Open Access Article
Open Access ArticleDigital discovery and the new experimental frontier
S. Hessam
M. Mehr

School of Chemistry, University of Glasgow, Advanced Research Centre, 11 Chapel Lane, Glasgow, G11 6EW, UK. E-mail: Hessam.Mehr@glasgow.ac.uk
First published on 11th March 2025
Abstract
The digitisation of chemistry has had a profound effect on the field by boosting the efficiency of information retrieval and data recording, and by automating repetitive laboratory operations. Increasingly complex molecules — both known and de novo — can be rapidly accessed with unprecedented speed and reproducibility. Despite progress as measured by these quantitative productivity metrics, a qualitative transformation in the design and structure of experimentation has yet to materialise. Here, we explore digitisation's role in a larger paradigm shift in experimental chemistry not just as a means of automated execution of procedures but dynamically sensing, interpreting, and manipulating chemical processes in real-time. This paradigm shift is characterised by transitioning from single-point measurements to continuous observation; from homogeneous to spatially organised systems; and from fixed linear experimental procedures to dynamic, branched “programs” that can unfold based on real-time feedback. This shift will enable new types of objectives in experimental chemistry, such as responsiveness, adaptability and persistence, expanding beyond static quantities like product structure, yield and purity. We explore the innovations needed to enable these transitions; the open questions they raise; and how digitisation can catalyse chemistry's evolution beyond its existing confines.
Introduction
Recent years have seen rapid progress in the development of general-purpose chemical automation1–4 and high bandwidth analytical methods,5 accompanied by accelerated computing and a revolution in machine learning methods.6 These advances have equipped chemistry with powerful new tools to tackle previously impenetrable problems. Self-driving labs using a range of robotic technology have automated repetitive, error-prone and hazardous manual procedures.7 Investigations can be run on a massive scale without the physical and cognitive burden of setting up and monitoring myriad parallel reactions. Computational prediction of structure and properties can be scaled up to molecules of immense size and complexity. Literature descriptions of properties and reactivity can be parsed and formalised into databases8,9 and mined as a starting point for further discoveries.10 The question arises whether the full impact of digitisation lies purely in the downstream efficiency gains of these advances or if extra leaps remain to fully unlock its potential for conceptually reshaping experimental chemistry.Early deployments of new technology frequently focus on applications as a drop-in replacement for existing techniques. Unanticipated impacts, often dwarfing the original use-case, are only gradually realised through iterative cycles of creative applications and improvements. It is not surprising that keyboards and character displays dominated the early development of personal computers, given their initial introduction to businesses as a replacement for typewriters and fax machines.11,12 Ultimately, the transition from static documents to dynamic, manipulatable data emerged as the main effect of this technology, transforming not just the act of writing documents but defining new relationships with information.
Likewise, sophisticated lab automation systems today rely on robotic adaptations of current manual setups, even using robotic chemists approximating human experimenters in order to maximise interoperability with existing labware and ease adaptation of literature procedures.4,13 Current applications of AI in chemistry similarly follow this pattern, largely focusing on interpolation of results or automation of decision-making within traditional experimental frameworks.14,15 One wonders how further abstraction of the human experimenter will enable new modes of experimentation that would be impossible to conceptualise or execute through traditional human-centric frameworks. In this opinion, we will reflect on the limitations of prevalent classical experimental frameworks within which digitisation is currently deployed, before exploring axes of innovation — and open questions posed by them — for experimentalists to best capture the benefits conferred by robotics, automation and AI. We examine three distinct facets of this transformation: the transition from discrete to continuous observation of chemical processes; the algorithmic specialisation of general procedures; and the reimagining of experimental design from tabular to graph-based representations. Each of these transitions represents an opportunity where digitisation could fundamentally reshape how we conceptualise and execute chemical experimentation, rather than merely accelerating existing practices.
Escaping single point chemistry
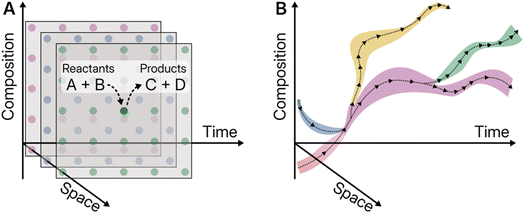
In the classical conceptual framework, chemical transformations are idealised as a single point. Prevalent experimental paradigms seek to reduce both their spatial extents (via stirring to enforce homogeneity), temporal dimension (favouring a picture where reactants are converted to products in a steady state), and variability in composition (purification applied after each step to enforce a single composition), Fig. 1A. The addition of automation and parallel execution does not alter this picture fundamentally, instead replicating it over time (automated sequences) and space (parallel execution). The de facto realisation of the single-point paradigm has been the homogeneous system. Homogeneous systems are describable using a handful of state variables, and any general system can be broken down into a set of roughly homogeneous ones.In contrast, considering the full extents of a chemical system not only in time and space, but also simultaneously composition, gives a richer picture, whereby transformations can be conceived of as trajectories connecting regions of this space. In this framework, we are not limited to the study of phenomena with single fixed beginning and end points; convergent and bifurcating paths-are both possible, Fig. 1B. Within these rich, transient systems, short-lived molecules can form, their respective chains of causation can be tracked and patterns of behaviour discerned and linked to their causal chains without being silenced by a dominant overall phenomenon. Much can be learned from systems biology's progress towards continuous monitoring of metabolites within biochemical networks, made interpretable via computational modelling,16,17 exemplified by techniques like 13C metabolic flux analysis.18
This transition from considering transformations as single points to regions inherently requires tackling more information — from single state variables to distributions. Data collection throughout time and space is key to interrogating these systems, with emergent commercial and academic systems tailored to this, leveraging both real-time imaging and spatially resolved spectroscopy.19,20 There is increasing access to physical tools for instantiating inhomogeneous systems in a reproducible way — from pipetting robots to microfluidics and droplet-on-demand dispensing systems — along with analytical methods that can interrogate them in real-time with excellent spatial resolution (e.g. chemical imaging). Open challenges include interpreting data obtained from these systems, and adapting mainstream protocols to these media.
Algorithmic specialisation of general procedures
The corpus of chemistry is built on the assumption that reusable experimental building blocks can be deployed in a multipurpose fashion. This concept is multi-faceted: from the composable design of laboratory glassware facilitated by standard joints; to reusable reactions, such as the Suzuki coupling; and reusable auxiliary protocols, such as phase separation and column chromatography. The utility of these blocks is contingent on a robust range of validity, i.e. reproducibility across a range of compositions and conditions, at odds with hyper-specific “smarter” systems with higher selectivity such as molecular recognition and enhanced regioselectivity. A symptom of this disparity is the proliferation of protecting groups, evidence that there is a large gap between the specificity of readily available methods and what is needed to achieve precise transformations.There is a chasm between the universe of general procedures and specialised protocols, with no mechanisms for smooth transition between the two modalities, i.e. no systematic way to specialise a general-purpose procedure on-demand. An example could be evolving a general metal-catalysed reaction to one conserving a specific stereocentre. Current procedures lack built-in chemical ‘programmability’ for on-demand adaptation, making specialisation a resource-intensive process of trial and error that often outweighs the benefits for bespoke transformations. Efforts to bridge this chasm are fertile ground for expansion of digital chemistry; specifically providing an algorithmic specialisation of general procedure to specific instances. A rudimentary solution to this challenge is the development of various implementations of closed-loop optimisers: with the advantage of being rooted in real, reproducible experimentation, they are nevertheless time-consuming and potentially wasteful. A second iteration of this is AI-powered prediction of optimal conditions and reagents to achieve a targeted transformation: a promising approach but hamstrung by its data-intensive nature and reliance on wealth of comparable examples.21 Subsequent iterations will require ever-closer collaboration between chemists, process specialists and computer scientists.
Transitioning from tabular to graph-based experimental representation
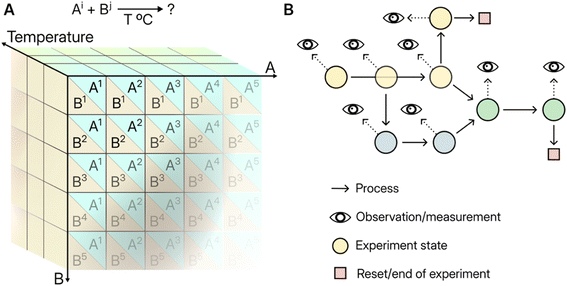
Classical machine learning models rely on tabular data for training. Tabular structures can bias exploration in the laboratory by evoking experiments with fixed “slots” for inputs and outcomes, Fig. 2A. Automation also thrives on tabulated programmes — whether executed sequentially or in large parallel batches — as this simplifies both programming and anticipating anomalies by the roboticist. The full repercussions of an infinitely variable operation sequences do not need to be accounted for a priori. One can hypothesise that the convenience of tabular structures comes with a rigid structure that precludes dynamic exploration or on-the fly optimisation because the invariable next step is pre-ordained. The tabular structure's “stateful” counterpart — to borrow a computer science term referring to the embedding of each action within a sequence of precedents — is the graph, in which operations can be specified with reference to previous starting state and can be the basis for sub-experiments or “children”, Fig. 2B.An upcoming challenge for the deployment of machine intelligence in chemistry will be to embrace and reason about experimentation within this graph-based formalism. These graph-based experimental structures are intractable in the absence of computer-aided reasoning. They offer an exciting new frontier for digital chemistry, where experiments can retain memory, be observed at intermediate time-points, and accumulate effects over multiple steps. Parallels may be drawn to persistent experimental paradigms in biology, e.g. the long-term evolution experiment on E. coli,22 where branching experimental structures enable observation of emergent evolutionary phenomena. One possible chemical equivalent could involve populations of surface-bound oligomers competing for surface sites while undergoing constant modification via successive localised addition of monomers and coupling/cleaving agents — any “evicted” molecules are removed by these washes, hence excluded from persisting. Microfluidics, inhomogeneous systems in general, and digital microfluidics all allow multiple instantiations in parallel and may be suitable physical substrates for implementing this paradigm.
A thought experiment to illustrate one possible evolution of the laboratory in response to the above: imagine a Petri dish containing small chemical “nuggets” — solid particles, liquid droplets, gas bubbles — dispensed robotically into a fluid medium. Chemicals are allowed to diffuse and mix naturally, but localised mechanical agitation and heating are also applied as needed. The system is constantly monitored using a range of imaging techniques as well as mass spectrometry via open-port sampling. A computational model of chemical interactions — formulated by the chemist and seeded with a corpus of known reactivity — is continuously queried to spot anomalies in the observed data (potential discoveries), and experiments with uncertain outcomes (to augment the model and learn new chemistry) that feed into the formulation of follow-up interventions to perform on the system. Additional intervention types include dispensing of new reagents and localised mass transfer from one part of the system to the other. One goal can be to keep the experiment running for as long as possible whilst maintaining a stable ratio of knowns and unknowns. Only when the world model is overwhelmed by unknowns, ideally after many exploratory steps, would the reactor be flushed, thereby erasing its memory to start a fresh experiment.
Conclusion
Chemistry's fundamental quest is to understand and control any desired molecular process, at any scale, anywhere in the universe. Far from an intellectual exercise, this universal remit is essential in a world where the field is tasked not just with furnishing raw molecular building blocks, but opening new avenues in precision therapeutics, smart materials, and new substrates for computation. The challenge areas included here aim to highlight the richness of matter's properties and behaviour outside chemistry's predominant area of focus and beyond established boundaries to adjacent fields, both incidentally arenas where deployment of digitisation will have the largest impact.Data availability
There is no data associated with this opinion piece.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Leverhulme Trust (ECF-2021-298) and the University of Glasgow Lord Kelvin–Adam Smith Leadership Fellowship. SHMM would like to thank Dr K. Al-Hourani for academic discussions in the preparation of this manuscript.References
- J. P. McMullen and K. F. Jensen, An automated microfluidic system for online optimization in chemical synthesis, Org. Process Res. Dev., 2010, 14(5), 1169–1176 Search PubMed.
- J. Li, S. G. Ballmer, E. P. Gillis, S. Fujii, M. J. Schmidt and A. M. E. Palazzolo, et al., Synthesis of many different types of organic small molecules using one automated process, Science, 2015, 347(6227), 1221–1226 CrossRef CAS PubMed.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda and G. Keenan, et al., Organic synthesis in a modular robotic system driven by a chemical programming language, Science, 2019, 363(6423), eaav2211 Search PubMed.
- B. Burger, P. M. Maffettone, V. V. Gusev, C. M. Aitchison, Y. Bai and X. Wang, et al., A mobile robotic chemist, Nature, 2020, 583(7815), 237–241 Search PubMed.
- X. Xu, D. Valavanis, P. Ciocci, S. Confederat, F. Marcuccio and J. F. Lemineur, et al., The new era of high-throughput nanoelectrochemistry, Anal. Chem., 2023, 95(1), 319–356 CrossRef CAS PubMed.
- M. Mehr SH, D. Caramelli and L. Cronin, Digitizing chemical discovery with a Bayesian explorer for interpreting reactivity data, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(17), e2220045120 CrossRef PubMed.
- B. P. MacLeod, F. G. L. Parlane, T. D. Morrissey, F. Häse, L. M. Roch and K. E. Dettelbach, et al., Self-driving laboratory for accelerated discovery of thin-film materials, Sci. Adv., 2020, 6(20), eaaz8867 CrossRef CAS PubMed.
- S. Rohrbach, M. Šiaučiulis, G. Chisholm, P. A. Pirvan, M. Saleeb and S. H. M. Mehr, et al., Digitization and validation of a chemical synthesis literature database in the ChemPU, Science, 2022, 377(6602), 172–180 CrossRef CAS PubMed.
- P. Kumar, S. Kabra and J. M. Cole, A database of stress-strain properties auto-generated from the scientific literature using ChemDataExtractor, Sci. Data, 2024, 11(1), 1273 Search PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Materials synthesis insights from scientific literature via text extraction and machine learning, Chem. Mater., 2017, 29(21), 9436–9444 CrossRef CAS.
- P. E. Ceruzzi, A History of Modern Computing, MIT Press, 2n edn, 2003, p. 468 Search PubMed.
- I. S. MacKenzie and K. Tanaka-Ishii, Text Entry Systems: Mobility, Accessibility, Universality, Elsevier, 2010, p. 343 Search PubMed.
- F. Yang and J. E. Hein, Training a robotic arm to estimate the weight of a suspended object, Device, 2023, 1(1), 100011 Search PubMed.
- C. Chen, Y. Zuo, W. Ye, X. Li, Z. Deng and S. P. Ong, A critical review of machine learning of energy materials, Adv. Energy Mater., 2020, 10(8), 1903242 CrossRef CAS.
- Z. W. Zhao, M. del Cueto and A. Troisi, Limitations of machine learning models when predicting compounds with completely new chemistries: possible improvements applied to the discovery of new non-fullerene acceptors, Digital Discovery, 2022, 1(3), 266–276 Search PubMed.
- U. Sauer, Metabolic networks in motion: 13C-based flux analysis, Mol. Syst. Biol., 2006, 2(1), 62 Search PubMed.
- P. Dvořák, B. Burýšková, B. Popelářová, B. E. Ebert, T. Botka and D. Bujdoš, et al., Synthetically-primed adaptation of Pseudomonas putida to a non-native substrate D-xylose, Nat. Commun., 2024, 15(1), 2666 Search PubMed.
- C. P. Long and M. R. Antoniewicz, High-resolution 13C metabolic flux analysis, Nat. Protoc., 2019, 14(10), 2856–2877 Search PubMed.
- K. W. Feindel, Spatially resolved chemical reaction monitoring using magnetic resonance imaging, Magn. Reson. Chem., 2016, 54(6), 429–436 Search PubMed.
- L. J. A. Macedo and F. N. Crespilho, Multiplex infrared spectroscopy imaging for monitoring spatially resolved redox chemistry, Anal. Chem., 2018, 90(3), 1487–1491 CrossRef CAS PubMed.
- A. Kristiadi, F. Strieth-Kalthoff, M. Skreta, P. Poupart, A. Aspuru-Guzik, and G. Pleiss, A sober look at LLMs for material discovery: are they actually good for bayesian optimization over molecules?, arXiv, 2024, preprint, arXiv:2402.05015, DOI:10.48550/arXiv.2402.05015.
- R. E. Lenski, M. R. Rose, S. C. Simpson and S. C. Tadler, Long-term experimental evolution in escherichia coli. i. adaptation and divergence during 2,000 generations, Am. Nat., 1991, 138(6), 1315–1341 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |