Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microsensor systems for cell metabolism – from 2D culture to organ-on-chip (2019–2024)
Johannes
Dornhof
 ,
Jochen
Kieninger
,
Jochen
Kieninger
 ,
Stefan J.
Rupitsch
and
Andreas
Weltin
,
Stefan J.
Rupitsch
and
Andreas
Weltin
 *
*
Laboratory for Electrical Instrumentation and Embedded Systems, IMTEK – Department of Microsystems Engineering, University of Freiburg, Georges-Köhler-Allee 103, 79110 Freiburg, Germany. E-mail: weltin@imtek.de; Tel: +49 761 203 7263
First published on 8th January 2025
Abstract
Cell cultures, organs-on-chip and microphysiological systems become increasingly relevant as in vitro models, e.g., in drug development, disease modelling, toxicology or cancer research. It has been underlined repeatedly that culture conditions and metabolic cues have a strong or even essential influence on the reproducibility and validity of such experiments but are often not appropriately measured or controlled. Here we review microsensor systems for cell metabolism for the continuous measurement of culture conditions in microfluidic and lab-on-chip platforms. We identify building blocks, features and essential advantages to underline the relevance of these systems and to derive appropriate requirements for development and practical use. We discuss different formats and geometries of cell culture, microfluidics and the resulting consequences for sensor placement, as the prerequisite for understanding the various approaches and classification of the systems. The major chemical and biosensors based on electrochemical and optical principles are discussed for general understanding and to contextualize current developments. We then review selected recent sensor systems with real-world implementations of sensing in cell cultures and organs-on-chip, employing a helpful characterization. That includes formats and cell models, microfluidic systems and sensor types applied in static and dynamic monitoring of 2D and 3D cell cultures, as well as single spheroids. We discuss notable advances, particularly with respect to sensor performance and the demonstration of long-term continuous measurements. We outline current approaches to system fabrication technologies, material choice, and interfacing, and comment on recent trends. Finally, we conclude with critical remarks on the current state of sensors in cell culture monitoring and identify avenues for future improvements for both developers and users of such systems, which will lead to better and more predictive in vitro models.
Introduction
Background and motivation
Organs-on-chip, 3D cell cultures, spheroids or microtissues are increasingly relevant in vitro models in biology and biomedicine. Classical 2D monolayer cell cultures still remain a cornerstone of cell biological and biomedical research. Countless microfluidic and lab-on-a-chip systems exist for growing and handling cell cultures and organs-on-chip, and many have integrated sensors to monitor some form of cellular state or the culture conditions. In general, the cellular state can range from simple general parameters such as viability, cell growth or cellular adhesion, over the basic energy metabolism to more complex signalling molecules and biomarkers. At the same time, fabrication technologies, miniaturized sensors and transduction principles continuously advance through progress in biotechnology, materials science and micro-/nanofabrication. Interestingly, even basic sensor integration into cell culture systems and organs-on-chip is not as widespread despite the promise of robust and powerful applications. Many aspects of cell cultures and organs-on-chip will strongly benefit from sensor integration, ranging from basic requirements, such as standardization of culture conditions, to ambitious future perspectives, such as personalized treatment in cancer therapy. The overall fields of application span from drug discovery and screening, over basic cancer research and therapy, disease modelling and personalized medicine, toxicology and the replacement/3R of animal models, to the assessment of food and environmental safety, or specific use cases such as research in spaceflight. Beyond the culture of human and other mammalian cells, e.g., plant cell models on-chip will become more relevant and can equally benefit from sensor integration.It has been clear for over a century that even the basic aspects of cellular energy metabolism, such as the availability of oxygen and glucose, can strongly determine the cell state, ranging from the simple survival of the cells in culture, to playing a central role, e.g., in both tumour progression and therapeutic resistance.1 In light of this importance, it is surprising how many cell culture systems to this day do not control or verify the respective metabolite levels in the cell culture, particularly not in the direct vicinity of the cells.2 Klein, et al., vividly called out a substantial lack of environmental control in mammalian cell culture altogether, putting reproducibility of results at risk.3 In a meta-analysis, Al-Ani, et al., reported a fully reproducible description of oxygenation conditions in only 6% of all published works.4 On a more anecdotal but far-reaching level: a reviewer for one of our cell culture monitoring systems argued that oxygen sensors for cell cultures are fundamentally unnecessary because all the relevant values are set at the incubator door. Naturally, we could not disagree more: the incubator atmosphere in the gas phase is certainly controlled, even though primarily by mixing air with carbon dioxide in a fixed ratio, and already non-atmospheric oxygen levels require more advanced hypoxia setups. It should be considered that the mass transport of metabolites and signaling molecules to and from cells primarily relies on diffusion. It has been shown that even under normoxic incubator culture conditions, highly metabolizing cancer cell cultures can easily reach hypoxic or even anoxic pericellular oxygen concentrations within the timeframe between typical medium exchange.5,6
From a historical perspective, the 1990s saw the rise of many chip-based cell culture monitoring systems, then predominantly called microphysiometers, that allowed cell culture on-chip with several integrated sensors for extracellular acidification, cellular respiration, adhesion and more.7–11 These systems promised high-content drug screening, and several products were commercially available. Most of these platforms have since disappeared for various reasons, such as high cost, limited option for parallelization and possibilities for high throughput screening, handling difficulties and incompatibility with routine workflow in cell labs, but possibly also for being too early for the surge in organoid and organ-on-chip use. In contrast, today's state-of-the-art microfluidic organ-on-chip platforms allow, e.g., the chip-based parallel phenotypic screening of a library of 1537 individual drug candidate compounds using a 3D angiogenesis assay in 64-chip microtiter plates and optical read-out of cell stainings.12
For sensor integration in cell cultures, these examples raise the questions that we aim to trace throughout this review: how can sensor integration in cell cultures and organ-on-chip systems be motivated; which challenges have to be met for the various culture formats and sensor types; what is the recent progress, and what is holding back sensor integration; and which points have to be addressed in the future?
Scope of the article, terminology, and state-of-the-art
This review continues our review on microsensors for cell metabolism in 2018.13 Here, we continue to explore the integration of sensors for cell metabolism into cell culture and organ-on-chip systems by putting recent developments and trends into the context of the existing literature.For the scope of this review, we generally prefer the more classical sensor definition of a self-integrated device, either within the cell culture platform or a discrete device that generates a typically quantitative, marker-free, time-dependent and reversible signal.14–17 In the cell culture monitoring application, the aforementioned sensor properties are often understood as key selling points because sensors for living cells and organs-on-chip can enable the continuous recording of real-time or near real-time signals about the cell state while preferably interfering with the cells as minimally as possible. Reversibility of the signal and, thus, the ability to show dynamics and recovery effects of cell metabolism sets sensors apart from classical end-point analyses. The term monitoring typically implies both real-time readout and the reversibility of the signal. There certainly exist grey areas with respect to strict classifications, starting with optical sensors based on sensor patches with the optically active material in the cell culture and an external read-out, to particles and markers embedded within cellular structures such as spheroids, to bioengineered markers directly expressed by the cells. All of these approaches can fulfill the above-mentioned requirements for sensors and have tremendous merits in their respective application scenarios.
In this review, we consider sensors for cell metabolism, in line with the above-mentioned criteria, that measure biochemical quantities continuously and in situ that are either produced or consumed by the cells in the culture. That encompasses primarily small molecules of cellular energy metabolism, such as oxygen, glucose, lactate, pH, neurotransmitters, reactive species and signalling molecules. We do not cover purely electrical signals, such as impedance, or transendo-/epithelial electrical resistance (TEER), which have been summarized recently,18 or electrical activity such as measured by microelectrode array chips for the neurosciences,19 although sensing capabilities have been shown.20,21 With respect to the scope of this journal, we focus on sensor-integrated, lab-on-a-chip scale devices that are miniaturized and share to some degree the prospect of automated or mass fabrication. We do not cover industrial-grade macroscopic sensors, such as those used in classical bioprocess monitoring, e.g., in batch (micro-)reactors, even though this is a highly relevant field. Nonetheless, some sensor principles, design criteria, and challenges in sensor performance may be of similar relevance across the fields.
The industry perspective on microphysiological systems was discussed by Ewart and Roth, reflecting both the positions of organ-on-chip device manufacturers and pharma end users.30 Despite strong claims in the technology-centric literature, they confirm that up to then, no regulatory submission ever included data from microphysiological systems. With the FDA Modernization Act 2.0 from 2022, the FDA has now allowed organs-on-chips as a potential alternative to animal experiments at the end of 2022.31 Vulto and Joore gave another perspective on the industry adoption of organ-on-chip-systems.32 Sensors in cell culture systems and organs-on-chip have recently been reviewed,33–35 while others focused on the materials,36 or reviewed the gap between tumours-on-chip and clinical practice.37 Recently, Reyes, et al., gave an interesting perspective on the standardization in microphysiological systems with an overview of stakeholders and available standards and guidelines that are relevant for a broad adoption of such systems.38
In this article, we outline the relevance of sensors in cell cultures and organs-on-chip. We discuss the requirements for sensor integration in such systems, taking into account the different cell culture formats and the resulting challenges for sensor integration. Various sensor types and parameters are briefly introduced and classified. We then review selected sensor systems for 2D, 3D, and single spheroid/organoid culture to discuss the recent advances in sensing with a focus on sensor performance and actual demonstration by long-term measurements. We outline current fabrication techniques and materials and comment on current trends. Finally, we conclude with a summary of recent developments and a critical evaluation of challenges that must be addressed.
Cell culture models and sensor integration
Relevance, principles, requirements and challenges
To understand and motivate sensor integration in such systems, the generic relevance and function of sensors in organ-on-chip and cell culture monitoring systems is summarized in Fig. 1, along with building blocks, challenges and feedback loops. The essential building blocks include the cell model, the microstructures and, optionally, the microfluidics. The cell model is imposed by the desired application and can be broadly classified based on the origin. This ranges from established cell lines, over patient-derived stem cells, to organoid and tissue models usually derived from stem cells. The cells often need to be confined by microstructures, such as barrier structures to confine artificial extracellular matrices, to trap or agglomerate cells and microtissues, and to form microchannels with the appropriate interfaces between matrix and liquid medium. The resulting microfluidics can then perform a variety of functions. One of the most basic requirements is to periodically or continuously supply the cells with fresh medium containing the nutrients/metabolites and to remove waste products of cellular metabolism. The medium exchange can be also used to supply drugs and other stimuli, and with the help of appropriate layouts, dynamic or static concentration gradients of both nutrients/metabolites can be generated. Besides, the removed medium can be used for sample collection and external analysis, e.g., of signaling molecules.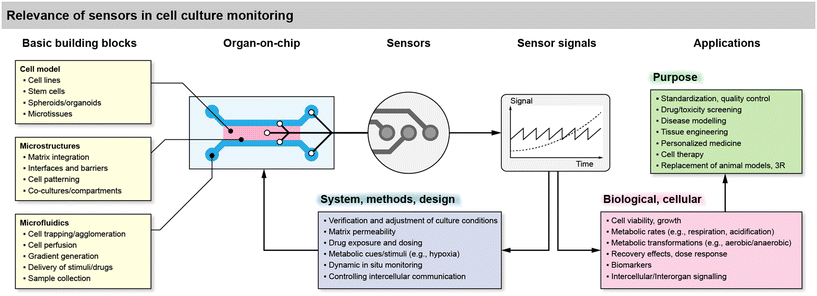 | ||
| Fig. 1 Overview of the relevance of sensor integration in cell culture systems and organs-on-chip, including basic building blocks, benefits for the system side or the biological side, and use cases. | ||
Microsensors can be integrated directly in the cell culture area or volume, in the microchannels in direct vicinity to the cells, in a downstream microchannel on-chip, or in the microfluidic circuit but off-chip in a modular approach. A modular approach may be practically beneficial, but short-lived or lowly concentrated analytes require a sensor placement as close as possible. The sensors typically generate a continuous, time-dependent output signal from which cellular metabolic rates, e.g., oxygen consumption or acidification can be derived.
The sensor signals can be used for various purposes in cell culture systems on the side of the system, e.g., verification of the methods and the experimental design such as the culture conditions, microfluidic regimes or matrix permeability in 3D cultures. In combination with the microfluidic flow, they can be used to modulate metabolic cues and stimuli, such as hypoxia or tailored drug exposure and dosing. Adjustment of the flow, in combination with sensor feedback, can be used for real-time in situ monitoring under dynamic conditions or even to control intercellular communication by interrupting diffusion pathways with the flow.
On the side of the biological relevance, sensor signals derived from cells can be simple indicators of cell viability and growth. If a stop/flow regime is applied, metabolic rates can be extracted, but only in the temporal resolution of the stop/flow phases because each cycle typically leads to one rate. True real-time measurements of metabolism are also possible but may be challenging due to the boundary conditions of the mass transport situation. These transient metabolic data can then be used to reveal metabolic transformations, e.g., the shift from aerobic to anaerobic metabolism, or the pharmacodynamics during drug exposure, including recovery effects thanks to the time-continuous signals, together with the option to dynamically switch back and forth between blank medium and drug exposure. More complex biomarkers or signaling molecules are challenging to measure in situ and in real-time. For such substances microsensors can also be placed in downstream microfluidics or used with samples of the medium.
Cell culture system formats and sensor integration
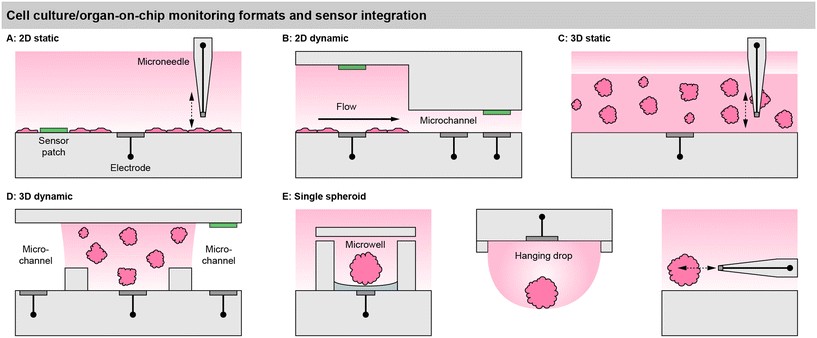
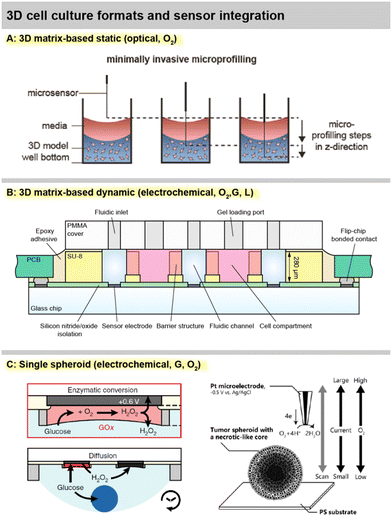
Sensor access to cell cultures and sensor integration into the systems must be understood in the context of the cell culture format. This is particularly important because all sensor signals are dependent on the mass transport of the analyte to the sensor. Fig. 2 summarizes different spatial and fluidic concepts of cell cultures, surrounding microsystems and microfluidics, as well as the positioning of the sensors within the lab-on-chip or as external devices.For meaningful sensor readings, it is essential to find a suitable sensor position considering the development of strong gradients. Most critical is the formation of gradients for oxygen, and a look into the underlying physics and metrics of oxygen levels is highly recommended.39 Those gradients can range from atmospheric dissolved oxygen concentration (approx. 19–21%) at the top of the medium in a flask and almost anoxic conditions (<1%) near cancer cells under a few millimetres of medium.6,40 Therefore, sensors in 2D cultures should ideally be integrated at the bottom, next to the cells (“pericellular”), to measure meaningful pericellular concentrations. A gradient in the vertical direction can be accessed either by a vertically positioned sensor array or a moving needle-type sensor probe.
In matrix-based, static 3D cultures [Fig. 2C], the situation is more complex because, radial gradients towards the cell agglomerates can develop additionally. Sensors at fixed positions can give representative values only if a homogenous concentration distribution is assumed, which is even more critical inside a gel with slower diffusion. For example, with the incubator atmosphere above being the only oxygen source, it has to be considered that cells at the top of the 3D layer are closer to the source. Here, vertically movable, needle-type sensor probes can give a more accurate representation.41
In 2D models, the perfusion channel itself or a dedicated larger chamber acts as the culture surface [Fig. 2C].42 If it is expanded with integrated pores, 3D cell agglomerates may also be embedded in it.44 In both cases, the medium flows over the cells. This fluid movement may induce shear stress, which can be intentional (e.g., polarization of endothelial cells) but can also lead to undesired cell detachment. The microsensors can be positioned between the cells, similar as in static systems to obtain pericellular values. Due to the lower height of the medium column above the cells, the sensor placement above the cell culture at the ceiling of the channel [Fig. 2C] can also lead to a meaningful measurement.45 Alternatively, sensors can be placed in the downstream microchannel as well as upstream for reference. The spatial separation of the sensor and cell culture may be necessary if sensors are incompatible with cells or if sensors need to be replaced or calibrated independently of the cell culture. How well the sensor signals represent the pericellular concentrations, has to be carefully considered. It has been shown that undiluted media can be transported to sensors with microfluidics,42 but dispersion by diffusion will occur over longer distances and times. Short-lived analytes may decompose, and the in- or outflux of volatile substances such as dissolved oxygen has to be considered.
In matrix-based 3D cultures, the perfusion channel must be separated from the culture volume [Fig. 2D]. Typically, this is achieved by barrier structures comprising free-standing posts,46 phaseguides,47 or a combination of both.43 The barrier structures confine and hold the artificial extracellular matrix gel, while the interface allows substance exchange from the gel to the liquid in the channel. Sensors can be either integrated into the channels or directly in the cell compartments. Sensors in the compartments can directly provide pericellular concentrations. With the placement in the channel, the diffusional transport out of the gel has to be considered to draw conclusions about pericellular levels. Even fast diffusing molecules will take several minutes to diffuse through submillimetre-scale dimensions and generate a meaningful particle flux to the sensor or to the cells.
Ideally, measurements inside spheroids are also attractive, but can be achieved only with the smallest footprint needle-type sensors, and may unfavourably affect the cellular behaviour, in addition to the challenging and cumbersome handling. External microneedles may be used to determine spheroid metabolism at close distance and with the smallest spatial resolution [Fig. 2E]. Hanging droplets have been established as a capable technique for handling spheroid cell cultures, and sensor-equipped systems exist where the sensor electrodes are placed at the ceiling of the droplet and access the single spheroid metabolism [Fig. 2E].56,57
Basic sensor principles
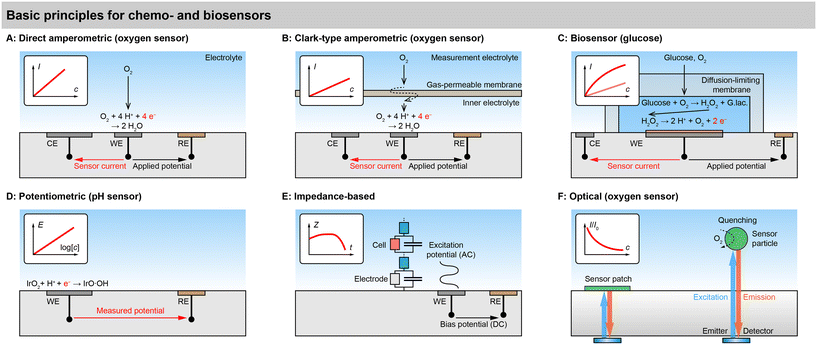
An overview of basic sensor principles for metabolic monitoring in cell cultures and organs-on-chip is shown in Fig. 3. Electrochemical and optical principles are predominant in the biochemical monitoring of cell cultures. These principles describe almost all of today's chemical and biochemical sensors in cell culture systems directly or can be deduced from them with minor adaptations.Electrochemical sensors
Direct amperometric sensors. Fig. 3A shows a typical example of a direct amperometric electrochemical sensor.58 In this principle, the working electrode (WE) is polarized to an appropriate potential with respect to the reference electrode (RE) by a potentiostat. The resulting current through the working and counter electrode (CE) is proportional to the electrochemical reaction that occurs at the working electrode. In case of a direct amperometric oxygen sensor, the current is generated by the electrochemical reduction of the oxygen molecule in a 4-electron process at a platinum electrode. If the mass transfer of the oxygen is limited by diffusion, e.g., by the use of an appropriate membrane, the transfer function of current over concentration becomes linear, as governed by Fick's first law. Such amperometric sensors need to be calibrated in almost any case because no absolute quantitative relationship between current and concentration can be formulated. Direct amperometric sensors are often found also for other electroactive small molecules, such as reactive oxygen and nitrogen species, hydrogen peroxide, or catecholamine neurotransmitters such as dopamine.
General advantages of electrochemical sensors are the linearity across the measurement range with low offset, the many options to tune sensitivity, linear range and selectivity for a variety of analytes, the high temporal and spatial resolution and the straightforward multiplexing. Challenges are the inherent analyte consumption and possible interference with the medium. Achieving selectivity and stability in complex media includes stabilizing the biointerface. The requirement for appropriate electrochemical instrumentation limits large-scale parallelization.
Clark-type principle. The principle of the direct amperometric oxygen sensor is sometimes called “pseudo-Clark-type”, as opposed to a true Clark-type sensor shown in Fig. 3B. Importantly, the Clark-type oxygen sensor has a gas-permeable membrane, such as a silicone, that separates the inner electrolyte from the solution. The oxygen molecule has to permeate from the outer liquid through the solid membrane into the inner liquid. In line with Clark and Lyons original formulations, the membrane is ion impermeable, and the inner electrolyte is therefore electrically and ionically isolated.59 It should be noted that the original design by Clark did not have a noble metal cathode and used a 2-electrode setup. The third electrode, which can regenerate at the anode the oxygen that is consumed by the sensing cathode, was later introduced by Ross.60 Early microfabricated 2- and 3-electrode Clark-type sensors followed the concept of a silicone membrane.61,62
Advantages of the Clark principle are good selectivity, especially in biological environments with many interfering molecules, and good stability. The transfer function is ideally linear, as for the direct amperometric sensor, but with lower sensitivity due to a stronger mass transfer limitation. Miniaturization of this principle is challenging because of the confinement and proper isolation of the inner electrolyte and, thus, sealing and integrity of the membrane, as well as keeping the pH of the inner electrolyte stable. Recently, there have been confusing implementations in the literature, which seemingly omit the inner electrolyte, use water/ion-permeable membranes, or have one electrode outside the inner electrolyte, raising the question about aqueous electrochemistry at the electrode, the current path, or the true implementation of the principle. Several examples certainly would not have worked if they had been properly fabricated as described.
The general advantage of biosensors is their ability to detect non-electroactive or non-optically active species that would be impossible to access without the help of the biological recognition element. The immobilization of the recognition element is challenging, and stability, especially with respect to temperature, but also chemical degradation can be limited. A long-term loss in sensitivity is not uncommon.
Advantages of potentiometric sensors are the fact that they do not consume the analyte because the electrode is in equilibrium and their relatively simple instrumentation, which requires only an amplifier without a feedback loop. However, achieving a high enough input impedance to fulfil the zero-current premise is critical, and leakage currents can lead to the disturbance of the equilibrium of the electrode, changes in electrode properties or unwanted accumulation of charge, which can all negatively affect sensor performance. Insufficiently high input-impedance is especially critical for microsensors.
Advantages of optical sensors are that they do not consume oxygen and do not interfere with the medium, and that they do not require physical contact between the transducer and read-out unit, which means one read-out unit can easily address multiple sensor points in sequence. Challenges are the inherent non-linearity, especially at low concentrations, the need for optical transparency, and the stability of the indicator in cell culture environment. The quenching process leads to an excited state of the analyte molecule, potentially causing harm to the cells. Therefore, dyes are typically applied in combination with a scavenger or embedded in a membrane, ensuring a high enough diffusion length to prevent the short-lived state from reaching the cells.
Selected cell culture sensor systems and sensor performance
In this section, we will review recent selected sensor systems for cell cultures and organ-on-chip, focusing on their sensor performance. We will largely adhere to the aforementioned classifications but will discuss systems beyond the scope if relevant advances, e.g., in approach, design, sensing or fabrication technology were made. A useful categorization of cell culture monitoring systems is challenging due to the complex interplay of cell model, platform format and integrated sensors, which makes grouping difficult because overlaps and exceptions exist. However, we consider the following key aspects for the grouping formats, in which all types of sensor integration can be found:• Static cell culture conditions (labware, simple vessels, stagnant medium) vs. dynamic cell culture conditions (microfluidics, perfusion systems)
• 2D monolayer cell culture (mostly adherent to a rigid surface) or bilayer co-culture (mostly on a membrane) vs. 3D agglomerates as, e.g., spheroids, matrix-based cultures and other organotypic models. Here, the lines are often blurred between cells simply distributed in a 3D environment and 3D agglomerates that actually recapitulate 3D structures or organotypic functions
• Monitoring of single isolated 3D agglomerates or spheroids
Different systems are summarized in Tables 1 and 2 according to these categories, and their properties include:
• The exact cell model
• The dimension of the cell model and its format
• The realization of microfluidics, including the number of individually addressable channels
• Sensor parameters
• Sensor principle
• The time-scale on which sensor-based measurements of cell metabolism are demonstrated
• Notable achievements of the presented system, including, e.g., sensor principles, sensor performance and biological relevance
| Author, year | Cell model | Dim./format | μFluidics | Parameter | Principle | Time-scale | Notable achievements | Ref. |
|---|---|---|---|---|---|---|---|---|
| EC = electrochemical, OPT = optical, TEER = transepithelial electrical resistance, G = glucose, L = lactate. | ||||||||
| Static 2D | ||||||||
| Kieninger, 2018 | Breast/brain cancer (T-47D/T98G) | 2D monolayer | No | O2, pH | EC | 5 d | Integration in standard labware, 2-parameter long-term | 40 |
| Marzioch, 2019 | Breast cancer (T-47D) | 2D monolayer | No | O2 | EC | 5 d | First photodynamic therapy on-chip | 64 |
| Tanumihardja, 2021 | Endothelial (HUVEC) | 2D monolayer | No | O2, pH | EC | 3 d | Two principles using the same electrode material | 65 |
| Tanumihardja, 2021 | Pluripotent stem cell-derived cardiomyocytes (hPSC CM) | 2D monolayer | No | NO | EC | <1 h | Demonstration of selectivity and cellular NO release | 66 |
| Liebisch, 2020 | Breast cancer (T-47D) | 2D monolayer | No | O2 | EC | 8 d | True Clark-type, dry fabrication, zero consumption | 67 |
| Hsueh, 2021 | (Bacteria only) | — | No | O2 | EC | 1 h | True Clark-type, dry fabrication | 68 |
| Static 3D | ||||||||
| Eggert, 2021 | Breast cancer (MDA-MB-231) | 3D matrix | No | O2 | OPT | 28 d | 96-Well format, sequential vertical microprofiling of O2 | 41 |
| Dynamic 2D | ||||||||
| Müller, 2021 | Lung cancer (A549) | 2D monolayer | Yes | O2, pH | OPT | 6.5 h | Optical pH sensor calibration data | 69 |
| Zirath, 2021 | Lung/colorectal cancer (A549/Caco-2), HUVEC | 2D monolayer | Yes | O2 | OPT | 3 h | Four dual-channels, nanoparticle exposure | 70 |
| Fuchs, 2022 | Induced pluripotent stem cells (hiPSC) | 2D monolayer | Yes | O2, pH | OPT | <3 h | Use of pluripotent stem cells | 71 |
| Fuchs, 2023 | Supernatant only | — | Yes | G | OPT | 5 d | Long-term optical glucose sensing | 72 |
| Busche, 2022 | Primary hepatocytes (PHH) | Unspecified | Yes | O2 | OPT | <1 d | Use of liver-on-chip model | 73 |
| Bouquerel, 2022 | Lung cancer (A549) | 2D monolayer | Yes | O2 | OPT | 1 h | Advanced perfusion system for O2 control | 74 |
| Cognetti, 2022 | Bronchial epithelial | 2D monolayer | Yes | Cytokines | Photonic | <6 h | Cytokine monitoring, photonic chip in cell culture | 75 |
| Dynamic bilayer/3D | ||||||||
| Alexander, 2018 | Hepatocyte (HepG2) | 3D multi-spheroid | Yes | O2, pH | EC | 2 d | Multi-parametric respiration/acidification, multi-spheroid insert for microfluidic sensor chip | 76 |
| Azizgolshani, 2021 | Kidney proximal tubule epithelial (hRPTEC), | 2D bilayer | Yes, 2-ch | O2, TEER | OPT | 250 min | 96-Well automated platform, 2 microchannels | 77 |
| Kann, 2022 | Retinal microvascular endothial (RMVEC) | 2D monolayer | Yes, 2-ch | O2 | OPT | <10 min | Cellular consumption rates, drug exposure | 78 |
| Kann, 2023 | 2D bilayer | Yes, 2-ch | O2 | OPT | 11 d | Automated high-throughput long-term | 79 | |
| Kreß, 2022 | Adipose-derived mesench. stem cells (adMSC), fibroblast (NHDF), keratinocyte (HaCaT) | 3D matrix | Yes | O2, pH, G, L | OPT, EC | 5 d | Microperfusion of 3D matrix and downstream sensing | 80 |
| Dornhof, 2022 | Breast cancer stem cell (BCSC1) | 3D matrix | Yes, 3-ch | O2, G, L | EC | 8.5 d | Microfluidic matrix-based culture, long-term 3-parameter | 43 |
| Scheinpflug, 2023 | Osteoblasts in scaffolds | 3D matrix | Yes | O2 | OPT | 1 d | Oxygen regulation, control of mechanical load | 81 |
| Schlünder, 2024 | Pancreatic pseudo-islets | 3D matrix | Yes | O2 | OPT | 4 h | O2 consumption at various glucose levels | 82 |
| Author, year | Spheroid cell model | Param. | Format | Principle/electrode Ø | Time-scale | Notable achievements | Ref. |
|---|---|---|---|---|---|---|---|
| EC = electrochemical. | |||||||
| Rousset, 2022 | Colorectal cancer (H116) | Glucose | Hanging drop network | EC, 400 μm | 2.5 h | Spheroid glucose consumption, deduction of intra-spheroid situation | 83 |
| Mukomoto, 2020 | Breast cancer (MCF-7) | O2 | — | EC, 20 μm | 2 h | Spheroid respiration rates, correlation with necrotic core | 84 |
| Nashimoto, 2023 | Lung fibroblast (hLF), HUVEC | O2 | Microfluidic perfusion | EC, 20 μm | <1 h | Spheroid respiration rates dependent on vascular perfusion, drug exposure | 85 |
| Nashimoto, 2023 | Colorectal cancer (HCT116/HT29), CRC-derived organoids (KUC16) | O2 | Conical wells | EC, 1–10 μm | — | Cell lines vs. patient-derived, variability in respiration rates of subpopulations | 86 |
| Dornhof, 2022 | Breast cancer (MCF-7) | O2 | Sensor wells | EC, 200 μm | 6 h | Parallel bioprinting onto sensor chip, respiration in minutes, drug exposure | 52 |
Monitoring of 2D cell cultures
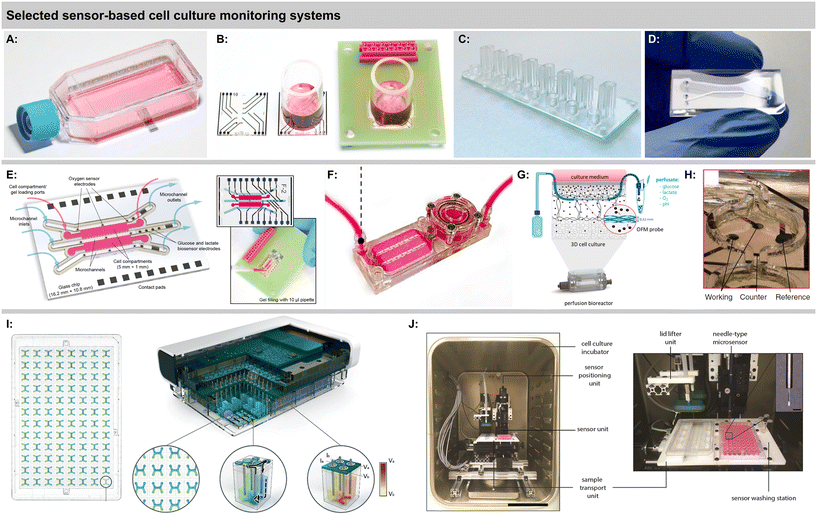
Sensor integration in standard labware was shown by Kieninger, et al., with an electrochemical multiparametric sensor chip for oxygen and pH in a standard cell culture flask, including six sensor electrodes in one flask and a multiplexer rack for four flasks in parallel [Fig. 6A].40 Amperometric platinum-based oxygen sensors and potentiometric iridium oxide-based pH sensors allowed precise measurements of cell respiration and acidification over up to 5 days in brain or breast cancer cells. Strong hypoxic conditions were found for both normoxic cultures at atmospheric incubator conditions and hypoxic cultures at 4% incubator oxygen. These results further underline the need for in situ monitoring of culture conditions directly at the cellular level because strong gradients develop in standard cell culture practice.
Marzioch, et al., used a chip-based electrochemical oxygen sensor platform to measure the metabolic effects of photodynamic therapy (PDT) on cancer cells [Fig. 6B].64 The transparent chip allowed illumination with a fibre optical system to trigger a photosensitizer in the culture medium. The application of PDT led to a reduced cellular respiration, and a repopulation effect was shown even for the treated cells during continuous measurements up to 5 days. Further, the spatial efficacy of PDT could be demonstrated by simply shading half the cell culture area during PDT and using a line array of four sensors, underlining the capabilities of simultaneous multi-channel chip-based pericellular monitoring.87 The same platform was also used to rapidly determine patient-derived brain cancer cell respiration rates within minutes by temporarily reducing the medium volume with an insert.88
A multiparametric electrochemical sensor platform for cell cultures was developed by Tamnumihardja, et al.65,66 pH and oxygen sensors by ruthenium oxide electrodes were established using potentiometric and amperometric principles, respectively, having the advantage of using the same electrode material for both methods and parameters.65 Oxygen consumption rates of cardiomyocytes in a transwell-style setup were determined over approx. 3 days, but no sensor stability data were shown, and electrodes were fairly large at 3 mm diameter. The ruthenium oxide nanorod electrode was also applied as a sensor for nitric oxide, but data are limited to durations below 1 h.66 Notably, the comparison with Pt electrodes was shown, and selectivity, e.g. against nitrite, was demonstrated.
A true Clark-type oxygen microsensor system integrated in a cell culture flask was developed by Liebisch, et al.67 A silicone membrane sealed a hydrogel-supported inner electrolyte pre-impregnated with the buffer salts and hydrated via water vapour through the membrane. The sealing of the gas-permeable membrane was verified by impedance measurements and sensor operation in deionized water was possible. Notably, the Ross-principle was implemented and verified, in which the counter electrode regenerates the oxygen that is consumed during the measurement and therefore enables net-zero consumption in the inner electrolyte, further underlining the long-term stability of the electrochemical cell in the isolated inner electrolyte. Monitoring oxygen for more than a week was demonstrated using a breast cancer cell line. Hsueh, et al., fabricated a miniaturized Clark-type sensor in a similar approach for bacterial cultures, but used a 2-electrode setup and demonstrated stability over 12 h [Fig. 4A].68 The work was extended by a bipolar Clark-type array, in which oxygen was measured by electrochemiluminescence (ECL) through the reaction in a confined chamber at the anode.89 Another ECL approach was presented by Hiramoto, et al. A potential step electrochemical protocol was applied to gold electrodes to generate hydrogen peroxide from the oxygen reduction as a sensitizer for the luminophore, which was then used for mapping oxygen around spheroids over time with good oxygen sensitivity and spatial resolution.90
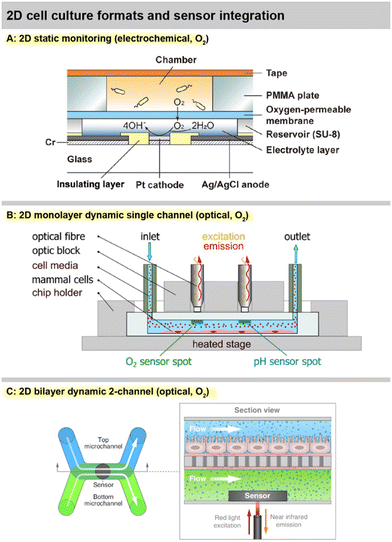 | ||
| Fig. 4 Practical implementations of 2D cell cultures with integrated sensors. A: 2D static monitoring with the Clark-type electrochemical O2 sensor principle68 (reproduced with permission from the American Chemical Society). B: 2D monolayer dynamic monitoring with optical O2 and pH sensors69 (Copyright: the authors, reproduced under the CC BY 4.0 license). C: 2D bilayer dynamic monitoring with a membrane separating two channels and optical O2 sensors78 (Copyright: the authors, reproduced under the CC BY 4.0 license). | ||
A number of systems incorporating fibreoptical oxygen, pH and glucose sensors from Pyroscience with direct involvement or from associated research groups have been introduced in the last years. Different thermoplastic-based microfluidic platforms were introduced [Fig. 4B and 6C and D].69,70 Optical oxygen and pH sensors were integrated at the ceiling of the channels, while cells grew in a monolayer at the channel bottom. Stop flow experiments allowed determination of respiration rates and extracellular acidification, including alterations due to nanoparticle exposure. Oxygen scavenging by a tailored material on-chip was demonstrated in an ischemic stroke model.91 Fuchs, et al., used a comparable setup and monitored respiration and acidification of human induced pluripotent stem cells (iPS).71 The same group reported the realization of an optical glucose sensor for microfluidic cell cultures.72 The enzymatic principle features immobilized glucose oxidase and is based on the measurement of the oxygen consumption of the enzymatic reaction. Stability over 5 days with a drift of 3% per day was reported. The linear range up to 10–30 mM covers the relevant range. This results in comparably high detection limits of 200–700 μM, and sensor spots at 1 mm are large. Cell measurements were reported from supernatants only. The need for advanced oxygen control was addressed by Bouquerel, et al., using commercial microfluidic chips and optical oxygen sensors from Pyroscience and Presens in the chip and both up- and downstream.74 The adjustment of oxygen levels under elaborate perfusion conditions and the influence of materials were shown. Bussooa, et al., used a Presens fibreoptical oxygen sensor to evaluate material properties of thermoplastics in microfluidic systems.92 The paper compares the oxygen dynamics of PDMS-based systems to cyclic-olefin-based systems.
A largely automated, 96-well-format organ-on-chip platform was introduced by Azizgolshani, et al., and further applied and characterized by Kann, et al. [Fig. 4C and 6H].77–79 The microfluidic plate features 96 2-channel microfluidic systems that are vertically separated by a membrane. The channels feature optical oxygen sensors from Pyroscience, integrated TEER-based sensors, automated fluidic control, and transparency allows advanced optical analysis. Oxygen readings from each channel pair were acquired sequentially every 30 min over 2.5 h at static and dynamic conditions.77 Transient oxygen levels were measured in a stop/flow regime, and corresponding cellular consumption rates were modelled and determined in a membrane-bilayer format, including drug exposure and alteration of respiration rates.78 In a nephrotoxicity cell model, oxygen consumption rates over 11 d were measured, and a 5 d dose response for a chemotherapeutic was successfully demonstrated.79 Time- and dose-dependent oxygen measurements showed more robust results than classical colorimetric read-out, especially at later time points of compound exposure.
A photonic chip with ring resonators was employed for the first time in cell cultures to measure four different inflammatory cytokines such as interleukin-6 and interleukin-1β.75 An 8-fibre array on a waveguide chip detected cytokine secretion from bronchial epithelial cells in a microfluidic setup. Non-linear sensor performance with real-time detection in the 1–100 ng mL−1 range was achieved for up to 6 h.
Monitoring 3D cell cultures
Microprofiling of 3D cell cultures using a fibreoptical oxygen sensor from Presens and a robotic stage in an incubator was demonstrated by Eggert, et al., [Fig. 5A and 6I].41 The fibre was scanned vertically through single wells of a 96-well plate, which contained a 60 μL/500-μm-thick 3D breast cancer construct at the bottom and around 4 mm of medium supernatant on top. A vertical scanning step size of 250 μm revealed strong vertical gradients in oxygen and hypoxic conditions at the well bottom for different cell densities and over multiple days of cell growth. This highlights the inherent limitation of static 3D cultures regarding metabolite transport. A chemotherapeutic dose response with six compound doses was successfully performed using 54 wells in parallel, and data up to 35 days are shown, although not continuously measured.
 | ||
| Fig. 5 Practical implementations of 3D cell cultures with integrated sensors. A: 3D matrix-based culture with static monitoring by vertically profiling the culture wells with optical O2 sensors41 (reproduced with permission from the American Chemical Society). B: 3D matrix-based microfluidic culture for dynamic monitoring with electrochemical O2 sensors43 (Copyright: the authors, reproduced under the CC BY 4.0 license). C: Single spheroid electrochemical glucose monitoring in hanging droplets (left)83 (Copyright: the authors, modified and reproduced under the CC BY 4.0 license). Probing of tumor spheroid microenvironment with needle-type electrochemical oxygen sensors (right)84 (reproduced with permission from the Royal Society of Chemistry). | ||
 | ||
| Fig. 6 Sensor-based cell culture monitoring systems. A: Sensing cell culture flask for electrochemical O2 and pH monitoring40 (Copyright: the authors). B: Sensing cell culture well for electrochemical O2 monitoring64 (Copyright: the authors, modified and reproduced under the CC BY 4.0 license). C: Thermoplastic multi-channel device for optical O2 monitoring70 (Copyright: the authors, modified and reproduced under the CC BY-NC 3.0 license). D: Thermoplastic device for optical O2 monitoring82 (Copyright: the authors, reproduced under the CC BY 4.0 license). E: Microfluidic organ-on-chip platform for matrix-based cultures and electrochemical O2, glucose and lactate monitoring43 (Copyright: the authors). F: Microfluidic platform for monitoring scaffold-based bone formation with integrated optical O2 sensors81 (Copyright: the authors, modified and reproduced under the CC BY-NC 3.0 license). G: Bioreactor with microperfusion setup and downstream sensing (Copyright: the authors, modified and reproduced under the CC BY 4.0 license). H: Hanging droplet network platform with integrated electrochemical glucose sensors83 (Copyright: the authors, modified and reproduced under the CC BY 4.0 license). I: Automated 96-well plate format system for bilayer culture with integrated optical O2 monitoring77 (Copyright: the authors, modified and reproduced under the CC BY-NC 3.0 license). J: Setup for automated vertical profiling of 3D cultures in microwells with optical O2 sensors41 (reproduced with permission from the American Chemical Society). | ||
The above-mentioned optical Pyroscience-based systems include only 2D monolayers at the channel bottom or on a membrane. To address more complex organ-on-chip models, Busche, et al., integrated similar oxygen sensors into their liver-on-chip to monitor primary hepatocytes.73,93 Scheinpflug, et al., developed a bone formation model by embedding osteoblasts inside scaffolds into a microfluidic system [Fig. 6F].81 The complex system features, gas and liquid perfusion loops, a perfusion chamber for cell laden scaffolds and optical oxygen sensors. Continuous oxygen sensing at approx. 7% oxygen was shown over 24 h. The influence of different oxygen levels and mechanical loads on the cell-laden scaffolds was investigated. An advanced microphysiological pancreas-on-chip system with integrated optical oxygen sensors was introduced by Schlünder, et al.82 The system includes a vascular-like perfusion channel, separated by a membrane with spheroid trapping holes from the tissue channel, where spheroids are embedded in a hydrogel, potentially also allowing co-culture. The gas-tight system enabled the determination of oxygen consumption at different glucose levels, substantially enhancing the offline analysis of insulin secretion. Rivera, et al., demonstrated another optical oxygen measurement system in microfluidics with a phosphorescence-based principle.94 Cells were embedded in a hydrogel which was covered by medium and a gas-exchange channel above. Gas-tightness and simple respiration measurements from undefined 3D agglomerates were shown. Besides discrete sensor patches or fibres, recent approaches also successfully demonstrated time-transient oxygen measurements by optical sensor beads embedded in perfused 3D.95,96
Sánchez-Salazar, et al., have recently pursued the interesting approach of using a commercially available continuous glucose monitoring system designated for subcutaneous application in cell culture measurements.97 This has the advantage of an already established, accurate, autonomous and stable sensor system, together with an easy read-out via a smartphone. Continuous data over 14 days at approx. 3–5 mM glucose concentrations were shown in microfluidic colorectal cancer cell-laden microsphere culture. Another interesting concept is using a perfusion bioreactor for 3D culture and embedding an openflow microperfusion probe centrically in the matrix by layering it between cell-laden collagen fleeces [Fig. 6G].80 Microscale optical pH and oxygen, as well as electrochemical glucose and lactate sensors were located downstream to analyse the perfusate. All four parameters were measured continuously over 5 days highlighting the discrepancy between medium supernatant and interstitial fluids for different cells.
A platform for multi-parametric monitoring of 3D breast cancer spheroids with microfluidics, matrix-based 3D culture, and electrochemical sensors in both channels and cell chambers, integrated on one chip was demonstrated by Dornhof, et al., [Fig. 5B and 6E].43 Cancer organoids developed in microfluidic culture in Matrigel compartments over days from single patient-derived triple-negative breast cancer stem cells. Electrochemical oxygen sensor stability in medium was demonstrated over one week of continuous measurement with no detectable drift, and respiration rates could be determined in both the cell compartments and the adjacent channels with similar validity. Enzymatic glucose and lactate sensors allowed reproducible determination of cellular metabolic rates at relevant concentrations throughout the entire measurement. The system is gas-tight, which allows the adjustment of oxygenation in hypoxic cultures purely via medium perfusion.
Monitoring of single spheroids and organoids
The monitoring of single spheroids and organoids aims at monitoring the metabolism of a single 3D cell agglomerate. Modern organoids and microtissues can reflect target tissue properties very well even at small size. The inherent challenge is the low cell number that mandates small and sensitive sensors, as well as good control of the geometry, and often also the handling of small media volumes. If these challenges are met, accurate deductions of cell metabolism are possible, because the absolute cell number is more easily controlled and determined, as are the boundary conditions of the metabolite transport.Rousset, et al., expanded on the group's previous work on sensor-equipped hanging droplet networks56,57 by measuring glucose dynamics of single cancer spheroids in hanging droplet networks.83 Eight interconnected droplets included electrochemical glucose-oxidase-based and blank microsensors [Fig. 5C and 6H]. Sensors were tuned for high sensitivity with a limit of detection below 1 μM glucose and a consequently rather low linear range. The elaborate modelling of glucose mass transport and sensor signals allowed insight into the situation inside the spheroid via external sensors and statements on the fraction of metabolically active cells within the spheroid. A downside is that the selected enzyme immobilization is not very long-term stable, but is acceptable if spheroids consume the available glucose in a droplet within 20–40 min.
A microneedle-type electrochemical oxygen sensor to probe the surroundings of spheroids was demonstrated by Mukomoto, et al., [Fig. 5C].84,85 The platinum microdisk electrode inside a glass capillary from a scanning electrochemical microscope (SECM) setup had a 20 μm diameter. Consumption rates of breast cancer spheroids were determined and a correlation to the development of a necrotic core could be made with the conclusion that the core contributes only minimally to oxygen consumption.84 Vascularized spheroids on a micropore membrane and perfused from below were investigated similarly.85 Differences in extracellular oxygen gradients upon substance exposure of cancer organoids could be resolved depending on the perfusion of the vasculature. Metabolic differences in subpopulations of patient-derived spheroids were later also shown, underlining metabolic heterogeneity in cancer.86
Dornhof, et al., demonstrated for the first time the bioprinting of a single spheroid into a sensor platform to directly measure single spheroid metabolism.52 Breast cancer spheroids were dispensed in droplets into a gel-filled microwell that featured an electrochemical microsensor at the bottom. Oxygen consumption rates could be accurately determined from a 55 nl volume within minutes by gas-tight sealing of the well. Linear sensitivity with negligible offset <2% and a relative error <2.5% across the entire atmospheric range was demonstrated. The sensor's oxygen consumption, however, is not negligible compared to that of the spheroid and needs to be subtracted, which is only possible with accurate and stable sensors.
Trends in fabrication technologies, materials and interfacing of cell culture monitoring and organ-on-chip platforms
In the past, advances in miniaturization and microfabrication technologies have largely driven the material choices and technologies for sensors and sensor platforms in cell culture. Since the purely technological limitations and the reliance on microelectronic and microsystems manufacturing have been largely overcome, fabrication and material choices have noticeably shifted towards more application-oriented routes for fabrication of cell culture monitoring and organs-on-chip. In addition, on the one hand, there is a justifiable desire for a facile and decentralized fabrication with limited investment costs, which, e.g., stimulates additive manufacturing techniques. On the other hand, the demand for parallelization and automation in biology and compliance with standard manufacturing practices drives the adherence to highly scalable standard processes such as found in standard labware.| Author, year | System type | μFluidics | Materials | Gas-tight | Notable features | Ref. |
|---|---|---|---|---|---|---|
| 2D and 3D cell culture | ||||||
| Marzioch, 2019, Weltin, 2023 | Sensing cell culture well chip | No | Glass/SU-8 barrier/PMMA well | No/yes w. insert | Glass-based chip with PMMA tube, optional PMMA insert | 64, 88 |
| Gehre, 2020 | Multichannel microfluidic system | Yes | PMMA/adhesive tape/glass | Yes | PMMA channel plate, PDMS cell carriers, glass cover | 95 |
| Zirath, 2021 | Multichannel microfluidic system | Yes | Glass/adhesive tape/glass or COC | Yes | Xurographic rapid prototyping approach, transfer to injection molding | 70 |
| Fuchs, 2022 | Multichannel microcapillary system | Yes | Acrylic | Yes | Commercial microfluidic chips outfitted with optical sensors | 71 |
| Bouquerel, 2022 | Microfluidic chip | Yes | Glass/COC | Yes | Commercial glass or COC chips outfitted with optical sensors | 74 |
| Busche, 2020, 2022 | 24-Well plate with channels | Yes | COP plate/COP foil | Yes | 24-Well plate format, integrated electrophoresis electrodes | 93, 73 |
| Azizgolshani, 2021 | Microfluidic membrane-based bilayer system | Yes | COC/COP/FEP multilayer laminate | Yes | Advanced 96-well plate scale multilayer lamination of microchannel, fluidic port and optical layers | 77 |
| Kreß, 2022 | Bioreactor with microperfusion and downstream sensing | Yes | Resin by formlabs (dental SG, clear) | Yes | Additive manufacturing by stereolithography of parts in contact with cells | 80 |
| Dornhof, 2022 | Microfluidic 3D-matrix monitoring chip | Yes | Glass chip/SU-8 barrier/PMMA cover | Yes | Advanced barriers for Matrigel integration, demonstration of gas-tightness, adjustment of hypoxia via flow only | 43 |
| Scheinpflug, 2023 | Perfused 3D scaffold platform | Yes | COC | Yes | Injection molded COC for main system, 3D printed peripherals | 81 |
| Schlünder, 2024 | Membrane-bilayer system | Yes | PC/PMMA | Yes | Thermally bonded, laser-structured PMMA and PC layers | 82 |
| Single spheroid | ||||||
| Rousset, 2022 | Single spheroid, hanging drop | Yes | Glass chip/SU-8 barrier | No | Interconnected fluidic sensor-integrated hanging droplet network | 83 |
| Dornhof & Zieger, 2022 | Single spheroid, sensor well chip | No | Glass chip/SU-8 barrier/glass cover | Yes | Gas-tight sensor well for respiration monitoring in single spheroids | 52 |
| Nashimoto, 2023 | Single spheroid, needle sensor | Yes | PDMS | No | Perfused spheroid platform | 85 |
Sterility and low bioburden are often not discussed in the literature but have utmost practical relevance. In a laboratory or research environment, ethanol disinfection and/or UV exposure may be sufficient for successful cell cultures. However, in standard industry biotechnological and biomedical applications, biological manufacturing practices simply demand non-negotiable sterilization procedures such as gamma-ray/ionizing irradiation or autoclaving and similar heat/steam treatments. While this requirement may not be a factor for standard labware, it should be considered that both sensor elements and electronic components may not withstand these procedures. Particularly, biological sensor components, e.g., enzymes or antibodies, but also materials such as polymers will not necessarily withstand high heat or high doses of ionizing irradiation. This limitation extends to the point at which it may not be feasible at all to combine the cell culture area or vessel and sensor device, and instead have the sterile barrier in between. Consequently, this limits the perspective of single-chip solutions for real-world applications.
Classical, commercial 3D printers allow the facile fabrication of microfluidic structures such as channels and chambers. Differentiated evaluations of common 3D printing modalities are rarely found.103,104 The shortcomings of the materials and processes range from limited feature size, dimensional accuracy, the surface quality and optical clarity, potential absorption of molecules, to unclear biocompatibility of the materials, hindering cell culture applications.28 In contrast, both mass fabricated polymeric platforms and standard cleanroom techniques can easily avoid many of these drawbacks. For example, microfabricated barrier structures for contact angle pinning, which require smooth surfaces and sharp edges are not easily realized via additive manufacturing, but easily moulded and/or fabricated directly via clean room processing. Regarding sensor integration, two common limitations of additive manufacturing stand out in particular. First, it is difficult to co-integrate electrodes in order to seamlessly include electrical and electrochemical sensors. Second, optical clarity of the materials is often not sufficient, which is detrimental for both high quality imaging and the integration of optical sensors. Additive manufacturing of novel material classes, such as the 3D printing of glass105,106 or metals,107 and even their potential co-integration are highly promising. Glass, in contrast to polymers, offers excellent chemical and thermal stability, low absorption of chemicals, a larger variety of surface modifications, excellent optical clarity, and defined optical properties, in addition to a different level of sustainability. At the same time, advanced additive manufacturing processes allow geometries and shapes that otherwise cannot be realized at all, e.g., hollow, porous and multilayer structures.
Conclusions
Sensors can be very useful in cell cultures because even basic environmental parameters are extremely determinant for the experimental outcome. The need for standardization and control of cultures conditions, and thereby reproducibility and biological relevance of in vitro models, has been identified and recently emphasized in various papers from the biological perspective. Numerous studies with continuous sensor data have underlined that assumed culture conditions and the actual pericellular microenvironment may differ strongly from each other. Other measurements have shown how metabolic cues can significantly influence cellular behaviour and are often the key to the validity of the in vitro model. Therefore, this overall challenge can barely be addressed without applying sensors in the cell culture. While the cost for sensor integration may appear prohibitive, lowering the cost and increasing accessibility of extremely expensive therapies, such as cell and gene therapy, is also imperative and well justifies sensor integration.Nonetheless, in situ sensors and sensor systems are still far from being standard in cell culture practice, particularly not in normal routines in the laboratory or industrial applications. The reasons for that are numerous: the difficulty of sensor integration in cell culture and handling and manufacturing practices, lack of straightforward parallelization and automation, lack of stability over relevant timeframes, but also the apparent complexity of the data and low drive for adoption and understanding of their relevance from the end user side. In light of these observations, we formulate the following concluding remarks for the future development of sensors and sensor-equipped platforms for cell cultures and organs-on-chip:
In sensor principles, optical sensors see an apparent increase in popularity due to their non-invasiveness, lack of on-chip electrical components and somewhat better options for multiplexed read-out, but are largely limited to oxygen and pH. Electrochemical sensors are advantageous if sensors need to be small, which is why they dominate in single spheroid monitoring, and a larger variety of parameters exists. For glucose and lactate as the primary energy metabolites, or glutamine and glutamate to cover the nitrogen metabolism, optical techniques are fairly limited, and electro-enzymatic sensors are the standard or the only alternative. Interestingly, a detailed look at real-world long-term performance data of solid implementations does not hint at substantial performance difference between the two principles, particularly not a clear advantage of optical sensors, as often claimed. More direct comparison studies of both principles under comparable conditions would be attractive. Despite decades of sensor development, the stability of biochemical sensors over a timeframe of, e.g., four weeks as desired for meaningful in vitro models, is still far from standard. With respect to the much desired need for parallelization in biology, there is also an argument for the development of more affordable and more multi-channel instrumentation, such as embedded hardware, independent of the individual sensor principle or platform.
Most cell culture microsystems still feature 2D adherent cell cultures because they allow the easiest sensor access, certainly not because they have the highest biological relevance. Extracting meaningful signals from 3D cultures is hard, especially from those growing heterogeneously in a matrix, which has arguably the highest in vivo relevance. There are still very few systems that can measure from matrix-based 3D culture, because culture integration is challenging, and mass transfer is complex, resulting in high demands for sensor location and performance. It is still not widely acknowledged that biochemistry overall and sensing of metabolites in particular are inevitably linked to the understanding and study of the molecule mass transfer. A solid understanding, modelling and verification of the mass transfer in cell culture monitoring systems are essential and should be emphasized more. Interestingly, oxygen control and the relevance of hypoxia have taken hold in system design, and gas-tightness is an increasingly implemented system property that also affects material choice.
The trends in materials point towards simpler, disposable materials and easier, more efficient fabrication processes with generally fewer electrical or electronic components. Almost all recent systems are fully transparent for optical access, such as high content imaging, and made from gas-impermeable polymers or glass. Silicone, e.g., PDMS, for microfluidics is being replaced due to its oxygen permeability and unfavourable absorption of various compounds, as is silicon as a chip material for not being transparent. The usage of thermoplastics and relatively easily scalable fabrication techniques is increasing. Materials and processes are preferably close to those of the standard labware. Despite other claims, additive manufacturing plays little to no role in advanced embodiments of sensor-equipped cell culture systems, due to low scalability and fabrication efficiency, questionable material properties, lack of optical clarity, and difficult electrical sensor integration.
The technological community should, therefore, overcome their own endless technology hype cycles that include sensor materials and fabrication processes, in combination with a substantial tendency to overestimate the relevance of technology in contrast to the constraints and requirements imposed by the biological model. Despite a high number of apparent innovations in sensor materials and technology, the number of translations to meaningful cell experiments is surprisingly low and relatively conservative. The focus should particularly be directed towards a more realistic assessment of sensor performance, which includes more long-term data up to the relevant time-scales of weeks under real-world conditions. We encourage reviewers for journals but also for funding bodies to reconsider and re-evaluate their criteria. A shift has to be made from apparent novelty as an end in itself and from, often unnecessary, technological complexity, towards a more goal-oriented approach. A better understanding and honest assessment of needs, capabilities and limitations from both the technological and biological side will facilitate a more widespread application of sensors in cell cultures and organs-on-chip, lead to better and more predictive in vitro models, and will improve transdisciplinary connectivity and collaboration.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
JD: literature review, writing – review and editing; JK: conceptualization, writing – review and editing; SJR: writing – review and editing; AW: conceptualization, literature review, writing – original draft, writing – review and editing, visualization.Conflicts of interest
The authors declare no conflict of interest.References
- D. C. Singleton, A. Macann and W. R. Wilson, Nat. Rev. Clin. Oncol., 2021, 18, 751–772 Search PubMed.
- V. Palacio-Castañeda, N. Velthuijs, S. Le Gac and W. P. R. Verdurmen, Lab Chip, 2022, 22, 1068–1092 Search PubMed.
- S. G. Klein, S. M. Alsolami, A. Steckbauer, S. Arossa, A. J. Parry, G. Ramos Mandujano, K. Alsayegh, J. C. Izpisua Belmonte, M. Li and C. M. Duarte, Nat. Biomed. Eng., 2021, 5, 787–792 Search PubMed.
- A. Al-Ani, D. Toms, D. Kondro, J. Thundathil, Y. Yu and M. Ungrin, PLoS One, 2018, 13, e0204269 Search PubMed.
- E. O. Pettersen, L. H. Larsen, N. B. Ramsing and P. Ebbesen, Cell Proliferation, 2005, 38, 257–267 Search PubMed.
- J. Kieninger, K. Aravindalochanan, J. A. Sandvik, E. O. Pettersen and G. A. Urban, Cell Proliferation, 2014, 47, 180–188 Search PubMed.
- D. G. Hafeman, J. W. Parce and H. M. Mcconnell, Science, 1988, 240, 1182–1185 Search PubMed.
- F. Hafner, Biosens. Bioelectron., 2000, 15, 149–158 Search PubMed.
- S. E. Eklund, D. E. Cliffel, E. Kozlov, A. Prokop, J. Wikswo and F. Baudenbacher, Anal. Chim. Acta, 2003, 496, 93–101 Search PubMed.
- E. Thedinga, A. Kob, H. Holst, A. Keuer, S. Drechsler, R. Niendorf, W. Baumann, I. Freund, M. Lehmann and R. Ehret, Toxicol. Appl. Pharmacol., 2007, 220, 33–44 Search PubMed.
- P. Mestres and A. Morguet, Expert Opin. Drug Discovery, 2009, 4, 785–797 Search PubMed.
- C. Soragni, K. Queiroz, C. P. Ng, A. Stok, T. Olivier, D. Tzagkaraki, J. Heijmans, J. Suijker, S. P. M. de Ruiter, A. Olczyk, M. Bokkers, F. Schavemaker, S. J. Trietsch, H. L. Lanz, P. Vulto and J. Joore, Angiogenesis, 2024, 27, 37–49 Search PubMed.
- J. Kieninger, A. Weltin, H. Flamm and G. A. Urban, Lab Chip, 2018, 18, 1274–1291 Search PubMed.
- X. D. Wang and O. S. Wolfbeis, Anal. Chem., 2016, 88, 203–227 Search PubMed.
- X. D. Wang and O. S. Wolfbeis, Anal. Chem., 2020, 92, 397–430 Search PubMed.
- D. R. Thévenot, K. Toth, R. A. Durst and G. S. Wilson, Biosens. Bioelectron., 2001, 16, 121–131 Search PubMed.
- A. Weltin, J. Kieninger and G. A. Urban, Anal. Bioanal. Chem., 2016, 408, 4503–4521 Search PubMed.
- M. A. Holzreuter and L. I. Segerink, Lab Chip, 2024, 24, 1121–1134 Search PubMed.
- X. Yuan, A. Hierlemann and U. Frey, IEEE J. Solid-State Circuits, 2021, 56, 2466–2475 Search PubMed.
- J. Dragas, V. Viswam, A. Shadmani, Y. Chen, R. Bounik, A. Stettler, M. Radivojevic, S. Geissler, M. E. J. Obien, J. Müller and A. Hierlemann, IEEE J. Solid-State Circuits, 2017, 52, 1576–1590 Search PubMed.
- J. Abbott, A. Mukherjee, W. Wu, T. Ye, H. S. Jung, K. M. Cheung, R. S. Gertner, M. Basan, D. Ham and H. Park, Lab Chip, 2022, 22, 1286–1296 Search PubMed.
- M. Hofer and M. P. Lutolf, Nat. Rev. Mater., 2021, 6, 402–420 Search PubMed.
- B. L. LeSavage, R. A. Suhar, N. Broguiere, M. P. Lutolf and S. C. Heilshorn, Nat. Mater., 2022, 21, 143–159 Search PubMed.
- J. Drost and H. Clevers, Nat. Rev. Cancer, 2018, 18, 407–418 Search PubMed.
- A. Sontheimer-Phelps, B. A. Hassell and D. E. Ingber, Nat. Rev. Cancer, 2019, 19, 65–81 Search PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 Search PubMed.
- L. A. Low, C. Mummery, B. R. Berridge, C. P. Austin and D. A. Tagle, Nat. Rev. Drug Discovery, 2021, 20, 345–361 Search PubMed.
- C. M. Leung, P. de Haan, K. Ronaldson-Bouchard, G. A. Kim, J. Ko, H. S. Rho, Z. Chen, P. Habibovic, N. L. Jeon, S. Takayama, M. L. Shuler, G. Vunjak-Novakovic, O. Frey, E. Verpoorte and Y. C. Toh, Nat. Rev. Methods Primers, 2022, 2, 33 Search PubMed.
- J. Rogal, K. Schlünder and P. Loskill, ACS Biomater. Sci. Eng., 2022, 8, 4643–4647 Search PubMed.
- L. Ewart and A. Roth, Nat. Rev. Drug Discovery, 2021, 20, 327–328 Search PubMed.
- J. J. Han, Artif. Organs, 2023, 47, 449–450 Search PubMed.
- P. Vulto and J. Joore, Nat. Rev. Drug Discovery, 2021, 20, 961–962 Search PubMed.
- M. M. Modena, K. Chawla, P. M. Misun and A. Hierlemann, ACS Chem. Biol., 2018, 13, 1767–1784 Search PubMed.
- S. Fuchs, S. Johansson, A. Tjell, G. Werr, T. Mayr and M. Tenje, ACS Biomater. Sci. Eng., 2021, 7, 2926–2948 Search PubMed.
- S. M. Grist, K. L. Bennewith and K. C. Cheung, Annu. Rev. Anal. Chem., 2022, 15, 221–246 Search PubMed.
- H. Kavand, R. Nasiri and A. Herland, Adv. Mater., 2022, 34, 2107876 Search PubMed.
- C. Bouquerel, A. Dubrova, I. Hofer, D. T. T. Phan, M. Bernheim, S. Ladaigue, C. Cavaniol, D. Maddalo, L. Cabel, F. Mechta-Grigoriou, C. Wilhelm, G. Zalcman, M. C. Parrini and S. Descroix, Lab Chip, 2023, 23, 3906–3935 Search PubMed.
- D. R. Reyes, M. B. Esch, E. Lorna, R. Nasiri, A. Herland, K. E. Sung, M. Piergiovanni, C. Lucchesi, J. Vukasinovic, J. T. Shoemaker, H. Nakae, J. Hickman, K. Pant, A. M. Taylor, N. Heinz and N. Ashammakhi, Lab Chip, 2023, 24, 1076–1087 Search PubMed.
- R. Wenger, V. Kurtcuoglu, C. Scholz, H. Marti and D. Hoogewijs, Hypoxia, 2015, 35 Search PubMed.
- J. Kieninger, Y. Tamari, B. Enderle, G. Jobst, J. A. Sandvik, E. O. Pettersen and G. A. Urban, Biosensors, 2018, 8, 1–11 Search PubMed.
- S. Eggert, M. S. Gutbrod, G. Liebsch, R. Meier, C. Meinert and D. W. Hutmacher, ACS Sens., 2021, 6, 1248–1260 Search PubMed.
- A. Weltin, K. Slotwinski, J. Kieninger, I. Moser, G. Jobst, M. Wego, R. Ehret and G. A. Urban, Lab Chip, 2014, 14, 138–146 Search PubMed.
- J. Dornhof, J. Kieninger, H. Muralidharan, J. Maurer, G. A. Urban and A. Weltin, Lab Chip, 2022, 22, 225–239 Search PubMed.
- D. Bavli, S. Prill, E. Ezra, G. Levy, M. Cohen, M. Vinken, J. Vanfleteren, M. Jaeger and Y. Nahmias, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2231–E2240 Search PubMed.
- H. Zirath, M. Rothbauer, S. Spitz, B. Bachmann, C. Jordan, B. Müller, J. Ehgartner, E. Priglinger, S. Mühleder, H. Redl, W. Holnthoner, M. Harasek, T. Mayr and P. Ertl, Front. Physiol., 2018, 9, 1–12 Search PubMed.
- Y. Wang, L. Wang, Y. Zhu and J. Qin, Lab Chip, 2018, 18, 851–860 Search PubMed.
- S. J. Trietsch, G. D. Israëls, J. Joore, T. Hankemeier and P. Vulto, Lab Chip, 2013, 13, 3548–3554 Search PubMed.
- C. Seydel, Nat. Methods, 2021, 18, 1452–1456 Search PubMed.
- K. Vandereyken, A. Sifrim, B. Thienpont and T. Voet, Nat. Rev. Genet., 2023, 24, 494–515 Search PubMed.
- F. Danzi, R. Pacchiana, A. Mafficini, M. T. Scupoli, A. Scarpa, M. Donadelli and A. Fiore, Signal Transduction Targeted Ther., 2023, 8, 137 Search PubMed.
- A. Weltin, S. Hammer, F. Noor, Y. Kaminski, J. Kieninger and G. A. Urban, Biosens. Bioelectron., 2017, 87, 941–948 Search PubMed.
- J. Dornhof, V. Zieger, J. Kieninger, D. Frejek, R. Zengerle, G. A. Urban, S. Kartmann and A. Weltin, Lab Chip, 2022, 22, 4369–4381 Search PubMed.
- A. Kalmykov, C. Huang, J. Bliley, D. Shiwarski, J. Tashman, A. Abdullah, S. K. Rastogi, S. Shukla, E. Mataev, A. W. Feinberg, K. Jimmy Hsia and T. Cohen-Karni, Sci. Adv., 2019, 5, eabf9153 Search PubMed.
- M. C. Lefevre, G. Dijk, A. Kaszas, M. Baca, D. Moreau and R. P. O'Connor, npj Flexible Electron., 2021, 5, 1–9 Search PubMed.
- Y. Park, C. K. Franz, H. Ryu, H. Luan, K. Y. Cotton, J. U. Kim, T. S. Chung, S. Zhao, A. Vazquez-Guardado, D. S. Yang, K. Li, R. Avila, J. K. Phillips, M. J. Quezada, H. Jang, S. S. Kwak, S. M. Won, K. Kwon, H. Jeong, A. J. Bandodkar, M. Han, H. Zhao, G. R. Osher, H. Wang, K. H. Lee, Y. Zhang, Y. Huang, J. D. Finan and J. A. Rogers, Sci. Adv., 2021, 7, eabf9153 Search PubMed.
- O. Frey, P. M. Misun, D. A. Fluri, J. G. Hengstler and A. Hierlemann, Nat. Commun., 2014, 5, 4250 Search PubMed.
- P. M. Misun, J. Rothe, Y. R. F. Schmid, A. Hierlemann and O. Frey, Microsyst. Nanoeng., 2016, 2, 16022 Search PubMed.
- J. Kieninger, Electrochemical Methods for the Micro- and Nanoscale, De Gruyter, Berlin/Boston, 2022 Search PubMed.
- L. C. Clark and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29–45 Search PubMed.
- J. W. Ross, US Pat., 3260656A, 1966 Search PubMed.
- M. Koudelka, Sens. Actuators, 1986, 9, 249–258 Search PubMed.
- G. Jobst, G. Urban, A. Jachimowicz, F. Kohl, O. Tilado, I. Lettenbichler and G. Nauer, Biosens. Bioelectron., 1993, 8, 123–128 Search PubMed.
- N. Arroyo-Currás, J. Somerson, P. A. Vieira, K. L. Ploense, T. E. Kippin and K. W. Plaxco, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 645–650 Search PubMed.
- J. Marzioch, J. Kieninger, A. Weltin, H. Flamm, K. Aravindalochanan, J. A. Sandvik, E. O. Pettersen, Q. Peng and G. A. Urban, Lab Chip, 2018, 18, 3353–3360 Search PubMed.
- E. Tanumihardja, R. H. Slaats, A. D. Van Der Meer, R. Passier, W. Olthuis and A. Van Den Berg, ACS Sens., 2021, 6, 267–274 Search PubMed.
- E. Tanumihardja, A. Paradelo Rodríguez, J. T. Loessberg-Zahl, B. Mei, W. Olthuis and A. van den Berg, Sens. Actuators, B, 2021, 334, 129631 Search PubMed.
- F. Liebisch, A. Weltin, J. Marzioch, G. A. Urban and J. Kieninger, Sens. Actuators, B, 2020, 322, 128652 Search PubMed.
- A. J. Hsueh, S. Park, T. Satoh, T. Shimizu, K. Koiwai, M. Nakashima, Y. Morimoto, M. Kinoshita and H. Suzuki, Anal. Chem., 2021, 93, 5577–5585 Search PubMed.
- B. Müller, P. Sulzer, M. Walch, H. Zirath, T. Buryška, M. Rothbauer, P. Ertl and T. Mayr, Sens. Actuators, B, 2021, 334, 129664 Search PubMed.
- H. Zirath, S. Spitz, D. Roth, T. Schellhorn, M. Rothbauer, B. Müller, M. Walch, J. Kaur, A. Wörle, Y. Kohl, T. Mayr and P. Ertl, Lab Chip, 2021, 21, 4237–4248 Search PubMed.
- S. Fuchs, R. W. J. van Helden, M. Wiendels, M. N. S. de Graaf, V. V. Orlova, C. L. Mummery, B. J. van Meer and T. Mayr, Mater. Today Bio, 2022, 17, 100475 Search PubMed.
- S. Fuchs, V. Rieger, A. Tjell, S. Spitz, K. Brandauer, R. Schaller-Ammann, J. Feiel, P. Ertl, I. Klimant and T. Mayr, Biosens. Bioelectron., 2023, 237, 115491 Search PubMed.
- M. Busche, D. Rabl, J. Fischer, C. Schmees, T. Mayr, R. Gebhardt and M. Stelzle, EXCLI J., 2022, 21, 144–161 Search PubMed.
- C. Bouquerel, W. César, L. Barthod, S. Arrak, A. Battistella, G. Gropplero, F. Mechta-Grigoriou, G. Zalcman, M. C. Parrini, M. Verhulsel and S. Descroix, Lab Chip, 2022, 22, 4443–4455 Search PubMed.
- J. S. Cognetti, M. T. Moen, M. G. Brewer, M. R. Bryan, J. D. Tice, J. L. McGrath and B. L. Miller, Lab Chip, 2022, 23, 239–250 Search PubMed.
- F. Alexander, S. Eggert and J. Wiest, Cytotechnology, 2018, 70, 375–386 Search PubMed.
- H. Azizgolshani, J. R. Coppeta, E. M. Vedula, E. E. Marr, B. P. Cain, R. J. Luu, M. P. Lech, S. H. Kann, T. J. Mulhern, V. Tandon, K. Tan, N. J. Haroutunian, P. Keegan, M. Rogers, A. L. Gard, K. B. Baldwin, J. C. de Souza, B. C. Hoefler, S. S. Bale, L. B. Kratchman, A. Zorn, A. Patterson, E. S. Kim, T. A. Petrie, E. L. Wiellette, C. Williams, B. C. Isenberg and J. L. Charest, Lab Chip, 2021, 21, 1454–1474 Search PubMed.
- S. H. Kann, E. M. Shaughnessey, J. R. Coppeta, H. Azizgolshani, B. C. Isenberg, E. M. Vedula, X. Zhang and J. L. Charest, Microsyst. Nanoeng., 2022, 8, 109 Search PubMed.
- S. H. Kann, E. M. Shaughnessey, X. Zhang, J. L. Charest and E. M. Vedula, Analyst, 2023, 148, 3204–3216 Search PubMed.
- S. Kreß, R. Schaller-Ammann, J. Feiel, J. Wegener, J. Priedl, W. Dietrich, C. Kasper and D. Egger, Cell, 2022, 11, 1–14 Search PubMed.
- J. Scheinpflug, C. T. Höfer, S. S. Schmerbeck, M. Steinfath, J. Doka, Y. A. Tesfahunegn, N. Violet, K. Renko, K. Gulich, T. John, M. R. Schneider, E. Wistorf, G. Schönfelder and F. Schulze, Lab Chip, 2023, 23, 3405–3423 Search PubMed.
- K. Schlünder, M. Cipriano, A. Zbinden, S. Fuchs, T. Mayr, K. Schenke-Layland and P. Loskill, Lab Chip, 2024, 24, 2080–2093 Search PubMed.
- N. Rousset, R. L. Sandoval, M. M. Modena, A. Hierlemann and P. M. Misun, Microsyst. Nanoeng., 2022, 8, 14 Search PubMed.
- R. Mukomoto, Y. Nashimoto, T. Terai, T. Imaizumi, K. Hiramoto, K. Ino, R. Yokokawa, T. Miura and H. Shiku, Analyst, 2020, 145, 6342–6348 Search PubMed.
- Y. Nashimoto, R. Mukomoto, T. Imaizumi, T. Terai, S. Shishido, K. Ino, R. Yokokawa, T. Miura, K. Onuma, M. Inoue and H. Shiku, Biosens. Bioelectron., 2023, 219, 114808 Search PubMed.
- Y. Nashimoto, S. Shishido, K. Onuma, K. Ino, M. Inoue and H. Shiku, Front. Bioeng. Biotechnol., 2023, 11, 1–7 Search PubMed.
- J. Marzioch, J. Kieninger, A. Weltin and G. A. Urban, in 2019 20th International Conference on Solid-State Sensors, Actuators and Microsystems and Eurosensors XXXIII, TRANSDUCERS 2019 and EUROSENSORS XXXIII, 2019, pp. 968–970 Search PubMed.
- A. Weltin, J. Dornhof, J. Kieninger, S. J. Rupitsch, J. Beck and K. Joseph, 2023 IEEE Biosens. Conf., 2023, pp. 1–4 Search PubMed.
- Y. Shirato, A. J. Hsueh, N. A. Ab Mutalib, Y. Deng, R. Suematsu, A. Kato, B. M. Kearney, M. Kinoshita and H. Suzuki, ACS Omega, 2024, 9, 10825–10833 Search PubMed.
- K. Hiramoto, K. Ino, K. Komatsu, Y. Nashimoto and H. Shiku, Biosens. Bioelectron., 2021, 181, 113123 Search PubMed.
- D. Sticker, M. Rothbauer, J. Ehgartner, C. Steininger, O. Liske, R. Liska, W. Neuhaus, T. Mayr, T. Haraldsson, J. P. Kutter and P. Ertl, ACS Appl. Mater. Interfaces, 2019, 11, 9730–9739 Search PubMed.
- A. Bussooa, E. Tubbs, F. Revol-Cavalier, A. Chmayssem, M. Alessio, M. L. Cosnier and N. Verplanck, Biosens. Bioelectron.:X, 2022, 11, 4–11 Search PubMed.
- M. Busche, O. Tomilova, J. Schütte, S. Werner, M. Beer, N. Groll, B. Hagmeyer, M. Pawlak, P. D. Jones, C. Schmees, H. Becker, J. Schnabel, K. Gall, R. Hemmler, M. Matz-Soja, G. Damm, S. Beuck, T. Klaassen, J. Moer, A. Ullrich, D. Runge, K. Schenke-Layland, R. Gebhardt and M. Stelzle, Lab Chip, 2020, 20, 2911–2926 Search PubMed.
- K. R. Rivera, V. A. Pozdin, A. T. Young, P. D. Erb, N. A. Wisniewski, S. T. Magness and M. Daniele, Biosens. Bioelectron., 2019, 123, 131–140 Search PubMed.
- C. Gehre, M. Flechner, S. Kammerer, J. H. Küpper, C. D. Coleman, G. P. Püschel, K. Uhlig and C. Duschl, Sci. Rep., 2020, 10, 1–12 Search PubMed.
- M. F. Wesseler, M. N. Johansen, A. Kızıltay, K. I. Mortensen and N. B. Larsen, Lab Chip, 2022, 22, 4167–4179 Search PubMed.
- M. G. Sánchez-Salazar, R. Garza-Garza, R. Crespo-López Oliver, V. S. Jerezano-Flores, S. Gallegos-Martínez, S. Ramos-Meizoso, M. Verduzco-Valenzuela, G. Trujillo-de Santiago and M. M. Alvarez, Front. Lab. Chip. Technol., 2024, 3, 1–14 Search PubMed.
- A. A. Rowe, A. J. Bonham, R. J. White, M. P. Zimmer, R. J. Yadgar, T. M. Hobza, J. W. Honea, I. Ben-Yaacov and K. W. Plaxco, PLoS One, 2011, 6, e23783 Search PubMed.
- M. D. M. Dryden and A. R. Wheeler, PLoS One, 2015, 10, 1–17 Search PubMed.
- D. Bill, M. Jasper, A. Weltin, G. A. Urban, S. J. Rupitsch and J. Kieninger, Anal. Chem., 2023, 95, 13003–13009 Search PubMed.
- C. Mota, S. Camarero-Espinosa, M. B. Baker, P. Wieringa and L. Moroni, Chem. Rev., 2020, 120, 10547–10607 Search PubMed.
- A. C. Daly, M. E. Prendergast, A. J. Hughes and J. A. Burdick, Cell, 2021, 184, 18–32 Search PubMed.
- F. Li, N. P. Macdonald, R. M. Guijt and M. C. Breadmore, Lab Chip, 2019, 19, 35–49 Search PubMed.
- L. A. Milton, M. S. Viglione, L. J. Y. Ong, G. P. Nordin and Y. C. Toh, Lab Chip, 2023, 23, 3537–3560 Search PubMed.
- F. Kotz, K. Arnold, W. Bauer, D. Schild, N. Keller, K. Sachsenheimer, T. M. Nargang, C. Richter, D. Helmer and B. E. Rapp, Nature, 2017, 544, 337–339 Search PubMed.
- F. Kotz, N. Schneider, A. Striegel, A. Wolfschläger, N. Keller, M. Worgull, W. Bauer, D. Schild, M. Milich, C. Greiner, D. Helmer and B. E. Rapp, Adv. Mater., 2018, 30, 1–5 Search PubMed.
- M. Luitz, M. Lunzer, A. Goralczyk, M. Mader, S. Bhagwat, A. Warmbold, D. Helmer, F. Kotz and B. E. Rapp, Adv. Mater., 2021, 33, 1–7 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |