Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Supplementation of low-protein diets with plant protein and probiotics enhances muscle health by regulating gut microbiota and metabolomic profiles in SAMP8 mice†
Ruyi
Han‡
a,
Limin
Ouyang‡
b,
Chenggang
Yin‡
a,
Long
Cai
a,
Qiming
Wu
b,
Liang
Chen
 b,
Jun
Du
b,
Xilong
Li
a,
Zhigang
Zhu
*b and
Yu
Pi
b,
Jun
Du
b,
Xilong
Li
a,
Zhigang
Zhu
*b and
Yu
Pi
 *a
*a
aKey Laboratory of Feed Biotechnology of Ministry of Agriculture and Rural Affairs, Institute of Feed Research, Chinese Academy of Agricultural Sciences, Beijing 100081, China. E-mail: piyu@caas.cn; Tel: +86-010-82108134
bNutrilite Health Institute, Amway (Shanghai) Innovation & Science Co., Ltd, Shanghai, 201203, China. E-mail: joe.zhu@amway.com
First published on 1st May 2025
Abstract
Muscle health is crucial, especially for aging populations. This study investigates how plant protein and probiotic supplementation in a low-protein (LP) diet affects the gut microbiota, metabolome, and muscle health in aging SAMP8 mice, emphasizing the gut–muscle axis in older populations. Twenty-four 8-month-old male SAMP8 mice were divided into four groups: control (CON, standard diet), low-protein (LP), LP supplemented with plant protein (LP + P), and LP supplemented with plant protein and probiotics (LP + P + B). The experimental treatment lasted 8 weeks. Results showed that, by week 6, body weight increased significantly in all LP groups, with a trend toward higher body fat. Protein utilization and muscle strength improved significantly in the supplemented groups compared to the LP group (P < 0.05). The LP group showed elevated levels of inflammatory cytokines (IL-12p70, IL-6, TNF-α) (P < 0.05) in serum, which were reduced in the supplemented groups, especially in the LP + P + B group (P < 0.05). Gene expression related to muscle protein synthesis (mTOR, S6K1) and oxidative stress (CAT, Nrf2) was upregulated in the LP group but downregulated in the supplemented groups (P < 0.05). Immune-related genes followed similar patterns, with inflammation markers being significantly reduced after supplementation (P < 0.05). After supplementation, fecal beneficial bacteria (Bifidobacterium, Roseburia) and metabolites (butyric acid, indole-3-propionic acid, kyotorphin) increased, indicating enhanced gut health (P < 0.05). Specific bacterial species were correlated with metabolites and immune markers, highlighting their role in immune modulation (P < 0.05). These results show that supplementing plant protein and probiotics into an LP diet improved muscle strength, reduced inflammation, optimized gut microbiota, and boosted beneficial metabolism. The findings suggest that plant protein and probiotics can maintain muscle health and regulate immune responses in the elderly.
Introduction
Muscle health is a critical aspect of overall well-being, particularly in aging populations where sarcopenia and muscle atrophy are prevalent concerns.1 Sarcopenia, the age-related loss of muscle mass and function, significantly contributes to increased frailty, reduced quality of life, and higher mortality rates in the elderly.2 Understanding the underlying mechanisms that govern muscle health and identifying effective interventions to mitigate muscle decline are therefore of paramount importance.The gut–muscle axis, an emerging concept in nutritional and physiological research, highlights the intricate relationship between gut microbiota and muscle function.3 The gut microbiota plays a pivotal role in maintaining metabolic balance, modulating immune responses, and producing beneficial metabolites, such as short-chain fatty acids (SCFAs), all of which are essential for muscle health.4 In aging models, such as the senescence-accelerated mouse prone 8 (SAMP8) model, disturbances in gut microbiota have been linked to impaired muscle function, increased systemic inflammation, and oxidative stress, thereby exacerbating muscle deterioration.5,6
Dietary protein plays a critical role in maintaining muscle health, as adequate protein intake is essential for muscle protein synthesis, maintenance, and repair.7 The estimated average requirement (EAR) for protein is 60 g day−1 for male adults and 50 g day−1 for female adults, both aged 18 years or older (0.9 g kg−1 day−1). The recommended nutrient intake (RNI) is slightly higher, at 65 g day−1 for men and 55 g day−1 for women (1.0 g kg−1 day−1). A recent survey shows that protein intake among elderly Chinese individuals has declined over 28 years, dropping from 63.3 g day−1 to 57.8 g day−1. During this time, the proportion of individuals consuming less than the EAR for protein significantly increased, while the proportion exceeding the RNI declined across all population subgroups.8 This suggests that many elderly Chinese are now at risk of insufficient dietary protein intake. Recent research indicates that low-protein (LP) diets result in diminished amino acid (AA) levels, induce DNA damage, and intensify inflammation within the intestinal tissues of male mice affected by inflammatory bowel disease (IBD).9 In addition, LP diets, often recommended for specific health conditions such as chronic kidney disease or to reduce renal load in aging populations, carry significant risks. Chief among these is the potential for muscle loss due to inadequate AA availability. This risk is compounded by the fact that elderly individuals often experience reduced digestive efficiency and anabolic resistance, which make it harder for them to preserve muscle mass even when protein intake meets or exceeds recommended levels. Furthermore, LP diets can lead to an imbalance in the gut microbiota, reducing the production of beneficial metabolites like SCFAs, which are vital for maintaining muscle and metabolic health.10 Ensuring sufficient protein intake, particularly in older populations, is therefore essential for maintaining overall health and preventing muscle deterioration.
To address this challenge, recent research has focused on the potential of dietary supplementation with plant protein and probiotics to enhance muscle health through the gut–muscle axis.5,11,12 Plant proteins, derived from sources such as soy, pea, and rice, not only provide essential AAs but also contain bioactive compounds that have been shown to exert anti-inflammatory and antioxidant effects, which are beneficial for gut and muscle health.13,14 Probiotics, which are live microorganisms that confer health benefits to the host, have been demonstrated to positively influence gut microbiota composition, enhance nutrient absorption, and reduce gut-derived inflammation,15 all of which can contribute to improved muscle function. However, the synergistic effects of plant proteins combined with probiotics remain unclear.
In this study, we investigate the combined effects of plant protein and probiotics as a supplementation strategy in an LP diet on muscle health in SAMP8 mice. We hypothesize that an LP diet may exacerbate muscle inflammation in elderly mice, while supplementation with plant protein and probiotics will improve muscle function by modulating the gut–muscle axis. This may lead to enhanced gut microbiota composition, increased production of beneficial microbial metabolites, improved nutrient utilization, and reduced systemic inflammation. This research aims to provide new insights into the role of diet in managing muscle health, particularly in aging populations and individuals on protein-restricted diets, and to identify potential dietary strategies for clinical applications.
Materials and methods
Probiotics and plant protein
The probiotics consist of three probiotic strains, including Bifidobacterium animalis subsp. lactis UABla-12™, Bifidobacterium animalis subsp. lactis BB-12®, and Lactobacillus acidophilus La-5 (Novonesis, Denmark). Probiotic freeze-dried powder is added to water, at a dosage of 3.75 × 109 colony forming units (CFUs) of Bifidobacterium animalis subsp. lactis UABla-12™, 7.5 × 108 CFUs of Bifidobacterium animalis subsp. lactis BB-12® and 7.5 × 108 CFUs of Lactobacillus acidophilus La-5 per milliliter of sterile water. The probiotic freeze-dried powder is stored in a 4 °C refrigerator, and the probiotic solution is freshly prepared daily. After preparation, the solution is poured into a customized 50 mL water bottle for the mice to drink. The amount of the solution added to the bottle is determined based on the approximate daily water intake of the mice, ensuring that the entire volume of the solution is consumed by the mice each day. The plant protein used in the present study is provided by Nutrilite Health Institute (Shanghai, China). The plant protein, which is derived from soybeans and peas, has its main nutritional composition presented in Table S1.†Animal and experiment design
Twenty-four male 8-month-old SAMP8 mice were purchased from SPF Biotechnology Co., Ltd (Beijing, China) and housed in an environmentally controlled room (temperature, 25 ± 2 °C; relative humidity, 45 to 60%; lighting cycle, 12 h light [L]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h dark [D]) with ad libitum access to food and water throughout the study. All mice were housed in individual cages. The experimental protocols involving animals in this research were approved by the Institutional Animal Care and Use Committee (IACUC) at the Institute of Feed Research, Chinese Academy of Agricultural Sciences (Approval No. IFR-CAAS20230520). After 1 week of adaptation, the mice were randomly allocated to four groups (n = 6 per group): the control group (CON) was fed the AIN-93M diet, the low-protein group (LP) was fed a low-protein diet based on the AIN-93M diet, the low-protein supplementation with plant protein group (LP + P) was fed the low-protein diet supplemented with plant protein, and the low-protein supplementation with plant protein and probiotics group (LP + P + B) was fed the low-protein diet supplemented with plant protein and probiotics administered through drinking water. The composition and nutritional components of the feed used in the experiment are shown in Table S2.† The trial period is 8 weeks in total. During the experiment, the body weight, food intake, and water consumption of the mice were checked weekly. The experimental scheme is shown in Fig. 1A. During the experiment, one mouse in the CON group suddenly died for unknown reasons. As a result, only five samples in the CON group were available for further research.
12 h dark [D]) with ad libitum access to food and water throughout the study. All mice were housed in individual cages. The experimental protocols involving animals in this research were approved by the Institutional Animal Care and Use Committee (IACUC) at the Institute of Feed Research, Chinese Academy of Agricultural Sciences (Approval No. IFR-CAAS20230520). After 1 week of adaptation, the mice were randomly allocated to four groups (n = 6 per group): the control group (CON) was fed the AIN-93M diet, the low-protein group (LP) was fed a low-protein diet based on the AIN-93M diet, the low-protein supplementation with plant protein group (LP + P) was fed the low-protein diet supplemented with plant protein, and the low-protein supplementation with plant protein and probiotics group (LP + P + B) was fed the low-protein diet supplemented with plant protein and probiotics administered through drinking water. The composition and nutritional components of the feed used in the experiment are shown in Table S2.† The trial period is 8 weeks in total. During the experiment, the body weight, food intake, and water consumption of the mice were checked weekly. The experimental scheme is shown in Fig. 1A. During the experiment, one mouse in the CON group suddenly died for unknown reasons. As a result, only five samples in the CON group were available for further research.
During the final week of the experiment, all feces were meticulously collected to accurately determine the apparent digestibility of protein and energy. To prevent any potential contamination, only fresh fecal samples were carefully gathered from the rectum for subsequent intestinal microbiota testing. At the end of the experimental period, the mice were fasted for 12 hours and euthanized after collecting blood samples by extracting eyeballs. Approximately 1 mL of blood was allowed to coagulate at room temperature for at least 2 hours. Subsequently, the samples were centrifuged at 3000g for 10 minutes to isolate the serum. The obtained serum was then stored at −80 °C for subsequent assessments. The middle colon tissue and gastrocnemius muscle were collected from each mouse and then immediately stored at −80 °C for future analysis.
Muscle strength and aerobic endurance measurement
The 4-limb hanging test and weight test were conducted with minor modifications based on a previous study.16 For the 4-limb hanging test, mice were placed on a wire grid for 1 minute to acclimate. The grid and mouse were then gently inverted, and the time until the mouse fell off was recorded. Each mouse was tested 3 times with at least a 5-minute break between trials, and the final result was the average of the three tests.For the weight test, the apparatus consisted of seven weights, each made from a ball of tangled fine-gauge stainless steel wire and steel chain links. The weights ranged from 15.2 g to 60.8 g. During the test, each mouse was held by the base of its tail and lowered to grasp the first weight (15.2 g) with its forepaws. The mouse was then lifted, and if it held the weight for 3 seconds, it proceeded to the next heavier weight. If it dropped the weight before 3 seconds, the time was recorded, and the mouse was given two more attempts. If it failed all three attempts, the heaviest weight successfully held was recorded. The total score was calculated as the product of the number of links in the heaviest chain held for 3 seconds, multiplied by the time held. For example, a mouse holding a 5-link weight for 3 seconds, but dropping the 6-link weight, would score (5 × 3) = 15. If it held the 6-link weight for 1 second, the score would be (5 × 3) + 1 = 16.
Body fat percentage analysis
Body fat percentage was determined using a body composition analysis and imaging system (MesoQMR, Shanghai Neway Electronic Technology Co., Ltd, Shanghai, China) three days before sacrifice. The data were analyzed using the software provided by the manufacturer.Chemical analysis and determination of apparent digestibility of nutrients
Following the methodologies outlined in the referenced literature,17 we conducted analyses of dry matter (DM; method 930.15) and crude protein (CP; method 990.03) in diet and fecal samples. The gross energy (GE) content of the diet and fecal samples was assessed using an automatic oxygen and nitrogen calorimeter. The apparent total tract digestibility (ATTD) of CP and GE were calculated by [(total CP or GE intake – total CP or GE in stool)/total CP or GE intake].Measurements of cytokine and chemokine levels
Serum samples (25 μL) were analyzed using the ProcartaPlex Multiplex Immunoassay, a 36-plex mouse cytokine/chemokine magnetic bead panel (Thermo Fisher), following the specific protocols provided with the kit. The analytes were quantified using the Magpix analytical instrument, employing a standard curve based on recombinant cytokine and chemokine standards. This process utilizes xMAP technology (Luminex Co., Austin, USA) and xPONENT 4.2 software (Luminex). The results were reported in pg mL−1.RNA extraction and quantitative RT-PCR
Total RNA was isolated from the colonic and gastrocnemius tissues using TRIzol reagent (Takara Bio, Otsu, Shiga, Japan). The concentration of total RNA and its A260/A280 ratio were measured using an Epoch microplate spectrophotometer. Only samples with an A260/A280 ratio falling within the range of 1.8 to 2.0 were selected for subsequent analysis. One microgram of RNA was reverse transcribed using the PrimeScript® RT Reagent Kit and gDNA Eraser (Takara Bio Inc., Dalian, China). Quantitative PCR (qPCR) was carried out on an ABI 7300 Real-Time PCR System (Applied Biosystems, Foster, CA, USA) using gene-specific primers (Table S3†) and SYBR Green Master Mix (Takara Bio), following the protocol previously described.18 Relative mRNA expression levels were determined using the ΔΔCt method.1816S rRNA gene sequencing, and data analysis of fecal samples
Microbial genomic DNA from fecal samples was extracted using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany). The V3–V4 regions of the 16S rRNA gene were amplified with primers 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), following previously described protocols.19 The resulting PCR products were purified, quantified, and combined in equal concentrations for paired-end 300 bp sequencing on an Illumina MiSeq system (Illumina, San Diego, USA), following the protocols established by Majorbio Bio-Pharm Technology Co. Ltd (Shanghai, China). Raw sequence reads were analyzed using QIIME 2 on the Majorbio I-Sanger Cloud Platform (https://www.i-sanger.com/).20 DADA2 was employed for quality control and denoising, using default settings, to generate amplicon sequence variants (ASVs), and only ASVs with at least two reads across more than one sample were included. A phylogenetic tree was constructed using the SEPP algorithm with reference to the Silva 138 database. α-Diversity analyses were performed using Mothur (version 1.30.2). β-diversity analyses were performed using the vegan package (version 3.3.1). Principal coordinates analysis (PCoA) based on Bray–Curtis distances, weighted UniFrac distance, and unweighted UniFrac distance was conducted to evaluate community differences. Differential taxa were determined using LEfSe.Non-targeted metabolomic analysis of fecal samples
A 50 mg fecal sample was precisely weighed, and metabolites were extracted using 400 μL of a methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water solution (4
water solution (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) containing 0.02 mg mL−1 L-2-chlorophenylalanine as an internal standard. The mixture was left to settle at −10 °C and processed using the high-throughput tissue crusher Wonbio-96c (Shanghai Wonbio Biotechnology Co., Ltd) at 50 Hz for 6 minutes, followed by sonication at 40 kHz for 30 minutes at 5 °C. The samples were then stored at −20 °C for 30 minutes to precipitate proteins. After centrifugation at 13000g for 15 minutes at 4 °C, the supernatant was carefully transferred to vials for LC-MS/MS analysis.
1, v/v) containing 0.02 mg mL−1 L-2-chlorophenylalanine as an internal standard. The mixture was left to settle at −10 °C and processed using the high-throughput tissue crusher Wonbio-96c (Shanghai Wonbio Biotechnology Co., Ltd) at 50 Hz for 6 minutes, followed by sonication at 40 kHz for 30 minutes at 5 °C. The samples were then stored at −20 °C for 30 minutes to precipitate proteins. After centrifugation at 13000g for 15 minutes at 4 °C, the supernatant was carefully transferred to vials for LC-MS/MS analysis.
The LC-MS analysis was performed using the Thermo Fisher Scientific UHPLC-Q Exactive system. Once the mass spectrometry was completed, the raw LC-MS data were preprocessed using Progenesis QI software (Waters Corporation, Milford, USA). Internal standard peaks and false positive peaks, such as noise, column bleed, and derivatized reagent peaks, were removed from the data matrix, followed by deduplication and peak pooling. Metabolite identification was performed using the HMDB (https://www.hmdb.ca/), Metlin (https://metlin.scripps.edu/), and Majorbio databases. The processed data were uploaded to the Majorbio cloud platform (https://cloud.majorbio.com) for analysis.20 Metabolic features detected in at least 80% of any sample set were retained. For samples with metabolite levels below the lower limit of quantification, minimum values were imputed, and each metabolic feature was normalized using sum normalization. To minimize errors from sample preparation and instrument variability, the mass spectrum peak intensity of each sample was normalized using sum normalization. The normalized data matrix was generated, with variables showing a relative standard deviation (RSD) > 30% in QC samples removed. Log10 transformation was applied to create the final data matrix for further analysis.
Variance analysis was performed on the preprocessed data matrix. Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were conducted using the R package ropls (Version 1.6.2). Additionally, Student's t-test and fold-change analysis were performed. Significantly different metabolites were identified based on Variable Importance in Projection (VIP) from the PLS-DA model and p-values from Student's t-test. Metabolites with VIP > 1 and P < 0.05 were considered significantly different. These differential metabolites were mapped to biochemical pathways through enrichment and pathway analysis, using databases like KEGG (https://www.genome.jp/kegg/). The metabolites were classified based on the pathways they were involved in or their functional roles. Enrichment analysis was used to determine whether a set of metabolites appeared within a functional node. Instead of annotating a single metabolite, this analysis focused on groups of metabolites. Fisher's exact test, implemented using the scipy.stats module (Python packages) (https://docs.scipy.org/doc/scipy/), was used to identify statistically significant enriched pathways.
Statistical analysis
All statistical analyses, except for the fecal microbiome and metabolome data, were conducted using one-way analysis of variance (ANOVA), followed by post hoc Tukey's test for pairwise comparisons using SPSS 20 software (IBM, Armonk, NY, USA). For fecal microbiome analysis, α-diversity was assessed using the Kruskal–Wallis test, and PCoA results were evaluated using ANOSIM. The relative abundance of fecal microbiota and fecal metabolites and the levels of serum cytokines/chemokines were analyzed using the Kruskal–Wallis test and adjusted using FDR correction. Data were presented as mean ± SEM, with statistical significance set at P < 0.05.Results
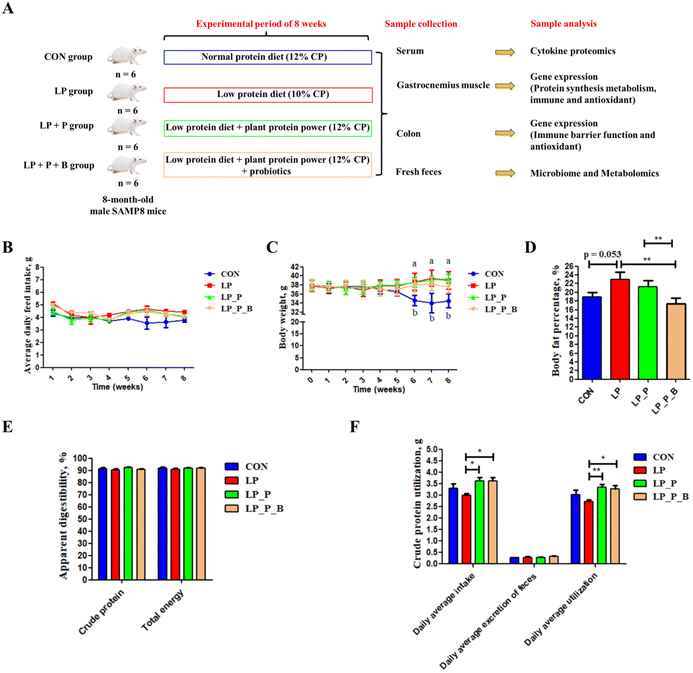
Weight, body composition, and nutrient digestibility
As shown in Fig. 1, there were no significant differences in feed intake among the experimental groups (Fig. 1B), indicating that supplementation with plant protein and probiotics did not affect feed consumption. By the 6th week of the experiment, body weight significantly increased in the LP, LP + P, and LP + P + B groups compared to the CON group (Fig. 1C). Regarding body fat percentage, no significant differences were observed among the LP, LP + P, and LP + P + B groups (Fig. 1D). However, the LP group showed a trend toward increased body fat percentage (P < 0.10), while supplementation with plant protein and probiotics reduced body fat percentage, aligning it more closely with the control group, though without significant differences. In terms of nutrient digestibility, there were no significant changes in the apparent digestibility of crude protein and energy across the groups (Fig. 1E). However, protein utilization was significantly improved in both the LP + P and LP + P + B groups compared to the LP group (Fig. 1F), suggesting that supplementation with plant protein and probiotics enhanced the bioavailability of dietary protein under LP conditions, potentially benefiting muscle health.Muscle strength
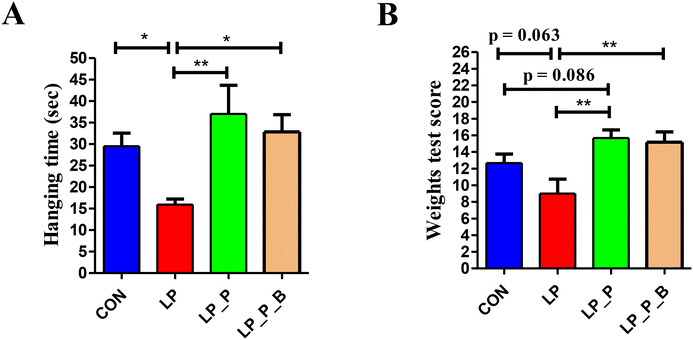
Muscle endurance was measured by the time spent in the four-limb hanging test. As shown in Fig. 2A, compared to the CON group, the LP group exhibited a significantly reduced hanging time, while there was no significant difference between the LP + P and LP + P + B groups and the CON group. However, the hanging time of the LP + P and LP + P + B groups was longer than that of the LP group. Muscle strength was measured using the weight test. As shown in Fig. 2B, compared to the CON group, the LP group showed a decrease in strength score (P < 0.10), while both the LP + P and LP + P + B groups showed significantly increased strength scores compared to the LP group. These results indicate that protein deficiency in aged mice is detrimental to muscle health.Serum cytokine levels
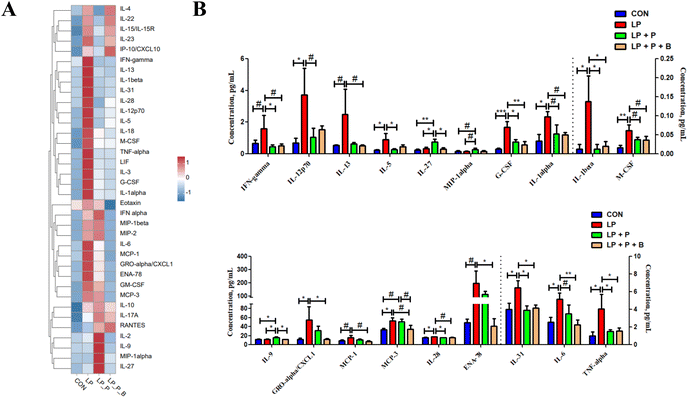
Serum cytokines can reflect the inflammatory state of the body. As shown in the heatmap of serum cytokine levels (Fig. 3A), overall, the cytokine levels in the LP group were higher compared to the CON group, while supplementation with plant protein and probiotics had a mitigating effect on cytokine levels. Further differential analysis revealed that (Fig. 3B), compared to the CON group, the LP group showed significantly increased levels of cytokines such as IL-12p70, IL-5, G-CSF, IL-1alpha, IL-1beta, M-CSF, CXCL1, MCP-3, IL-28, IL-31, IL-6, and TNF-alpha, along with a trend of increasing IFN-gamma, IL-13, MCP-1, and ENA-78 (P < 0.10). After supplementing with plant protein and probiotics, these cytokine levels decreased. Compared to the LP group, the LP + P group showed significantly reduced levels of IFN-gamma, IL-12p70, IL-5, G-CSF, IL-1beta, IL-28, IL-31, and TNF-alpha, along with a trend of decreasing IL-1alpha, M-CSF, and IL-6 (P < 0.10), while significantly increasing IL-27 and IL-9, with a trend of increasing MIP-1alpha (P < 0.10). In comparison with the LP group, the LP + P + B group showed significantly reduced levels of G-CSF, IL-1beta, CXCL1, ENA-78, IL-31, IL-6, and TNF-alpha, along with a trend of decreasing IFN-gamma, IL-13, IL-1alpha, M-CSF, MCP-1, and MCP-3 (P < 0.10). Furthermore, compared to the LP + P group, the LP + P + B group showed significantly reduced IL-17 and IL-9 levels and a trend of decreasing MCP-3 (P < 0.10). These results indicate that protein deficiency increases the inflammatory state in aged mice, while supplementation with plant protein and probiotics can alleviate this change to some extent.Gene expression related to protein metabolism, immune response, and oxidative stress in muscle
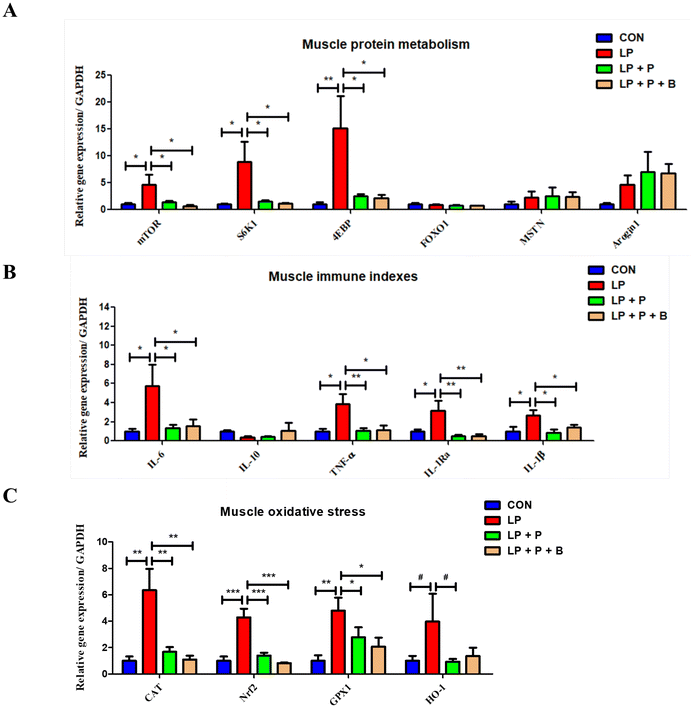
As shown in Fig. 4A, regarding protein metabolism, compared to the CON group, the LP group showed significantly upregulated expression of genes related to protein synthesis, including mTOR, S6K1, and 4EBP. However, when plant protein and probiotics were added, these significant changes disappeared. Compared to the LP group, the expression of these genes in the LP + P and LP + P + B groups was significantly downregulated. There were no significant changes in the expression of FOXO1, MSTN, and Arogin1 across the groups. These results suggest that under protein deficiency, there may be negative feedback regulation of protein synthesis genes in aged muscles, and supplementation with plant protein and probiotics can alleviate this state.Regarding immune-related genes (Fig. 4B), the LP group showed significantly upregulated expression of pro-inflammatory cytokine genes such as IL-6, TNF-alpha, IL-1Ra, and IL-1beta compared to the CON group. However, when plant protein and probiotics were added, these significant changes disappeared. Furthermore, compared to the LP group, the expression of these genes was significantly downregulated in both the LP + P and LP + P + B groups. These results suggest that protein deficiency in aged individuals may lead to an increase in muscle inflammation, while plant protein and probiotics supplementation can mitigate this inflammatory response.
In terms of oxidative stress-related genes (Fig. 4C), the LP group showed significantly upregulated expression of antioxidant genes, including CAT, Nrf2, and GPX1, compared to the CON group. However, after supplementation with plant protein and probiotics, these significant changes were no longer observed. Compared to the LP group, the expression of these genes was significantly downregulated in the LP + P and LP + P + B groups. These findings suggest that under protein deficiency, there may be negative feedback regulation of antioxidant gene expression in aged muscles, and plant protein and probiotics supplementation can help alleviate this oxidative stress state.
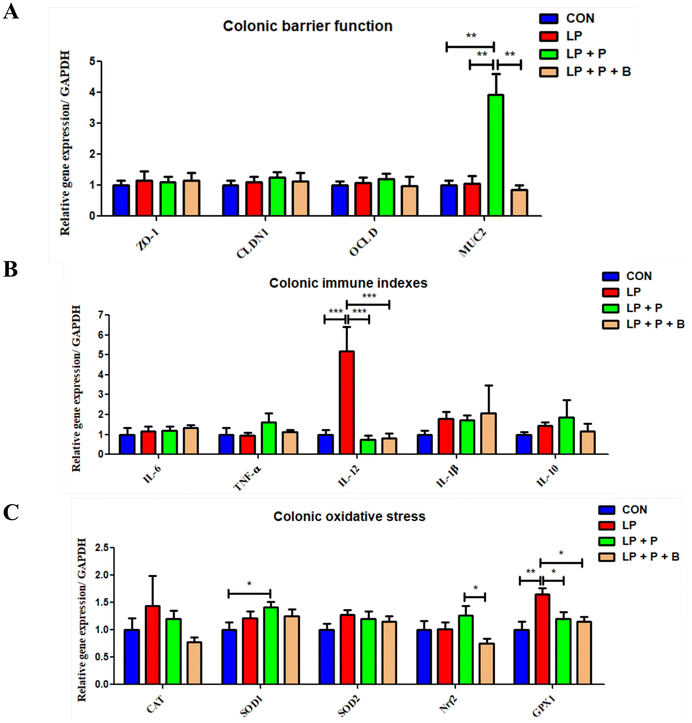
Gene expression related to barrier function, immune response, and oxidative stress in the colon
The gut is a critical organ for digestion and immunity. As shown in Fig. 5, regarding intestinal barrier function, there were no significant differences in the expression levels of ZO-1, CLDN1, and OCLD genes between groups. However, compared to the CON, LP, and LP + P + B groups, the expression of the MUC2 gene was significantly upregulated in the LP + P group (Fig. 5A). For immune-related genes (Fig. 5B), the expression levels of IL-6, TNF-α, IL-1β, and IL-10 showed no significant differences between the groups. In contrast, for the pro-inflammatory cytokine IL-12, the LP group showed a significant upregulation compared to the CON group. However, when plant protein and probiotics were added, these significant changes disappeared, and the expression of IL-12 was significantly downregulated in the LP + P and LP + P + B groups compared to the LP group. In terms of oxidative stress-related genes (Fig. 5C), there were no significant differences in the expression of CAT and SOD2 between groups. However, GPX1 expression was significantly upregulated in the LP group compared to the CON group. Again, with the addition of plant protein and probiotics, these significant changes disappeared, and GPX1 expression was significantly downregulated in the LP + P and LP + P + B groups compared to the LP group. Moreover, the LP + P group showed a significant upregulation of SOD1 expression compared to the CON group, while the LP + P + B group showed a significant downregulation of Nrf2 expression compared to the LP + P group. These results suggest that in a protein-deficient state, the colon of elderly individuals may experience inflammation and a negative feedback regulation of antioxidant gene expression. Supplementation with plant protein and probiotics can help alleviate this condition.Fecal microbiota composition
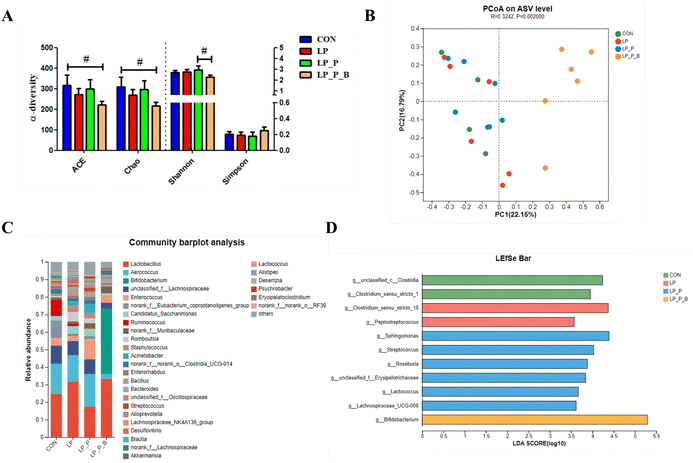
Gut microbiota plays a crucial role in regulating host health. To further investigate the differences in microbiota composition, 16S rRNA gene sequencing was performed on fecal bacterial DNA. The rarefaction curves based on the sobs index indicated that the sequencing depth approached the saturation plateau (Fig. S1†). For α-diversity, indices such as ACE, Chao, Shannon, and Simpson showed no significant differences between the CON, LP, and LP + P groups. However, compared to the CON group, the LP + P + B group exhibited a trend toward increased ACE and Chao indices (P < 0.10) (Fig. 6A). Additionally, the LP + P + B group showed a significant reduction in the Simpson index compared to the LP + P group. In terms of β-diversity, clear segregation of the fecal microbiota structure was observed among treatment groups, as demonstrated by the Bray–Curtis dissimilarity index (Fig. 6B), weighted UniFrac distance (Fig. S2A†), and unweighted UniFrac distance (Fig. S2B†) (ANOSIM, P < 0.05). The microbiota was primarily dominated by three phyla—Firmicutes, Bacteroidota, and Actinobacteriota (Fig. S3A†)—and five genera, including Lactobacillus, Aerococcus, Bifidobacterium, unclassified_f_Lachnospiraceae, and Enterococcus (Fig. 6C). Further analysis at the phylum level revealed no significant differences between the CON, LP, and LP + P groups. However, the LP + P + B group exhibited a significantly lower relative abundance of Firmicutes and a significantly higher relative abundance of Actinobacteriota compared to the other groups (Fig. S3B†). At the genus level, LEfSe analysis (LDA > 3.5) (Fig. 6D) identified notable differences in specific taxa. The LP group showed an increase in the relative abundance of Clostridium_sensu_stricto_18 and Peptostreptococcus, while the LP + P group was enriched in Sphingomonas, Streptococcus, Roseburia, unclassified_f__Erysipelotrichaceae, Lactococcus, and Lachnospiraceae_UCG-006. The LP + P + B group demonstrated a significant increase in the relative abundance of Bifidobacterium. These findings suggest that the addition of plant protein and probiotics can modulate the gut microbiota composition, enhancing diversity and promoting the growth of beneficial bacteria, which may help improve gut health under protein-deficient conditions.Fecal metabolome
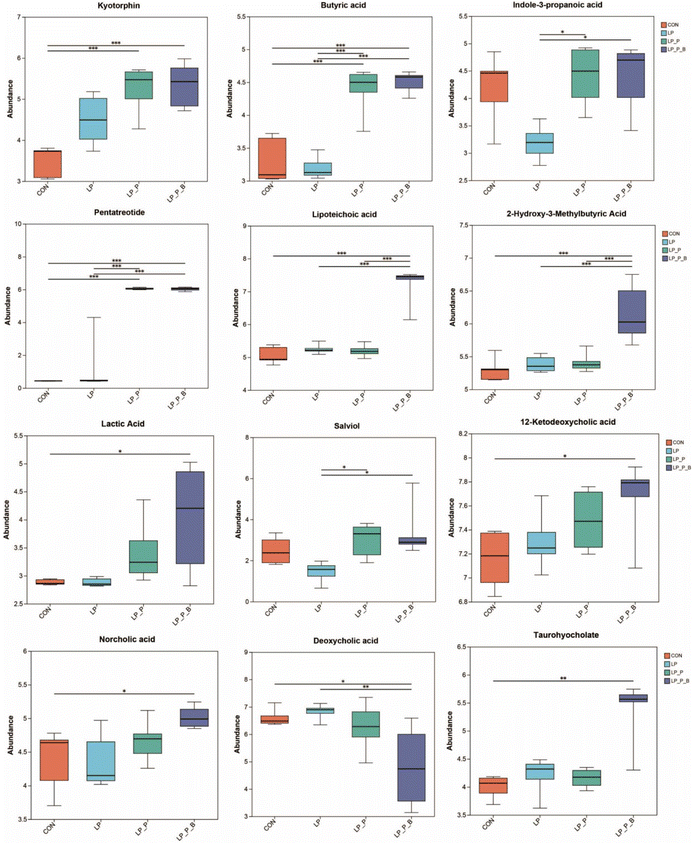
To determine the changes in fecal metabolic profiles, an untargeted metabolomic analysis was conducted. Overall differences in metabolic composition were evaluated using PCA (Fig. S4A and S4B†) and PLS-DA (Fig. S4C and S4D†). The PLS-DA plot revealed a clear separation among the LP + P, LP + P + B, and LP/CON groups, while the distinction between the CON and LP groups was less obvious. A total of 3224 metabolites were detected in the entire metabolome. From the Venn plot (Fig. S4E†), it can be seen that 2776 metabolites were shared among the four groups, accounting for 83.64% of the total metabolites. Among the top 30 metabolites based on their VIP value (VIP > 1, P < 0.05), compared to the CON group, 7 metabolites were upregulated and 23 were downregulated in the LP group (Fig. S5A†). Compared to the LP group, 30 metabolites were upregulated in the LP + P group (Fig. S5B†). Compared to the LP group, 29 metabolites were upregulated and 1 was downregulated in the LP + P + B group (Fig. S5C†). Furthermore, compared to the LP + P group, 24 metabolites were upregulated and 6 were downregulated in the LP + P + B group (Fig. S5D†). Further analysis using the Kruskal–Wallis H test (Fig. 7) revealed that, compared to the CON group, the LP group showed no significant changes. However, the LP + P group exhibited a significant increase in the abundance of kyotorphin, butyric acid, and pentatreotide. The LP + P + B group exhibited significantly increased abundance of kyotorphin, butyric acid, pentatreotide, lipoteichoic acid, 2-hydroxy-3-methylbutyric acid, lactic acid, 12-ketodeoxycholic acid, norcholic acid, and taurohyocholate, while exhibiting a significantly reduced abundance of deoxycholic acid. Compared to the LP group, the LP + P group showed significantly increased abundance of butyric acid, indole-3-propanoic acid, pentatreotide, and salviol, while the LP + P + B group showed a significantly increased abundance of butyric acid, pentatreotide, lipoteichoic acid, 2-hydroxy-3-methylbutyric acid, and salviol, with a significant reduction in deoxycholic acid. Finally, compared to the LP + P group, the LP + P + B group showed significantly increased abundance of lipoteichoic acid and 2-hydroxy-3-methylbutyric acid. KEGG analysis revealed that, compared to the CON group, the LP group showed a significant downregulation of the PPAR signaling pathway, alpha-linolenic acid metabolism, and linoleic acid metabolism, while showing a significant upregulation of the tryptophan metabolism pathway (Fig. S6A†). Compared to the LP group, the LP + P group showed significantly upregulated pathways including those involved in tryptophan metabolism, protein digestion and absorption, alpha-linolenic acid metabolism, and linoleic acid metabolism (Fig. S6B†). Similarly, compared to the LP group, the LP + P + B group showed significantly upregulated pathways including those involved in tryptophan metabolism, protein digestion and absorption, alpha-linolenic acid metabolism, and linoleic acid metabolism, while showing a significant downregulation of the nucleotide metabolism pathway (Fig. S6C†). Compared to the LP + P group, the LP + P + B group showed significantly downregulated purine metabolism and nucleotide metabolism pathways (Fig. S6D†). This analysis suggests that plant protein and probiotic supplementation significantly alters key metabolic pathways and the fecal metabolome, increasing beneficial metabolites related to gut health while reducing harmful ones, particularly under protein-deficient conditions.Correlation between significantly changed fecal microbiota, fecal metabolites, and serum cytokines
To further clarify the relationships between gut microbiota, fecal metabolome, and serum cytokines, Pearson correlation analysis was performed (Fig. 8). The main results showed the following: Roseburia was significantly positively correlated with IL-9 and MIP-1alpha, and significantly negatively correlated with butyric acid and pentatreotide. Unclassified_c__Clostridia was significantly negatively correlated with kyotorphin. Lachnospiraceae_ UCG-006 was significantly negatively correlated with indole-3-propanoic acid and salviol. Clostridium_sensu_stricto_1 was significantly negatively correlated with 12-ketodeoxycholic acid and taurohyocholate. Lactococcus was significantly positively correlated with MCP-3 and significantly negatively correlated with taurohyocholate. Peptostreptococcus was significantly positively correlated with MCP-3 and deoxycholic acid, and significantly negatively correlated with taurohyocholate. Streptococcus was significantly positively correlated with MCP-3 and significantly negatively correlated with butyric acid. Unclassified_f__Erysipelotrichaceae was significantly negatively correlated with butyric acid and pentatreotide. Clostridium_sensu_stricto_18 was significantly positively correlated with G-CSF, MCP-3, and IL-6, and significantly negatively correlated with lipoteichoic acid and taurohyocholate. Bifidobacterium was significantly positively correlated with 2-hydroxy-3-methylbutyric acid, lipoteichoic acid, taurohyocholate, 12-ketodeoxycholic acid, lactic acid, pentatreotide, butyric acid, kyotorphin, and norcholic acid, and significantly negatively correlated with deoxycholic acid. In terms of metabolites: deoxycholic acid was significantly positively correlated with IL-27 and IL-6. Lipoteichoic acid and taurohyocholate were significantly negatively correlated with IL-6. Pentatreotide was significantly positively correlated with IL-5, IL-28, TNF-alpha, and G-CSF. Butyric acid was significantly positively correlated with IL-5 and significantly negatively correlated with IL-9. Kyotorphin was significantly positively correlated with M-CSF, IL-28, IL-12p70, TNF-alpha, and G-CSF. Indole-3-propanoic acid was significantly negatively correlated with IL-27, IL-9, and MIP-1alpha. Salviol was significantly negatively correlated with IL-27 and IL-9. These findings suggest a complex network of interactions between gut microbiota, metabolites, and immune markers, indicating that specific gut bacteria and their metabolites may play critical roles in modulating immune responses and maintaining host health.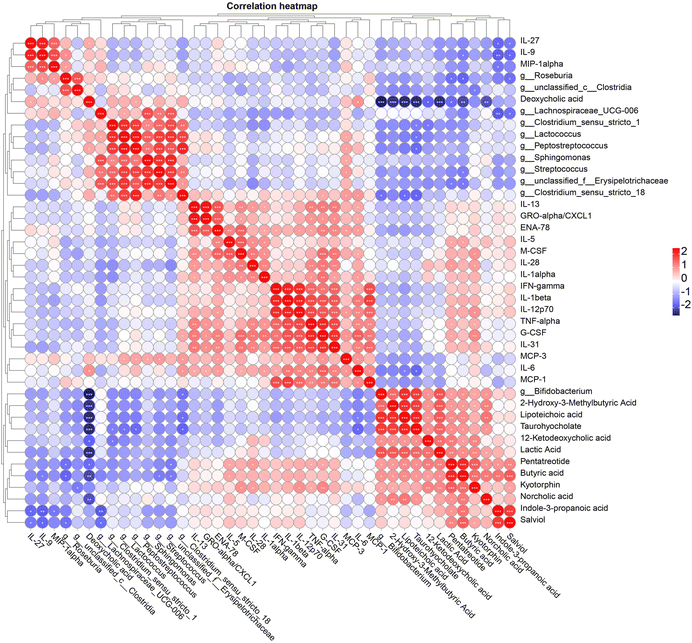 | ||
| Fig. 8 Pearson correlation analysis between significantly changed fecal microbiota at the genus level, fecal metabolites, and serum cytokines; *P < 0.05; **P < 0.01; ***P < 0.001. | ||
Discussion
Aging populations frequently face protein deficiencies, which are closely linked to declines in muscle mass, immune function, and overall health.21–23 To address these challenges, the role of plant protein and probiotics in mitigating protein deficiency is gaining attention, as these dietary interventions may enhance nutrient absorption, promote gut health, and reduce inflammation.24 Our study evaluated the effects of supplementing LP diets with plant protein and probiotics on aged SAMP8 mice. The results indicate significant improvements in nutrient metabolism, body composition, gut microbiota, immune regulation, muscle health, and systemic inflammation, all of which underscore the therapeutic potential of these dietary interventions in maintaining muscle function and systemic health via the gut–muscle axis.Body weight, composition, and nutrient absorption
Despite similar food intake, the LP, LP + P, and LP + P + B groups showed significant weight gain, indicating that the LP diet and its supplementation may promote weight gain. The LP group exhibited a trend towards increased body fat percentage, potentially as a compensatory response to protein deficiency.21 In aged mouse models, the mechanisms behind LP diet-induced weight gain, particularly fat accumulation, may include muscle loss and reduced metabolic efficiency, as studies indicate that the absence of essential AAs (e.g., methionine, lysine) in LP diets significantly impairs muscle synthesis in aged mice, while supplementation improves growth performance, indirectly highlighting the role of AA deficiency in exacerbating muscle breakdown,25 as supported by the compensatory upregulation of muscle protein synthesis-related genes observed in this study. Additionally, age-related metabolic adaptations, such as mitochondrial dysfunction and increased oxidative stress, may reduce energy utilization efficiency, predisposing aged mice to fat storage rather than expenditure, with LP diets potentially worsening metabolic inefficiency by further lowering NAD+ levels.26 Moreover, LP diets may disrupt circadian rhythms of serum insulin and hepatic lipid metabolism, leading to persistently lower daytime insulin levels and increased nighttime fat synthesis,27 indirectly promoting fat accumulation, while also enhancing hepatic lipogenesis through upregulated expression of enzymes like acetyl-CoA carboxylase 1 (ACC1), as evidenced by elevated ACC1 mRNA and protein levels in the livers of LP-fed mice, resulting in hepatic triglyceride accumulation.27 However, supplementation with plant protein and probiotics mitigated fat accumulation, likely by improving nutrient metabolism and microbiome health, consistent with previous findings that LP diets can increase fat storage.28–30 Nutrient absorption analysis revealed no significant differences in crude protein and energy digestibility among groups. Still, the LP + P and LP + P + B groups demonstrated higher protein utilization than the LP group, suggesting enhanced protein bioavailability due to supplementation. Probiotics likely contributed to improved nutrient absorption through gut microbiota modulation, highlighting the critical role of the gut–muscle axis in muscle health maintenance.24,31 This finding is particularly relevant for elderly populations, who often face inadequate protein intake and for whom gut health is crucial for maintaining overall metabolic balance.Regarding protein metabolism-related genes, the LP group exhibited upregulation of genes involved in protein synthesis in muscle, such as mTOR, S6K1, and 4EBP, likely as an adaptive response to protein deficiency. However, supplementation with plant protein and probiotics normalized these gene expressions, indicating their potential to alleviate metabolic stress associated with protein deficiency. Additionally, oxidative stress-related genes such as CAT, Nrf2, GPX1, and HO-1 were upregulated in the LP group, reflecting increased oxidative stress levels; supplementation led to their downregulation, indicating reduced oxidative stress and improved muscle health.32 This is particularly relevant for elderly populations, who typically experience increased oxidative stress and muscle mass decline, highlighting the benefits of dietary supplements.
Muscle strength, endurance, and systemic inflammation
Protein deficiency has well-documented adverse effects on muscle performance, especially in aging populations.33 Consistent with this, the LP group showed significantly reduced muscle endurance and strength. However, supplementation with plant protein and probiotics restored muscle function in the LP + P and LP + P + B groups, suggesting that these supplements effectively counteract muscle degradation by improving nutrient utilization and reducing inflammation. This finding is critical for elderly populations, as muscle function deterioration is closely related to protein metabolism imbalances, and appropriate dietary interventions can significantly mitigate these issues.34,35Studies have shown that supplementation with Lactobacillus casei Shirota or a combination of Bifidobacterium bifidum and Lactobacillus paracasei can enhance muscle function through the gut–muscle axis in aged mice.5,36 This finding underscores the significant role of probiotics in improving muscle health in the elderly. In contrast, although our study did not use probiotics alone, we employed a combination of probiotics and plant protein. Remarkably, the restoration of muscle function in the LP + P and LP + P + B groups was far more pronounced. This could be attributed to the synergistic effect between the plant protein and the multi-strain probiotic blend employed in our study. Such a combination might exert a more extensive influence on gut health and muscle metabolism.
In terms of protein sources, at similar protein intakes, studies have reported a lower ability of plant-based protein sources to stimulate protein synthesis at the skeletal muscle level and induce muscle mass gain compared to animal-based protein sources, especially in older people.37 However, other research has shown that plant-based protein supplementation attenuated age-related sarcopenia by inducing antioxidant enzymes and SCFAs via gut microbiota regulation.12 Our study further supports the latter view. In our study, although there was no comparison with animal-based protein in the experiment, the combination of plant protein and probiotics not only addresses the potential limitations of using plant proteins alone but also enhances the positive impacts on muscle function through the interaction with probiotics. The underlying mechanism of this interaction requires further investigation.
Elderly individuals often experience increased systemic inflammation, particularly under nutrient-deficient conditions such as LP diets.38,39 In our present study, serum cytokine analysis revealed elevated levels of pro-inflammatory cytokines (e.g., IL-1β, IL-6, and TNF-α) in the LP group, indicating exacerbated systemic inflammation under LP conditions. Supplementation significantly reduced these cytokines, with the LP + P + B group showing the most pronounced anti-inflammatory effects. The modulation of inflammation and cytokine levels further emphasizes the interplay between gut health and muscle function, highlighting the importance of the gut–muscle axis in maintaining muscle health. This inflammation regulation is crucial for elderly individuals, often suffering from chronic low-grade inflammation affecting overall functionality.24,40 Similar to our findings, a study also reported a reduction in pro-inflammatory cytokines after probiotic supplementation in elderly subjects with chronic inflammation.5 However, their study did not incorporate protein supplementation. Our results suggest that the combination of plant protein and probiotics may have a more potent anti-inflammatory effect, which could be related to the improved gut microbiota composition and enhanced nutrient metabolism resulting from the combined intervention.
The overall impact on gut health
The integrity of the intestinal barrier is crucial for preventing harmful substances from entering the bloodstream and maintaining overall gut health.41 This study evaluated several key genes related to barrier function, including ZO-1, CLDN1, and OCLD, which are involved in tight junction formation and epithelial barrier integrity. No significant differences were observed in the expression of these genes across the experimental groups, indicating that neither the LP diet nor the supplementation significantly altered tight junction integrity at the gene expression level. However, MUC2, a gene responsible for mucin production—a key component of the protective mucus layer in the gut—was significantly upregulated in the LP + P group compared to the CON, LP, and LP + P + B groups. This suggests that plant protein supplementation alone may stimulate mucus production, potentially enhancing the gut's protective barrier function.42,43 The upregulation of MUC2 could be a compensatory mechanism to reinforce mucosal defense in response to an LP diet.Another key finding of the study was the modulation of immune-related gene expression in the colon. Although the expression levels of pro-inflammatory cytokines such as IL-6, TNF-α, and IL-1β showed no significant differences between the groups, IL-12, a pro-inflammatory cytokine that promotes T-helper cell differentiation,44 was significantly upregulated in the LP group compared to the CON group. This indicates that an LP diet might elevate specific inflammatory markers in the gut. Notably, this pro-inflammatory response was attenuated by supplementation with plant protein and probiotics. Both the LP + P and LP + P + B groups exhibited significantly downregulated IL-12 expression compared to the LP group, suggesting that supplementation effectively reduced inflammation in the colon.45,46 This aligns with the systemic anti-inflammatory effects observed in muscle tissue and serum, emphasizing the potential of plant protein and probiotics to modulate immune responses both systemically and locally in the gut.
Oxidative stress is another important factor affecting colon health, especially under conditions of nutrient deficiency.47,48 The study evaluated the expression of several oxidative stress-related genes, including CAT, SOD1, SOD2, GPX1, and Nrf2. No significant differences were found in the expression of CAT and SOD2, indicating that the general oxidative stress response was not markedly altered across groups. However, GPX1, an important gene involved in detoxifying reactive oxygen species, was significantly upregulated in the LP group compared to the control group, suggesting that an LP diet induced oxidative stress in the colon.49 Supplementation with plant protein and probiotics reversed this effect, with both the LP + P and LP + P + B groups showing significantly downregulated GPX1 expression compared to the LP group. This suggests that the supplements helped alleviate oxidative stress in the colon, potentially contributing to the overall health benefits observed in the supplemented groups.
Interestingly, the LP + P group showed a significant upregulation of SOD1, a gene involved in superoxide dismutation, compared to the CON group, while the LP + P + B group showed a significant downregulation of Nrf2 expression compared to the LP + P group. These findings suggest that plant protein supplementation alone might activate certain antioxidant pathways, as evidenced by the upregulation of SOD1, whereas the combination of plant protein and probiotics might exert a more balanced regulatory effect by downregulating Nrf2, a key regulator of antioxidant responses.50 This nuanced regulation of oxidative stress pathways indicates that probiotics, in combination with plant protein, might provide a more targeted approach to managing oxidative stress in the gut, particularly under conditions of protein deficiency.
The results from the gene expression analysis in the colon suggest that an LP diet can induce localized inflammation and oxidative stress in the gut, even in the absence of significant changes in systemic inflammation. The upregulation of the pro-inflammatory cytokine IL-12 and oxidative stress marker GPX1 in the LP group indicates that protein deficiency can negatively affect gut health, potentially contributing to systemic dysfunction.51 However, supplementation with plant protein and probiotics effectively mitigated these negative effects by downregulating both inflammatory and oxidative stress markers. Additionally, the specific upregulation of MUC2 and SOD1 in the LP + P group highlights the role of plant protein in enhancing the gut's barrier and antioxidant defenses.
Intestinal microbiota composition and diversity
16S rRNA sequencing revealed significant alterations in the gut microbiota composition across treatment groups. Although α-diversity indices such as ACE, Chao, Shannon, and Simpson did not differ significantly between the CON, LP, and LP + P groups, there was an increasing trend in ACE and Chao indices in the LP + P + B group, suggesting enhanced microbial richness. This trend aligns with previous studies showing that probiotics can improve gut microbial diversity.12 β-Diversity analysis demonstrated distinct microbial communities across groups, indicating that both plant protein and probiotics play a role in shaping the gut microbiota structure. Increased gut microbial diversity not only aids in better digestion but also positively impacts overall health. At the phylum level, the LP + P + B group exhibited a significant decrease in Firmicutes and an increase in Actinobacteriota, notably Bifidobacterium. Bifidobacterium is known for its beneficial effects on gut health, including SCFA production and pathogen inhibition.52 We found that Bifidobacterium was positively correlated with anti-inflammatory metabolites, including butyric acid and kyotorphin, further supporting the immune-modulating effects of probiotics. Thus, the increased abundance of Bifidobacterium further supports the beneficial role of probiotics in improving gut health and underscores the importance of gut microbiota in systemic inflammation regulation.Fecal metabolome and key pathways
Untargeted metabolomic analysis showed significant changes in fecal metabolites between groups. Compared to the CON group, the LP group exhibited reduced metabolite diversity, highlighting the detrimental effects of protein deficiency. However, the LP + P and LP + P + B groups showed upregulation of several key metabolites, including butyric acid, indole-3-propanoic acid, and kyotorphin, which are associated with improved gut barrier function and reduced inflammation.53–55 The LP + P + B group also demonstrated increased levels of lipoteichoic acid and lactic acid, alongside specific changes in bile acid levels. Notably, there was an increase in 12-ketodeoxycholic acid, norcholic acid, and taurohyocholate, and a decrease in deoxycholic acid in this group. Bile acids play a crucial role in intestinal anti-inflammatory and immune regulation processes and are closely related to the intestinal microbiota.56–58 These findings indicate a link between immune modulation and gut homeostasis, further emphasizing the benefits of these supplements under protein-deficient conditions.Pathway analysis revealed that the LP group downregulated pathways related to PPAR signaling and fatty acid metabolism, which are critical for maintaining metabolic homeostasis. In contrast, the LP + P and LP + P + B groups upregulated pathways involved in tryptophan metabolism, protein digestion, and alpha-linolenic acid metabolism, suggesting that plant protein and probiotics help restore key metabolic pathways disrupted by protein deficiency.37,59 The restoration of these pathways is particularly important for elderly populations who often suffer from poor protein metabolism, with appropriate dietary supplements helping improve metabolic functions.
Limitations of this study
Despite standardizing the experimental conditions, the variability in the baseline microbiota among mice, coupled with individual differences such as their responses to external stressors, might have confounded our results. For future research, it is recommended to characterize the gut microbiota composition at the beginning of the experiment to ensure the consistency of the baseline microbiota and to strictly control all experimental conditions, including animal handling, housing conditions, and dietary changes, which are external factors that can induce stress in mice. Additionally, although the present study employs an aging mouse model, there are considerable challenges in generalizing the findings to the elderly human population. In humans, long-term dietary patterns, medication usage, underlying health conditions, and a wide spectrum of dietary preferences all contribute to high variability in gut microbiota. Moreover, there are significant disparities in dietary habits between humans and mice. This variability in gut microbiota may lead to different responses to the dietary interventions of plant-based proteins and probiotics compared to the mice in this study. Consequently, personalized research approaches that consider individual gut microbiota profiles are crucial for human-centered investigations. Furthermore, the sample size of animals in this study was relatively limited. For future research, a larger sample size is indispensable to enhance the reliability and representativeness of the results. The lack of a single probiotic treatment group also compromises the comprehensiveness of the study's findings. Finally, while our research has identified correlations among gut microbiota, metabolites, and muscle health, the underlying mechanistic relationships are still speculative. Further experiments, such as fecal microbiota transplantation or supplementation with targeted metabolites (e.g., butyric acid, indole-3-propionic acid, kyotorphin, etc.), could provide evidence to establish causal relationships.Conclusion
This study demonstrates that supplementing a low-protein diet with plant protein and probiotics significantly affects gut microbiota composition, fecal metabolite profiles, immune responses, and muscle health in aged mice. The findings suggest that these dietary interventions could enhance protein utilization, reduce systemic inflammation, and improve muscle function. Future research should focus on identifying specific probiotic strains and examining their long-term effects on human health, particularly in aging populations and under conditions of protein malnutrition. These results highlight the potential of plant protein and probiotics as effective dietary strategies for promoting gut and muscle health, especially in elderly individuals with limited protein intake. The interplay between gut health, nutrient metabolism, and muscle function presents a promising avenue for improving health outcomes in protein-deficient conditions through targeted dietary interventions.Author contributions
Ruyi Han: writing – original draft, investigation, and data curation; Limin Ouyang: methodology, investigation, and data curation; Chenggang Yin: visualization and formal analysis; Long Cai: validation and methodology; Qiming Wu: methodology and writing – review & editing; Liang Chen: resources, conceptualization, and writing – review & editing; Jun Du: methodology and writing – review & editing; Xilong Li: resources and writing – review & editing; Zhigang Zhu: writing – review & editing, resources, and conceptualization; Yu Pi: writing – review & editing, supervision, project administration, funding acquisition, and conceptualization.Data availability
All data used in this study are presented in the illustrated figures, and the raw data will promptly be made available upon request.Conflicts of interest
Limin Ouyang, Qiming Wu, Liang Chen, Jun Du, and Zhigang Zhu are employees of Amway (Shanghai) Innovation & Science Co., Ltd with long-term research and commercial interest in plant protein. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. None of the other authors declared any conflicts of interest.Acknowledgements
This study was financially supported by the China Postdoctoral Science Foundation (2023M730594), the Agricultural Science and Technology Innovation Program of the Feed Research Institute of the Chinese Academy of Agricultural Sciences (CAAS-IFR-ZDRW202402), the Central Public-interest Scientific Institution Basal Research Fund (1610382023011), and an Investigated Program (Am20230115RD) funded by Amway (China) Limited Company.References
- T. Dao, A. E. Green, Y. A. Kim, S. J. Bae, K. T. Ha, K. Gariani, M. Lee, K. J. Menzies and D. Ryu, Sarcopenia and Muscle Aging: A Brief Overview, Endocrinol. Metab., 2020, 35, 716–732 CrossRef CAS PubMed.
- L. Larsson, H. Degens, M. Li, L. Salviati, Y. I. Lee, W. Thompson, J. L. Kirkland and M. Sandri, Sarcopenia: aging-related loss of muscle mass and function, Physiol. Rev., 2019, 99, 427–511 CrossRef PubMed.
- J. Zhao, Y. Huang and X. Yu, A narrative review of gut-muscle axis and sarcopenia: The potential role of gut microbiota, Int. J. Gen. Med., 2021, 1263–1273 CrossRef PubMed.
- J. Frampton, K. G. Murphy, G. Frost and E. S. Chambers, Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function, Nat. Metab., 2020, 2, 840–848 CrossRef CAS PubMed.
- L. H. Chen, S. S. Chang, H. Y. Chang, C. H. Wu, C. H. Pan, C. C. Chang, C. H. Chan and H. Y. Huang, Probiotic supplementation attenuates age–related sarcopenia via the gut–muscle axis in SAMP8 mice, J. Cachexia Sarcopenia Muscle, 2022, 13, 515–531 CrossRef PubMed.
- L.-H. Chen, M.-F. Wang, C.-C. Chang, S.-Y. Huang, C.-H. Pan, Y.-T. Yeh, C.-H. Huang, C.-H. Chan and H.-Y. Huang, Lacticaseibacillus paracasei PS23 effectively modulates gut microbiota composition and improves gastrointestinal function in aged SAMP8 mice, Nutrients, 2021, 13, 1116 CrossRef CAS PubMed.
- C. Putra, N. Konow, M. Gage, C. G. York and K. M. Mangano, Protein source and muscle health in older adults: a literature review, Nutrients, 2021, 13, 743 CrossRef CAS PubMed.
- Y. F. Ouyang, T. Y. Tan, X. Y. Song, F. F. Huang, B. Zhang, G. Q. Ding and H. J. Wang, Dietary Protein Intake Dynamics in Elderly Chinese from 1991 to 2018, Nutrients, 2021, 13, 3806 CrossRef CAS PubMed.
- H. Ming, J. Tan, S.-Y. Cao, C.-P. Yu, Y.-T. Qi, C. Wang, L. Zhang, Y. Liu, J. Yuan and M. Yin, NUFIP1 integrates amino acid sensing and DNA damage response to maintain the intestinal homeostasis, Nat. Metab., 2025, 1–17 CAS.
- H. Masuoka, W. Suda, E. Tomitsuka, C. Shindo, L. Takayasu, P. Horwood, A. R. Greenhill, M. Hattori, M. Umezaki and K. Hirayama, The influences of low protein diet on the intestinal microbiota of mice, Sci. Rep., 2020, 10, 17077 CrossRef CAS PubMed.
- R. Qaisar, A. Burki, A. Karim, M. S. Iqbal and F. Ahmad, Probiotics Supplements Improve the Sarcopenia-Related Quality of Life in Older Adults with Age-Related Muscle Decline, Calcif. Tissue Int., 2024, 114, 583–591 CrossRef CAS PubMed.
- S. S. Chang, L. H. Chen, K. C. Huang, S. W. Huang, C. C. Chang, K. W. Liao, E. C. Hu, Y. P. Chen, Y. W. Chen, P. C. Hsu and H. Y. Huang, Plant-based polyphenol rich protein supplementation attenuated skeletal muscle loss and lowered the LDL level via gut microbiota remodeling in Taiwan's community-dwelling elderly, Food Funct., 2023, 14, 9407–9418 RSC.
- H. Yuan, Z. Luo, Z. Ban, R. J. Reiter, Q. Ma, Z. Liang, M. Yang, X. Li and L. Li, Bioactive peptides of plant origin: distribution, functionality, and evidence of benefits in food and health, Food Funct., 2022, 13, 3133–3158 RSC.
- M. Bagherniya, A. Mahdavi, N. Shokri-Mashhadi, M. Banach, S. Von Haehling, T. P. Johnston and A. Sahebkar, The beneficial therapeutic effects of plant-derived natural products for the treatment of sarcopenia, J. Cachexia Sarcopenia Muscle, 2022, 13, 2772–2790 CrossRef PubMed.
- M. E. Sanders, D. J. Merenstein, G. Reid, G. R. Gibson and R. A. Rastall, Probiotics and prebiotics in intestinal health and disease: from biology to the clinic, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 605–616 CrossRef PubMed.
- R. M. J. Deacon, Measuring the Strength of Mice, J. Visualized Exp., 2013, 76, 2610 Search PubMed.
- N. Thiex, L. Novotny and A. Crawford, Determination of ash in animal feed: AOAC official method 942.05 revisited, J. AOAC Int., 2012, 95, 1392–1397 CrossRef CAS PubMed.
- C. J. Zhang, M. Yu, Y. X. Yang, C. L. Mu, Y. Su and W. Y. Zhu, Differential effect of early antibiotic intervention on bacterial fermentation patterns and mucosal gene expression in the colon of pigs under diets with different protein levels, Appl. Microbiol. Biotechnol., 2017, 101, 2493–2505 CrossRef CAS PubMed.
- Y. Pi, X. Y. Zhang, Y. J. Wu, Z. Y. Wang, Y. Bai, X. Y. Liu, D. D. Han, J. B. Zhao, I. Tobin, J. C. Zhao, G. L. Zhang and J. J. Wang, Alginate Alleviates Dextran Sulfate Sodium-Induced Colitis by Promoting Bifidobacterium animalis and Intestinal Hyodeoxycholic Acid Synthesis in Mice, Microbiol. Spectrum, 2022, 10, e02979–22 Search PubMed.
- Y. Ren, G. Yu, C. Shi, L. Liu, Q. Guo, C. Han, D. Zhang, L. Zhang, B. Liu and H. Gao, Majorbio Cloud: A one–stop, comprehensive bioinformatic platform for multiomics analyses, iMeta, 2022, 1, e12 CrossRef CAS PubMed.
- J. Bollwein, R. Diekmann, M. J. Kaiser, J. M. Bauer, W. Uter, C. C. Sieber and D. Volkert, Distribution but not amount of protein intake is associated with frailty: a cross-sectional investigation in the region of Nürnberg, Nutr. J., 2013, 12, 109 CrossRef CAS PubMed.
- J. Bauer, G. Biolo, T. Cederholm, M. Cesari, A. J. Cruz-Jentoft, J. E. Morley, S. Phillips, C. Sieber, P. Stehle, D. Teta, R. Visvanathan, E. Volpi and Y. Boirie, Evidence-Based Recommendations for Optimal Dietary Protein Intake in Older People: A Position Paper From the PROT-AGE Study Group, J. Am. Med. Dir. Assoc., 2013, 14, 542–559 CrossRef PubMed.
- A. J. Cruz-Jentoft, J. P. Baeyens, J. M. Bauer, Y. Boirie, T. Cederholm, F. Landi, F. C. Martin, J.-P. Michel, Y. Rolland and S. M. Schneider, Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in Older People, Age Ageing, 2010, 39, 412–423 CrossRef PubMed.
- H. Tilg and A. R. Moschen, Microbiota and diabetes: an evolving relationship, Gut, 2014, 63, 1513–1521 CrossRef CAS PubMed.
- E. S. Riddle, M. H. Stipanuk and A. E. Thalacker-Mercer, Amino acids in healthy aging skeletal muscle, Front. Biosci. (Elite Ed.), 2016, 8, 326–350 Search PubMed.
- M. Romani, V. Sorrentino, C. M. Oh, H. Li, T. I. de Lima, H. Zhang, M. Shong and J. Auwerx, NAD(+) boosting reduces age-associated amyloidosis and restores mitochondrial homeostasis in muscle, Cell Rep., 2021, 34, 108660 CrossRef CAS PubMed.
- S. I. Yokota, K. Nakamura, M. Ando, A. Haraguchi, K. Omori and S. Shibata, A low-protein diet eliminates the circadian rhythm of serum insulin and hepatic lipid metabolism in mice, J. Nutr. Biochem., 2019, 63, 177–185 CrossRef CAS PubMed.
- R. M. Daly, S. L. O'Connell, N. L. Mundell, C. A. Grimes, D. W. Dunstan and C. A. Nowson, Protein-enriched diet, with the use of lean red meat, combined with progressive resistance training enhances lean tissue mass and muscle strength and reduces circulating IL-6 concentrations in elderly women: a cluster randomized controlled trial, Am. J. Clin. Nutr., 2014, 99, 899–910 CrossRef CAS PubMed.
- S. A. de França, M. P. dos Santos, M. A. R. Garófalo, L. C. Navegantes, I. D. Kettelhut, C. F. Lopes and N. H. Kawashita, Low protein diet changes the energetic balance and sympathetic activity in brown adipose tissue of growing rats, Nutrition, 2009, 25, 1186–1192 CrossRef PubMed.
- D. G. Le Couteur, S. Solon-Biet, V. C. Cogger, S. J. Mitchell, A. Senior, R. de Cabo, D. Raubenheimer and S. J. Simpson, The impact of low-protein high-carbohydrate diets on aging and lifespan, Cell. Mol. Life Sci., 2016, 73, 1237–1252 CrossRef CAS PubMed.
- M. Giron, M. Thomas, D. Dardevet, C. Chassard and I. Savary-Auzeloux, Gut microbes and muscle function: can probiotics make our muscles stronger?, J. Cachexia Sarcopenia Muscle, 2022, 13, 1460–1476 CrossRef PubMed.
- I. Liguori, G. Russo, F. Curcio, G. Bulli, L. Aran, D. Della-Morte, G. Gargiulo, G. Testa, F. Cacciatore and D. Bonaduce, Oxidative stress, aging, and diseases, Clin. Interventions Aging, 2018, 757–772 CrossRef CAS.
- N. A. Burd, C. F. McKenna, A. F. Salvador, K. J. M. Paulussen and D. R. Moore, Dietary Protein Quantity, Quality, and Exercise Are Key to Healthy Living: A Muscle-Centric Perspective Across the Lifespan, Front. Nutr., 2019, 6, 83 CrossRef PubMed.
- F. Landi, R. Calvani, M. Tosato, A. M. Martone, E. Ortolani, G. Savera, E. D’Angelo, A. Sisto and E. Marzetti, Protein Intake and Muscle Health in Old Age: From Biological Plausibility to Clinical Evidence, Nutrients, 2016, 8, 295 CrossRef PubMed.
- K. Prokopidis, M. M. Cervo, A. Gandham and D. Scott, Impact of Protein Intake in Older Adults with Sarcopenia and Obesity: A Gut Microbiota Perspective, Nutrients, 2020, 12, 2285 CrossRef CAS PubMed.
- J. S. Baek, Y. J. Shin, X. Y. Ma, H. S. Park, Y. H. Hwang and D. H. Kim, Bifidobacterium bifidum and Lactobacillus paracasei alleviate sarcopenia and cognitive impairment in aged mice by regulating gut microbiota-mediated AKT, NF-κB, and FOXO3a signaling pathways, Immun. Ageing, 2023, 20, 56 CrossRef CAS PubMed.
- I. Berrazaga, V. Micard, M. Gueugneau and S. Walrand, The Role of the Anabolic Properties of Plant- versus Animal-Based Protein Sources in Supporting Muscle Mass Maintenance: A Critical Review, Nutrients, 2019, 11, 1825 CrossRef CAS PubMed.
- H. Fang, S. Ghosh, L. C. Sims, K. P. Stone, C. M. Hill, D. Spires, D. V. Ilatovskaya, C. D. Morrison, T. W. Gettys and K. Stadler, FGF21 prevents low-protein diet-induced renal inflammation in aged mice, Am. J. Physiol.: Renal Physiol., 2021, 321, F356–F368 CrossRef CAS PubMed.
- J. H. Guo, H. Y. Wang, Q. Zhang, M. Feng and L. J. Zhang, The low protein diet affects the nonspecific inflammatory response of middle-aged and old mice through mTOR, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 7551–7561 Search PubMed.
- S. M. Phillips, Dietary protein requirements and adaptive advantages in athletes, Br. J. Nutr., 2012, 108, S158–S167 CrossRef CAS PubMed.
- J. König, J. Wells, P. D. Cani, C. L. García-Ródenas, T. MacDonald, A. Mercenier, J. Whyte, F. Troost and R. J. Brummer, Human Intestinal Barrier Function in Health and Disease, Clin. Transl. Gastroenterol., 2016, 7, e196 CrossRef PubMed.
- P. Paone and P. D. Cani, Mucus barrier, mucins and gut microbiota: the expected slimy partners?, Gut, 2020, 69, 2232–2243 CrossRef CAS PubMed.
- Y. Liu, X. J. Yu, J. X. Zhao, H. Zhang, Q. X. Zhai and W. Chen, The role of MUC2 mucin in intestinal homeostasis and the impact of dietary components on MUC2 expression, Int. J. Biol. Macromol., 2020, 164, 884–891 CrossRef CAS PubMed.
- G. Trinchieri, Proinflammatory and immunoregulatory functions of interleukin-12, Int. Rev. Immunol., 1998, 16, 365–396 CrossRef CAS PubMed.
- C. Eftychi, R. Schwarzer, K. Vlantis, L. Wachsmuth, M. Basic, P. Wagle, M. F. Neurath, C. Becker, A. Bleich and M. Pasparakis, Temporally Distinct Functions of the Cytokines IL-12 and IL-23 Drive Chronic Colon Inflammation in Response to Intestinal Barrier Impairment, Immunity, 2019, 51, 367–380 CrossRef CAS PubMed.
- S. Chikano, K. Sawada, T. Shimoyama, S. I. Kashiwamura, A. Sugihara, K. Sekikawa, N. Terada, K. Nakanishi and H. Okamura, IL-18 and IL-12 induce intestinal inflammation and fatty liver in mice in an IFN-γ dependent manner, Gut, 2000, 47, 779–786 CrossRef CAS PubMed.
- B. L. Tan, M. E. Norhaizan and W.-P.-P. Liew, Nutrients and oxidative stress: friend or foe?, Oxid. Med. Cell. Longevity, 2018, 2018, 9719584 CrossRef PubMed.
- A. Bardelcíková, J. Soltys and J. Mojzis, Oxidative Stress, Inflammation and Colorectal Cancer: An Overview, Antioxidants, 2023, 12, 901 CrossRef PubMed.
- R. Quintana-Cabrera, S. Fernandez-Fernandez, V. Bobo-Jimenez, J. Escobar, J. Sastre, A. Almeida and J. P. Bolaños, γ-Glutamylcysteine detoxifies reactive oxygen species by acting as glutathione peroxidase-1 cofactor, Nat. Commun., 2012, 3, 718 CrossRef PubMed.
- J. Kim and Y. S. Keum, NRF2, a Key Regulator of Antioxidants with Two Faces towards Cancer, Oxid. Med. Cell. Longevity, 2016, 2746457 CrossRef PubMed.
- T. Tian, Z. Wang and J. Zhang, Pathomechanisms of oxidative stress in inflammatory bowel disease and potential antioxidant therapies, Oxid. Med. Cell. Longevity, 2017, 2017, 4535194 CrossRef PubMed.
- D. A. Russell, R. P. Ross, G. F. Fitzgerald and C. Stanton, Metabolic activities and probiotic potential of bifidobacteria, Int. J. Food Microbiol., 2011, 149, 88–105 CrossRef CAS PubMed.
- P. Louis, G. L. Hold and H. J. Flint, The gut microbiota, bacterial metabolites and colorectal cancer, Nat. Rev. Microbiol., 2014, 12, 661–672 CrossRef CAS PubMed.
- K. Conceiçao, P. R. Magalhaes, S. R. R. Campos, M. M. Domingues, V. G. Ramu, M. Michalek, P. Bertani, A. M. Baptista, M. Heras, E. R. Bardaji, B. Bechinger, M. L. Ferreira and M. A. R. B. Castanho, The anti-inflammatory action of the analgesic kyotorphin neuropeptide derivatives: insights of a lipid-mediated mechanism, Amino Acids, 2016, 48, 307–318 CrossRef PubMed.
- D. X. Ming, X. C. Xu, X. R. Jiang, Y. P. Li, W. J. Sun, J. B. Xiang, M. Y. Huang, Y. Pi and X. L. Li, Indole-3-propionic acid enhances growth performance and reduces diarrhea via modulating redox status and intestinal inflammation in weaned piglets, Anim. Nutr., 2024, 19, 240–247 CrossRef CAS PubMed.
- I. Mohanty, C. Allaband, H. Mannochio-Russo, Y. El Abiead, L. R. Hagey, R. Knight and P. C. Dorrestein, The changing metabolic landscape of bile acids - keys to metabolism and immune regulation, Nat. Rev. Gastroenterol. Hepatol., 2024, 21, 493–516 CrossRef PubMed.
- Y. Pi, Y. J. Wu, X. Y. Zhang, D. D. Lu, D. D. Han, J. C. Zhao, X. J. Zheng, S. Y. Zhang, H. Ye, S. Lian, Y. Bai, Z. Y. Wang, S. Y. Tao, D. J. Ni, X. H. Zou, W. Jia, G. L. Zhang, D. F. Li and J. J. Wang, Gut microbiota-derived ursodeoxycholic acid alleviates low birth weight-induced colonic inflammation by enhancing M2 macrophage polarization, Microbiome, 2023, 11, 19 CrossRef CAS PubMed.
- S. L. Collins, J. C. Stine, J. E. Bisanz, C. D. Okafor and A. D. Patterson, Bile acids and the gut microbiota: metabolic interactions and impacts on disease, Nat. Rev. Microbiol., 2023, 21, 236–247 CrossRef CAS PubMed.
- R. P. Singh, A. Shadan and Y. Ma, Biotechnological Applications of Probiotics: A Multifarious Weapon to Disease and Metabolic Abnormality, Probiotics Antimicrob. Proteins, 2022, 14, 1184–1210 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fo01400j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |