORCA input files.
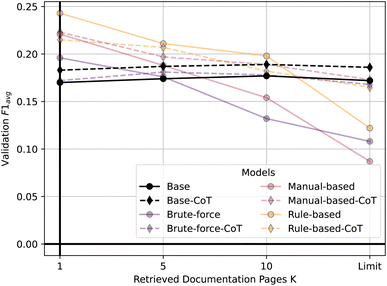 | ||
| Fig. 7 Validation performance of all models using RAG as a function of retrieved documentation pages (K). | ||
Base model configurations
Table 2 summarizes the performance of the base models ( and
and  ) in all possible configurations with RAG and CoT. A detailed analysis of the corresponding ORCA errors encountered when executing the generated inputs is provided in the ESI Materials.†
) in all possible configurations with RAG and CoT. A detailed analysis of the corresponding ORCA errors encountered when executing the generated inputs is provided in the ESI Materials.†
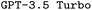 and
and 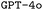 . F1avg is the mean of F1keywords, F1options, and F1setting. When CoT is not used, ‘basic’ prompt engineering is used instead. For all metrics, the best performance across the
. F1avg is the mean of F1keywords, F1options, and F1setting. When CoT is not used, ‘basic’ prompt engineering is used instead. For all metrics, the best performance across the 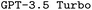 and the
and the 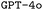 models, respectively, is bolded
models, respectively, is bolded
| Base model | CoT | RAG | % executable | F1total | F1avg | F1keywords | F1options | F1settings |
|---|---|---|---|---|---|---|---|---|
| GPT-3.5 Turbo | ✗ | ✗ | 3.401 | 0.217 | 0.182 | 0.344 | 0.171 | 0.029 |
| ✗ | ✓ | 11.905 | 0.242 | 0.194 | 0.374 | 0.176 | 0.031 | |
| ✓ | ✗ | 3.741 | 0.229 | 0.193 | 0.366 | 0.177 | 0.035 | |
| ✓ | ✓ | 14.626 | 0.229 | 0.188 | 0.359 | 0.166 | 0.037 | |
| GPT-4o | ✗ | ✗ | 5.872 | 0.378 | 0.279 | 0.545 | 0.227 | 0.066 |
| GPT-4o | ✗ | ✓ | 10.884 | 0.383 | 0.296 | 0.544 | 0.241 | 0.103 |
| GPT-4o | ✓ | ✗ | 7.570 | 0.382 | 0.281 | 0.558 | 0.222 | 0.063 |
| GPT-4o | ✓ | ✓ | 10.204 | 0.390 | 0.284 | 0.567 | 0.214 | 0.070 |
We find that the  base model performed poorly in ORCA input file synthesis, producing few functional files and struggling with accurate synthesis of settings and options. The relatively respectable F1keywords score is likely because the keywords are often specified literally in the prompt. This poor performance supports the notion that LLMs are not well-suited for DSL code synthesis due to insufficient exposure in their pre-training. When we combine the poor base model with RAG, we observe a higher fraction of executable files compared to the
base model performed poorly in ORCA input file synthesis, producing few functional files and struggling with accurate synthesis of settings and options. The relatively respectable F1keywords score is likely because the keywords are often specified literally in the prompt. This poor performance supports the notion that LLMs are not well-suited for DSL code synthesis due to insufficient exposure in their pre-training. When we combine the poor base model with RAG, we observe a higher fraction of executable files compared to the  baseline, even outperforming the
baseline, even outperforming the  base model in this regard. Intuitively, this makes sense as we provide external information about ORCA inputs including numerous functional examples. Using RAG also leads to slight improvements in the F1 scores. While improvements are insignificant for predicted options and settings, the additional context helped the model to improve keyword prediction, likely by referencing a broader range of valid keywords from the added information. We observe a similar impact of RAG on
base model in this regard. Intuitively, this makes sense as we provide external information about ORCA inputs including numerous functional examples. Using RAG also leads to slight improvements in the F1 scores. While improvements are insignificant for predicted options and settings, the additional context helped the model to improve keyword prediction, likely by referencing a broader range of valid keywords from the added information. We observe a similar impact of RAG on  , however, the performance increase is significantly smaller compared to
, however, the performance increase is significantly smaller compared to 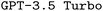 .
.
Employing CoT prompt engineering boosts F1 scores, however, the improvement is more modest compared to RAG, and the fraction of executable files hardly increases. Based on previous literature,36,51,52 we expected larger performance improvements. We explain this marginal improvement by the LLMs simply lacking knowledge about the desired chemistry simulation, making it unable to exploit the advantages of reasoning. Nevertheless, CoT is still valuable for its added explainability despite limited performance improvement. Combining RAG and CoT results in the highest fraction of executable files for 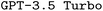 , but it only improves F1 scores for
, but it only improves F1 scores for 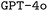 .
.
Finetuned model configurations
The performance of our finetuned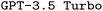 models, in all possible configurations, and our finetuned
models, in all possible configurations, and our finetuned 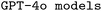 , in a few select configurations, is provided in Table 3. Again, we provide a detailed analysis of the different ORCA errors encountered when executing the corresponding input files in the ESI Materials.†
, in a few select configurations, is provided in Table 3. Again, we provide a detailed analysis of the different ORCA errors encountered when executing the corresponding input files in the ESI Materials.†
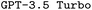 and
and 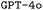 model configurations. F1avg is the mean of F1keywords, F1options, and F1setting. When CoT is not used, ‘basic’ prompt engineering is used instead. For all metrics, the best performance across the
model configurations. F1avg is the mean of F1keywords, F1options, and F1setting. When CoT is not used, ‘basic’ prompt engineering is used instead. For all metrics, the best performance across the 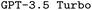 and the
and the  models, respectively, is bolded
models, respectively, is bolded
| Base model | CoT | RAG | Finetuning | % executable | F1total | F1avg | F1keywords | F1options | F1settings |
|---|---|---|---|---|---|---|---|---|---|
| GPT-3.5 Turbo | ✗ | ✗ | ✗ | 3.401 | 0.217 | 0.182 | 0.344 | 0.171 | 0.029 |
| ✗ | ✗ | Brute-force | 15.306 | 0.382 | 0.267 | 0.527 | 0.147 | 0.128 | |
| ✗ | ✗ | Manual-based | 10.544 | 0.390 | 0.261 | 0.535 | 0.133 | 0.113 | |
| ✗ | ✗ | Rule-based | 11.225 | 0.333 | 0.237 | 0.475 | 0.199 | 0.037 | |
| ✗ | ✓ | Brute-force | 25.122 | 0.221 | 0.177 | 0.310 | 0.128 | 0.112 | |
| ✗ | ✓ | Manual-based | 15.926 | 0.311 | 0.228 | 0.424 | 0.133 | 0.128 | |
| ✗ | ✓ | Rule-based | 10.884 | 0.282 | 0.206 | 0.407 | 0.184 | 0.028 | |
| ✓ | ✗ | Brute-force | 17.006 | 0.384 | 0.264 | 0.535 | 0.137 | 0.121 | |
| ✓ | ✗ | Manual-based | 21.769 | 0.426 | 0.286 | 0.588 | 0.145 | 0.125 | |
| ✓ | ✗ | Rule-based | 22.048 | 0.361 | 0.254 | 0.506 | 0.207 | 0.047 | |
| ✓ | ✓ | Brute-force | 26.191 | 0.282 | 0.205 | 0.326 | 0.106 | 0.105 | |
| ✓ | ✓ | Manual-based | 15.986 | 0.330 | 0.223 | 0.459 | 0.104 | 0.106 | |
| ✓ | ✓ | Rule-based | 15.646 | 0.298 | 0.205 | 0.425 | 0.158 | 0.032 | |
| GPT-4o | ✗ | ✗ | ✗ | 5.872 | 0.378 | 0.279 | 0.545 | 0.227 | 0.066 |
| GPT-4o | ✓ | ✗ | Manual-based | 18.027 | 0.513 | 0.330 | 0.691 | 0.157 | 0.140 |
| GPT-4o | ✓ | ✓ | Manual-based | 15.306 | 0.473 | 0.308 | 0.632 | 0.151 | 0.141 |
Looking at the  results, across all three finetuning datasets, we see consistent performance increase, solidifying our expectation that teaching the base model ORCA input synthesis is essential due to its limited pre-trained knowledge. Importantly, these improvements are achieved with only 500 synthetic ORCA input files, highlighting the large potential of synthetic datasets to enhance real-world DSL synthesis performance. Overall, the manual-based models perform best, likely because the corresponding training data includes keywords and input blocks commonly used in real-world files, as they are extracted from documentation examples.
results, across all three finetuning datasets, we see consistent performance increase, solidifying our expectation that teaching the base model ORCA input synthesis is essential due to its limited pre-trained knowledge. Importantly, these improvements are achieved with only 500 synthetic ORCA input files, highlighting the large potential of synthetic datasets to enhance real-world DSL synthesis performance. Overall, the manual-based models perform best, likely because the corresponding training data includes keywords and input blocks commonly used in real-world files, as they are extracted from documentation examples.
This is in marked contrast to the brute-force dataset, which shows lower F1 scores. We argue that this is because many of the randomly selected keywords and input blocks are not used in real-world calculations. Interestingly, the brute-force approach was able to generate more executable input files compared to the other datasets. This is surprising as this approach combines documentation in the most random way. However, as all our finetuning datasets only included executable files, the specific combinations of keywords, settings, and options do not need to be meaningful to increase performance on generating executable inputs.
Unexpectedly, the rule-based models underperformed compared to the manual-based approach, despite the rule-based dataset being designed to represent real-world data. This is likely due to the limited subset of calculations we employed, which resulted in less diverse keywords and settings, as characterized by the low variability and high skewness of this dataset (cf. ESI Materials†). Consequently, this translated to worse generalisation performance.
Whereas the base models showed little performance improvement with prompt engineering, finetuning combined with CoT yields the highest F1 scores across all three finetuning datasets. The percentage of executable files also increases across all datasets with CoT-based finetuning, with the largest increase seen in the rule-based and manual-based datasets. Notably, the manual-based finetuning dataset, when combined with CoT, achieves the highest F1 scores of all  models. We explain this performance improvement by the finetuning teaching the model how to make use of the CoT prompt, learn mappings of descriptions, and dissect the different parts of an ORCA input file. Moreover, we hypothesize that the finetuned model has more knowledge about ORCA to use in its reasoning, making CoT more effective.
models. We explain this performance improvement by the finetuning teaching the model how to make use of the CoT prompt, learn mappings of descriptions, and dissect the different parts of an ORCA input file. Moreover, we hypothesize that the finetuned model has more knowledge about ORCA to use in its reasoning, making CoT more effective.
Conversely, using RAG generally results in significantly lower F1 scores. Nevertheless, it improves the number of executable files for both the brute-force and the manual-based datasets, but not for the rule-based dataset. Combining RAG with CoT again generally degrades all performance metrics compared to using CoT alone. An exception is observed with the brute-force dataset, where the fraction of executable files increases significantly, reaching its highest value. This is in line with our findings from the hyperparameter optimization, where we found that, generally, the finetuned models performed worse as more documents were retrieved.
Finally, we also observe a substantial performance increase for  when finetuned with the manual-based dataset. While the resulting fraction of executable input files is slightly smaller compared to the corresponding finetuned
when finetuned with the manual-based dataset. While the resulting fraction of executable input files is slightly smaller compared to the corresponding finetuned 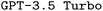 model, finetuning leads to the highest F1 scores, outperforming the best finetuned
model, finetuning leads to the highest F1 scores, outperforming the best finetuned 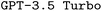 model by a significant margin. When looking at the bigger picture, we find very encouraging performance of our proposed model architecture. Our best finetuned
model by a significant margin. When looking at the bigger picture, we find very encouraging performance of our proposed model architecture. Our best finetuned 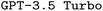 model outperforms the
model outperforms the 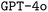 base model in all metrics significantly, except for the F1 score for input options. Both baseline models benefit substantially from finetuning with a very limited sample of computer-generated training data.
base model in all metrics significantly, except for the F1 score for input options. Both baseline models benefit substantially from finetuning with a very limited sample of computer-generated training data.
Limitations
As this study is a first systematic foray in DSL synthesis for quantum chemistry simulations, we identified several limitations of our approach. Here, we discuss the most important ones. A full overview is provided in the ESI Materials.†First, the performance of our best models, while outperforming the baselines both with respect to accuracy and the generation of executable input files, are still insufficient for real-world deployment, especially when facing inexperienced users. At the current level, the majority of the generated input files are not executable with ORCA. While users could try to simply submit the same prompt and rely on the probabilistic nature or submit a modified prompt, this does not necessarily lead to an executable input file. Perhaps the most promising action users could take is to provide the ORCA output as additional information in the prompt together with the previously generated input to allow for the LLM to self-correct. However, this is not a user-friendly solution and, therefore, at the current performance level, these models are not practical yet. Further work is needed to reach an appropriate performance level to avoid cumbersome workarounds and really reduce the time researcher spend on input file generation.
Second, we did not test the performance of all the implemented prompt engineering techniques on finetuned models. Despite the low impact of prompt engineering beyond the ‘Basic’ approach on the base model, we suspect that performance differences would likely be more pronounced on finetuned models. Hence, follow-up work should investigate the impact of different prompt engineering approaches more comprehensively. Nevertheless, we do not expect substantial performance improvements from alternative prompt engineering techniques.
Additionally, as mentioned before, we observed that the internal loss used by 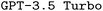 was a poor indication of final model performance. This is particularly problematic as the model parameters are updated based on the gradient of the loss, potentially even moving away from better model parameters during finetuning. While both
was a poor indication of final model performance. This is particularly problematic as the model parameters are updated based on the gradient of the loss, potentially even moving away from better model parameters during finetuning. While both 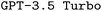 and
and 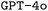 still provided us with strong base models that permit finetuning, open-source models would provide more insight regarding the loss, and even enable us to modify it.
still provided us with strong base models that permit finetuning, open-source models would provide more insight regarding the loss, and even enable us to modify it.
Furthermore, we used a limited amount of synthetic data for this first case study. Notably, it has been shown for the BERT model53 that the size of the finetuning dataset can impact performance drastically,54 which we did not take advantage of. The corresponding data generation methods could have been exploited further to create more synthetic training data. This would provide additional insights about the change in model performance with training samples. Combining data from multiple generation methods is also a promising avenue towards further performance improvements.
Finally, all the user prompts used for quantitative evaluation were synthetic. Given the novelty of our approach, the absence of such data is natural. While we believe that they are sufficiently realistic, future work is necessary to collect prompts from actual users.
Conclusions
In this study, we proposed a model and developed a Python package for code synthesis of domain specific languages, and applied it to the quantum chemistry program ORCA. We devised three methods for synthetic data generation of increased sophistication. We explored the effects of finetuning, prompt engineering, and retrieval-augmented generation on the performance of both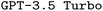 and
and 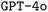 in synthesizing ORCA input files and showed that our best models were able to outperform the pristine base models significantly.
in synthesizing ORCA input files and showed that our best models were able to outperform the pristine base models significantly.
Our results highlight the importance of finetuning to improve performance, even when using small synthetically generated datasets. While chain-of-thought prompting does not substantially outperform basic prompt engineering, its combination with finetuning leads to synergistic improvements. In contrast, we find that RAG, while beneficial as a standalone enhancement, can degrade performance when used with finetuned models, highlighting the importance of careful method integration.
While our best-performing model is still insufficient for an end user product, our study is a first-of-its-kind and provides a comprehensive general framework for synthetic data generation and model setup, paving a clear path towards the goal of an LLM for DSL code generation in chemistry. There are several limitations to be addressed in future research. We believe that the most promising avenue is increasing the size and diversity of synthetic datasets. Additionally, applying our methodology to open-source LLMs, perhaps even models specialized on code synthesis, would allow to implement a meaningful loss function and ultimately lead to better models. Furthermore, exploring advanced RAG techniques, which incorporate mechanisms allowing for ranking and filtering the information provided to the LLM, or integrating context during finetuning present promising future avenues. Finally, and perhaps most importantly, the approach we developed, while applied to ORCA, is general, and thus allows for straightforward expansion to other widely used quantum chemistry simulation programs such as ADF,55 Gaussian,56 or Q-Chem.57 This could reveal whether the finetuned knowledge is transferable across different DSLs, offering broader application of our methodology in chemistry and beyond. Accordingly, we encourage others to use our approach as foundation for tackling similar problems.
Data availability
The code used for generating the datasets and running the experiments presented in this article is available at our GitLab Repository (https://git.lwp.rug.nl/pollice-research-group/ORCAInputFileSynthesis). The version of the code employed for this study is found in commit . This repository also includes the specific brute-force, manual-based and rule-based datasets used for finetuning both
. This repository also includes the specific brute-force, manual-based and rule-based datasets used for finetuning both 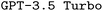 and
and 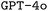 , the
, the  dataset used for appending coordinate blocks and the
dataset used for appending coordinate blocks and the  dataset used for evaluation of the model. The code and data were developed for ORCA version 5.0.4 and based on the corresponding documentation. Alternatively, the code can also be downloaded at https://doi.org/10.34894/WNRHA4.
dataset used for evaluation of the model. The code and data were developed for ORCA version 5.0.4 and based on the corresponding documentation. Alternatively, the code can also be downloaded at https://doi.org/10.34894/WNRHA4.
Author contributions
Pieter Floris Jacobs: data curation; formal analysis (lead); investigation; methodology (equal); project administration (equal); software (lead); validation (lead); visualization; writing – original draft (lead); writing – review & editing (equal). Robert Pollice: conceptualization; formal analysis (supporting); funding acquisition; methodology (equal); project administration (equal); software (supporting); resources; supervision; validation (supporting); writing – original draft (supporting); writing – review & editing (equal).Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank S. Chen and S. Tretiakov for providing their ORCA expertise and helping us construct our test and validation sets.Notes and references
- A. Fan, B. Gokkaya, M. Harman, M. Lyubarskiy, S. Sengupta, S. Yoo and J. M. Zhang, 2023 IEEE/ACM International Conference on Software Engineering: Future of Software Engineering (ICSE-FoSE), 2023, pp. 31–53 Search PubMed.
- F. Neese, F. Wennmohs, U. Becker and C. Riplinger, J. Chem. Phys., 2020, 152, 224108 CrossRef CAS PubMed.
- M. Z. Alom, T. M. Taha, C. Yakopcic, S. Westberg, P. Sidike, M. S. Nasrin, M. Hasan, B. C. Van Essen, A. A. Awwal and V. K. Asari, Electronics, 2019, 8, 292 CrossRef.
- K. Aitken, V. Ramasesh, Y. Cao and N. Maheswaranathan, Adv. Neural Inf. Process. Syst., 2021, 34, 22184–22195 Search PubMed.
- R. E. Noonan, Comput. Lang., 1985, 10, 225–236 CrossRef.
- X. Wang, I. Dillig and R. Singh, Proc. ACM Program. Lang., 2017, 2, 1–30 CAS.
- A. Desai, S. Gulwani, V. Hingorani, N. Jain, A. Karkare, M. Marron and S. Roy, Proceedings of the 38th International Conference on Software Engineering, 2016, pp. 345–356 Search PubMed.
- S. Gulwani, O. Polozov, R. Singh, et al., Foundations and Trends® in Programming Languages, 2017, vol. 4, pp. 1–119 Search PubMed.
- J. Jiang, F. Wang, J. Shen, S. Kim and S. Kim, arXiv, 2024, preprint, arXiv:2406.00515, DOI:10.48550/arXiv.2406.00515.
- F. F. Xu, U. Alon, G. Neubig and V. J. Hellendoorn, MAPS@PLDI 2022: 6th ACM SIGPLAN International Symposium on Machine Programming, San Diego, CA, USA, 2022, pp. 1–10 Search PubMed.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. Kaiser and I. Polosukhin, Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, December 4–9, 2017, Long Beach, CA, USA, 2017, pp. 5998–6008 Search PubMed.
- J. OpenAI, S. Adler, S. Agarwal, L. Ahmad, I. Akkaya, F. L. Aleman, D. Almeida, J. Altenschmidt, S. Altman, S. Anadkat, R. Avila, I. Babuschkin, S. Balaji, V. Balcom, P. Baltescu, H. Bao, M. Bavarian, J. Belgum, I. Bello, J. Berdine, G. Bernadett-Shapiro, C. Berner, L. Bogdonoff, O. Boiko, M. Boyd, A.-L. Brakman, G. Brockman, T. Brooks, M. Brundage, K. Button, T. Cai, R. Campbell, A. Cann, B. Carey, C. Carlson, R. Carmichael, B. Chan, C. Chang, F. Chantzis, D. Chen, S. Chen, R. Chen, J. Chen, M. Chen, B. Chess, C. Cho, C. Chu, H. W. Chung, D. Cummings, J. Currier, Y. Dai, C. Decareaux, T. Degry, N. Deutsch, D. Deville, A. Dhar, D. Dohan, S. Dowling, S. Dunning, A. Ecoffet, A. Eleti, T. Eloundou, D. Farhi, L. Fedus, N. Felix, S. P. Fishman, J. Forte, I. Fulford, L. Gao, E. Georges, C. Gibson, V. Goel, T. Gogineni, G. Goh, R. Gontijo-Lopes, J. Gordon, M. Grafstein, S. Gray, R. Greene, J. Gross, S. S. Gu, Y. Guo, C. Hallacy, J. Han, J. Harris, Y. He, M. Heaton, J. Heidecke, C. Hesse, A. Hickey, W. Hickey, P. Hoeschele, B. Houghton, K. Hsu, S. Hu, X. Hu, J. Huizinga, S. Jain, S. Jain, J. Jang, A. Jiang, R. Jiang, H. Jin, D. Jin, S. Jomoto, B. Jonn, H. Jun, T. Kaftan, Ł. Kaiser, A. Kamali, I. Kanitscheider, N. S. Keskar, T. Khan, L. Kilpatrick, J. W. Kim, C. Kim, Y. Kim, J. H. Kirchner, J. Kiros, M. Knight, D. Kokotajlo, Ł. Kondraciuk, A. Kondrich, A. Konstantinidis, K. Kosic, G. Krueger, V. Kuo, M. Lampe, I. Lan, T. Lee, J. Leike, J. Leung, D. Levy, C. M. Li, R. Lim, M. Lin, S. Lin, M. Litwin, T. Lopez, R. Lowe, P. Lue, A. Makanju, K. Malfacini, S. Manning, T. Markov, Y. Markovski, B. Martin, K. Mayer, A. Mayne, B. McGrew, S. M. McKinney, C. McLeavey, P. McMillan, J. McNeil, D. Medina, A. Mehta, J. Menick, L. Metz, A. Mishchenko, P. Mishkin, V. Monaco, E. Morikawa, D. Mossing, T. Mu, M. Murati, O. Murk, D. Mély, A. Nair, R. Nakano, R. Nayak, A. Neelakantan, R. Ngo, H. Noh, L. Ouyang, C. O'Keefe, J. Pachocki, A. Paino, J. Palermo, A. Pantuliano, G. Parascandolo, J. Parish, E. Parparita, A. Passos, M. Pavlov, A. Peng, A. Perelman, F. de Avila Belbute Peres, M. Petrov, H. P. de Oliveira Pinto, M. Pokorny, M. Pokrass, V. H. Pong, T. Powell, A. Power, B. Power, E. Proehl, R. Puri, A. Radford, J. Rae, A. Ramesh, C. Raymond, F. Real, K. Rimbach, C. Ross, B. Rotsted, H. Roussez, N. Ryder, M. Saltarelli, T. Sanders, S. Santurkar, G. Sastry, H. Schmidt, D. Schnurr, J. Schulman, D. Selsam, K. Sheppard, T. Sherbakov, J. Shieh, S. Shoker, P. Shyam, S. Sidor, E. Sigler, M. Simens, J. Sitkin, K. Slama, I. Sohl, B. Sokolowsky, Y. Song, N. Staudacher, F. P. Such, N. Summers, I. Sutskever, J. Tang, N. Tezak, M. B. Thompson, P. Tillet, A. Tootoonchian, E. Tseng, P. Tuggle, N. Turley, J. Tworek, J. F. C. Uribe, A. Vallone, A. Vijayvergiya, C. Voss, C. Wainwright, J. J. Wang, A. Wang, B. Wang, J. Ward, J. Wei, C. Weinmann, A. Welihinda, P. Welinder, J. Weng, L. Weng, M. Wiethoff, D. Willner, C. Winter, S. Wolrich, H. Wong, L. Workman, S. Wu, J. Wu, M. Wu, K. Xiao, T. Xu, S. Yoo, K. Yu, Q. Yuan, W. Zaremba, R. Zellers, C. Zhang, M. Zhang, S. Zhao, T. Zheng, J. Zhuang, W. Zhuk and B. Zoph, arXiv, 2024, preprint, arXiv:2303.08774, DOI:10.48550/arXiv.2303.08774.
- M. Reid, N. Savinov, D. Teplyashin, L. Dmitry, T. Lillicrap, J. baptiste Alayrac, R. Soricut, A. Lazaridou, O. Firat, J. Schrittwieser, I. Antonoglou, R. Anil, S. Borgeaud, A. Dai, K. Millican, E. Dyer, M. Glaese, T. Sottiaux, B. Lee, F. Viola, M. Reynolds, Y. Xu, J. Molloy, J. Chen, M. Isard, P. Barham, T. Hennigan, R. McIlroy, M. Johnson, J. Schalkwyk, E. Collins, E. Rutherford, E. Moreira, K. Ayoub, M. Goel, C. Meyer, G. Thornton, Z. Yang, H. Michalewski, Z. Abbas, N. Schucher, A. Anand, R. Ives, J. Keeling, K. Lenc, S. Haykal, S. Shakeri, P. Shyam, A. Chowdhery, R. Ring, S. Spencer, E. Sezener, L. Vilnis, O. Chang, N. Morioka, G. Tucker, C. Zheng, O. Woodman, N. Attaluri, T. Kocisky, E. Eltyshev, X. Chen, T. Chung, V. Selo, S. Brahma, P. Georgiev, A. Slone, Z. Zhu, J. Lottes, S. Qiao, B. Caine, S. Riedel, A. Tomala, M. Chadwick, J. Love, P. Choy, S. Mittal, N. Houlsby, Y. Tang, M. Lamm, L. Bai, Q. Zhang, L. He, Y. Cheng, P. Humphreys, Y. Li, S. Brin, A. Cassirer, Y. Miao, L. Zilka, T. Tobin, K. Xu, L. Proleev, D. Sohn, A. Magni, L. A. Hendricks, I. Gao, S. Ontanon, O. Bunyan, N. Byrd, A. Sharma, B. Zhang, M. Pinto, R. Sinha, H. Mehta, D. Jia, S. Caelles, A. Webson, A. Morris, B. Roelofs, Y. Ding, R. Strudel, X. Xiong, M. Ritter, M. Dehghani, R. Chaabouni, A. Karmarkar, G. Lai, F. Mentzer, B. Xu, Y. Li, Y. Zhang, T. L. Paine, A. Goldin, B. Neyshabur, K. Baumli, A. Levskaya, M. Laskin, W. Jia, J. W. Rae, K. Xiao, A. He, S. Giordano, L. Yagati, J.-B. Lespiau, P. Natsev, S. Ganapathy, F. Liu, D. Martins, N. Chen, Y. Xu, M. Barnes, R. May, A. Vezer, J. Oh, K. Franko, S. Bridgers, R. Zhao, B. Wu, B. Mustafa, S. Sechrist, E. Parisotto, T. S. Pillai, C. Larkin, C. Gu, C. Sorokin, M. Krikun, A. Guseynov, J. Landon, R. Datta, A. Pritzel, P. Thacker, F. Yang, K. Hui, A. Hauth, C.-K. Yeh, D. Barker, J. Mao-Jones, S. Austin, H. Sheahan, P. Schuh, J. Svensson, R. Jain, V. Ramasesh, A. Briukhov, D.-W. Chung, T. von Glehn, C. Butterfield, P. Jhakra, M. Wiethoff, J. Frye, J. Grimstad, B. Changpinyo, C. L. Lan, A. Bortsova, Y. Wu, P. Voigtlaender, T. Sainath, S. Gu, C. Smith, W. Hawkins, K. Cao, J. Besley, S. Srinivasan, M. Omernick, C. Gaffney, G. Surita, R. Burnell, B. Damoc, J. Ahn, A. Brock, M. Pajarskas, A. Petrushkina, S. Noury, L. Blanco, K. Swersky, A. Ahuja, T. Avrahami, V. Misra, R. de Liedekerke, M. Iinuma, A. Polozov, S. York, G. van den Driessche, P. Michel, J. Chiu, R. Blevins, Z. Gleicher, A. Recasens, A. Rrustemi, E. Gribovskaya, A. Roy, W. Gworek, S. M. R. Arnold, L. Lee, J. Lee-Thorp, M. Maggioni, E. Piqueras, K. Badola, S. Vikram, L. Gonzalez, A. Baddepudi, E. Senter, J. Devlin, J. Qin, M. Azzam, M. Trebacz, M. Polacek, K. Krishnakumar, S. yiin Chang, M. Tung, I. Penchev, R. Joshi, K. Olszewska, C. Muir, M. Wirth, A. J. Hartman, J. Newlan, S. Kashem, V. Bolina, E. Dabir, J. van Amersfoort, Z. Ahmed, J. Cobon-Kerr, A. Kamath, A. M. Hrafnkelsson, L. Hou, I. Mackinnon, A. Frechette, E. Noland, X. Si, E. Taropa, D. Li, P. Crone, A. Gulati, S. Cevey, J. Adler, A. Ma, D. Silver, S. Tokumine, R. Powell, S. Lee, K. Vodrahalli, S. Hassan, D. Mincu, A. Yang, N. Levine, J. Brennan, M. Wang, S. Hodkinson, J. Zhao, J. Lipschultz, A. Pope, M. B. Chang, C. Li, L. E. Shafey, M. Paganini, S. Douglas, B. Bohnet, F. Pardo, S. Odoom, M. Rosca, C. N. dos Santos, K. Soparkar, A. Guez, T. Hudson, S. Hansen, C. Asawaroengchai, R. Addanki, T. Yu, W. Stokowiec, M. Khan, J. Gilmer, J. Lee, C. G. Bostock, K. Rong, J. Caton, P. Pejman, F. Pavetic, G. Brown, V. Sharma, M. Lučić, R. Samuel, J. Djolonga, A. Mandhane, L. L. Sjösund, E. Buchatskaya, E. White, N. Clay, J. Jiang, H. Lim, R. Hemsley, Z. Cankara, J. Labanowski, N. D. Cao, D. Steiner, S. H. Hashemi, J. Austin, A. Gergely, T. Blyth, J. Stanton, K. Shivakumar, A. Siddhant, A. Andreassen, C. Araya, N. Sethi, R. Shivanna, S. Hand, A. Bapna, A. Khodaei, A. Miech, G. Tanzer, A. Swing, S. Thakoor, L. Aroyo, Z. Pan, Z. Nado, J. Sygnowski, S. Winkler, D. Yu, M. Saleh, L. Maggiore, Y. Bansal, X. Garcia, M. Kazemi, P. Patil, I. Dasgupta, I. Barr, M. Giang, T. Kagohara, I. Danihelka, A. Marathe, V. Feinberg, M. Elhawaty, N. Ghelani, D. Horgan, H. L. Walker, R. Tanburn, M. Tariq, D. Shrivastava, F. Xia, Q. Wang, C.-C. Chiu, Z. Ashwood, K. Baatarsukh, S. Samangooei, R. L. Kaufman, F. Alcober, A. Stjerngren, P. Komarek, K. Tsihlas, A. Boral, R. Comanescu, J. Chen, R. Liu, C. Welty, D. Bloxwich, C. Chen, Y. Sun, F. Feng, M. Mauger, X. Dotiwalla, V. Hellendoorn, M. Sharman, I. Zheng, K. Haridasan, G. Barth-Maron, C. Swanson, D. Rogozińska, A. Andreev, P. K. Rubenstein, R. Sang, D. Hurt, G. Elsayed, R. Wang, D. Lacey, A. Ilić, Y. Zhao, A. Iwanicki, A. Lince, A. Chen, C. Lyu, C. Lebsack, J. Griffith, M. Gaba, P. Sandhu, P. Chen, A. Koop, R. Rajwar, S. H. Yeganeh, S. Chang, R. Zhu, S. Radpour, E. Davoodi, V. I. Lei, Y. Xu, D. Toyama, C. Segal, M. Wicke, H. Lin, A. Bulanova, A. P. Badia, N. Rakićević, P. Sprechmann, A. Filos, S. Hou, V. Campos, N. Kassner, D. Sachan, M. Fortunato, C. Iwuanyanwu, V. Nikolaev, B. Lakshminarayanan, S. Jazayeri, M. Varadarajan, C. Tekur, D. Fritz, M. Khalman, D. Reitter, K. Dasgupta, S. Sarcar, T. Ornduff, J. Snaider, F. Huot, J. Jia, R. Kemp, N. Trdin, A. Vijayakumar, L. Kim, C. Angermueller, L. Lao, T. Liu, H. Zhang, D. Engel, S. Greene, A. White, J. Austin, L. Taylor, S. Ashraf, D. Liu, M. Georgaki, I. Cai, Y. Kulizhskaya, S. Goenka, B. Saeta, Y. Xu, C. Frank, D. de Cesare, B. Robenek, H. Richardson, M. Alnahlawi, C. Yew, P. Ponnapalli, M. Tagliasacchi, A. Korchemniy, Y. Kim, D. Li, B. Rosgen, K. Levin, J. Wiesner, P. Banzal, P. Srinivasan, H. Yu, Ç. Ünlü, D. Reid, Z. Tung, D. Finchelstein, R. Kumar, A. Elisseeff, J. Huang, M. Zhang, R. Aguilar, M. Giménez, J. Xia, O. Dousse, W. Gierke, D. Yates, K. Jalan, L. Li, E. Latorre-Chimoto, D. D. Nguyen, K. Durden, P. Kallakuri, Y. Liu, M. Johnson, T. Tsai, A. Talbert, J. Liu, A. Neitz, C. Elkind, M. Selvi, M. Jasarevic, L. B. Soares, A. Cui, P. Wang, A. W. Wang, X. Ye, K. Kallarackal, L. Loher, H. Lam, J. Broder, D. Holtmann-Rice, N. Martin, B. Ramadhana, M. Shukla, S. Basu, A. Mohan, N. Fernando, N. Fiedel, K. Paterson, H. Li, A. Garg, J. Park, D. Choi, D. Wu, S. Singh, Z. Zhang, A. Globerson, L. Yu, J. Carpenter, F. de Chaumont Quitry, C. Radebaugh, C.-C. Lin, A. Tudor, P. Shroff, D. Garmon, D. Du, N. Vats, H. Lu, S. Iqbal, A. Yakubovich, N. Tripuraneni, J. Manyika, H. Qureshi, N. Hua, C. Ngani, M. A. Raad, H. Forbes, J. Stanway, M. Sundararajan, V. Ungureanu, C. Bishop, Y. Li, B. Venkatraman, B. Li, C. Thornton, S. Scellato, N. Gupta, Y. Wang, I. Tenney, X. Wu, A. Shenoy, G. Carvajal, D. G. Wright, B. Bariach, Z. Xiao, P. Hawkins, S. Dalmia, C. Farabet, P. Valenzuela, Q. Yuan, A. Agarwal, M. Chen, W. Kim, B. Hulse, N. Dukkipati, A. Paszke, A. Bolt, K. Choo, J. Beattie, J. Prendki, H. Vashisht, R. Santamaria-Fernandez, L. C. Cobo, J. Wilkiewicz, D. Madras, A. Elqursh, G. Uy, K. Ramirez, M. Harvey, T. Liechty, H. Zen, J. Seibert, C. H. Hu, A. Khorlin, M. Le, A. Aharoni, M. Li, L. Wang, S. Kumar, N. Casagrande, J. Hoover, D. E. Badawy, D. Soergel, D. Vnukov, M. Miecnikowski, J. Simsa, P. Kumar, T. Sellam, D. Vlasic, S. Daruki, N. Shabat, J. Zhang, G. Su, J. Zhang, J. Liu, Y. Sun, E. Palmer, A. Ghaffarkhah, X. Xiong, V. Cotruta, M. Fink, L. Dixon, A. Sreevatsa, A. Goedeckemeyer, A. Dimitriev, M. Jafari, R. Crocker, N. FitzGerald, A. Kumar, S. Ghemawat, I. Philips, F. Liu, Y. Liang, R. Sterneck, A. Repina, M. Wu, L. Knight, M. Georgiev, H. Lee, H. Askham, A. Chakladar, A. Louis, C. Crous, H. Cate, D. Petrova, M. Quinn, D. Owusu-Afriyie, A. Singhal, N. Wei, S. Kim, D. Vincent, M. Nasr, C. A. Choquette-Choo, R. Tojo, S. Lu, D. de Las Casas, Y. Cheng, T. Bolukbasi, K. Lee, S. Fatehi, R. Ananthanarayanan, M. Patel, C. Kaed, J. Li, S. R. Belle, Z. Chen, J. Konzelmann, S. Põder, R. Garg, V. Koverkathu, A. Brown, C. Dyer, R. Liu, A. Nova, J. Xu, A. Walton, A. Parrish, M. Epstein, S. McCarthy, S. Petrov, D. Hassabis, K. Kavukcuoglu, J. Dean and O. Vinyals, arXiv, 2024, preprint, arXiv:2403.05530, DOI:10.48550/arXiv.2403.05530.
- M. Chen, J. Tworek, H. Jun, Q. Yuan, H. P. de Oliveira Pinto, J. Kaplan, H. Edwards, Y. Burda, N. Joseph, G. Brockman, A. Ray, R. Puri, G. Krueger, M. Petrov, H. Khlaaf, G. Sastry, P. Mishkin, B. Chan, S. Gray, N. Ryder, M. Pavlov, A. Power, L. Kaiser, M. Bavarian, C. Winter, P. Tillet, F. P. Such, D. Cummings, M. Plappert, F. Chantzis, E. Barnes, A. Herbert-Voss, W. H. Guss, A. Nichol, A. Paino, N. Tezak, J. Tang, I. Babuschkin, S. Balaji, S. Jain, W. Saunders, C. Hesse, A. N. Carr, J. Leike, J. Achiam, V. Misra, E. Morikawa, A. Radford, M. Knight, M. Brundage, M. Murati, K. Mayer, P. Welinder, B. McGrew, D. Amodei, S. McCandlish, I. Sutskever and W. Zaremba, arXiv, 2021, preprint, arXiv:2107.03374, DOI:10.48550/arXiv.2107.03374.
- S. Liu, W. Nie, C. Wang, J. Lu, Z. Qiao, L. Liu, J. Tang, C. Xiao and A. Anandkumar, Nat. Mach. Intell., 2023, 5, 1447–1457 CrossRef.
- S. Wang, Y. Guo, Y. Wang, H. Sun and J. Huang, Proceedings of the 10th ACM international conference on bioinformatics, computational biology and health informatics, 2019, pp. 429–436 Search PubMed.
- S. Chithrananda, G. Grand and B. Ramsundar, arXiv, 2020, preprint, arXiv:2010.09885, DOI:10.48550/arXiv.2010.09885.
- W. Ahmad, E. Simon, S. Chithrananda, G. Grand and B. Ramsundar, arXiv, 2022, preprint, arXiv:2209.01712, DOI:10.48550/arXiv.2209.01712.
- A. D. White, G. M. Hocky, H. A. Gandhi, M. Ansari, S. Cox, G. P. Wellawatte, S. Sasmal, Z. Yang, K. Liu, Y. Singh and W. J. Peña Ccoa, Digital Discovery, 2023, 2, 368–376 RSC.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Nature, 2023, 624, 570–578 CrossRef CAS PubMed.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Nat. Mach. Intell., 2024, 1–11 Search PubMed.
- G. M. Hocky and A. D. White, Digital Discovery, 2022, 1, 79–83 RSC.
- Z. Hong, Energy Mater. Adv., 2023, 4, 0026 CrossRef.
- J. C. Verduzco, E. Holbrook and A. Strachan, GPT-4 as an interface between researchers and computational software: improving usability and reproducibility, arXiv, 2023, preprint, arXiv:2310.11458, DOI:10.48550/arXiv.2310.11458.
- Y. Gao, Y. Xiong, X. Gao, K. Jia, J. Pan, Y. Bi, Y. Dai, J. Sun, M. Wang and H. Wang, arXiv, 2024, preprint, arXiv:2312.10997, DOI:10.48550/arXiv.2312.10997.
- J. Kaddour, J. Harris, M. Mozes, H. Bradley, R. Raileanu and R. McHardy, arXiv, 2023, preprint, arXiv:2307.10169, DOI:10.48550/arXiv.2307.10169.
- Y. Lu, M. Bartolo, A. Moore, S. Riedel and P. Stenetorp, Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics, Volume. 1: (Long Papers), Dublin, Ireland, 2022, pp. 8086–8098 Search PubMed.
- B. Chen, Z. Zhang, N. Langrené and S. Zhu, arXiv, 2023, preprint, arXiv:2310.14735, DOI:10.48550/arXiv.2310.14735.
- D. OBrien, S. Biswas, S. M. Imtiaz, R. Abdalkareem, E. Shihab and H. Rajan, Proceedings of the IEEE/ACM 46th International Conference on Software Engineering, 2024, pp. 1–13 Search PubMed.
- N. Dainese, M. Merler, M. Alakuijala and P. Marttinen, arXiv, 2024, preprint, arXiv:2405.15383, DOI:10.48550/arXiv.2405.15383.
- T. Ridnik, D. Kredo and I. Friedman, arXiv, 2024, preprint, arXiv:2401.08500, DOI:10.48550/arXiv.2401.08500.
- J. Li, G. Li, Y. Li and Z. Jin, arXiv, 2023, preprint, arXiv:2305.06599, DOI:10.48550/arXiv.2305.06599.
- N. Koul, Prompt Engineering for Large Language Models, https://books.google.at/books?id=e9_jEAAAQBAJ Search PubMed.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever and D. Amodei, Adv. Neural Inf. Process. Syst., 2020, 1877–1901 Search PubMed.
- C. Wang, Y. Yang, C. Gao, Y. Peng, H. Zhang and M. R. Lyu, Proceedings of the 30th ACM joint European software engineering conference and symposium on the foundations of software engineering, 2022, pp. 382–394 Search PubMed.
- J. Wei, X. Wang, D. Schuurmans, M. Bosma, b. ichter, F. Xia, E. Chi, Q. V. Le and D. Zhou, Adv. Neural Inf. Process. Syst., 2022, 24824–24837 Search PubMed.
- S. Dhuliawala, M. Komeili, J. Xu, R. Raileanu, X. Li, A. Celikyilmaz and J. Weston, Findings of the Association for Computational Linguistics ACL 2024, Bangkok, Thailand and virtual meeting, 2024, pp. 3563–3578 Search PubMed.
- M. Besta, N. Blach, A. Kubicek, R. Gerstenberger, M. Podstawski, L. Gianinazzi, J. Gajda, T. Lehmann, H. Niewiadomski, P. Nyczyk and T. Hoefler, Proc. AAAI Conf. Artif. Intell., 2024, 38, 17682–17690 Search PubMed.
- S. Yao, D. Yu, J. Zhao, I. Shafran, T. Griffiths, Y. Cao and K. Narasimhan, Adv. Neural Inf. Process. Syst., 2023, 11809–11822 Search PubMed.
- K. Weiss, T. M. Khoshgoftaar and D. Wang, J. Big Data, 2016, 3, 1–40 CrossRef.
- A. Nigam, R. Pollice, M. Krenn, G. d. P. Gomes and A. Aspuru-Guzik, Chem. Sci., 2021, 12, 7079–7090 RSC.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- RDKit: Open-source Cheminformatics, https://rdkit.org.
- P. Lewis, E. Perez, A. Piktus, F. Petroni, V. Karpukhin, N. Goyal, H. Küttler, M. Lewis, W.-T. Yih and T. Rocktäschel, et al. , Adv. Neural Inf. Process. Syst., 2020, 33, 9459–9474 Search PubMed.
- R. Bjornsson, ORCA input Library, 2024, https://sites.google.com/site/orcainputlibrary/home Search PubMed.
- S. Zhou, U. Alon, F. F. Xu, Z. Wang, Z. Jiang and G. Neubig, arXiv, 2023, preprint, arXiv:2207.05987, DOI:10.48550/arXiv.2207.05987.
- OpenAI, Embeddings – OpenAI API, https://platform.openai.com/docs/guides/embeddings Search PubMed.
- M. Douze, A. Guzhva, C. Deng, J. Johnson, G. Szilvasy, P.-E. Mazaré, M. Lomeli, L. Hosseini and H. Jégou, arXiv, 2024, preprint, arXiv:2401.08281, DOI:10.48550/arXiv.2401.08281.
- M. Álvarez-Moreno, C. de Graaf, N. Lopez, F. Maseras, J. M. Poblet and C. Bo, J. Chem. Inf. Model., 2015, 55, 95–103 CrossRef.
- J.-Y. Yao, K.-P. Ning, Z.-H. Liu, M.-N. Ning, Y.-Y. Liu and L. Yuan, arXiv, 2024, preprint, arXiv:2310.01469, DOI:10.48550/arXiv.2310.01469.
- J. Chen, L. Chen, H. Huang and T. Zhou, arXiv, 2023, preprint, arXiv:2304.03262, DOI:10.48550/arXiv.2304.03262.
- Z. Shao, Y. Gong, Y. Shen, M. Huang, N. Duan and W. Chen, International Conference on Machine Learning, 2023, pp. 30706–30775 Search PubMed.
- J. Devlin, M. Chang, K. Lee and K. Toutanova, Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2019, Minneapolis, MN, USA, June 2–7, 2019, Volume 1 (Long and Short Papers), 2019, pp. 4171–4186 Search PubMed.
- H. Mehrafarin, S. Rajaee and M. T. Pilehvar, Findings of the Association for Computational Linguistics: ACL 2022, Dublin, Ireland, May 22-27, 2022, 2022, pp. 228–238 Search PubMed.
- G. te Velde, F. M. Bickelhaupt, E. J. Baerends, C. Fonseca Guerra, S. J. A. van Gisbergen, J. G. Snijders and T. Ziegler, J. Comput. Chem., 2001, 22, 931–967 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussiañ16 Revision C.01, Gaussian Inc., Wallingford CT, 2016 Search PubMed.
- E. Epifanovsky, A. T. B. Gilbert, X. Feng, J. Lee, Y. Mao, N. Mardirossian, P. Pokhilko, A. F. White, M. P. Coons, A. L. Dempwolff, Z. Gan, D. Hait, P. R. Horn, L. D. Jacobson, I. Kaliman, J. Kussmann, A. W. Lange, K. U. Lao, D. S. Levine, J. Liu, S. C. McKenzie, A. F. Morrison, K. D. Nanda, F. Plasser, D. R. Rehn, M. L. Vidal, Z.-Q. You, Y. Zhu, B. Alam, B. J. Albrecht, A. Aldossary, E. Alguire, J. H. Andersen, V. Athavale, D. Barton, K. Begam, A. Behn, N. Bellonzi, Y. A. Bernard, E. J. Berquist, H. G. A. Burton, A. Carreras, K. Carter-Fenk, R. Chakraborty, A. D. Chien, K. D. Closser, V. Cofer-Shabica, S. Dasgupta, M. de Wergifosse, J. Deng, M. Diedenhofen, H. Do, S. Ehlert, P.-T. Fang, S. Fatehi, Q. Feng, T. Friedhoff, J. Gayvert, Q. Ge, G. Gidofalvi, M. Goldey, J. Gomes, C. E. González-Espinoza, S. Gulania, A. O. Gunina, M. W. D. Hanson-Heine, P. H. P. Harbach, A. Hauser, M. F. Herbst, M. Hernández Vera, M. Hodecker, Z. C. Holden, S. Houck, X. Huang, K. Hui, B. C. Huynh, M. Ivanov, A. Jász, H. Ji, H. Jiang, B. Kaduk, S. Kähler, K. Khistyaev, J. Kim, G. Kis, P. Klunzinger, Z. Koczor-Benda, J. H. Koh, D. Kosenkov, L. Koulias, T. Kowalczyk, C. M. Krauter, K. Kue, A. Kunitsa, T. Kus, I. Ladjánszki, A. Landau, K. V. Lawler, D. Lefrancois, S. Lehtola, R. R. Li, Y.-P. Li, J. Liang, M. Liebenthal, H.-H. Lin, Y.-S. Lin, F. Liu, K.-Y. Liu, M. Loipersberger, A. Luenser, A. Manjanath, P. Manohar, E. Mansoor, S. F. Manzer, S.-P. Mao, A. V. Marenich, T. Markovich, S. Mason, S. A. Maurer, P. F. McLaughlin, M. F. S. J. Menger, J.-M. Mewes, S. A. Mewes, P. Morgante, J. W. Mullinax, K. J. Oosterbaan, G. Paran, A. C. Paul, S. K. Paul, F. Pavošević, Z. Pei, S. Prager, E. I. Proynov, A. Rák, E. Ramos-Cordoba, B. Rana, A. E. Rask, A. Rettig, R. M. Richard, F. Rob, E. Rossomme, T. Scheele, M. Scheurer, M. Schneider, N. Sergueev, S. M. Sharada, W. Skomorowski, D. W. Small, C. J. Stein, Y.-C. Su, E. J. Sundstrom, Z. Tao, J. Thirman, G. J. Tornai, T. Tsuchimochi, N. M. Tubman, S. P. Veccham, O. Vydrov, J. Wenzel, J. Witte, A. Yamada, K. Yao, S. Yeganeh, S. R. Yost, A. Zech, I. Y. Zhang, X. Zhang, Y. Zhang, D. Zuev, A. Aspuru-Guzik, A. T. Bell, N. A. Besley, K. B. Bravaya, B. R. Brooks, D. Casanova, J.-D. Chai, S. Coriani, C. J. Cramer, G. Cserey, I. DePrince, A. Eugene, J. DiStasio, A. Robert, A. Dreuw, B. D. Dunietz, T. R. Furlani, I. Goddard, A. William, S. Hammes-Schiffer, T. Head-Gordon, W. J. Hehre, C.-P. Hsu, T.-C. Jagau, Y. Jung, A. Klamt, J. Kong, D. S. Lambrecht, W. Liang, N. J. Mayhall, C. W. McCurdy, J. B. Neaton, C. Ochsenfeld, J. A. Parkhill, R. Peverati, V. A. Rassolov, Y. Shao, L. V. Slipchenko, T. Stauch, R. P. Steele, J. E. Subotnik, A. J. W. Thom, A. Tkatchenko, D. G. Truhlar, T. Van Voorhis, T. A. Wesolowski, K. B. Whaley, I. Woodcock, H. Lee, P. M. Zimmerman, S. Faraji, P. M. W. Gill, M. Head-Gordon, J. M. Herbert and A. I. Krylov, J. Chem. Phys., 2021, 155, 084801 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: An explanation of our choice of ORCA, an overview of related work, a formal description of model inference and fine-tuning, details on how we combine finetuning with prompt engineering, our rationale for selecting the LLM and its architecture, the process of acquiring and processing test data, key dataset statistics, an explanation of our hyperparameters, an analysis of ORCA errors with synthesized inputs, qualitative performance of models on real-world prompts, a discussion on ORCA error analysis and performance on real-world prompts, study limitations, future research suggestions, and the exact prompts used in the Appendix. See DOI: https://doi.org/10.1039/d4dd00366g |
| This journal is © The Royal Society of Chemistry 2025 |