Open Access Article
Open Access ArticleEffects of acidic and alkaline electrolyzed water treatments on the volatilomics and proteomics changes in fresh-cut apple during storage†
Zinash A.
Belay
*ab,
Mbukeni
Nkomo
b,
Gadija
Mohamed
bc,
Makgafele L.
Ntsoane
 d and
Oluwafemi James
Caleb
d and
Oluwafemi James
Caleb
 *ace
*ace
aDepartment of Food Science, Faculty of AgriSciences, Stellenbosch University, Matieland 7602, South Africa. E-mail: caleboj@sun.ac.za
bPost-Harvest and Agro-Processing Technologies (PHATs), Agricultural Research Council (ARC) Infruitec-Nietvoorbij, Private Bag X5026, Stellenbosch 7599, South Africa. E-mail: BelayZ@arc.agric.za
cDepartment of Biotechnology, Life Science Building, University of the Western Cape, Robert Sobukwe Road, Bellville 7530, South Africa
dPostharvest Technology, Agricultural Research Council, Tropical and Subtropical Crops, Private Bag X11208, Nelspruit 1200, South Africa
eAgriFood BioSystems and Technovation Research Group, African Institute for Postharvest Technology, Faculty of AgriSciences, Stellenbosch University, Matieland 7602, South Africa
First published on 8th January 2025
Abstract
Electrolyzed water (EW) has shown potential to decontaminate and maintain the quality of fresh-cut apple; however, the underlying response of the product to this treatment remains unclear. Thus, this study aims to identify the possible quality regulation mechanisms of acidic electrolyzed water (AEW) and alkaline electrolyzed water (ALEW) treatments on fresh-cut ‘Granny Smith’ apples via volatile organic compound (VOC) and qualitative proteomics analysis during storage at 2 °C for 10 days. The results identified 43 VOCs, including 10 esters, 9 alcohols, 9 alkanes, 8 carboxylic acids, 6 ketones, and 1 aldehyde. The distribution of VOCs was significantly affected by the pretreatment conditions; fresh-cut apple treated with AEW was characterised by the highest number of esters, alcohols, and carboxylic acids, whereas samples treated with ALEW exhibited predominantly carboxylic acids, alcohols, and alkanes in comparison to control (untreated) samples. Ethyl dodecanoate, which was identified only in the ALEW samples on each sampling day, had the highest concentration among all the individual VOCs. The proteomics results showed that a total of 3434, 3401, and 3313 proteins were identified on day 3, 6, and 10, respectively, across all samples. Until day 6 of storage, no significant differences were observed among the samples. Notably, on day 6, “M16C_associated domain-containing protein” was shown to be unique to the control samples. KH type-2 domain-containing protein, methylenetetrahydrofolate reductase (MTHFR), and 1,4-alpha-glucan branching enzyme were unique proteins identified after AEW treatment at day 6 and 10 of storage. No unique protein was identified for the ALEW samples. These results provide the first report of the proteomic and volatilomic changes associated with EW-treated fresh-cut apple during storage. Data are available via ProteomeXchange with identifier PXD056621.
Sustainability spotlightElectrolyzed water (EW) treatment has emerged as a novel decontaminating washing step for fresh and fresh-cut horticultural produce and an effective alternative to chlorine-based solutions. Compared to conventional chemical sanitizing agents, electrolyzed water is considered to be an environmentally friendly, efficacious antimicrobial agent that is safe and capable of extending the storability or shelf-life of fresh and fresh-cut horticultural produce. These are critical factors that make a case for the sustainability of EW and its application, which ensures extended access to safe and nutritious ready-to-eat produce, thereby contributing to SDG #2 Zero Hunger. Therefore, understanding the underlying responses of fresh-cut apples to EW treatment as a case study will be critical to further optimization strategies for other fresh-cut produce. |
1 Introduction
Electrolyzed water (EW) treatment of fresh-cut fruit has been gaining considerable attention due to its effective decontamination and disinfection potential with minimal or no determinate effects on the overall quality of the fruit or on human health and the environment. Electrolyzed water is produced by mixing an electrolyte containing chlorine, generally sodium chlorite (NaCl) and/or hydrochloric acid (HCL), with tap water through an electrolyte cell.1 The production conditions of EW can produce three different types of EW, namely, acidic, alkaline, and neutral EW, which have different end-product pH, available chlorine concentration (ACC), and oxidation–reduction potential (ORP). Acidic electrolyzed water (AEW) produced on the anode side is charactered by low pH (2.3 to 2.7), high ORP (>1000 mV) and ACC (20 to 200 mg L−1) and the presence of hypochlorous acid and hypochlorite ions, whereas alkaline electrolyzed water (ALEW), which is generated on the cathode side, is charactered by high pH (11.0 to 13.5), low ORP (−800 to −900 mV) and ACC (20–60) and the presence of sodium hydroxide. On the other hand, neutral electrolyzed water (NEW) is generated without separation of the anode and the cathode side by a membrane, and has a pH of 5.0 to 8.5 and an ORP of 500–700 mV. The antimicrobial activity of EW is due to its high content of HOCl, high oxidation–reduction potential (ORP) and various pH values, which lead to oxidative damage to biomolecules.2Various studies have demonstrated the potential of different EWs as an alternative to sodium hypochlorite for treatment of fresh-cut apple. Gao et al.3 demonstrated the strong bactericidal effects and quality retention effects of 5 min of treatment with slightly acidic electrolyzed water (ACC = 21 mg L−1, pH = 6) on fresh-cut apple during storage at 4 °C. Similarly, Graça et al.4 reported the inactivation potential of AEW and NEW against various yeasts (Candida sake, Metschnikowia pulcherrima, Pichia fermentation, and Hanseniaspora uvarum) on fresh-cut apple (cv. Royal Gala) stored at 4 °C for 9 days. Furthermore, Plesoianu et al.5 demonstrated significant retention of firmness, total phenolic content, and antioxidant activity as well as reduced browning for fresh-cut apple (cv. Florina and Ionathan) treated with AEW during 14 days of storage at 8 °C. The preparation of fresh-cut apple impacts the structural integrity of the apple cells, resulting in the intercellular disruption that can lead to loss of nutrients and water, softening, and acceleration of decay and deterioration.5,6 Furthermore, both postharvest treatments and minimal processing of the fruit can induce the production of secondary metabolites and disrupt the biological, physiological responses and defence systems in fruit.7,8
Volatile compounds in apples mostly include esters, alcohols, aldehydes, ketones, and ethers. The concentrations and compositions of these VOCs differ in different cultivars; their production is also affected by several pre- and postharvest factors. Moreover, the biosynthesis of these compounds involves various metabolic pathways, and they are the main products of fatty acids and amino acids.9 Similarly, proteomic tools are efficient to understand structural and quantitative information related to the dynamics of all cellular proteins and the functional state of the cell. Available literature on proteomic studies for apples have mainly focused on the changes in the apples (cv. Ambrosia) due to delayed cooling during storage, changes associated with pre- and postharvest 1-MCP treatment on the quality of apple (cv. Honeycrisp), and the anti-browning mechanism of selenium in fresh-cut apple (cv. Fiji).10–12 In addition, studies of the EW treatment of apple have mainly focused on whole fruit,1,13 in addition to recent studies on the hurdle effect of EW on fresh-cut apple to preserve their quality.3,5,14 Despite these research efforts, more studies of the fundamental basis of EW treatments are required to provide an in-depth understanding of its potential to maintain quality. This study aims to investigate the effects of acidic and alkaline electrolyzed water treatments and storage duration on the changes in the volatile organic compounds (VOCs) and quantitative protein response. The goal is to identify significantly regulated proteins and metabolic pathways.
2 Materials and methods
2.1 Fruit material
Apple fruit (Malus domestica cv. Granny Smith) was harvested at commercial maturity at the Agricultural Research Council (ARC) Elgin Research Farm, Grabouw, South Africa. The harvest maturity index of the fruit was based on its total soluble solids (TSS) content of 10.7 ± 0.17 °Brix and titratable acidity of 0.9 ± 0.04 g 100 mL−1. Harvested fruit were then transported under cool conditions (4 °C) in well refrigerated trucks to the Agri-Food Systems and Omics Laboratory, ARC Infruitec-Nietvoorbij, Stellenbosch, South Africa. Only healthy fruit of uniform size and colour with no physical injuries or infection were selected for this experiment and were stored at 0.5 °C and 95% RH for further processing.2.2 Preparation of electrolyzed water
Electrolyzed water (EW) was generated by the electrolysis of NaCl solution using the ELA-12 000ANW system (ECA Technologies, Envirolyte, South Africa). The pH and ORP were determined immediately using a pH meter (D-22, Horiba, Kyoto, Japan) and ORP meter (HM-60V, TOA Electronics Ltd, Tokyo, Japan). For this study, two types of EW with different pH values and ORP levels were generated. The acidic EW (AEW) had an ACC of 500 mg L−1, a pH of 2–3 and an ORP of >1000 mV. The alkaline EW (ALEW) had an ACC of 500 mg L−1, an ORP of >−900 mV and a pH of 11–13. The ACC for both EWs were adjusted to 200 ppm. The ACC and ORP were obtained using an ORP meter (HM-60V, TOA Electronics Ltd, Tokyo, Japan) and pH meter (D-22, Horiba, Kyoto, Japan).2.3 Fruit treatment
The ‘Granny Smith’ apple fruit were cut into slices using a mechanical slicer, separated randomly into three groups, and dipped into the following treatments for 10 min in triplicate: (1) control group (no treatment), (2) AEW (fruit treated with acidic EW), and (3) ALEW (fruit treated with alkaline EW). The sliced apple was immersed in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (w/v) in the respective treatment conditions at 15 °C. The samples were then packed in rigid polypropylene containers with lids and stored in a cold atmosphere (2 °C and 95% RH) for 10 days. Sampling for secondary metabolites and proteomics analysis was conducted on day 0, 3, 6 and 9. On each sampling day, packs were randomly pulled out from the cold storage, immediately immersed in liquid nitrogen, and stored at −20 °C until further analysis.
5 (w/v) in the respective treatment conditions at 15 °C. The samples were then packed in rigid polypropylene containers with lids and stored in a cold atmosphere (2 °C and 95% RH) for 10 days. Sampling for secondary metabolites and proteomics analysis was conducted on day 0, 3, 6 and 9. On each sampling day, packs were randomly pulled out from the cold storage, immediately immersed in liquid nitrogen, and stored at −20 °C until further analysis.
2.4 Volatile organic compounds
To quantify the relative abundance of volatile organic compounds in the fresh-cut ‘Granny Smith’ apple, apple aliquots of approximately 5 mL were pipetted into 20 mL solid-phase micro-extraction (SPME) vials and hermetically sealed. Each vial was allowed to equilibrate for 5 min at 50 °C in the CTC auto-sampler incubator at 250 rpm. Volatile compounds trapped in the headspace of the vials were extracted using the static headspace solid-phase micro-extraction (SHS-SPME) technique as previously described by Martínez et al.15 Subsequently, an SPME fibre (50/30 μm) coated with divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) was automatically inserted into the sample headspace for 10 min at 50 °C. After volatile extraction from the SHS of the vials, desorption of the adsorbed compounds from the fibre coating was carried out in the injection port of the gas chromatography-mass spectrometry (GC-MS) instrument for 10 min. The injector temperature was maintained at 250 °C. Separation and quantification of the volatile compounds was performed using an Agilent 6890 N GC (Agilent, Pablo Alto, CA, USA) coupled with an MS-detector Agilent 5975B MS (Agilent, Palo Alto, CA, USA). Separation of the volatiles was performed on a DB-WaxEtr (30 m, 0.25 mm ID, 0.25 μm film thickness) capillary column (Agilent Technologies Inc., Palo Alto, CA, USA). Helium was used as the carrier gas for these analyses at a flow of 1 mL min−1 with a nominal initial pressure of 196 kPa and an average velocity of 36 cm s−1.The oven temperature program was set as follows: 40 °C for 6 min, thereafter ramped to 260 °C at a rate of 8 °C min−1, and held for 3 min. The MSD was operated in full scan mode, and the ion source as well as the quadrupole temperature were maintained at 230 °C and 150 °C, respectively. The transfer line temperature was maintained at 250 °C. The mass spectrometer was operated in electron impact (EI) mode at an ionization energy of 70 eV, scanning from 30 to 700 m/z. Compounds were identified by comparison of their retention time (RT) and retention index (RI) with those registered in the National Institute of Standards and Technology (NIST v.05, Gaithersburg, MD, USA) and the WHILEY 275 mass spectral libraries with similarity above 90%. Only compounds that were consistently identified in all treated and control samples were considered for analysis.
2.5 Proteomics analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 3 min at 4 °C. The resultant pellet was rinsed with 2 mL of cold 80% acetone containing 5 mM dithiothreitol (DTT) and centrifuged for 3 min at 16
000×g for 3 min at 4 °C. The resultant pellet was rinsed with 2 mL of cold 80% acetone containing 5 mM dithiothreitol (DTT) and centrifuged for 3 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g at 4 °C. The acetone rinse steps were repeated three times until a white pellet was obtained. The protein pellet was air-dried at room temperature and further used for protein extraction.
000×g at 4 °C. The acetone rinse steps were repeated three times until a white pellet was obtained. The protein pellet was air-dried at room temperature and further used for protein extraction.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 30 min at 4 °C, and the resulting pellet was washed once with cold methanol, followed by an 80% cold acetone wash. The sample matrix can interfere with analysis, decreasing both accuracy and sensitivity. This can lead to false positives or negatives and the disruption of the injector or separation column. Hence, the additional cleanup step is crucial.
000×g for 30 min at 4 °C, and the resulting pellet was washed once with cold methanol, followed by an 80% cold acetone wash. The sample matrix can interfere with analysis, decreasing both accuracy and sensitivity. This can lead to false positives or negatives and the disruption of the injector or separation column. Hence, the additional cleanup step is crucial.
The final pellet was partially air-dried and redissolved in 30 μL protein solubilization buffer (4 M urea, 2% SDS, 50 mM Tris–HCl, pH 8.0). Samples were thoroughly vortexed for 15 min at RT prior to centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 5 min at 4 °C. The protein concentration of the resultant supernatant (total soluble proteins) was determined using the Pierce microplate BCA protein assay kit (Thermo Scientific, Rockford, IL, USA) according to the manufacturer's instructions with bovine serum albumin used as a standard.
000×g for 5 min at 4 °C. The protein concentration of the resultant supernatant (total soluble proteins) was determined using the Pierce microplate BCA protein assay kit (Thermo Scientific, Rockford, IL, USA) according to the manufacturer's instructions with bovine serum albumin used as a standard.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio with 4× Laemmli sample buffer (250 mM Tris–HCl, pH 6.8; 4% SDS; 30% glycerol; 350 mM β-mercaptoethanol; 0.02% bromophenol blue) and boiled at 70 °C for 10 min. The proteins were then resolved according to their molecular weight on 12% polyacrylamide gels under a constant 100 V with the aid of the Mini-Protean III® Cell gel casting system (Bio-Rad Laboratories Ltd, Rosebank, Johannesburg, South Africa) until the bromophenol blue reached the bottom of the gel. After electrophoresis, the proteins were visualized using the Acqua Stain, and the gels were captured using Quantity One software on the Molecular Imager PharosFX Plus System (Bio-Rad Laboratories Ltd, Rosebank, Johannesburg, South Africa).
3 ratio with 4× Laemmli sample buffer (250 mM Tris–HCl, pH 6.8; 4% SDS; 30% glycerol; 350 mM β-mercaptoethanol; 0.02% bromophenol blue) and boiled at 70 °C for 10 min. The proteins were then resolved according to their molecular weight on 12% polyacrylamide gels under a constant 100 V with the aid of the Mini-Protean III® Cell gel casting system (Bio-Rad Laboratories Ltd, Rosebank, Johannesburg, South Africa) until the bromophenol blue reached the bottom of the gel. After electrophoresis, the proteins were visualized using the Acqua Stain, and the gels were captured using Quantity One software on the Molecular Imager PharosFX Plus System (Bio-Rad Laboratories Ltd, Rosebank, Johannesburg, South Africa).
The protein samples (50 μg) were resuspended in 50 mM ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA) before reduction with 10 mM dithiothreitol (DTT) (Sigma) for 30 min at room temperature. The reduced proteins were further alkylated with 30 mM iodoacetamide at room temperature in the dark for 30 min and diluted with an equal volume of binding buffer (200 mM ammonium acetate, pH 4.5, 30% acetonitrile). The protein solution was added to pre-equilibrated MagResyn HILIC magnetic beads (Resyn Biosciences (Pty), Ltd Gauteng, South Africa) prepared according to the manufacturer's instructions and incubated for 16 h at 4 °C. The magnetic beads (with bound protein) were then placed in a magnetic rack and the supernatant was removed. The protein-bound magnetic beads were then washed two times with 200 μL of 95% acetonitrile before resuspension in 50 mM ammonium bicarbonate containing sequencing-grade modified trypsin (New England Biolabs®, Ipswich, UK) to a final enzyme–substrate ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. Following digestion at 37 °C for 16 h, the beads were placed in a magnetic rack, and the supernatants containing tryptic peptides were transferred to new tubes and acidified at a final concentration of 0.5% (v/v) trifluoroacetic acid (TFA). Residual digestion reagents were removed from the peptide samples using custom laboratory-made StageTips prepared from Octadecyl C18 solid-phase extraction disks (Empore™, 66883-U). Eluted peptides were evaporated to dryness in a speed vacuum and conserved at −20 °C until further processing.
50. Following digestion at 37 °C for 16 h, the beads were placed in a magnetic rack, and the supernatants containing tryptic peptides were transferred to new tubes and acidified at a final concentration of 0.5% (v/v) trifluoroacetic acid (TFA). Residual digestion reagents were removed from the peptide samples using custom laboratory-made StageTips prepared from Octadecyl C18 solid-phase extraction disks (Empore™, 66883-U). Eluted peptides were evaporated to dryness in a speed vacuum and conserved at −20 °C until further processing.
Peptide and protein validation were carried out using the Peptide and Protein Prophet algorithms. Protein fold changes between the experimental conditions and the baseline were calculated using Student's t-test. Proteins were deemed significant if they exhibited a fold change greater than 2 or a p-value of <0.05. Significant proteins were highlighted to differentiate between upregulated and downregulated features, and the plot was annotated to emphasize key significant proteins, aiding in their biological interpretation and subsequent analysis. For positive protein identification, a minimum of two unique peptides, a protein identification probability of at least 95%, and a percentage sequence coverage greater than zero was used. The proteins identified under each condition were compiled using the FunRich Multi-Analysis software package (version 3.1.3) to identify common and unique proteins for each treatment.
2.6 Statistical analyses
For volatile organic compounds, multiple factor analysis (MFA) was conducted using XLSTAT and Metabo Analysi was used to discriminate the significant VOCs between different treatment types and storage durations. The data set was further subjected to principal component analysis (PCA) to find the distribution features of the data. Furthermore, the partial least discriminant analysis (PLS-DA) was employed to determine the variable importance in projection (VIP) of the significant VOCs. Significant differences between the treatment types were evaluated by p values of ≤0.05 and VIP values of >1.5. For the volcano plot analysis, the proteomics data from both baseline and experimental conditions were normalized. The volcano plot was generated using software Prism (ver. 10). This transformation facilitates a clear visualization of both the magnitude and significance of changes.Venn diagrams and volcano plots were employed to visualize and interpret complex proteomics data. For positive protein identification, we established the following criteria: a minimum of two unique peptides, a protein identification probability of at least 95%, and greater than zero percentage sequence coverage. These criteria facilitated the elucidation of relationships between protein sets by highlighting overlaps among significantly altered proteins under various experimental conditions compared to baseline samples. Proteins identified across conditions (base, control, KCN treatment, and NaCl treatment) were compiled using the FunRich Multi-Analysis software package (version 3.1.3) to identify common and unique proteins for each treatment, resulting in distinct lists for base vs. control, base vs. KCN, and base vs. NaCl. For the volcano plot analysis, we normalized proteomics data from baseline and experimental conditions to correct for systematic biases. We calculated the fold change of each protein between the experimental conditions (control, KCN treatment, and NaCl treatment) and the baseline, assessing statistical significance using Student's t-test, which provided p-values for each protein. The volcano plot was generated using the latest version of the software package Prism (version 10), displaying log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-transformed fold change on the x-axis and negative log
2-transformed fold change on the x-axis and negative log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10-transformed p-values on the y-axis. This transformation enabled clear visualization of both the magnitude and significance of changes. Proteins were considered significant if they exhibited a fold change greater than 2 or less than 0.5, with a p-value below 0.05. Significant proteins were highlighted to distinguish between upregulated and downregulated features, and the plot was annotated to emphasize key significant proteins, aiding in their biological interpretation and subsequent analysis.
10-transformed p-values on the y-axis. This transformation enabled clear visualization of both the magnitude and significance of changes. Proteins were considered significant if they exhibited a fold change greater than 2 or less than 0.5, with a p-value below 0.05. Significant proteins were highlighted to distinguish between upregulated and downregulated features, and the plot was annotated to emphasize key significant proteins, aiding in their biological interpretation and subsequent analysis.
3 Results and discussion
3.1 Volatile organic compounds
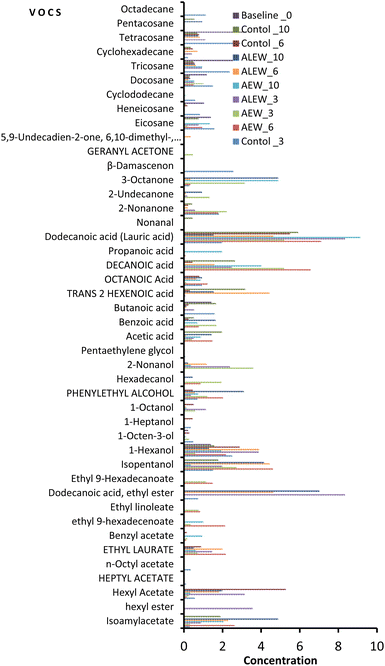
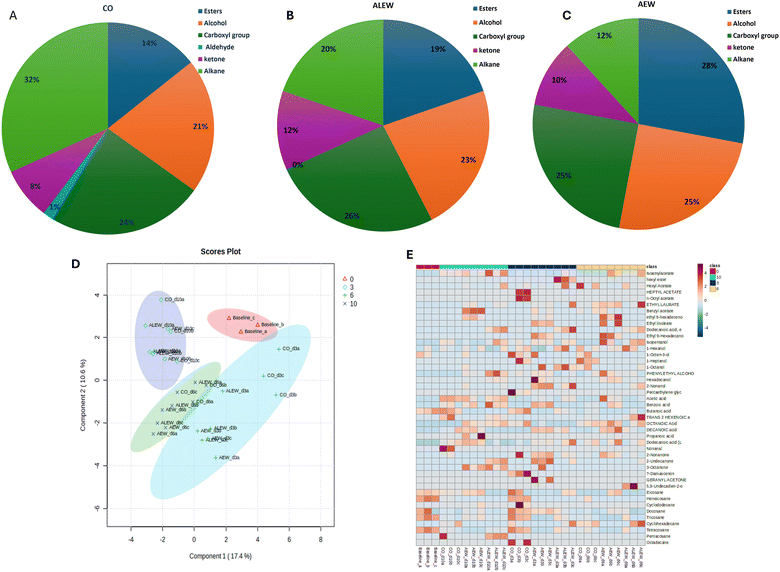
The analysis of VOCs showed that the aromatic profile of fresh-cut apple changed according to the treatment type and storage duration (Fig. 1). In total, 43 compounds including 10 esters, 9 alcohols, 9 alkanes, 8 carboxylic acids, 6 ketones, and 1 aldehyde were identified. In the baseline analysis, only alkane (eicosane, heneicosane, docosane, tricosane and tetracosane), alcoholic (1-hexanol and 1-octen-3-ol), and carboxylic (butanoic acid) VOCs were identified. From this analysis, it was evident that alkanes predominated compared to other VOCs at the beginning of the study.However, a significant number of VOCs were identified with continued storage at day 3, with 25 compounds being identified in the control and AEW samples and 22 compounds being identified for the ALEW samples. On day 6, the AEW samples had the highest number of ester compounds (33%) compared to the control and ALEW samples. In fact, the number of esters in the AEW samples was consistently higher on each sampling day. Similarly, AEW had the highest count (28%) of alcohols on day 10, whereas the control samples had the highest number of alkanes throughout the study. By the end of storage on day 10, the number of identified VOCs compounds had decreased slightly, reaching 18 in the control samples, 22 in the AEW samples and 21 in the ALEW samples. Relatively, dodecanoic acid (lauric acid) consistently had the highest concentration among all the identified VOCs, being present in all treated and control samples throughout the storage period. However, ethyl dodecanoate, which was identified only in the ALEW samples on each sampling day, had the highest concentration of all the VOCs. In general, for the control samples, the dominant VOCs at the end of storage were alkanes, whereas the treated samples exhibited more alcohols and esters. Under normal maturity and ripening conditions, the profile of apple volatile compounds at the beginning is predominated by aldehydes and with increasing maturation the profile changes to alcohols, whereas esters predominate at end.17 In the current study, the trend in the normal synthesis of volatile organic compounds was observed in the fruit treated with AEW.
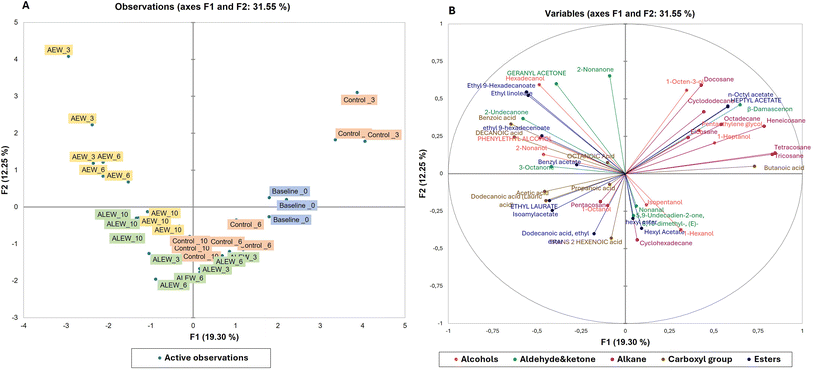
Principal component analysis was used to demonstrate the relationship between the volatile organic compounds and the treatment conditions in fresh-cut apple fruit (Fig. 2). As presented in the score plot (Fig. 2A), the AEW samples moved negatively along the PC2 axis and positively along the PC1 axis with longer storage, whereas the ALEW samples moved positively along the PC2 axis and negatively along the PC1 axis with longer storage duration.
Fig. 2B presents the biplot for the first two principal components, which demonstrates that the EW treatments influenced the emission of diverse VOCs. The PCA accounts for 33% of the total variance in the dataset; specifically, PC1 explained 12.5% and PC2 explained 19.3% of the variance. As can be seen in Fig. 2A, for the first component, the baseline and control samples (on day 3), and for the second component, the AEW samples (day 3 and day 6), were distributed in the positive region. For the first component, the control (day 6) and ALEW (day 6) samples, and for the second component, the ALEW (day 10) and (AEW day 10) samples were distributed in the negative region. According to the biplot result (Fig. 2B), the VOCs in the positive region of the first component were mostly alkanes (docosane, cyclododecane), an alcohol (1-octen-3-ol), esters (n-octyl acetate, heptyl acetate), aldehyde and ketones (β-damascenone) that can be related to the VOCs in the baseline and control samples.
Comparing the concentrations of the emitted VOCs, overall, carboxylic acids and alcohols predominated, while aldehydes and ketones remained very low throughout storage in all samples. In general, the control fruit was characterized by the highest concentration of alkanes, followed by carboxylic acids, alcohols and a lower number of esters and ketones. In contrast, the fruit treated with AEW showed the highest number of esters, alcohols, and carboxylic acids but the lowest number of alkane VOCs (Fig. 3A and B). One the other hand, fruit treated with ALEW exhibited a relatively average number of VOCs, predominantly carboxylic acids, alcohols, and alkanes (Fig. 3C). The observed VOC profile corresponds to the microbial growth pattern in different samples (data not shown). Higher microbial growth is evident in the control samples due to the presence of high alkanes in the control sample, as alkanes are indicative of an active microbial community involved in the breakdown of complex substrates that increase microbial biomass. In contrast, AEW resulted in significantly lower microbial counts, which correlates with the greater presence of ester, alcohol, and carboxylic acid VOCs. These VOCs could have exhibited antimicrobial properties, contributing to the reduction in yeast, mold, and aerobic mesophilic bacteria counts in the AEW-treated samples.
The positive region of the second component contains compounds such as 2-nonanone, hexadecanol, ethyl-9-hexadecanoate and ethyl linoleate, which are associated with esters and alcohols.18 In general, the compounds presented in the biplot are closely related to the treatment condition applied and could be used as VOC biomarkers for discriminating among the treatment conditions and storage time in fresh-cut apple. Moreover, PLS-DA calculations were performed on the secondary metabolite data between the control and treated fruit during storage, as shown in Fig. 3D; PCA1 accounted for 15% of the variance and PCA2 accounted for 15%. Separation between samples over the course of storage was observed, with slight overlapping between the VOCs on day 3 and 6, showing the influence of the storage duration on the emission of different VOCs in addition to the treatments. The VOC heat map (Fig. 3E) for the treatment and control samples during storage was obtained after normalization in the program MetaboAnalyst (ESI Fig. 1†).
3.1.1.1 Ester volatile organic compounds. At the beginning of the study, no ester compounds were detected in either the treated or control samples. Ester compounds are known for their ‘fruity’ note and result from esterification reactions; their low abundance at the beginning suggested a low level of esterification. However, it became clear that both storage duration and the EW treatments had a significant impact on ester compounds during storage. The highest number of esters was identified in samples treated with AEW, followed by those treated with ALEW and then the control (untreated). The observed substantial increase of esters after 3 days of storage mainly indicated fruity characteristics in the fresh-cut apples.
Ester VOCs such as isoamylacetate, hexyl acetate and ethyl laurate were the most abundant esters in all treated and controlled samples during storage. Other VOCs, such as heptyl acetate and n-octyl acetate, were only identified in the control samples at day 3, whereas ethyl 9-hexadecenoate and ethyl linoleate were only identified in AEW samples throughout storage. On the other hand, hexyl ester was only emitted for the ALEW samples on day 3. Hexyl acetate is an important volatile organic compound that gives apples their characteristic flavor. It is emitted though the LOX pathway from hexanol by the enzyme alcohol acyltransferase (AAT).19 Alcohol acyltransferase (AAT) catalyzes the last step of volatile ester biosynthesis. In this study, considerably high concentrations of hexyl acetate were observed for both control and treated fresh-cut apple during storage; however, its concentration was below the detection limit for the control samples at the end of storage. In general, the lowest concentration of ester was observed for the control samples throughout storage compared with the treated samples, which could be due to low activity of AAT in the control samples. According to Defilippi et al.,20 AAT catalyzes the final linkage of an acyl moiety and an alcohol to form esters and is thus directly responsible for producing esters.
The ALEW treatment significantly inhibited the emission of benzyl acetate throughout the storage. Benzyl acetate is responsible for the sweet and pleasant aroma of the fruit. Its disappearance after ALEW treatment could affect the aroma of the fruit. However, a high concentration of benzyl acetate was observed for the AEW samples during storage. Furthermore, of all ester compounds, the highest concentration was observed for dodecanoic acid, ethyl ester (ethyl dodecanoate), and it was identified only under ALEW for all sampling days. Ethyl dodecanoate, also known as ethyl laurate, is a fatty acid ethyl ester of lauric acid formed by esterification between ethanol and laurate and has a role as a metabolite. It can be found in many fruits including apples, apricot, guava and lemon and provides a fruity flavor.9,19
Principal component analysis using multiple factor analysis was performed to demonstrate the correlation between the treatment types and the emitted ester VOCs. The PCA analysis resulted in a clear separation among the different VOCs emitted across the different treatments (ESI Fig. 2†) accounting for 47% of total variance, with components one and two accounting for 26% and 20% of the variance, respectively. In the biplot results, ethyl linoleate and ethyl 9-hexadeconate are found in the positive PC1 region, and heptyl and n-octyl acetate are observed in the positive PC2 region.
3.1.1.2 Alcohols. The major volatile organic compounds quantitatively represented based on their concentration distribution that contributed to the aroma of fresh-cut apple are alcohol compounds, prominently 2-nonanol, 1-octen-3-ol, 1-heptanol, 1-octanol, 1-hexanol, isopentanol and phenyl ethyl alcohol, which are known for their sweet, fruity aroma. Among the identified alcohol functional group compounds, 1-hexanol was uniformly identified in all the control and EW treated samples throughout the storage duration. However, compounds such as 1-heptanol and pentaethylene glycol were only abundant in the control samples, whereas 2-nonanol and hexadecanol were predominant in the treated samples. Among all the alcohol compounds, the highest concentration was recorded for isopentanol and 1-hexanol in the ALEW samples.
The high concentrations of 1-hexanol are associated with the high enzyme activity of ADH, which can be related to increased self-defense mechanisms of the fruit induced by the treatment.19 Alcohol VOCs are formed by the reduction of the corresponding aldehydes through the action of alcohol dehydrogenase (ADH);20 this was evident in the treated samples, in which the occurrence of aldehydes was completely inhibited while higher alcohol VOCs were dominant at the end of storage. Based on the biplot result (ESI Fig. 3†), most of the alcohol VOCs were present in the positive region of the first component, which corresponds to the control and ALEW samples, whereas the samples treated with AEW were found in the negative region for both components.
3.1.1.3 Carboxylic acids and alkanes. Among the carboxylic acids, only butanoic acid was identified in the baseline analysis, whereas during storage all the carboxylic groups were identified in ALEW samples except for propanoic acid, which was only identified in AEW samples. On the other hand, butanoic acid and trans-2-hexenoic acid were the two carboxylic acids that were not identified in the AEW group. Dodecanoic acid (lauric acid) had the highest emitted concentration in all the control and treated samples during storage. Comparing all the VOC functional groups, carboxyl VOCs were uniformly emitted across the treatments, as also shown in the PCA analysis (ESI Fig. 4†).
Unlike the other VOCs, alkane compounds were the most abundant in the baseline analysis, and their concentration was significantly reduced during storage for all samples. Among the alkane groups, octadecane was not identified initially and was only emitted by the control fruit during storage. Heneicosane was only identified in the baseline and control samples during storage, whereas cyclohexadecane and pentacosane were only significantly emitted by the ALEW and CO samples. The significant effects of the treatments on the emission of alkane VOCs were also shown by the PCA analysis, in which the PCA accounted for 59.95% of the variance, indicating a clear separation (ESI Fig. 5†).
3.1.1.4 Aldehydes and ketones. In this study, the only identified aldehyde was nonanal, and its abundance was only recorded for control samples at the end of storage (day 10). Aldehyde VOCs predominate in immature apples. As the fruit matures, their concentration decreases and the concentrations of alcohol and esters increase, according to Espino-Díaz et al.9 This could be evidence that the apples were stored for a significant period before being processed into fresh-cut product and could be at their maximum maturity, which coincided well with the number of alcohols and esters. There were no ketones identified in the baseline analysis; furthermore, only a few ketones were identified for the control fruit during storage. In addition, in the PCA analysis, the first two components explained the 47% of the total variability (ESI Fig. 6†), presenting a separation between the control samples in the positive region of the second component and the treated samples, showing a significant difference among the treated and control sample.
Although 2-nonanone, 3-octanone and β-damascenone were predominant on day 3 in the control samples, their concentrations dropped below the detection limit as the storage duration progressed, except in the case of 2-nonanone, which was emitted continuously throughout the storage period. In contrast, when comparing all ketones, a significantly higher concentration of 3-octanone was identified only for the treated samples. Notably, most of the alkane groups were predominant across all treatment types and storage conditions. However, β-damascenone was only detected in the control fruit, and geranyl acetone was exclusively identified under AEW conditions. Additionally, 5,9-undecadien-2-one, 6,10-dimethyl-, (E)- was identified solely under ALEW conditions.
3.2 Proteomics changes of fresh-cut apple during storage after different electrolyzed water treatments
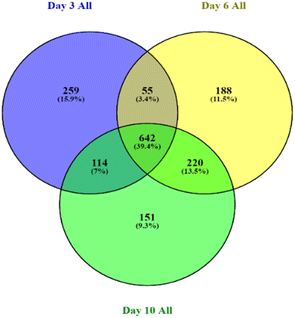
On day 3, day 6, and day 10, 3434, 3401, and 3313 proteins were identified, respectively, with an FDR of 0.9%. To ensure high-quality and accurate protein identification, a set of threshold criteria was established. To be considered positively identified, a protein must have a true molecular weight, a missing protein exclusive unique peptide count of at least 2, a protein identification probability of at least 95% (0.95), and a percentage sequence coverage greater than zero. Using these selection and identification parameters, we identified 1069 proteins on day 3 after the exclusion of 305 redundant proteins (see ESI S1†). On day 6, we identified 1417 proteins, of which 312 were redundant, and on day 10, we identified 1436 proteins, of which 309 were redundant. Fig. 4 presents the non-redundant proteins for each day in a Venn diagram, resulting in a total of 1070, 1105, and 1127 non-redundant proteins. Out of these, 642 (39.4%) were conserved across all three days.The proteomics results showed that there were differences between the protein expression levels of the different treatments and the baseline samples for day 6. When comparing the control, AEW, and ALEW samples individually to the baseline samples for day 6 (Fig. 5A–C), the Venn diagrams show that baseline vs. control had more conserved proteins (691) compared to baseline vs. AEW (605) and baseline vs. ALEW (597). Comparing all four treatments, a total of 538 conserved proteins were found, with 115 unique proteins only present for the baseline group, and 36 and 31 unique proteins in the control and AEW groups, respectively.
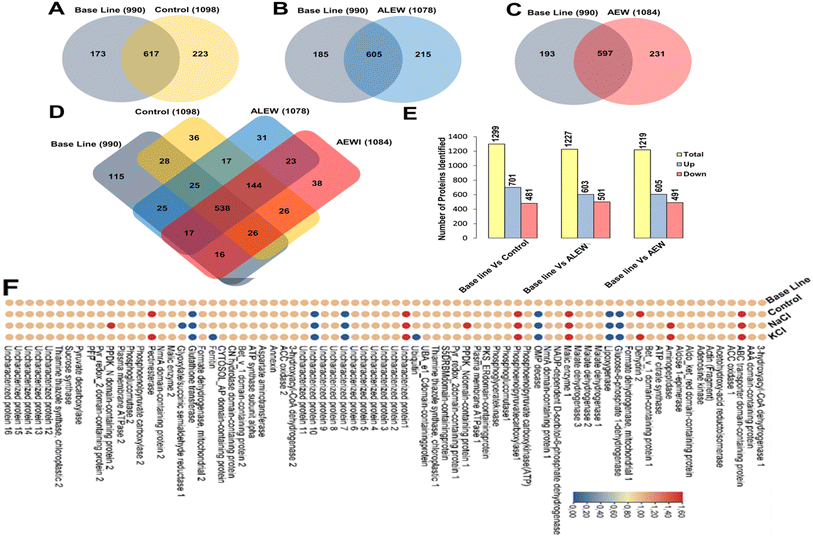 | ||
Fig. 5 Venn diagrams and a heatmap depicting the differential protein analysis on day 6 after treatment with ALEW and AEW compared to their respective control and baseline samples. (A)–(C) Venn diagrams representing the number of proteins identified in baseline samples compared to the (A) control, (B) ALEW, and (C) AEW treatments. (D) Venn diagram illustrating the numbers of conserved and differential unique proteins induced by the control, ALEW, and AEW treatments compared to the baseline group. (E) Total number of differential proteins identified in baseline samples compared to the control, ALEW, and AEW samples. (F) Heatmap showing the relative expression levels (fold change after log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 transformation) of the 75 most significantly upregulated proteins identified in the baseline samples compared to the control, ALEW, and AEW treatments. Detailed descriptions of these proteins can be found in ESI Table S1.† 10 transformation) of the 75 most significantly upregulated proteins identified in the baseline samples compared to the control, ALEW, and AEW treatments. Detailed descriptions of these proteins can be found in ESI Table S1.† | ||
Further analysis of the protein expression levels for proteins (Fig. 5A–C) showed that there were more upregulated proteins than downregulated proteins in all the comparisons. For baseline vs. control, 701 proteins were upregulated and 481 were downregulated, while for baseline vs. AEW, 605 were upregulated and 491 were downregulated. Similarly, for baseline vs. ALEW, 603 proteins were upregulated and 501 were downregulated.
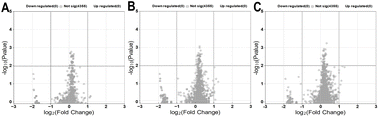
Volcano plots were used as a visualization tool to present the findings for the various storage durations (3, 6 and 10). Volcano plot selection criteria for assessing DEPs were defined as those with a −log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (p-value) of ≥2 and a fold change (FC) of ≥1 or ≤−1, with a false discovery rate (FDR) of 0.01. Proteins meeting these criteria were categorized as upregulated for those with FC ≥1 or downregulated for FC ≤−1. Fig. 6 shows a total of 4355 proteins that were identified in all treatments after 3 days of exposure to AEW and ALEW. However, comparing the protein expression changes for baseline vs. control, baseline vs. AEW, and baseline vs. ALEW, none of the 4355 proteins exhibited significant upregulation or downregulation relative to their baseline samples (Fig. 6A–C).
10 (p-value) of ≥2 and a fold change (FC) of ≥1 or ≤−1, with a false discovery rate (FDR) of 0.01. Proteins meeting these criteria were categorized as upregulated for those with FC ≥1 or downregulated for FC ≤−1. Fig. 6 shows a total of 4355 proteins that were identified in all treatments after 3 days of exposure to AEW and ALEW. However, comparing the protein expression changes for baseline vs. control, baseline vs. AEW, and baseline vs. ALEW, none of the 4355 proteins exhibited significant upregulation or downregulation relative to their baseline samples (Fig. 6A–C).
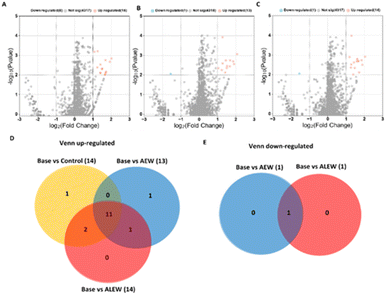
At the day 6 time point, a total of 4332 proteins were identified in the three samples (control, AEW and ALEW). Among these, when comparing baseline vs. control, 4317 proteins showed no significant differences, while the remaining 15 exhibited upregulation (Fig. 7A). In the comparison of baseline vs. AEW, 4318 proteins were identified, with 13 being upregulated and 1 downregulated (Fig. 7B).
A similar pattern was observed in the comparison of baseline vs. ALEW, with 1 downregulated protein and 14 upregulated proteins, while a larger number (4317 proteins) displayed no significant response (Fig. 7C). Overall, 41 proteins were upregulated at the 6th day after treatment comparing the baseline samples to the control, AEW and ALEW samples after 6 days. Of these, 41 proteins that were upregulated 68.8% (11) were shown to be conserved across all treatments (baseline vs. control, baseline vs. AEW and baseline vs. ALEW, respectively); of those 11 proteins that were upregulated, 3 of the proteins were classified as uncharacterised while the remaining 8 were classified (Fig. 7D). The “M16C_associated domain-containing protein” was shown to be unique to baseline vs. control on day 6.
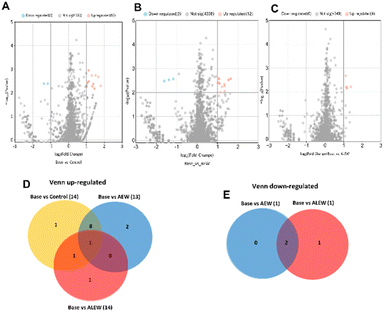
Baseline vs. AEW also had 1 unique protein, “KH type-2 domain-containing protein,” which was found on day 6 after treatment, while baseline vs. ALEW had no unique protein. For the day 10 time point, a total of 4353 proteins were identified in the three samples (control, AEW, and ALEW). Among these, when comparing baseline vs. control, 4339 proteins showed no significant differences, while the remaining 14 showed significant changes (Fig. 8A). Of the 14 proteins that showed significant changes, 2 were significantly downregulated, while the remaining 12 were significantly upregulated (Fig. 8A). In the comparison of baseline vs. AEW, 4353 proteins showed no significant difference, with 12 being upregulated and 3 downregulated (Fig. 8B). For baseline vs. ALEW, only 4 upregulated proteins were identified, while a larger number (4349 proteins) displayed no significant response (Fig. 8C).
Overall, 14 proteins were upregulated at the 10th day after treatment when comparing the baseline samples to the control, AEW and ALEW samples after 10 days. Out of these 14 proteins that were upregulated, 57.1% (8) were shown to be conserved across all treatments. Baseline vs. AEW had 2 unique proteins (methylenetetrahydrofolate reductase and 1,4-alpha-glucan branching enzyme), baseline vs. control had 1 unique protein (diadenosine tetraphosphate synthetase), and baseline vs. ALEW had one unique protein that was classified as uncharacterised.
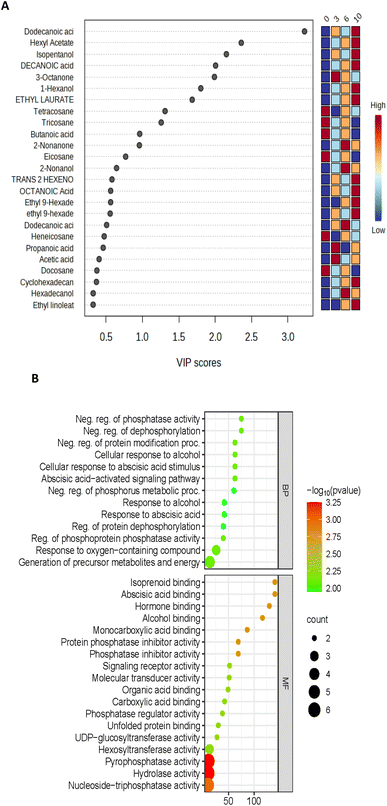
Using the VOC data, variable importance in the projection (VIP) was generated; the higher VIP score generated from the PLS-DA model ranked individual compounds for their potential to discriminate VOCs of importance (Fig. 9A). On day 10, AEW had 2 unique proteins; methylenetetrahydrofolate reductase (MTHFR) and 1,4-alpha-glucan branching enzyme were identified only for the AEW samples. Methylenetetrahydrofolate reductase (MTHFR) catalyses the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate using flavin adenine dinucleotide (FAD) as a cofactor.24 According to the literature, MTHFR is the least-understood enzyme involved in folate-mediated one-carbon metabolism in plants.25 It is clear from this figure that dodecanonic acid, hexyl acetate, isopentanol, decanoic acid, 3-octanone, 1-hexanol, ethyl laurate, tetracosane, and tricosane were the main contributors to the metabolic differences between the control and treated samples (VIP >1.5). The differences in the metabolic activities were identified using the software Funrich for analysis of the 24 identified upregulated proteins; upon removal of redundant proteins, these 24 upregulated proteins were clustered by biological process and molecular functions using the software ShinyGo 0.77 (Fig. 9B). The biological processes contained 13 top pathways and molecular functions presented 18 pathways. The highest rich factor corresponded to isoprenoid, abscisic acid, and alcohol binding pathways associated with molecular function. Moreover, the two top DEPs with lowest reach factor and the highest protein counts corresponded to pyrophosphate and hydrolase activity related pathways. There is no existing study with which to compare the findings of the current study; however, according to He et al.,26 combined metabolome and volatilome analysis demonstrated that slightly acidic EW ice maintained the contents of umami- and sweetness-related amino acids while inhibiting the accumulation of undesirable spoilage-related biomarkers such as lactic acid, 2,3-butanediol, 2-ethyl-1-hexanol and 2-methyl-butanal in shrimp. It is important to note that the studies differed in the biological materials used, as well as the type of EW applied.
4 Conclusions
Both volatilomics and proteomics analyses were shown to be effective to understand the molecular basis of changes in response to electrolyzed water treatments during the storage of fresh-cut apple. The profile of the VOCs changed significantly during storage and in response to the treatments, as indicated by the increase of alcohol and ester compounds in the EW-treated samples, which could contribute to the aroma of the fresh-cut apple. Similarly, the proteomics results showed that there were differences in the protein expression levels between the different treatments; however, significant difference was observed only after day 6. In general, 1 unique protein “KH type-2 domain-containing protein” on day 6 and 2 unique proteins (methylenetetrahydrofolate reductase and 1,4-alpha-glucan branching enzyme) in day 10 were identified for AEW. Similarly, the control sample had 1 unique M16C_associated domain-containing protein on day 6 and another unique protein (diadenosine tetraphosphate synthetase) on day 10. However, ALEW had no unique proteins on day 6 and one unique protein at day 10 that was classified as uncharacterised. These results demonstrate that EW treatment could contribute to different expression and molecular activities that contribute to the retention of quality.Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD056621.Author contributions
Z. A. Belay: conceptualization, writing – original draft, methodology, investigation and validation. M. Nkomo: writing – original draft, methodology, data analysis. G. G. Mohamed: methodology, data analysis, resources, validation. M. L. Ntsoane: methodology, data analysis, validation. Oluwafemi J. Caleb: conceptualization, review, methodology, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is based upon research supported wholly by the National Research Foundation (NRF), South Africa (Grant No. 137990) awarded to Dr O. J. Caleb and (138128) Dr Z. A. Belay. The research grant provided by Technology Innovation Agency (TIA) of South Africa for electrolyzed water application (2023/FUN132D/AA) to Dr O. J. Caleb is gratefully acknowledged.References
- N. E. Nyamende, Z. A. Belay, Z. Keyser, A. B. Oyenihi and O. J. Caleb, Impacts of alkaline-electrolyzed water treatment on physicochemical, phytochemical, antioxidant properties and natural microbial load on ‘Granny Smith’ apples during storage, Int. J. Food Sci. Technol., 2022, 57(1), 447–456 CrossRef CAS.
- N. E. Nyamende, Z. A. Belay and O. J. Caleb, Recent advances in electrolyzed water treatments: Mechanisms of action and its effect on browning, bioactive compounds, and disinfection of fresh-cut fruit and vegetables-A review, Food Chem. Adv., 2023, 3, 100569 CrossRef.
- Q. Gao, Z. Yang, B. Bi and J. He, Effects of slightly acidic electrolyzed water on the quality of fresh-cut apple, Foods, 2022, 12(1), 39 CrossRef PubMed.
- A. Graça, D. Santo, P. Pires-Cabral and C. Quintas, The effect of UV-C and electrolyzed water on yeasts on fresh-cut apple at 4 C, J. Food Eng., 2020, 282, 110034 CrossRef.
- A. M. Plesoianu, V. Nour, F. Tutulescu and M. E. Ionica, Quality of fresh-cut apples as affected by dip wash treatments with organic acids and acidic electrolyzed water, Food Sci. Technol., 2021, 42, e62620 CrossRef.
- H. K. Malahlela, Z. A. Belay, R. R. Mphahlele, G. O. Sigge and O. J. Caleb, Recent advances in activated water systems for the postharvest management of quality and safety of fresh fruits and vegetables, Compr. Rev. Food Sci. Food Saf., 2024, 23(2), e13317 CrossRef CAS PubMed.
- W. Hu, Sarengaowa, Y. Guan and K. Feng, Biosynthesis of phenolic compounds and antioxidant activity in fresh-cut fruits and vegetables, Front. Microbiol., 2022, 13, 906069 CrossRef PubMed.
- Z. A. Belay and O. J. Caleb, Role of integrated omics in unravelling fruit stress and defence responses during postharvest: A review, Food Chem.: Mol. Sci., 2022, 5, 100118 CAS.
- M. Espino-Díaz, D. R. Sepúlveda, G. González-Aguilar and G. I. Olivas, Biochemistry of apple aroma: A review, Food Technol. Biotechnol., 2016, 54(4), 375 Search PubMed.
- H. Luo, J. Song, P. Toivonen, Y. Gong, C. Forney, L. C. Palmer, S. Fillmore, X. Pang and Z. Zhang, Proteomic changes in ‘Ambrosia’ apple fruit during cold storage and in response to delayed cooling treatment, Postharvest Biol. Biotechnol., 2018, 137, 66–76 CrossRef CAS.
- Y. Gong, J. Song, J. DeEll, M. Vinqvist-Tymchuk, L. Campbell-Palmer, L. Fan, S. Fillmore, G. Lum and Z. Q. Zhang, Proteomic changes in association with storage quality of ‘Honeycrisp’ apples after pre and postharvest treatment of 1-MCP, Postharvest Biol. Biotechnol., 2023, 201, 112362 CrossRef CAS.
- X. Wang, X. Zhang, P. Jia, H. Luan, G. Qi, H. Li and S. Guo, Transcriptomics and metabolomics provide insight into the anti-browning mechanism of selenium in freshly cut apples, Front. Plant Sci., 2023, 14, 1176936 CrossRef PubMed.
- L. Sheng, X. Shen, O. Ulloa, T. V. Suslow, I. Hanrahan and M. J. Zhu, Evaluation of JC9450 and neutral electrolyzed water in controlling Listeria monocytogenes on fresh apples and preventing cross-contamination, Front. Microbiol., 2020, 10, 3128 CrossRef PubMed.
- A. Graca, M. Abadias, M. Salazar and C. Nunes, The use of electrolyzed water as a disinfectant for minimally processed apples, Postharvest Biol. Biotechnol., 2011, 61(2–3), 172–177 CrossRef CAS.
- A. Martínez, A. Hernandez, C. Moraga, P. Tejero, M. de Guía Córdoba and A. Martín, Detection of volatile organic compounds associated with mechanical damage in apple cv. ‘Golden Delicious’ by headspace solid-phase microextraction (HS-SPME) and GC-MS analysis, LWT--Food Sci. Technol., 2022, 172, 114213 CrossRef.
- Q. Zheng, J. Song, K. Doncaster, E. Rowland and D. M. Byers, Qualitative and quantitative evaluation of protein extraction protocols for apple and strawberry fruit suitable for two-dimensional electrophoresis and mass spectrometry analysis, J. Agric. Food Chem., 2007, 55, 1663–1673 CrossRef CAS PubMed.
- J. Speirs, E. Lee, K. Holt, K. Yong-Duk, S. N. Steele, B. Loveys and W. Schuch, Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols, Plant Physiol., 1998, 117(3), 1047–1058 CrossRef CAS PubMed.
- M. K. Hasan and L. J. Brady, Nucleic acid-binding KH domain proteins influence a spectrum of biological pathways including as part of membrane-localized complexes, J. Struct. Biol.: X, 2024, 10, 100106 CAS.
- N. Spaho, F. Gaši, E. Leitner, M. Blesić, A. Akagić, S. O. Žuljević, M. Kurtović, D. Đ. Ratković, M. S. Murtić and M. F. Akšić, et al., Characterization of volatile compounds and flavor in spirits of old apple and pear cultivars from the Balkan region, Foods, 2021, 10(6), 1258 CrossRef CAS PubMed.
- B. G. Defilippi, A. A. Kader and A. M. Dandekar, Apple aroma: alcohol acyltransferase, a rate limiting step for ester biosynthesis, is regulated by ethylene, Plant Sci., 2005, 168(5), 1199–1210 CrossRef CAS.
- L. F. Thatcher, L. G. Kamphuis, J. K. Hane, L. Onate-Sanchez and K. B. Singh, The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress, PLoS One, 2015, 10(5), e0126978 CrossRef PubMed.
- T. Chen, P. Cui, H. Chen, S. Ali, S. Zhang and L. Xiong, A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis, PLoS Genet., 2013, 9(10), e1003875 CrossRef PubMed.
- W. Wang, S. Wang, W. Gong, L. Lv, L. Xu, J. Nie and L. Huang, Valsa mali secretes an effector protein VmEP1 to target a K homology domain-containing protein for virulence in apple, Mol. Plant Pathol., 2022, 23(11), 1577–1591 CrossRef CAS PubMed.
- M. Kasap, A. Sazci, E. Ergul and G. Akpinar, Molecular phylogenetic analysis of methylenetetrahydrofolate reductase family of proteins, Mol. Phylogenet. Evol., 2007, 42(3), 838–846 CrossRef CAS PubMed.
- S. Roje, H. Wang, S. D. McNeil, R. K. Raymond, D. R. Appling and Y. Shachar-Hill, et al., Isolation, characterization, and functional expression of cDNAs encoding NADH-dependent methylenetetrahydrofolate reductase from higher plants, J. Biol. Chem., 1999, 274(51), 36089–36096 CrossRef CAS PubMed.
- Y. He, Z. Xie, Y. Xu, C. Guo, X. Zhao and H. Yang, Effect of slightly acid electrolysed water ice on metabolite and volatilome profile of shrimp (Penaeus vannamei) during cold storage, Food Control, 2023, 145, 109421 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fb00318g |
| This journal is © The Royal Society of Chemistry 2025 |