Open Access Article
Open Access ArticleAn introduction to machine learning tools for the analysis of microplastics in complex matrices
Brian R.
Coleman
 *
*
Metrology Research Centre, National Research Council Canada, Ottawa, Ontario, Canada. E-mail: brian.coleman@nrc-cnrc.gc.ca; Tel: +1 613-990-0954
First published on 18th November 2024
Abstract
As microplastic (MP) particles continue to spread globally, their pervasive presence is increasingly problematic. Analyzing MPs in matrices as varied as soil, river water, and biosolid fertilizers is critical, as these matrices directly impact the food sources of plants, animals, and humans. Current analytical methods for quantifying and identifying MPs are limited due to labor-intensive extraction processes and the time and effort required for counting and analysis. Recently, Machine Learning (ML) has been introduced to the analysis of MPs in complex matrices, significantly reducing the need for extensive extraction and increasing analysis speeds. This work aims to illuminate various ML techniques for new researchers entering this field. It highlights numerous examples in the application of these models, with a particular focus on spectroscopic techniques such as infrared and Raman spectroscopy; tools which are used to quantify and identify MPs in complex matrices. By demonstrating the effectiveness of these computer-based tools alongside the hands-on techniques currently used in the field, we are confident that these ML methodologies will soon become integral to all aspects of microplastic analysis in the environmental sciences.
Environmental significanceThe work presented herein presents the use of Machine Learning (ML) tools in the analysis of microplastics (MPs) in terrestrial and aquatic matrices. Microplastics have been known to cause harm to both fauna and flora and, as more and more plastics are produced each day everyday, our ability to count and identify these plastics with speed and precision becomes essential. ML represents an emerging tool for analyzing MPs, but oftentimes those that study computer science and environmental science do no overlap. This review paper intends to introduce ML tools to environmental scientists, allowing them to add rapid counting and identification methods to their tool kit. |
1 Introduction
For better or worse, plastics have become ubiquitous in our society. From bottles to tires to airbags and pacemakers, the benefits of plastic cannot be overstated. But with their ubiquity comes a number of unintended consequences. Microplastics (MPs) have been found everywhere from the Antarctic ice sheets1 to bottled water2 to the inside of our very lungs.3 From a human perspective, the prevalence of MPs in agricultural soil and riverine systems has raised a number of concerns about how MPs may be affecting agricultural practices and toxicity towards microbes,4–6 animals,7–10 plants,11–14 and humans.15,16 These MPs have also been found to be co-carriers of known carcinogens, such as PFAS17,18 or heavy metals.19,20 Specific emphasis in recent years has been placed on examining the role of biosolids as vectors for MP propagation in terrestrial environments such as agricultural soil,21,22 where biosolids collected from wastewater treatment plants are often amended to the soil as a fertilizer. This direct application of MPs has raised a number of concerns about how this affects agriculture and the food we eat.23 Given that human health may be at risk, the pervasive nature of MPs must be given the attention it deserves.The various matrices in which MPs are found present unique issues when it comes to identifying and quantifying the MPs found within, as they are incredibly complex. Plastic-free glassware and laboratory equipment are essential for analysis, regardless of the matrix, as they present easy sources of contamination. Less complex matrices, such as ocean and river samples, will undergo numerous filtering and sieving steps, often augmented with manual collection of particulates to cumulate plastic and debris.24 Sand, soil, and biosolid matrices display even greater complexity as they vary drastically between geographic locations. They contain different mixtures of inorganic (silt, sand, clay) and organic (plant and animal matter) components affecting the density, relative humidity, pH, and myriad other properties of the soil. More recently, the use of synthetic fertilizer and the application of biosolids25,26 to agricultural fields has increased the amount of plastic that percolates into the soil. Adhikari et al.27 found that soils that had been amended with biosolids over a 23 year period contained a mean concentration of 500 plastic particles per kg of dry soil, up to 3 times higher than in non-biosolid amended soils. Corradini et al.28 found that after 10 years of biosolid application in Chilean field soils, the fields they studied had anywhere from 1100 to 3500 plastic particles per kg soil, up to 20x higher than expected. As agricultural practices are actively adding MPs to soil, it becomes necessary to isolate the MPs from the major inorganic and organic soil components in order to identify and quantify the MPs. Techniques ranging from magnetic extraction,29,30 centrifugation,31 and solvent extraction separation32,33 have been employed to carry out the separation, but density separation techniques34,35 have become the most prevalent due to their ease of use, cost, and effectiveness. In this scenario, the soil is placed into a salt solution of high density, such as NaI (1.8 g cm−3). Denser inorganic material will sink, while lighter organic material and polymers will rise, allowing the two layers to be separated. Finally, the organic material is broken down using acids,36 enzymes,37,38 or peroxide solutions39–41 that leave the MPs unscathed, providing us with just the plastics to be characterized. It should be noted that these same techniques can be applied to concentrate the MPs found in marine and ocean samples which can also include soil and organic matter, but on the whole, terrestrial matrices have a much larger component of their collected bulk comprising non-MP elements.
One of the first steps in the process of analyzing MPs is the classification of the particles. What are these MPs made of? What shape are they in? What colour are they? Identification of polymers is often performed using spectroscopic techniques such as IR42–46 or Raman47–49 spectroscopy, often combined with an imaging method, such as microscopy. For example, Chouchene et al. used microscopy to select MPs based on size, colour, and morphology for future identification by FTIR.50 Sobhani et al. extracted MPs from Australian garden soil, using Raman to identify the plastics.51 Liu et al. extracted soils from Shanghai and not only identified the plastics using μFTIR, but also noted the variation in size and concentration of MPs when looking at top soil versus soil collected deeper in the earth (>6 cm).52 As most polymers have a unique spectroscopic fingerprint, these techniques are highly regarded for their easy identification of MPs, often through comparison with libraries. These processes unfortunately require either time-consuming mapping scans of the specimen, or countless individual scans throughout the sample. The library searches can also have difficulties identifying individual polymers when the samples are a mixture of polymers, or when samples contain contaminants from the soil.
It is also necessary to quantify the number of MPs found in a given sample. As the MPs may eventually reach our bodies through the food we eat, understanding how saturated a water or soil sample is with MPs can give us clues about the risk presented. Unfortunately, the best current technique is to manually count all the microplastics in the sample through a microscope. For example, Fakour et al.53 used a stereomicroscope to count the number of particles found in agricultural fields in Taiwan, as well as examining their colour, shape and morphology. Zhang et al.54 paired a camera with a simple light microscope to perform their counting. Jia et al.55 attempted to automate the process by using a Laser Directed Infrared (LDIR) Spectrometer, but admits that there are limitations on identifying particles below 100 μm with great accuracy. As one parses the literature, it becomes clear that new tools are needed to speed up the counting rate compared to a human, as well as improve the collection of data for MPs as they move from the micro to the nanoscale.
As the characterization of MPs has become a time consuming and laborious affair, scientists have once again turned to computing power to find a solution. Giant leaps in computing power and the maturation of the field of Machine Learning (ML) has opened up unforeseen avenues of exploration in a multitude of scientific fields, and MP analysis is no exception. This work hopes to untangle some of the mysteries of ML for those without computer science backgrounds, and show how these in silica approaches are being applied in soil samples.
2 Machine learning
ML isn't merely a single approach, but refers to a collection of various computational models that can learn from a given data set in order to extrapolate and eventually make predictions about new information that is provided. There are a number of different ML models that can be employed, and often it is best to apply multiple algorithms to the problem and compare the results. In the case of environmental samples, goals tend to focus on classification and quantification, and many different ML algorithms can achieve these goals. In this section, some of the basics of ML will be discussed, as well as several commonly used models and how they work. This is by no means an exhaustive list of techniques, but serves to show the many ways ML can attempt to solve the same problem. As the terminology in this field can often be confusing for new computer scientists, a glossary of terms can be found in Table 1.| Term | Description |
|---|---|
| Bias | The error introduced in a machine learning model by simplifying assumptions, leading to underfitting and poor accuracy on training data |
| Classification | A machine learning task where the goal is to categorize data into predefined classes or labels |
| Decision tree | A flowchart-like model used for classification and regression that splits data into branches based on feature values, leading to decision nodes or leaf nodes representing outcomes |
| Hyperplane | A boundary in support vector machines that separates classes by maximizing the margin between them for optimal classification |
| Latent variable | A hidden factor derived from the data that captures shared variation between predictors and responses, used to reduce dimensionality in partial least squares |
| Machine learning | A field of artificial intelligence where algorithms learn patterns from data to make predictions or decisions without being explicitly programmed for specific tasks |
| Regression | A machine learning task focused on predicting continuous numerical values based on input data |
| Supervised learning | A type of machine learning where the model is trained on labeled data, learning to predict outputs from known inputs |
| Unsupervised learning | A machine learning approach where the model learns patterns and structure from unlabeled data without specific guidance on outputs |
| Variance | The model's sensitivity to small changes in the training data, which can cause overfitting and poor generalization to new data |
2.1 Classification vs. regression
ML is often employed to solve one of two problems: classification or regression. Regression problems tend to focus on making a prediction based on historical data. For example, if one has a data set that shows the pH of the soil and the total amount of microplastics in that soil, a best fit line can be created to show the relationship between these two features. That best fit line can then be employed to predict the amount of microplastics in a soil sample if the pH is known.A classification problem is one in which an algorithm is employed to provide labels to a data set based on the training data. For example, if a series of IR spectra are measured for a set of plastics, a classification algorithm can be used to label each of the unknown polymers. When examining MPs in environmental matrices, this type of problem is most common.
2.2 Supervised vs. unsupervised learning
Another key distinction to make when discussing ML models is that of supervised vs. unsupervised learning. In the case of supervised learning, the training data provided to the algorithm has labels that help the model understand what the correct answer should be. For MPs, the data may be labelled as polystyrene (PS) or polypropylene (PP) so that the model knows that any spectra with these specific peak locations belong to one of those categories. In unsupervised learning, the training data is not labelled, so the model will have to make its own connections and identify patterns without help. In the case of MPs, supervised learning is far more common as much of the training data has been analyzed quite thoroughly.2.3 How ML works
When setting up a ML model, a large data set related to the eventual input data is needed for training and testing of the model. For example, if the purpose of the model is to classify the different types of polymers in a soil mixture using Raman spectroscopy, then a large collection of Raman spectra of various polymers will be needed in preparation. A general rule of thumb is that 10 data points are needed for each feature you wish to examine. For example, if one wishes to examine Raman spectra of aged polyethylene (PE), ideally 10 Raman spectra of PE are required. This is by no means a hard and fast rule. Generally, more training data is better than less, but the complexity and application of the ML model will often dictate of how many data points are needed.From here, the data needs to be processed for it to be in a useable format for ML algorithms. This often involves three steps: baseline correction, smoothing, and normalization. For Raman spectroscopy data in particular, the baseline is often distorted by background fluorescence, causing a great deal of drift along the baseline. Corrections can be performed using polynomial fitting,55 least squares smoothing,56 or wavelet transformations,57 among others. Smoothing of the data is generally needed as Raman spectra can often have a number of noisy signals that obscure and obfuscate the main spectral peaks. Therefore, smoothing techniques such as Savitzky–Golay filters are often used to smooth out the data.58 Finally, normalization is applied to the data so that all data is comparable on a 0–1 intensity scale, allowing data from different sources to be compared on an even playing field. The combination of these three processes will generally allow multiple data sources to be used in a machine learning algorithm.
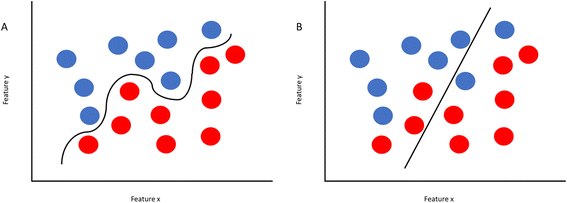
Once the data set is compiled and processed, the data is split up, with about 80% set aside for training and the remaining 20% set aside for testing. The model will then be trained on the first batch to learn trends and connections, and then it will test what it learned on the training set. From there, parameters are shifted in the algorithm in order to reduce bias and variance. Bias refers to the error between the average model prediction and the truth. High bias does not match the training data set well, while low bias will match the training set too closely, making the model unable to analyze new data that differs too greatly from the training set. Variance refers to the ability of a model to adjust to a data set. If a model is overfitting the data (Fig. 1A), that means the model is trying too hard to make the input data look like the training set. If you are underfitting (Fig. 1B), the model is doing a poor job of capturing what the data looks like, including outliers. By performing these training and testing steps, the model can be optimized to find the best fit.
3 Commonly used ML models
3.1 Partial least squares (PLS)
PLS functions best in situations where each data point has a large number of identifiable features. For example, a Raman spectrum of a mixed soil/microplastic sample (a data point) may have a large number of visible peaks (predictors), which the PLS can use to identify the types of plastics within it. In a classification problem, it may produce a response variable to indicate whether polymers are visible and measurable in the soil. PLS looks at the available information and transforms the data into what are known as latent variables, which are essentially summaries of the data that capture the most important information from the predictors. It will group these latent variables together and reduce the dimensionality of the data, focusing only on the most important and predictive elements. The important elements are selected by determining which combinations of predictors have the highest covariance with the response variables (if the prevalence of one predictor increases, so does the likelihood of identifying a microplastic). Once these elements have been determined, the model can use them to produce the response variable defined by the classifier.593.2 Support vector machine
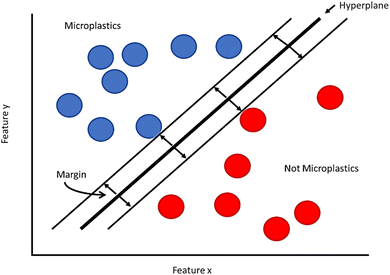
Support Vector Machines (SVMs) are classification tools that attempt to separate data into classes by examining features of that data. This type of model was developed to focus on binary classifications (yes/no systems), informing the user whether an object is or is not what they are looking for. In the case of MPs, this can be used to determine if a particle in a microscope image is an MP, or simply leftover debris from the extraction process.An SVM works by plotting the data set based on two or more features (these make up the axes of your plot). Features in the case of MPs may refer to physical dimensions like size or aspect ratio, or for spectroscopic data sets the features may be spectral peaks known to correlate with a given functional group. The data set is then plotted on this set of feature axes (Fig. 2). The next step is for the algorithm to create a boundary line between through the data set called a hyperplane. As there are a number of hyperplanes that can be drawn through a dataset, SVM maximizes the margin between different classes, making it clear that there is a true distinction between each side of the hyperplane. One advantage SVM has over similar models like PLS is that the hyperplane can be drawn through three-dimensional space using kernel functions, improving the classification by adding extra features. Looking at our example in Fig. 2, the side of the hyperplane that the data is found on will determine if the object is or is not a MP. For many cases, it may be obvious whether the data shows an MP or not. Many data points will not be so clear, and that's why having the algorithm find the correct hyperplane is so important and why having an appropriate training set can greatly improve the results of your analysis. SVMs tend to be used more for classification problems than regression problems.60
3.3 K-nearest neighbour (KNN)
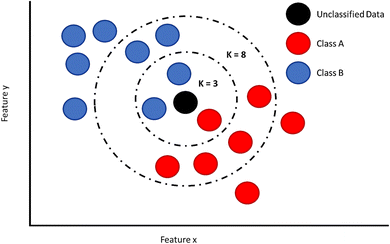
KNN models also look to classify a data point based on its similarity and proximity to known datasets. For example, a known dataset has been classified into two groups based on a set of features, and the model has placed each data point into one of two separate groups (Fig. 3). As a new data point is introduced into the model, it will be sorted based on the assigned features. But what if it falls between classifications? In the case of K-Nearest Neighbour, its classification will be based upon its proximity to one of the other classifications (generally based on a Euclidean distance function). The distance values will be sorted from shortest distance to longest distance. A K value is then selected. This refers to the selected population size (starting with the smallest distance), which will be averaged. High K values lead to underfitting, meaning that the model is too simple to represent the true relationship between the input (new data point) and output (classification). Low K values can lead to overfitting, as the model becomes too sensitive to variations in the data, making it difficult to properly present the trend of the data, especially when it varies from the training data. Therefore, a Goldilocks zone needs to be found for k.613.4 Random forest (RF)
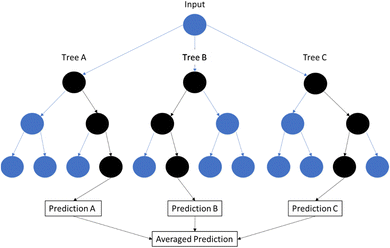
A Random Forest (RF) is a machine learning algorithm that builds on the concept of a decision tree (Fig. 4). A decision tree is a method of determining an output based on a series of binary choices. For example, if one is trying to determine what type of plastic is under investigation, there are a series of choices to be made that can help make that determination? Does the IR spectrum have a C–H stretch at 2900 cm−1? If no, then it's probably not PS. Does it have an N–H stretch at 2930 cm−1? If yes, then it may be a nylon. In ML, an RF model will take a number of decision trees and run them simultaneously. This is done to minimize overfitting to a training set, or reduce bias that is caused by errors built into each individual tree. For a classification RF model, the classification is based on the majority vote of the individual decision trees. For a regression analysis, the output is the average output of all the individual decision trees. In this way, RF models help to generalize the output and produce more accurate results.623.5 Neural networks (NN)
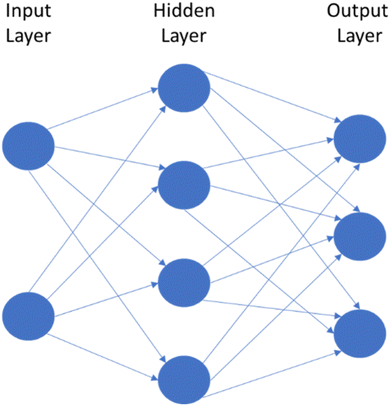
NNs are designed to act like neurons in the brain, taking an input value and weighing it against known information to determine the correct output.63 NNs use a series of weighted nodes to evaluate the input information. The internal layers, known as hidden layers, are made up of a series of nodes that evaluate one characteristic of the input (Fig. 5). Is this particle round? Is it blue? Does it fluoresce? The model then compares the input data against the training data, and passes the information onto the next layer. Each of these decisions have an associated weight based on how important that characteristic is to determining the output. More weight may be put on a particle having a blue or green colour versus whether a particle is round or square. Once the data has passed through a specified set of hidden layers, it reaches the output layer where it makes its final determination based on the appraisal of the data. For a classification model, the output layer may be a single node that makes a binary decision; yes, this is a MP or no, it is not. It's important to note that NNs move in the forward direction (called a feed forward network). Newer models64 can institute what is known as a recurrent neural network, where the output of some nodes can be used to affect later inputs into the same nodes, acting as an internal learning process. Other neural network models continue to appear as the field matures.3.6 Choosing the best algorithm
This section has shown that there a vast number of ML algorithms available for researchers to use, and breadth of the field is a feature not a bug. In line with Wolpert's “No Free Lunch Theorem”,65 these is no single machine learning approach that performs best across all possible problems. Therefore, to find the best model, one must simply test several and find the best for yourself. Thankfully, Kotsiantsis66 has put together an excellent review of a number of models that demonstrates their pros and cons (see Table 2). If overall accuracy is a priority, maybe an SVM is the best model, even if it learns at a slow pace. Maybe classification speed matters most, so a NN is the ideal model. Overall, Kotsiantis provides us an excellent starting point, but there is no substitute for hard work.| Decision trees | Neural networks | KNN | SVM | |
|---|---|---|---|---|
| Accuracy in general | ** | *** | ** | **** |
| Speed of learning | *** | * | **** | * |
| Speed of classification | **** | **** | * | ** |
| Tolerance to missing values | *** | * | ** | **** |
| Tolerance to irrelevant attributes | *** | * | ** | **** |
| Tolerance to highly interdependent attributes | ** | *** | * | *** |
| Dealing with discrete/binary/continuous attributes | **** | *** | *** | ** |
| Tolerance to noise | ** | ** | * | ** |
| Dealing with danger of overfitting | ** | * | *** | ** |
| Attempts for incremental learning | ** | *** | **** | ** |
| Explanation ability | **** | * | ** | * |
| Model parameter handling | *** | * | *** | * |
4 Application of ML techniques to MPs in various matrices
4.1 Identification of MPs
MP identification typically relies on spectroscopic techniques (such as IR or Raman spectroscopy), which analyze sample composition. IR and Raman spectroscopy offer distinct advantages, providing unique fingerprints of the targeted specimen. When combined with microscopic techniques, they enable individual particle analysis, enhancing characterization capabilities. These techniques are also non-destructive, allowing for further analysis of the particles by other means, such as TGA or Py-GCMS. On the other hand, this analysis is quite time-consuming, and identifying uncommon substances (that may have undergone degradation) can be tricky if they fall outside of known libraries. This is why ML has begun to be paired with these techniques to both speed up and improve the accuracy of identification.The above examples were performed using synthetic or pristine polymers, free from environmental matrices. However, when dealing with soil, marine sediments, peat, or wastewater biosolid, classification becomes significantly more challenging. One approach to tackle this is to pair the ML with human expertise. Weber et al.72 collected sediment samples from a series of wastewater treatment plants in Germany, and extracted the MPs via density separation in sodium polytungstate before using hydrogen peroxide and sodium hypochlorite to tackle the organic components still dwelling amongst the MPs. They would then generate more than sixty thousand Raman spectra from the samples, which were painstakingly classified by humans based on a set of criteria. This resulted in a high rate of false positives in classification of MPs, though it had a higher overall precision (higher percent of MPs were categorized and classified). Next, the data was input into a deep-learning NN, where it was found to have higher rate of true positives in classification of MPs than the human only methodology, but had a precision rate lower than the human approach. They then chose to combine the human and machine, first applying the ML algorithm to collect all possible MP signals, and then using the human expert to remove the false positives. Along with a major reduction in the time needed to perform the analysis, this combined method led to a recall of >98% (compared to human only value of roughly 80%) and a precision of around 97% (compared to machine-only 90%). These results show that ML can act as a powerful complementary technique to current classification methods.
What happens when the MPs are still mixed within the matrix? Li et al.73 chose to combine Raman imaging, Raman spectroscopy, and a PLS method in order to examine MPs found in river sand without extraction or digestion of mineral and organic matter. A volume maximizer (AVmax)74 is used to unmix spectral signals and maximize features of interest before using an unconstrained form of PLS to predict which components in an image are MPs, along with their identity. Li was able to discern PE that was mixed into river sand samples, without any physical preprocessing or filtering of the samples.
The addition of a polymer spike adds certainty to the analysis, but in real world samples the concentration of MPs is expected to be much lower. In the case of marine and ocean samples, large volumes of water need to be processed to analyze measurable quantities of MPs. Tian et al.80 collected large volumes of river water samples from the German Rhine and Meus rivers in order to classify the weathered MPs found in the water. After extensive sieving, chemical treatment, and density filtration, the resulting particulate was examined using LDIR, which uses a Quantum Cascade Laser as its infrared source. Using both a KNN model and Decision Tree model, the researchers were able to classify the unknown polymers with 89.9% accuracy (KNN) and 77.1% accuracy (decision tree). They then went one step further, using a non-supervised model (Density-Based Spatial Clustering of Applications with Noise, or DBSCAN) to help determine the remaining unknowns (which are likely eroded and weathered versions of the known polymers). DBSCAN, as an unsupervised technique, will simply group together data points with similar features. If one happens to know what some of those data points are, and they are clustered with a series of unknowns, it is reasonable to believe some of those unknowns fall into the same category. While this technique cannot truly confirm the identity of the unknown data points, it allows a rough estimate of which of the remaining points may be MPs, and which are simply outliers.
Water samples and soil samples are unlikely to have similar weathering processes, so its important to examine soil as well. Hufnagl et al.81 chose to examine MPs within soil, wastewater treatment plant outlet, deep sediment, and compost samples, which required the soil to be sieved and separated using mechanical and chemical techniques. Once the MPs were isolated, they used an RF model was used to identify polymers from focal plane array-based micro-Fourier transform infrared (FPA-μFTIR) imaging (Fig. 6). This IR technique creates chemical images by recording thousands of IR spectra. The RF model would then predict the identity of the particles by comparing the measured spectra against known reference or training data. This technique was able to distinguish up to 20 different polymers on a 1000 × 1000-pixel image in less than 10 minutes, while also measuring length, width, aspect ratio, area, and orientation. This technique was shown to work in a wide variety of matrices, including sediment, soil, compost, and sewage sludge.
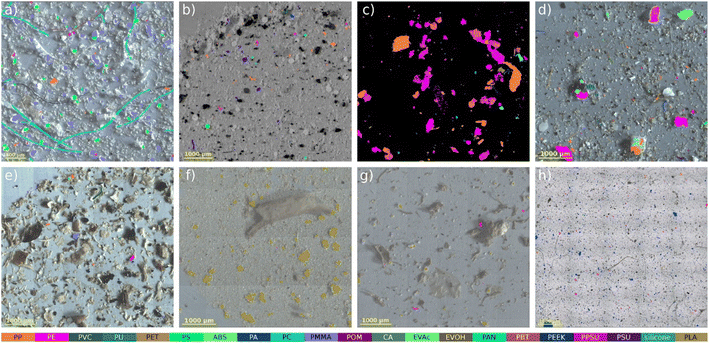 | ||
| Fig. 6 Application examples for different matrices. (a–c). Plankton samples (d) wastewater treatment plant outlet, (e) deep sediment sample, (f) soil sample, (g) compost sample, and (h) sea salt sample. Adapted with permission under a Creative Commons Attribution 3.0 Unported License from Hufnagl et al.81 Copyright 2022, The Royal Society of Chemistry. | ||
4.2 Quantification of MPs
Quantifying the number of MPs in a sample can be just as important as knowing which kind of MPs are in the sample. The ability to determine the number of MPs in soil or water can help monitor the effectiveness of pollution mitigation efforts, and inform researchers whether the situation is improving or deteriorating. By combining ML with spectroscopic techniques, such as IR, Raman, and NIR, ML can be used to determine if a sample reaches a certain threshold of MP concentration, and can be even used to estimate the number of MPs in the given sample.Quantification of the number of particles (without identification), thus far, has seen most of its progress focused on simpler samples, such as pure microplastics82 or water samples that have very little in the way of matrix that obscures and obfuscates the MPs.83 Tan et al.84 chose an in-silica approach to count the number of MPs in a sample using mass measurements obtained from existing datasets, including those taken from beach sands, seawater, and seabirds. They computationally “sieved” the particles into different size ranges and determine the total particle weight in each size range. Using particle density and size measurements for rubber particles, films, beads, fibres, and organic components in the known datasets, they could train an RF model and a linear regression model (known as a Kernel Ridge Regression)85 to estimate the number of MP particles on the basis of the aggregate particle weight measurements. This allowed the researchers to predict the number of MPs in a single population or in mixed MP samples. Results showed the model would perform better than humans for large and homogeneous mixtures, and that organic material that can be often difficult to remove was not a major source of error, as it represented such a small component of the mixtures overall mass.
Moving on from simulation and testing, more complex samples and matrices would become the focus of study. Using Australian soil samples spiked with PET and low-density PE, Ng et al.86 combined NIR spectroscopy with a NN to classify samples based on the percentage of microplastics found within the sample. They were able to sort soil samples into batches that were less than 1% MPs, between 1% and 3% MPs, and above 3% MPs based on the NIR images with relative accuracy, although it had trouble separating situations of no MP concentration and low MPs concentration. Wu et al.87 also went the spiked-polymer route, adding PE, PS, PP, and PVC to waste incineration ash. The samples were examined with NIR, and care was taken to exclude spectral information from the ash using background subtractions from pure ash samples. The application of an SVM model was able to predict the quantity of plastics within the ash with a greater than 89% accuracy for each of the polymers. Each of these studies show that spectroscopic techniques combined with ML are able to parse through matrices in order to complete quantification experiments, particularly when the concentration is high, as one would expect from a spiked sample.
Spiked samples have a known number of MPs to measure, but this is not the case with environmental samples. Lorenzo-Navarro et al.88 collected beach sands from the Canary Islands archipelago. Even after pre-treatment via mechanical sieving and density-based separation techniques, they were unable to completely separate polymer pellets and fibres from tar and organic particles. In order to properly count the MPs, they had to first introduce a classifier that would separate the pellets and the fibres from the tar and organic material based off microscope imagery. A series of ML models were then employed to contrast and compare, including a KNN, an RF, and an SVM. Fibres and pellets proved easy to classify based on geometric properties (fibres have high aspect ratios, pellets are spherical), while the remaining components would require more properties (colour, texture, etc.) to be further classified. Once the models could identify the particles by class, it could then carry out the job of counting. SVM was found to be the best of the techniques, with a recall rate of 88%.
While soil itself is a complex matrix, this also means that it has more features that can be analyzed and measured. Tran et al.89 used this to their advantage, as they went about predicting MP quantities in peatland sediments based on soil pH, salinity, and composition, amongst others. Using peatland soil from the three industrial regions around the Mekong Delta of Vietnam, they measured 7 different physicochemical properties for 300 samples, and, after performing sieving and density filtration, counted the number of MPs found in each sample. This information was used as training and testing data for RF and SVM models (although the large differences in the region made mixing the data from different areas too monumental of a task). Their results showed that both pH and total organic carbon (TOC) had a positive correlation with the number of MPs in a sample from a given area. Salinity and electrical conductivity were found to have a negative correlation. Overall, the correlations were not considered strong enough to stand on their own (the models couldn't predict samples with high numbers of MPs due to the weak correlations), but it has merit as a complimentary tool to the many spectroscopic and microscopic tools seen thus far.
4.3 Methodological comparison
The studies discussed above show that ML techniques can be applied to MPs derived from myriad environmental sources, whether provided as pristine polymer spikes or weathered and eroded samples found in situ. Each of these studies remains reliant on the ability to extract the MPs from the matrix diligently enough for spectroscopic and microscopic techniques to be applied. New techniques, such as those demonstrated by Ng et al.86 and Tran et al.,89 allow for analysis to occur with very little preprocessing of the matrix, or by using the matrix to improve the analysis. By focusing on these kinds of techniques, the difficult extraction step can be minimized, allowing for more reliable and reproducible data between groups.This work has previously discussed the important decision of which ML model to select for the task and a quick perusal of the above studies show how Wolpert's “No Free Lunch Theorem” rings true. Table 3 shows a comparison of all the studies in this work, with a particular emphasis on the ML models used in each study and their final accuracy determination. Almost all studies used multiple ML models and, while each study had one model that worked best, no particular model stands head and shoulders above the rest across the board. When it comes to applying ML tools in this field, it is simply a necessity to roll up one's sleeves and try several algorithms to find the best one. It should be noted that Weber et al.’s72 combination of a neural network with a human “expert” led to not only high classification accuracy, but also minimized false positive and false negatives, suggesting that removing the human element from machine learning remains unwise.
| Study | Analysis technique | Matrix | Goal | ML technique | Accuracy of technique |
|---|---|---|---|---|---|
| Lei et al.66 | Raman | Commercial plastics | Classification | RF | >95% |
| KNN | >95% | ||||
| Luo et al.67 | Raman | Water samples | Classification | NN | 99% |
| SVM | 94% | ||||
| BPNN | 81% | ||||
| Ramanna et al.68 | Raman | Weathered plastics | Classification | RF | 93.81% |
| NN | 71.13% | ||||
| SVM | 73.19% | ||||
| DT | 69.07% | ||||
| KNN | 73.19% | ||||
| Ren et al.71 | Raman | Weathered plastics | Classification | CNN | 95% |
| Weber et al.72 | Raman | Wastewater sediments | Classification | NN/human | 98% |
| Li et al.73 | Raman | River sand | Classification | PLS | >94% |
| Zou et al.76 | NIR | Sand | Classification | PLS | >95% (coloured) |
| >80% (colourless) | |||||
| Shan et al.77 | NIR | Soil | Classification | SVM | 76% (black) |
| 77% (white) | |||||
| Paul et al.78 | NIR | Soil | Classification | SVM | 92% |
| PLS | 83% | ||||
| Chen et al.79 | SWIR | Soil | Classification | PLS | >95% (InGaAs) |
| >92% (MCT) | |||||
| >95% (InGaAs) | |||||
| >92% (MCT) | |||||
| Tian et al.80 | LDIR | River water | Classification | KNN | 89.9% |
| DT | 77.1% | ||||
| Hufnagl et al.81 | μFTIR | Soil | Classification | RF | >90% |
| Ng et al.86 | Vis-NIR | Soil | Quantification | CNN | 78.5% |
| Wu et al.87 | NIR | Waste incineration ash | Quantification | SVM | 89% |
| Lorenzo-Navarro et al.88 | Microscopy | Beach sand | Quantification | KNN | 72.1% |
| RF | 81.9% | ||||
| SVM | 88% |
5 Beyond spectroscopy
While this work has placed an emphasis on the synthesis of spectroscopic techniques and ML, it is important to note that a number of other techniques have been used to study MPs, particularly thermometric techniques such as Thermogravimetric Analysis (TGA)90,91 and Pyrolysis Gas Chromatography Mass Spectrometry (Py-GCMS).92–94 There has recently been a push to utilize ML alongside these techniques. Chowdhury et al. used a NN alongside TGA to predict the heat deterioration of PET particles.95 Christian et al. tested a number of ML algorithms to examine PET degradation amongst modified and aged PET particles.96 Zhang et al. tested RF and SVM models to predict the presence of PE and PP based on decomposition data produced by Py-GCMS.97 Lastly, Forbes et al. was able to characterize MPs as pristine, weathered, or in mixtures by combining Py-GCMS with a series of ML models.98 Each of these studies show how ML can work in conjunction with thermal methods to analyze MPs. What each of these studies lack is the application of these techniques to MPs in environmental samples. Particularly with soil, the field remains very much in its infancy, with direct application of ML on MPs in soil samples yet to be studied at this time.996 Limitations and conclusions
The use of ML to study soil samples has shown amazing promise in analyzing MPs in complex matrices, but limitations still remain. All ML models require large sets of data for training and validation, which can be difficult to obtain. Many of the above examples were required to pull data from multiple databases (often derived from different sources and methods), or even produce synthetic data to be trained upon. These datasets are also not easily accessible. Many datasets may be found behind paywalls or held back for proprietary reasons, making it difficult to get a strong dataset to train on. The field is desperately calling out for more easily accessible datasets, and, even more ideally, a single open access database where it can all be archived and made available for future researchers.Another major limitation is the complexity of the matrices themselves. Extracting MPs from different matrices involves multiple steps to remove the mineral and organic content, and often doesn't see complete separation or full recovery of the MPs. Remove too much of the soil, and one doesn't get an accurate measurement of MPs. Remove too little, and the MPs get buried in the noise. Add in the fact that soil or water varies heavily between regions in composition and usage, and it becomes difficult to apply a one-sized fits all approach to analysis. This may be solved through the creation of new standards and reference materials created from individual soil/MP mixtures, which may speed up and improve the accuracy of the training and testing procedure. There remains an opening for metrologists to add their own expertise to the field.
A final limitation of ML comes from the reproducibility of ML models, as sample preparation (choosing one chemical degradant over another), the type of spectrometer (such as variations in laser power and intensity), and even the data acquisition method (selection of range of interest, duration of laser application) can lead to variations in the data that is fed into the models. In order to combat this, full transparency of data and methodology will be key to demonstrating why and how models may vary. Both raw and processed data should be made available publicly, as well as any assumptions regarding why each data point was or was not included in the training/testing dataset. An advantage of making all the data available is that large databases can be built to create training data, improving future models. The ML code itself needs to be made readily available, so that the code can be evaluated and tested for errors and inaccuracies and even improved upon for future iterations. If the data and code are available for all to see, then reproducibility can be much improved, or, in the cases where there is poor reproducibility, the source of the issue can be laid bare for all to see.
Despite these limitations, the future remains open to optimism. As more data is produced, the available pool of training data increases, improving the reliability of the models. As new techniques and models come online, research can dig deeper into their data and finally gain a full picture of the MPs scattered throughout our world.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The author declares that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors appreciate the Ocean Program at the National Research Council Canada (NRC) and the Advancing a Circular Plastics Economy for Canada Program from the Government of Canada for financial support. The author would also like to acknowledge the contributions of Dr Shan Zou, Dr Daniel Prezgot, and Dr Adrian Pegoraro for their helpful suggestions to improve this work.References
- A. R. Aves, L. E. Revell, S. Gaw, H. Ruffell, A. Schuddeboom and N. E. Wotherspoon, et al., First evidence of microplastics in Antarctic snow, Cryosphere, 2022, 16(6), 2127–2145, DOI:10.5194/tc-16-2127-2022.
- I. Gambino, F. Bagordo, T. Grassi, A. Panico and A. De Donno, Occurrence of microplastics in tap and bottled water: Current knowledge, Int. J. Environ. Res. Public Health, 2022, 19(9), 5283, DOI:10.3390/ijerph19095283.
- L. C. Jenner, J. M. Rotchell, R. T. Bennett, M. Cowen, V. Tentzeris and L. R. Sadofsky, Detection of microplastics in human lung tissue Using μFTIR spectroscopy, Sci. Total Environ., 2022, 831, 154907, DOI:10.1016/j.scitotenv.2022.154907.
- Y. Fei, S. Huang, H. Zhang, Y. Tong, D. Wen and X. Xia, et al., Response of soil enzyme activities and bacterial communities to the accumulation of microplastics in an acid cropped soil, Sci. Total Environ., 2020, 707, 135634, DOI:10.1016/j.scitotenv.2019.135634.
- A. A. de Souza Machado, C. W. Lau, J. Till, W. Kloas, A. Lehmann and R. Becker, et al., Impacts of microplastics on the soil biophysical environment, Environ. Sci. Technol., 2018, 52(17), 9656–9665, DOI:10.1021/acs.est.8b02212.
- H. T. Wang, J. Ding, C. Xiong, D. Zhu, G. Li and X. Y. Jia, et al., Exposure to microplastics lowers arsenic accumulation and alters gut bacterial communities of earthworm Metaphire californica, Environ. Pollut., 2019, 251, 110–116, DOI:10.1016/j.envpol.2019.04.054.
- E. Huerta Lwanga, H. Gertsen, H. Gooren, P. Peters, T. Salánki and M. van der Ploeg, et al., Microplastics in the terrestrial ecosystem: Implications for lumbricus terrestris (oligochaeta, lumbricidae), Environ. Sci. Technol., 2016, 50(5), 2685–2691, DOI:10.1021/acs.est.5b05478.
- M. E. Hodson, C. A. Duffus-Hodson, A. Clark, M. T. Prendergast-Miller and K. L. Thorpe, Plastic bag derived-microplastics as a vector for metal exposure in terrestrial invertebrates, Environ. Sci. Technol., 2017, 51(8), 4714–4721, DOI:10.1021/acs.est.7b00635.
- M. O. Gaylor, E. Harvey and R. C. Hale, Polybrominated diphenyl ether (PBDE) accumulation by earthworms (eisenia fetida) exposed to biosolids-, polyurethane foam microparticle-, and penta-BDE-amended soils, Environ. Sci. Technol., 2013, 47(23), 13831–13839, DOI:10.1021/es403750a.
- S. Matthews, E. Genbo Xu, E. R. Dumont, V. Meola, O. Pikuda and S. Cheong R, et al., Polystyrene micro- and nanoplastics affect locomotion and daily activity of Drosophila melanogaster, Environ. Sci.: Nano, 2021, 8(1), 110–121, 10.1039/D0EN00942C.
- S. Pignattelli, A. Broccoli and M. Renzi, Physiological responses of garden cress (L. sativum) to different types of microplastics, Sci. Total Environ., 2020, 727, 138609, DOI:10.1016/j.scitotenv.2020.138609.
- Y. Qi, X. Yang, A. M. Pelaez, E. Huerta Lwanga, N. Beriot and H. Gertsen, et al., Macro- and micro- plastics in soil-plant system: Effects of plastic mulch film residues on wheat (Triticum aestivum) growth, Sci. Total Environ., 2018, 645, 1048–1056, DOI:10.1016/j.scitotenv.2018.07.229.
- M. Simonetta, L. Giorgetti, M. Corsini, G. Di Florio and L. Bellani, Nano and submicron fluorescent polystyrene particles internalization and translocation in seedlings of Cichorium endivia L, Environ. Sci.: Nano, 2022, 9(12), 4585–4598, 10.1039/D2EN00732K.
- S. E. Taylor, C. I. Pearce, K. A. Sanguinet, D. Hu, W. B. Chrisler and Y. M. Kim, et al., Polystyrene nano- and microplastic accumulation at Arabidopsis and wheat root cap cells, but no evidence for uptake into roots, Environ. Sci.: Nano, 2020, 7(7), 1942–1953, 10.1039/D0EN00309C.
- D. He, Y. Luo, S. Lu, M. Liu, Y. Song and L. Lei, Microplastics in soils: Analytical methods, pollution characteristics and ecological risks, TrAC, Trends Anal. Chem., 2018, 109, 163–172, DOI:10.1016/j.trac.2018.10.006.
- Q. Wang, J. Bai, B. Ning, L. Fan, T. Sun and Y. Fang, et al., Effects of bisphenol A and nanoscale and microscale polystyrene plastic exposure on particle uptake and toxicity in human Caco-2 cells, Chemosphere, 2020, 254, 126788, DOI:10.1016/j.chemosphere.2020.126788.
- J. Sun, H. Xiang, X. Jiang, X. Wang, X. Luo and J. Fu, et al., Effects of polyamide microplastics on the adsorption of perfluoroalkyl substances in soil, J. Hazard. Mater. Adv., 2024, 13, 100391, DOI:10.1016/j.hazadv.2023.100391.
- J. W. Scott, K. G. Gunderson, L. A. Green, R. R. Rediske and A. D. Steinman, Perfluoroalkylated substances (PFAS) associated with microplastics in a lake environment, Toxics, 2021, 9(5), 106, DOI:10.3390/toxics9050106.
- B. Liu, S. Zhao, T. Qiu, Q. Cui, Y. Yang and L. Li, et al., Interaction of microplastics with heavy metals in soil: Mechanisms, influencing factors and biological effects, Sci. Total Environ., 2024, 918, 170281, DOI:10.1016/j.scitotenv.2024.170281.
- Q. An, T. Zhou, C. Wen and C. Yan, The effects of microplastics on heavy metals bioavailability in soils: a meta-analysis, J. Hazard. Mater., 2023, 460, 132369, DOI:10.1016/j.jhazmat.2023.132369.
- J. Crossman, R. R. Hurley, M. Futter and L. Nizzetto, Transfer and transport of microplastics from biosolids to agricultural soils and the wider environment, Sci. Total Environ., 2020, 724, 138334, DOI:10.1016/j.scitotenv.2020.138334.
- S. Marchuk, S. Tait, P. Sinha, P. Harris, D. L. Antille and B. K. McCabe, Biosolids-derived fertilisers: A review of challenges and opportunities, Sci. Total Environ., 2023, 875, 162555, DOI:10.1016/j.scitotenv.2023.162555.
- L. Li, Y. Luo, R. Li, Q. Zhou, W. J. G. M. Peijnenburg and N. Yin, et al., Effective uptake of submicrometre plastics by crop plants via a crack-entry mode, Nat Sustainability, 2020, 3(11), 929–937, DOI:10.1038/s41893-020-0567-9.
- M. Kedzierski, M. Falcou-Préfol, M. E. Kerros, M. Henry, M. L. Pedrotti and S. Bruzaud, A Machine Learning Algorithm for High Throughput Identification of FTIR Spectra: Application on Microplastics Collected in the Mediterranean Sea, Chemosphere, 2019, 234, 242–251, DOI:10.1016/j.chemosphere.2019.05.113.
- B. N. Naderi, S. Karimifard, J. Gilley, T. Messer, A. Schmidt and S. Bartelt-Hunt, Higher concentrations of microplastics in runoff from biosolid-amended croplands than manure-amended croplands, Commun. Earth Environ., 2023, 4(1) DOI:10.1038/s43247-023-00691-y.
- A. Mohajerani and B. Karabatak, Microplastics and pollutants in biosolids have contaminated agricultural soils: An analytical study and a proposal to cease the use of biosolids in farmlands and utilise them in sustainable bricks, Waste Manage., 2020, 107, 252–265, DOI:10.1016/j.wasman.2020.04.021.
- K. Adhikari, C. I. Pearce, K. A. Sanguinet, A. I. Bary, I. Chowdhury and I. Eggleston, et al., Accumulation of microplastics in soil after long-term application of biosolids and atmospheric deposition, Sci. Total Environ., 2023, 1, 168883, DOI:10.1016/j.scitotenv.2023.168883.
- F. Corradini, P. Meza, R. Eguiluz, F. Casado, E. Huerta-Lwanga and V. Geissen, Evidence of microplastic accumulation in agricultural soils from sewage sludge disposal, Sci. Total Environ., 2019, 671, 411–420, DOI:10.1016/j.scitotenv.2019.03.368.
- J. Grbic, B. Nguyen, E. Guo, J. B. You, D. Sinton and C. M. Rochman, Magnetic Extraction of Microplastics from Environmental Samples, Environ. Sci. Technol. Lett., 2019, 6(2), 68–72, DOI:10.1021/acs.estlett.8b00671.
- F. Rhein, F. Scholl and H. Nirschl, Magnetic seeded filtration for the separation of fine polymer particles from dilute suspensions: Microplastics, Chem. Eng. Sci., 2019, 207, 1278–1287, DOI:10.1016/j.ces.2019.07.052.
- G. Grause, Y. Kuniyasu, M. F. Chien and C. Inoue, Separation of microplastic from soil by centrifugation and its application to agricultural soil, Chemosphere, 2022, 288, 132654, DOI:10.1016/j.chemosphere.2021.132654.
- E. D. Okoffo, F. Ribeiro, J. W. O'Brien, S. O'Brien, B. J. Tscharke and M. Gallen, et al., Identification and quantification of selected plastics in biosolids by pressurized liquid extraction combined with double-shot pyrolysis gas chromatography–mass spectrometry, Sci. Total Environ., 2020, 715, 136924, DOI:10.1016/j.scitotenv.2020.136924.
- J. La Nasa, G. Biale, M. Mattonai and F. Modugno, Microwave-assisted solvent extraction and double-shot analytical pyrolysis for the quali-quantitation of plasticizers and microplastics in beach sand samples, J. Hazard. Mater., 2021, 401, 123287, DOI:10.1016/j.jhazmat.2020.123287.
- R. Nakajima, M. Tsuchiya, D. J. Lindsay, T. Kitahashi, K. Fujikura and T. Fukushima, A new small device made of glass for separating microplastics from marine and freshwater sediments, PeerJ, 2019, 7 DOI:10.7717/peerj.7915.
- M. Chen, B. Coleman, L. Gaburici, D. Prezgot, Z. J. Jakubek and B. Sivarajah, et al., Identification of microplastics extracted from field soils amended with municipal biosolids, Sci. Total Environ., 2024, 907, 168007, DOI:10.1016/j.scitotenv.2023.168007.
- M. Claessens, L. Van Cauwenberghe, M. B. Vandegehuchte and C. R. Janssen, New techniques for the detection of microplastics in sediments and field collected organisms, Mar. Pollut. Bull., 2013, 70(1–2), 227–233, DOI:10.1016/j.marpolbul.2013.03.009.
- C. Palacios-Mateo, K. Meng, L. Legaz-Pol, E. Steen-Redeker, E. Huerta-Lwanga and L. M. Blank, Enzymes for microplastic-free agricultural soils, Ecotoxicol. Environ. Saf., 2023, 258, 114982, DOI:10.1016/j.ecoenv.2023.114982.
- B. Toto, A. Refosco, J. Dierkes and T. Kögel, Efficient extraction of small microplastic particles from rat feed and feces for quantification, Heliyon, 2023, 9(1), e12811, DOI:10.1016/j.heliyon.2023.e12811.
- Q. Li, J. Wu, X. Zhao, X. Gu and R. Ji, Separation and identification of microplastics from soil and sewage sludge, Environ. Pollut., 2019, 254, 113076, DOI:10.1016/j.envpol.2019.113076.
- S. Frei, S. Piehl, B. S. Gilfedder, M. G. J. Löder, J. Krutzke and L. Wilhelm, et al., Occurrence of microplastics in the hyporheic zone of rivers, Sci. Rep., 2019, 9(1) DOI:10.1038/s41598-019-51741-5.
- R. R. Hurley, A. L. Lusher, M. Olsen and L. Nizzetto, Validation of a method for extracting microplastics from complex, organic-rich, environmental matrices, Environ. Sci. Technol., 2018, 52(13), 7409–7417, DOI:10.1021/acs.est.8b01517.
- B. Thakur, J. Singh, J. Singh, D. Angmo and A. P. Vig, Identification and characterization of extracted microplastics from agricultural soil near industrial area: FTIR and X-ray diffraction method, Environ. Qual. Manag., 2023, 33(1), 173–181, DOI:10.1002/tqem.22035.
- B. Zhou, J. Wang, H. Zhang, H. Shi, Y. Fei and S. Huang, et al., Microplastics in agricultural soils on the coastal plain of Hangzhou Bay, east China: Multiple sources other than plastic mulching film, J. Hazard. Mater., 2020, 388, 121814, DOI:10.1016/j.jhazmat.2019.121814.
- S. Afrin, M. d. K. Uddin and M. d. M. Rahman, Microplastics contamination in the soil from urban landfill site, Dhaka, Bangladesh, Heliyon, 2020, 6(11), e05572, DOI:10.1016/j.heliyon.2020.e05572.
- M. Scheurer and M. Bigalke, Microplastics in Swiss floodplain soils, Environ. Sci. Technol., 2018, 52(6), 3591–3598, DOI:10.1021/acs.est.7b06003.
- Q. Li, J. Wu, X. Zhao, X. Gu and R. Ji, Separation and identification of microplastics from soil and sewage sludge, Environ. Pollut., 2019, 254, 113076, DOI:10.1016/j.envpol.2019.113076.
- C. F. Araujo, M. M. Nolasco, A. M. P. Ribeiro and P. J. A. Ribeiro-Claro, Identification of microplastics using Raman spectroscopy: Latest developments and future prospects, Water Res., 2018, 142, 426–440, DOI:10.1016/j.watres.2018.05.060.
- Y. Luo, C. T. Gibson, C. Chuah, Y. Tang, R. Naidu and C. Fang, Applying Raman imaging to capture and identify microplastics and nanoplastics in the garden, J. Hazard. Mater., 2022, 426, 127788, DOI:10.1016/j.jhazmat.2021.127788.
- H. B. El, L. El Fels, K. Quénéa, M. F. Dignac, C. Rumpel and V. K. Gupta, et al., Microplastics from lagooning sludge to composts as revealed by fluorescent staining- image analysis, Raman spectroscopy and pyrolysis-GC/MS, J. Environ. Manage., 2020, 275, 111249, DOI:10.1016/j.jenvman.2020.111249.
- K. Chouchene, T. Nacci, F. Modugno, V. Castelvetro and M. Ksibi, Soil contamination by microplastics in relation to local agricultural development as revealed by FTIR, ICP-MS and pyrolysis-GC/MS, Environ. Pollut., 2022, 303, 119016, DOI:10.1016/j.envpol.2022.119016.
- Z. Sobhani, Y. Luo, C. Gibson, Y. Tang, R. Naidu and M. Megharaj, et al., Collecting microplastics in gardens: Case study (i) of soil, Toxicology, Pollution, and the Environment, 2021 DOI:10.3389/fenvs.2021.739775.
- M. Liu, S. Lu, Y. Song, L. Lei, J. Hu and W. Lv, et al., Microplastic and mesoplastic pollution in farmland soils in suburbs of Shanghai, China, Environ. Pollut., 2018, 242, 855–862, DOI:10.1016/j.envpol.2018.07.051.
- H. Fakour, S. L. Lo, N. T. Yoashi, A. M. Massao, N. N. Lema and F. B. Mkhontfo, et al., Quantification and analysis of microplastics in farmland soils: Characterization, sources, and pathways, Agriculture, 2021, 11(4), 330, DOI:10.3390/agriculture11040330.
- (a) S. Zhang, X. Yang, H. Gertsen, P. Peters, T. Salánki and V. Geissen, A simple method for the extraction and identification of light density microplastics from soil, Sci. Total Environ., 2018, 616–617, 1056–1065, DOI:10.1016/j.scitotenv.2017.10.213; (b) W. Jia, A. Karapetrova, M. Zhang, L. Xu, K. Li and M. Huang, et al., Automated identification and quantification of invisible microplastics in agricultural soils, Sci. Total Environ., 2022, 844, 156853, DOI:10.1016/j.scitotenv.2022.156853.
- X. Y. Jiang, Q. Y. Wang, Q. X. Mu and J. Hao, An Improved Iterative Polynomial Fitting Algorithm for Baseline Correction in X-Ray Spectrum, Adv. Sci. Technol., 2021 DOI:10.4028/www.scientific.net/ast.105.90.
- Z.-M. Zhang, S. Chen and Y.-Z. Liang, Baseline Correction Using Adaptive Iteratively Reweighted Penalized Least Squares, Analyst, 2010, 135(5), 1138, 10.1039/b922045c.
- Y. Hu, T. Jiang, A. Shen, W. Li, X. Wang and J. Hu, A Background Elimination Method Based on Wavelet Transform for Raman Spectra, Chemom. Intell. Lab. Syst., 2007, 85(1), 94–101, DOI:10.1016/j.chemolab.2006.05.004.
- Y. Luo, W. Su, X. Xu, D. Xu, Z. Wang, H. Wu, B. Chen and J. Wu, Raman Spectroscopy and Machine Learning for Microplastics Identification and Classification in Water Environments, IEEE J. Sel. Top. Quantum Electron., 2023, 29(4: Biophotonics), 1–8, DOI:10.1109/jstqe.2022.3222065.
- H. Wang, X. Chu, P. Chen, J. Li, D. Liu and Y. Xu, Partial Least Squares Regression Residual Extreme Learning Machine (PLSRR-ELM) Calibration Algorithm Applied in Fast Determination of Gasoline Octane Number with Near-Infrared Spectroscopy, Fuel, 2022, 309, 122224, DOI:10.1016/j.fuel.2021.122224.
- scikit learn. 1.4, Support Vector Machines — Scikit-Learn 0.20.3 Documentation. Scikit-learn.Org, https://scikit-learn.org/stable/modules/svm.html Search PubMed.
- IBM, What is the K-Nearest Neighbors Algorithm?, IBM, https://www.ibm.com/topics/knn#:~:text=The-k%2Dnearest-neighbors-(KNN Search PubMed.
- IBM, What Is Random Forest?, IBM, https://www.ibm.com/topics/random-forest Search PubMed.
- IBM, What Are Neural Networks?, https://www.ibm.com/topics/neural-networks Search PubMed.
- J. Yang, X. Wang, R. Wang and H. Wang, Combination of convolutional neural networks and recurrent neural networks for predicting soil properties using vis–NIR spectroscopy, Geoderma, 2020, 380, 114616, DOI:10.1016/j.geoderma.2020.114616.
- D. H. Wolpert and W. G. Macready, No Free Lunch Theorems for Optimization, IEEE Trans. Evol. Comput., 1997, 1(1), 67–82, DOI:10.1109/4235.585893.
- (a) S. B. Kontsiantis, Supervised Machine Learning: A Review of Classification Techniques, Informatica, 2007, 31, 249–268 Search PubMed; (b) B. Lei, J. R. Bissonnette, Ú. E. Hogan, A. E. Bec, X. Feng and R. Smith, Customizable machine-learning models for rapid microplastic identification using Raman microscopy, Anal. Chem., 2022, 94(49), 17011–17019, DOI:10.1021/acs.analchem.2c02451.
- Y. Luo, W. Su, X. Xu, D. Xu, Z. Wang, H. Wu, B. Chen and J. Wu, Raman Spectroscopy and Machine Learning for Microplastics Identification and Classification in Water Environments, IEEE J. Sel. Top. Quantum Electron., 2023, 29(4: Biophotonics), 1–8, DOI:10.1109/jstqe.2022.3222065.
- S. Ramanna, D. Morozovskii, S. Swanson and J. Bruneau, Machine Learning of polymer types from the spectral signature of Raman spectroscopy microplastics data, Adv. Artif. Intell. Mach. Learn., 2023, 03(01), 647–668, DOI:10.48550/arXiv.2201.05445.
- SLoPP and SLoPP-E Raman Spectral Libraries for Microplastics Research. plasticactioncentre.ca, Available from: https://plasticactioncentre.ca/directory/slopp-and-slopp-e-raman-spectral-libraries-for-microplastics-research/ Search PubMed.
- M. Dong, Q. Zhang, X. Xing, W. Chen, Z. She and Z. Luo, A Raman Database of Microplastics Weathered under Natural Environments. Datamendeley.Com, 2020, vol. 2, https://data.mendeley.com/datasets/kpygrf9fg6/2 Search PubMed.
- L. Ren, S. Liu, S. Huang, Q. Wang, Y. Lu, J. Song and J. Guo, Identification of Microplastics Using a Convolutional Neural Network Based on Micro-Raman Spectroscopy, Talanta, 2023, 124611, DOI:10.1016/j.talanta.2023.124611.
- F. Weber, A. Zinnen and J. Kerpen, Development of a machine learning-based method for the analysis of microplastics in environmental samples using μ-Raman spectroscopy, Microplastics and Nanoplastics, 2023, 3(9) DOI:10.1186/s43591-023-00057-3.
- F. Li, D. Li, X. Guo, Z. Zhang, F. Martin and A. Lu, et al., Identification and visualization of environmental microplastics by Raman imaging based on hyperspectral unmixing coupled machine learning, J. Hazard. Mater., 2024, 465, 133336, DOI:10.1016/j.jhazmat.2023.133336.
- A. ArulMurugan, T. H. Chan, W. K. Ma and C. Y. Chi, A Robust Alternating Volume Maximization Algorithm for Endmember Extraction in Hyperspectral Images, Institute of Electrical and Electronics Engineers, 2010 Search PubMed.
- E. Choi, Y. Choi, H. Lee, J. Kim and O. Han Bin, Development of a machine-learning model for microplastic analysis in an FT-IR microscopy image, Bull. Korean Chem. Soc., 2024, 45(5), 379–481, DOI:10.1002/bkcs.12835.
- H. H. Zou, P. J. He, W. Peng, D. Y. Lan, H. Y. Xian and L. ü Fan, et al., Rapid detection of colored and colorless macro- and micro-plastics in complex environment via near-infrared spectroscopy and machine learning, J. Environ. Sci., 2023, 147, 512–522, DOI:10.1016/j.jes.2023.12.004.
- J. Shan, J. Zhao, L. Liu, Y. Zhang, X. Wang and F. Wu, A novel way to rapidly monitor microplastics in soil by hyperspectral imaging technology and chemometrics, Environ. Pollut., 2018, 238, 121–129, DOI:10.1016/j.envpol.2018.03.026.
- A. Paul, L. Wander, R. Becker, C. Goedecke and U. Braun, High-throughput NIR spectroscopic (NIRS) detection of microplastics in soil, Environ. Sci. Pollut. Res., 2018, 26(8), 7364–7374, DOI:10.1007/s11356-018-2180-2.
- H. Chen, T. Shin, B. Park, K. Ro, C. Jeong and H. Jeon, et al., Coupling hyperspectral imaging with machine learning algorithms for detecting polyethylene (PE) and polyamide (PA) in soils, J. Hazard. Mater., 2024, 471, 134346, DOI:10.1016/j.jhazmat.2024.134346.
- X. Tian, F. Been and P. S. Bäuerlein, Quantum Cascade Laser Imaging (LDIR) and Machine Learning for the Identification of Environmentally Exposed Microplastics and Polymers, Environ. Res., 2022, 212, 113569, DOI:10.1016/j.envres.2022.113569.
- B. Hufnagl, M. Stibi, H. Martirosyan, U. Wilczek, J. N. Möller and M. G. J. Löder, et al., Computer-assisted analysis of microplastics in environmental samples based on μFTIR Imaging in combination with machine learning, Environ. Sci. Technol. Lett., 2021, 9(1), 90–95, DOI:10.1021/acs.estlett.1c00851.
- B. Shi, M. Patel, D. Yu, J. Yan, Z. Li and D. Petriw, et al., Automatic quantification and classification of microplastics in scanning electron micrographs via deep learning, Sci. Total Environ., 2022, 825, 153903, DOI:10.1016/j.scitotenv.2022.153903.
- C. Massarelli, C. Campanale and V. F. Uricchio, A handy open-source application based on computer vision and machine learning algorithms to count and classify microplastics, Water, 2021, 13(15), 2104, DOI:10.3390/w13152104.
- S. Tan, J. A. Taylor and E. Passeport, Efficient prediction of microplastic counts from mass measurements, Environ. Sci. Technol., 2022, 2(2), 299–308, DOI:10.1021/acsestwater.1c00316.
- 1.1.3. Kernel Ridge Regression. Scikit-Learn, Available from: https://scikit-learn.org/stable/modules/kernel_ridge.html Search PubMed.
- W. Ng, B. Minasny and A. McBratney, Convolutional neural network for soil microplastic contamination screening using infrared spectroscopy, Sci. Total Environ., 2020, 702, 134723, DOI:10.1016/j.scitotenv.2019.134723.
- R. Wu, L. Hao, H. Tian, J. Liu, C. Dong and J. Xue, Qualitative Discrimination and Quantitative Prediction of Microplastics in Ash Based on Near-Infrared Spectroscopy, J. Hazard. Mater., 2024, 469, 133971, DOI:10.1016/j.jhazmat.2024.133971.
- J. Lorenzo-Navarro, M. Castrillon-Santana, E. Santesarti, M. De Marsico, I. Martinez and E. Raymond, et al., SMACC: A system for microplastics automatic counting and classification, IEEE Access, 2020, 8, 25249–25261 Search PubMed.
- H. T. Tran, M. Hadi, T. Thu, H. G. Hoang, M. K. Nguyen and K. N. Nguyen, et al., Machine learning approaches for predicting microplastic pollution in peatland areas, Mar. Pollut. Bull., 2023, 194, 115417, DOI:10.1016/j.marpolbul.2023.115417.
- R. Mansa and S. Zou, Thermogravimetric analysis of microplastics: A mini review, Environ. Adv., 2021, 5, 100117, DOI:10.1016/j.envadv.2021.100117.
- J. Yu, P. Wang, F. Ni, J. Cizdziel, D. Wu and Q. Zhao, et al., Characterization of microplastics in environment by thermal gravimetric analysis coupled with Fourier transform infrared spectroscopy, Mar. Pollut. Bull., 2019, 145, 153–160, DOI:10.1016/j.marpolbul.2019.05.037.
- F. Blancho, M. Davranche, H. E. Hadri, B. Grassl and J. Gigault, Nanoplastics identification in complex environmental matrices: Strategies for polystyrene and polypropylene, Environ. Sci. Technol., 2021, 55(13), 8753–8759, DOI:10.1021/acs.est.1c01351.
- N. Yakovenko, A. Carvalho and A. ter Halle, Emerging use thermo-analytical method coupled with mass spectrometry for the quantification of micro(nano)plastics in environmental samples, TrAC, Trends Anal. Chem., 2020, 131, 115979, DOI:10.1016/j.trac.2020.115979.
- E. S. Meredith and J. M. Lynch, Previous successes and untapped potential of pyrolysis–GC/MS for the analysis of plastic pollution, Anal. Bioanal. Chem., 2023, 415, 2873–2890, DOI:10.1007/s00216-023-04671-1.
- T. Chowdhury and Q. Wang, Study on thermal degradation processes of polyethylene terephthalate microplastics using the kinetics and artificial neural networks models, Processes, 2023, 11(2), 496, DOI:10.3390/pr11020496.
- E. E. Christian, E. O. Prosper, H. R. Mominul, Q. Wang and A. T. Mohammad, Thermal degradation evaluation of polyethylene terephthalate microplastics: Insights from kinetics and machine learning algorithms using non-isoconversional TGA data, J. Environ. Chem. Eng., 2024, 12(2), 111909, DOI:10.1016/j.jece.2024.111909.
- J. Zhang, T. Yuan, J. Wen and Q. Zhang, Machine learning assisted identification of polymers in coated paper using pyrolysis gas chromatography mass spectrometry, SSRN, 2023 DOI:10.2139/ssrn.4564903.
- T. P. Forbes, J. M. Pettibone, E. Windsor, J. M. Conny and R. A. Fletcher, Rapid chemical screening of microplastics and nanoplastics by thermal desorption and pyrolysis mass spectrometry with unsupervised fuzzy clustering, Anal. Chem., 2023, 95(33), 12373–12382, DOI:10.1021/acs.analchem.3c01897.
- P. Guo, Y. Wang, M. Parastoo, W. Meng, S. Wu and Y. Bao, Artificial intelligence-empowered collection and characterization of microplastics: A review, J. Hazard. Mater., 2024, 471, 134405, DOI:10.1016/j.jhazmat.2024.134405.
| This journal is © The Royal Society of Chemistry 2025 |