Open Access Article
Open Access ArticleAzo derivatives of monoterpenes as anti-Helicobacter pylori agents: from synthesis to structure-based target investigation†
Francesco
Melfi‡
a,
Marialuigia
Fantacuzzi‡
a,
Simone
Carradori
 *a,
Ilaria
D'Agostino
*a,
Ilaria
D'Agostino
 b,
Alessandra
Ammazzalorso
b,
Alessandra
Ammazzalorso
 a,
Noemi
Mencarelli
a,
Marialucia
Gallorini
a,
Mattia
Spano
a,
Noemi
Mencarelli
a,
Marialucia
Gallorini
a,
Mattia
Spano
 c,
Paolo
Guglielmi
c,
Mariangela
Agamennone
c,
Paolo
Guglielmi
c,
Mariangela
Agamennone
 a,
Sazan
Haji Ali
a,
Sazan
Haji Ali
 de,
Ali
Al-Samydai
f and
Francesca
Sisto
g
de,
Ali
Al-Samydai
f and
Francesca
Sisto
g
aDepartment of Pharmacy, “G. d'Annunzio” University of Chieti-Pescara, via dei Vestini 31, 66100 Chieti, Italy. E-mail: simone.carradori@unich.it
bDepartment of Pharmacy, Univerity of Pisa, Via Bonanno 6, 56126 Pisa, Italy
cDepartment of Drug Chemistry and Technology, “Sapienza” University of Rome, P.le Aldo Moro 5, 00185 Rome, Italy
dDepartment of Pharmaceutical Chemistry, College of Pharmacy, Hawler Medical University, Erbil 44000, Iraq
eDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Anadolu University, Eskişehir, 26470, Turkey
fPharmacological and Diagnostic Research Centre (PDRC), Faculty of Pharmacy, Al-Ahliyya Amman University, Amman-Jordan- Al Salt Road, Amman, 19328, Jordan
gDepartment of Biomedical, Surgical and Dental Sciences, University of Milan, Via Pascal 36, 20133 Milan, Italy
First published on 9th October 2024
Abstract
Helicobacter pylori (Hp) infection affects nearly half of the global population. Current therapeutic options include the administration of a combination of antibiotics and proton pump inhibitors, although antimicrobial resistance rise remains a big concern. Phenolic monoterpenes, e.g., eugenol, vanillin, carvacrol, and thymol, have always attracted researchers for their multifaced biological activities and the possibility to be easily derivatized. Thereby, herein we present the functionalization of such compounds through the conventional aryl diazotization reaction, generating a series of mono- and bis-azo derivatives (1–28). Also, to continue previous studies, we investigated the role of the free phenolic moiety of thymol with eight compounds (29–36). The compounds were tested against four Hp strains including three clinical isolates, finding some potent and selective inhibitors of bacterial growth. Thus, the representative compounds underwent in vitro cytotoxicity evaluation on two normal cell lines and putative target investigation by performing a structure-based approach based on docking calculations on some of the most studied pharmacological targets for Hp, e.g., urease, β-hydroxyacyl-acyl carrier protein dehydratase, glucose 6-phosphate dehydrogenase, and inosine 5′-monophosphate dehydrogenase.
1. Introduction
Helicobacter pylori (Hp) is a microaerophilic, Gram-negative, spiral-shaped bacterium infecting an estimated 50% of the global population.1 The prevalence of this infection is influenced by different factors related to geographic regions, hygiene conditions, temperature, and socioeconomic status.2 Recently, a meta-analysis estimated the overall prevalence of Hp infection worldwide, highlighting a downward trend in adults in the past three decades (52.6% before 1990, 43.9% from 2015 to 2022), and an upward fashion for children and adolescents (from 26.6% before 1990 to 35.1% from 2015 to 2022).3Hp colonizes the inhospitable gastric environment by several mechanisms including urease activity, motility, and adhesion.4 Even though the infection is usually asymptomatic, it is correlated with a series of diseases, including duodenal and gastric ulcers, and malignancies such as mucosa-associated lymphoid tissue (MALT) lymphoma and gastric cancer.1 As a matter of fact, the International Agency for Research on Cancer (IARC), a subordinate organization of the World Health Organization (WHO), classified Hp as a group 1 carcinogen.5 Currently, the eradication of the infection relies on treatments including the combination of antibiotics and strong acid suppressants.4 In particular, the first-line treatment is based on a triple therapy including proton pump inhibitors (PPIs), clarithromycin, and amoxicillin. However, if the treatment population has a clarithromycin resistance rate above 20%, clarithromycin-containing triple therapy is not indicated. Thus, bismuth-containing quadruple therapy (BQT) or non-bismuth quadruple therapy could be -used as a first line empirical treatment.4 Albeit a seven-day first-line treatment usually leads to eradication rates exceeding 90%, antibiotic resistance insurgence, insufficient acid suppression, and inadequate adherence to medications, can cause alarming failures,6–8 drawing attention to the importance of developing novel and effective anti-Hp therapies.9 However, only a few new antibiotics have received approval for the market in the past years and the search for new chemical entities is urgently required.10–13In recent years azo benzene compounds have been extensively explored given their broad spectrum of pharmaceutical and industrial applications and their cost-effective, easy, and highly reproducible synthetic procedure. Notably, azo derivatives from substituted anilines resulted in the development of derivatives endowed with a versatile biological profile, including antimicrobial activity.14–16
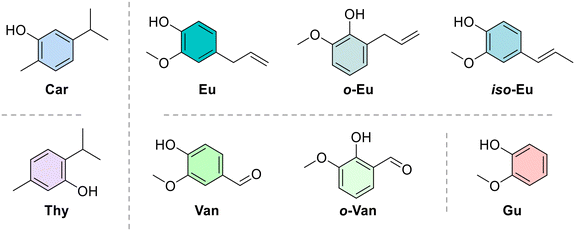
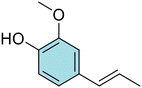
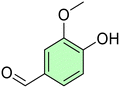
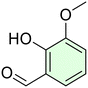
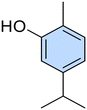
Conversely, to date, only a few researchers investigated the azo compounds as effective tools for the management of Hp and, among them, Jangra and colleagues reported a series of substituted 3-aryldiazenyl indoles targeting the Hp inosine 5′-monophosphate dehydrogenase (IMPDH, EC 1.1.1.205) enzyme.17 Furthermore, azobenzene derivatives can be degraded by intestinal microorganisms in vivo, as demonstrated by Chung et al., leading to carcinogenic and/or toxic metabolites, after azo-reduction by intestinal anaerobes.18 On the other hand, the wide polypharmacology of monoterpenes led to an in-depth investigation of carvacrol (Car, Fig. 1) and thymol (Thy, Fig. 1) along with eugenol (Eu, Fig. 1) and vanillin (Van, Fig. 1) and their regioisomers o-eugenol (o-Eu, Fig. 1), iso-eugenol (iso-Eu, Fig. 1), and o-vanillin (o-Van, Fig. 1) as antimicrobials. In this context, we recently reported the ability of Thy-, Eu-, and Car-based compounds to act as inhibitors of Hp growth.19–21
Also, by combining the two chemical features, Prof. Kantar's research group evaluated a series of azo compounds containing a Eu, Van, and guaiacol (Gu, Fig. 1) core, demonstrating promising inhibitory activity against Hp urease and antibacterial effectiveness.22–25
Pursuing our efforts in the development of novel effective anti-Hp drugs and inspired by the antimicrobial results observed for azo compounds, we designed a series of novel azobenzene derivatives based on the phenolic monoterpenes Thy, Car, Eu, and Van. The derivatives were synthesized through the conventional diazotization-coupling reaction, as depicted in Scheme 1. The antibacterial activity for these compounds along with a panel of commercially available phenolic monoterpenes and derivatives was assessed against Hp strains, including the genomically characterized NCTC 11637 and three other clinical isolates (F1, 23, and F4), and reported as minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC). Finally, two of the most active compounds (20 and 28) have been tested on healthy cell lines to assess their safety and subjected to a target investigation protocol by means of a molecular docking protocol employing a reverse docking approach to unravel the putative target(s) among the most extensively studied, including urease (urea amidohydrolase, EC 3.5.1.5), β-hydroxyacyl-acyl carrier protein (ACP) dehydratase (FabZ, EC 4.2.1.59), glucose 6-phosphate dehydrogenase (G6PD, EC 1.1.1.49), and inosine 5′-monophosphate dehydrogenase (IMPDH).
 | ||
| Scheme 1 Synthetic routes to azo compounds 1–36. Reagents and conditions: (i) NaNO2, 1.2 M HCl, 0 °C, 5 min; (ii) substituted phenol, 1.3 M NaOH, 0 °C, 5 min. | ||
2. Results and discussion
2.1. Design and synthesis of the monoterpene derivatives
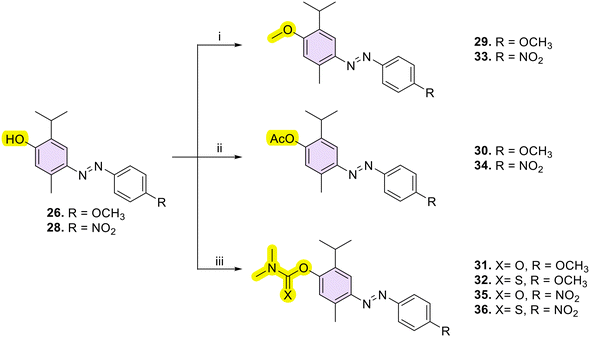
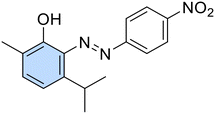
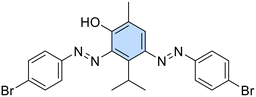
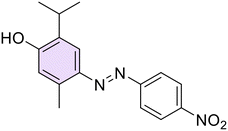
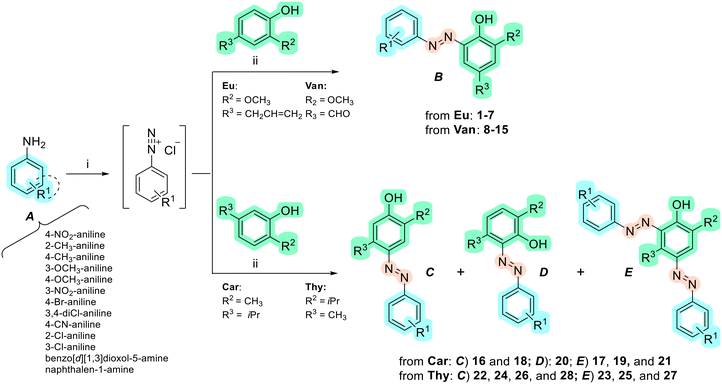
A new series of azobenzene derivatives (1–36) was obtained through the conventional diazotization-coupling reaction by using the appropriately substituted aromatic amine (A, Scheme 1) under acidic conditions in the presence of sodium nitrite at 0 °C, followed by the addition of the appropriate phenolic monoterpene in a basic environment (Scheme 1).Starting from Eu and Van, one azo compound (B, Scheme 1) was obtained, while interestingly, despite using the same experimental procedure involving equimolar amounts of the starting materials, in several cases, two products (C and E, Scheme 1) were isolated when Car and Thy were employed. In particular, the expected derivative characterized by the azo function at para-position to the phenolic OH moiety (C) was obtained as the major product, while the bis-azo compound E endowed with two azo functions at both ortho and para positions with respect to the phenolic group was isolated only in low yields. Moreover, only in one case, when Car was coupled with 4-nitroaniline (20), were we able to isolate only the ortho-azo product (D).
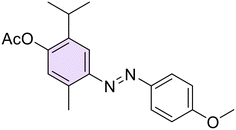
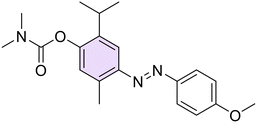
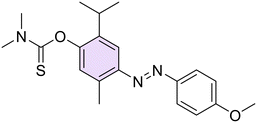
Then, with the aim to better understand the role of phenolic function in the anti-Hp activity, this function was chemically modified through O-derivatization reactions on two compounds (26 and 28, characterized by an electron-withdrawing or an electron-donating moiety, respectively), including methylation (29, 33), acetylation (30, 34), and conversion into dimethyl(thio)carbamates (31, 32, 35, 36) (Scheme 2).
O-Methylation of 26 and 28 was performed through an excess of dimethyl sulfate in the presence of K2CO3 at room temperature (r.t.) to afford the methoxy derivatives 29 and 33, respectively. Then, by singularly reacting 26 and 28 with sodium bicarbonate (NaHCO3) and an excess of acetic anhydride (Ac2O) at r.t., acetylated 30 and 34 were easily obtained. In the end, compounds 31, 32, 35, and 36 were obtained by reacting the suitable 26 or 28 with dimethyl (thio)carbamoyl chloride and potassium carbonate (K2CO3) as a base at r.t.
Interestingly, for all these compounds, the N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N double bond was found to be totally in the E configuration. In fact, the 1H NMR spectra showed the presence of just one species (ESI†), then confirmed to be the E isomer, as previously established by UV-vis experiments.26
N double bond was found to be totally in the E configuration. In fact, the 1H NMR spectra showed the presence of just one species (ESI†), then confirmed to be the E isomer, as previously established by UV-vis experiments.26
2.2. In vitro antibacterial activity evaluation against H. pylori and other bacterial species
The antibacterial profile of the commercial monoterpenes and their analogues, and the synthesized azo derivatives 1–36, was assessed on four Hp strains, including the reference NCTC 11637 strain and the clinical isolates F1, 23, and F4 from our laboratory by using metronidazole (MTZ), clarithromycin (CLR), and amoxicillin (AMX) as reference antibiotics (Table 1).The phenotypic diversity of resistant strains influences the biological results of both parent compounds and their derivatives. Indeed, this depends on the genotypic differences among resistant and sensitive strains. It is also known that the CLR resistance (F1 and F4) could be related to the mutation of the macrolide target, while the resistance to MTZ (like in NCTC 11637 and F4 strains) should be linked to the inactivation by mutation of the gene encoding an oxygen-independent NADPH nitroreductase.21
By analyzing the anti-Hp activities of the parent phenolic monoterpenes, we noticed that only o-Van, characterized by methoxy and aldehyde groups at the ortho position of the hydroxy group, and iso-Eu, bearing the methoxy and prop-1-en-1-yl moieties, at the ortho and para positions of the phenolic function, exhibited a promising antibacterial activity (Table 1). In particular, o-Van displayed a value between 4 and 16 μg mL−1 for both MIC and MBC, while iso-Eu reached a value of 32 μg mL−1 for MIC and MBC against all the tested strains. The other monoterpenes and analogues, including thymol blue, resulted in being almost or completely inactive on the four tested Hp strains (Table 1). In contrast, their azo analogues showed in some cases an anti-Hp activity with an MBC value of 2 μg mL−1, especially for Thy and Car azo derivatives (Table 1).
Eu derivatives 1–7 resulted in being inactive on the four strains, mainly exhibiting MIC and MBC values of around 128 μg mL−1, with the lipophilic/hydrophilic and electronic nature of substituents not affecting the activity profile (Table 1). Indeed, the most active compounds 4, bearing a para-nitro substituent, and 2, endowed with an ortho-methyl group, showed only in some cases MICs and MBCs of 64 μg mL−1 (Table 1).
Similarly, previously reported derivatives endowed with electron-donating substituents, i.e., para-methyl or para-methoxy groups, showed higher activity on the NCTC 11637 strain, with MIC values of 64 and 32 μg mL−1, respectively.21 Otherwise, electron-withdrawing substituents, i.e., para-cyano, para-bromo, and 3,4-dichloro substituents, were found to be almost inactive, with MIC values ≥128 μg mL−1.21 Interestingly, monochloro derivatives, with the halogen at the meta or ortho position, still remain the best-in-class Eu compounds, with MIC equal to 8–16 μg mL−1 on the tested strains.21
For Van derivatives 8–15, a linear trend for the biological results was not highlighted, even though the introduction of a substituted phenyl ring through the azo function generally ameliorated the anti-Hp profile of the monoterpene (Table 1). In detail, the insertion of the bromine at the para position as in 13 led to an increase of the activity (MIC range: 8–32 μg mL−1) especially on the F1 strain, while either its substitution with the other halogen, as for the 3-chlorine 9, or the presence of two chlorine atoms at the meta and para positions (15) decreased dramatically the activity against all the strains. Otherwise, single chlorine at the ortho position (8) was a better option for chloro-derivatives, especially against the F1 strain, with MIC and MBC values of 16 μg mL−1. Other electron-withdrawing groups like the cyano substituent placed at the para (14) or the nitro moiety at the meta position (10) showed better results with MBC values of 4 to 16 μg mL−1, respectively. The presence of electron-donating substituents in para led to different results depending on the magnitude of the electron-releasing effect. As a matter of fact, compound 12, endowed with a stronger electron-donating methoxy group, exhibited a weak or null activity, while compound 11, with the weaker methyl group, resulted to be the most potent Van derivative with both MIC and MBC values in the range of 2–8 μg mL−1, showing more efficacy against the F1 strain (Table 1).
Car and Thy derivatives (16–21 and 22–28, respectively) showed different trends: on the one hand, the mono-azo derivatives resulted to be the most potent ones against Hp overall, while the molecules that presented two azo functions in their structure, the bis-azo derivatives 17, 19, 21, 23, 25, and 27, were completely inactive, suggesting that an enlargement of the scaffold could lead to excessive bulkiness or lipophilicity, decreasing the antibacterial potency (Table 1). In terms of activity, there was only a slight difference between the para-methoxy Car-based compound 18 and its Thy-based regioisomer 26, with the former exhibiting MICs between 4 and 8 μg mL−1, whereas the latter showed better outcomes (2–4 μg mL−1) and also exhibiting a potent activity on the F4 strain (2 μg mL−1). Moreover, derivatives 16 and 24, the ortho-chloro azo derivatives of Car and Thy, respectively, showed a similar inhibitory profile, with the only exception of MIC and MBC values for F4 (2 μg mL−1) for compound 16 instead of 4 μg mL−1 as observed for 24. The para-methyl substituted azo derivative of Thy (22) exhibited 4 μg mL−1 for MIC and MBC on all the clinical isolates. Lastly, para-nitro derivatives 20 and 28 showed the best inhibitory profile on most strains: both compounds exhibited MIC and MBC values on F1, 23, and NCTC 11637 equal to 2 μg mL−1, while in the case of the F4 strain, the values were equal to 4 μg mL−1. Moreover, 20 resulted in strongly inhibiting the growth of strains 23 and NCTC 11637, while 28 was found to be active against the F1 strain (Table 1).
It is interesting to note that when the phenolic group of compounds 26 and 28 was derivatized as in 29–32 and 33–36, respectively, a dramatic decrease of activity independently of the chemical characteristics of the derivatization occurred (Table 1), suggesting that the free phenolic group is essential for the anti-Hp activity within this library of derivatives. This is in line with that observed for Car, Thy, and EuO-functionalization with alkyl, alkenyl, acetyl, and ester groups previously explored.19–21 Therefore, the chemically stable functionalization of the phenolic function is expected to increase the hydrophobicity of these compounds, and we could assume that the phenolic function of the azo derivatives influences the lipophilic/hydrophilic balance, affecting their antibacterial profile.
Furthermore, to shed light on the selectivity of the compounds' antibacterial effect, we tested the most potent anti-Hp derivatives 20 and 28 against three additional bacterial species: two Gram-negative, e.g., the lactose-fermenting Escherichia coli ATCC 25922 and the non-lactose-fermenting Pseudomonas aeruginosa ATCC 27853, and the Gram-positive Staphylococcus aureus ATCC 29213, and MIC and MBC values are reported in Table 2.
| Cpd | MIC/MBC (μg mL−1) | ||
|---|---|---|---|
| E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | S. aureus ATCC 29213 | |
| 20 | >128/>128 | >128/>128 | 2/32 |
| 28 | >128/>128 | >128/>128 | 2/32 |
By comparing data in Table 1, a lack of activity (MIC and MBC values >128 μg mL−1) on the Gram-negative bacteria E. coli and P. aeruginosa, with a consequent high selectivity for Hp (MIC and MBC values in the range 2–4 μg mL−1), was clear. Lower MIC values were obtained against the Gram-positive S. aureus (= 2 μg mL−1), whereas worse MBC values (= 32 μg mL−1) suggested a bacteriostatic effect for such compounds, as indicated by the MBC/MIC ratio greater than 4.
2.3. Molecular docking simulations
In the search for the pharmacological target of the azo derivatives and, in particular, of the two best-in-class compounds 20 and 28, a ligand-based target fishing approach was performed. Briefly, we conducted an in silico reverse screening in which a query compound was compared to ligands of specific targets.27,28 Different free web tools were used, like SwissTargetPrediction,29 PLATO,30 SuperPred,31 PPB2,32 and SEA Search.33 However, in almost all databases no Hp or any other bacterial targets were included. Thus, a structure-based approach was employed. A series of docking simulations were conducted on the most extensively studied targets associated with Hp, namely urease, FabZ, G6PD, and IMPDH.Urease is a nickel-dependent metalloenzyme that catalyzes the hydrolysis of urea to ammonia, thus neutralizing gastric acid and creating an alkaline local environment required for Hp survival.34,35 The 3D coordinates of Hp urease were retrieved by Protein Data Bank (PDB ID 6ZJA).36 Validation of the docking protocol was achieved by re-docking the cognate ligand DJM and obtaining a geometry with a docking score of −11.131 kcal mol−1 and an RMSD of 2.413 Å compared to the crystallographic ligand.
FabZ is a potent enzyme in fatty acid biosynthesis and catalyzes the dehydration of β-hydroxyacyl-ACP to trans-2-acyl-ACP.37 The discovery of Hp-FabZ inhibitors is of special interest in the treatment of various gastric diseases.38 The structure of the enzyme (PDB ID 3DOZ)38 was prepared and minimized before performing docking simulations. A fundamental water molecule was maintained in the active site for interaction with the crystallographic ligand (docking score −5.491 kcal mol−1 and RMSD 0.8322 Å).
Hp G6PD is a key enzyme in the metabolism of Hp, particularly in the pentose phosphate pathway that provides NADH+/ATP through glycolysis.39 As the 3D coordinates of the bacterial G6PD are still unknown, the homology modeling tool in Maestro40 was used to predict the structure of the enzyme. The primary sequence of the enzyme (strain ATCC 700392/26695) was used to identify the most suitable model in the Protein Data Bank41 using BLAST.42Leishmania donovani G6PD (PDB ID 7ZHV)43 with the best identity percentage (31.74%) was used as the template. The visual inspection of the docking poses of the azo derivatives in the active site of urease, FabZ, and G6PD and the obtained docking score values suggest that azo compounds do not interact with these targets.
IMPDH, a fundamental enzyme in the biosynthesis of purine nucleotides, is responsible for the oxidation of inosine 5′-monophosphate (IMP) to xanthosine 5′-monophosphate (XMP), which is further converted to guanosine 5′-monophosphate (GMP) by GMP synthase.44 Inhibition of IMPDH reduces the guanine nucleotide pool, thereby inhibiting microbial proliferation. Given the fundamental differences between prokaryotic and eukaryotic IMPDH in terms of their structural and kinetic characteristics, it is possible to obtain selective inhibitors of the bacterial enzyme (selective toxicity). Consequently, Hp-IMPDH inhibitors can be developed as valid antimicrobial agents.17 As the 3D structure of Hp-IMPDH is not yet known, it was modelled using the Homology Model tool of Maestro.40 The primary sequence of Hp-IMPDH (strain ATCC 700392/26695) was used to identify the best template in the Protein Data Bank41 using BLAST.42 The optimal model, with 60.2% identity, was identified as Campylobacter jejuni (Cj strain ATCC 700819 IMPDH (Cj-IMPD)), and the available 3D structure of Cj-IMPD (PDB ID 4MZ1)45 was employed as a template, including the IMP ligand. The modelled Hp-IMPDH was prepared and minimized before docking analysis. In the active site of Hp-IMPDH, the IMP ligand forms hydrogen bond contacts with Ser298, Asp333, Gly335, Gly356, Ser357, Met383, Gly384, and Glu410, as illustrated in Fig. 2a. This pattern of interactions is consistent with that observed in Cj-IMPD (PDB ID 4MZ1), as shown in Fig. 2b.
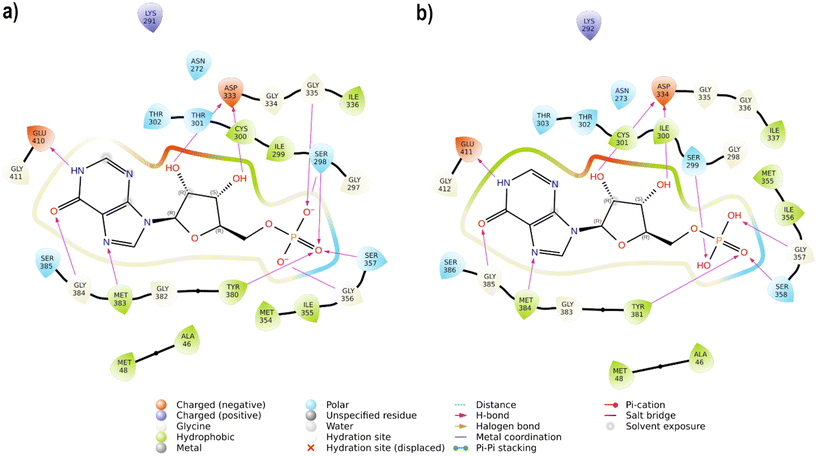 | ||
| Fig. 2 (a) 2D interaction of IMP in the active site of modelled Hp-IMPDH, (b) 2D interaction of IMP in the active site of Cj-IMPDH (PDB ID 4MZ1). | ||
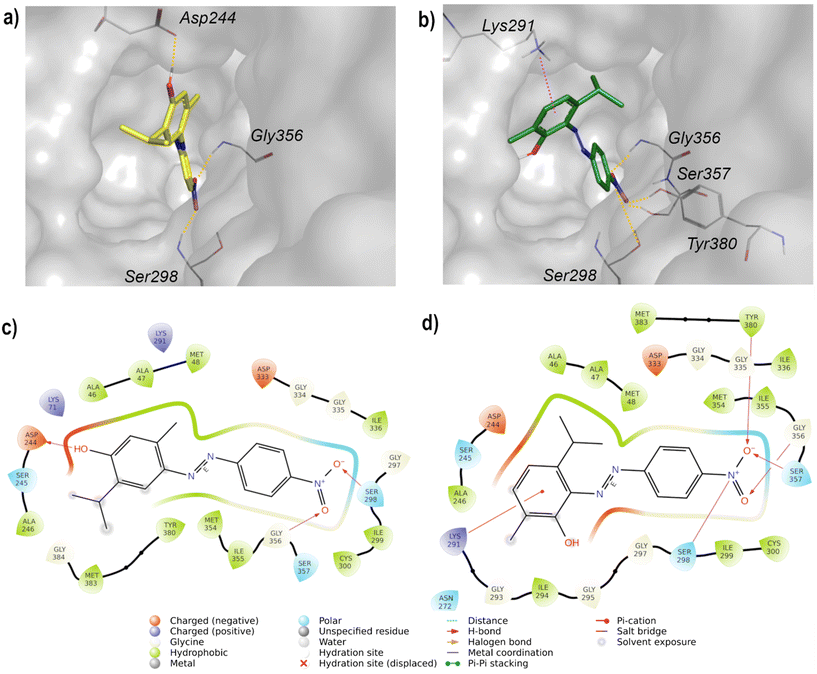
The best-docked pose of IMP possesses a docking score of −9.045 kcal mol−1 and an RMSD of 0.8322 Å between the docked pose and the crystallographic ligand, confirming the validation of the protocol. The docking study of compounds 20 (docking score of −5.967 kcal mol−1) and 28 (docking score of −6.370 kcal mol−1) revealed that they are deeply inserted into the active site of IMPH, establishing a pattern of interactions similar to those of IMP. The nitro group of 20 interacts with Ser298 and Gly356 through H-bonds, while the hydroxyl group forms H-bonds with Asp244 (Fig. 3a and c). As the hydroxyl group of 28 is at the ortho position to the azo group, it is unable to reach Asp244, while the aromatic ring is involved in a cation–π interaction with Lys291. In addition, the nitro group is H-bonded to Ser298, Gly356, Ser357, and Tyr380 (Fig. 3b and d).
The nitrophenyl rings of 20 and 28 occupy the area typically occupied by the phosphate group of IMP, but the rigidity of the azo linker prevents the benzene ring from arranging like the purine ring of the IMP, projecting the phenol on the opposite side (Fig. 4). Nevertheless, favorable interactions are still present.
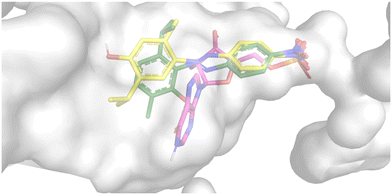 | ||
| Fig. 4 Superimposition of IMP (magenta stick), 20 (yellow stick), and 28 (green stick) in the active site of modelled Hp-IMPDH represented as culled surface. | ||
Given that the majority of IMPDH inhibitors act as non-competitive inhibitors,46 a docking study was conducted to investigate the non-competitive binding of the most active compounds in Hp-IMPDH. The grid was constructed with IMP in the protein and the grid box was centered on the crystallographic ligand of Cj-IMPD. The docking results indicated that 28 can form a π–π interaction between the phenol ring and the purine moiety of IMP and a cation–π interaction between the nitrophenyl ring and Lys71, in accordance with the findings of other non-competitive inhibitors (Fig. 5). In conclusion, the docking study of the most active compounds in the active site of Hp-IMPDH revealed that they can act as competitive and non-competitive inhibitors. The obtained results suggest that Hp-IMPDH is a possible target of the studied compounds in the Hp, but further experimental studies are necessary to confirm our hypothesis.
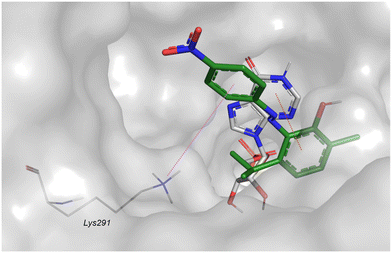 | ||
| Fig. 5 Best docked pose of 28 (green stick) as a non-competitive ligand. IMP (grey stick), dotted violet line for cation–π, and dotted red line for π–π. | ||
2.4. In silico physicochemical and pharmacokinetic properties
The most promising compounds (16, 18, 20, 26, and 28) were selected for a in silico analysis of pharmacokinetic properties. For these purposes, the pkCSM web tool47 was employed and the assessed chemical, ADME, drug-likeness, and toxicological properties are reported in Table 3.| Properties | Compounds | |||||
|---|---|---|---|---|---|---|
| 16 | 18 | 20 | 26 | 28 | ||
| Absorption | Water solubility (log mol/L) | −5.918 | −5.454 | −4.978 | −4.675 | −4.937 |
Caco-2 permeability expressed as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Papp in pkCSM 10−6 cm s−1 Papp in pkCSM 10−6 cm s−1 |
1.485 | 1.396 | 0.364 | 1.363 | 0.4 | |
| Intestinal absorption (human) (% adsorbed) | 87.887 | 91.908 | 88.159 | 91.621 | 88.823 | |
Skin permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KP) KP) |
−2.231 | −2.642 | −2.732 | −2.723 | −2.737 | |
| P-glycoprotein substrate | No | No | Yes | Yes | Yes | |
| P-glycoprotein I inhibitor | No | No | No | No | No | |
| P-glycoprotein II inhibitor | No | No | No | No | No | |
| Distribution | Volume of distribution at steady state (VDss) (human) (log L/kg) | 0.625 | 0.464 | 0.027 | 0.356 | 0.161 |
| Fraction unbound (human) (Fu) | 0 | 0.003 | 0 | 0.022 | 0 | |
Blood–brain barrier (BBB) permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BB) BB) |
0.002 | −0.14 | −0.167 | −0.184 | −0.264 | |
CNS permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PS) PS) |
−1.452 | −1.627 | −1.638 | −1.619 | −1.664 | |
| Metabolism | CYP2D6 substrate | No | No | No | No | No |
| CYP3A4 substrate | Yes | Yes | Yes | Yes | Yes | |
| CYP1A2 inhibitor | Yes | Yes | Yes | Yes | Yes | |
| CYP2C19 inhibitor | Yes | Yes | Yes | Yes | Yes | |
| CYP2C9 inhibitor | Yes | Yes | Yes | Yes | Yes | |
| CYP2D6 inhibitor | No | No | No | No | No | |
| CYP3A4 inhibitor | No | Yes | Yes | No | Yes | |
| Excretion | Total clearance (log mL min−1 Kg) | 0.104 | −0.106 | −0.095 | −0.108 | −0.111 |
| Renal organic cationic transporter (OCT2) substrate | No | No | No | No | No | |
All tested compounds exhibited high intestinal absorption (absorption >85%) as well as high permeability in human epithelial colorectal adenocarcinoma (Caco-2) cells, an index of intestinal permeability and efflux liability. Three of the selected compounds (16, 18, and 26) displayed values of logarithm of the apparent permeability coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Papp) higher than 0.90, which is considered as the threshold value for high permeability, whereas derivatives 20 and 28 displayed log
Papp) higher than 0.90, which is considered as the threshold value for high permeability, whereas derivatives 20 and 28 displayed log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Papp of 0.36 and 0.40, respectively, accounting for moderate absorption.
Papp of 0.36 and 0.40, respectively, accounting for moderate absorption.
Although a good blood–brain barrier (BBB) permeability for the compounds was found to be in the negative range, with logarithmic ratio of brain to plasma drug concentration (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BB) values <0.3, all analyzed derivatives were predicted to penetrate the central nervous system (CNS), resulting to be CNS-positive, with values of in vivo BBB permeability-surface area (log
BB) values <0.3, all analyzed derivatives were predicted to penetrate the central nervous system (CNS), resulting to be CNS-positive, with values of in vivo BBB permeability-surface area (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PS) ≥ −2. This discrepancy could be due to the different parameters considered to compute BBB permeability (measured in vivo as log
PS) ≥ −2. This discrepancy could be due to the different parameters considered to compute BBB permeability (measured in vivo as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BB) and CNS permeability (obtained from in situ brain perfusions with the compound directly injected into the carotid artery). On the other hand, considering the localization of the Hp in the stomach mucosa, the ability to penetrate the CNS is not a central issue for the anti-bacterial activity.
BB) and CNS permeability (obtained from in situ brain perfusions with the compound directly injected into the carotid artery). On the other hand, considering the localization of the Hp in the stomach mucosa, the ability to penetrate the CNS is not a central issue for the anti-bacterial activity.
With regard to the metabolism, these compounds could act as a substrate or inhibit cytochrome P450 (CYP) 3A4. In particular, the tested compounds resulted in being a substrate for the CYP3A4 isoform, whereas compounds 18, 20, and 28 were predicted to inhibit this enzyme. In contra, compounds 16 and 26 did not show any inhibitory activity against CYP3A4. Interestingly, none of these compounds were found to be CYP2D6 isoform substrates or inhibitors. As stated above, the currently employed standard therapy for Hp eradication consists of triple therapy including PPIs, CLR, and AMX;48 thereby, the development of drugs/drug candidates not interfering with the other drugs' metabolism could be useful to assess a potential co-administration in polytherapy.
2.5. Cell toxicity in normal cell lines
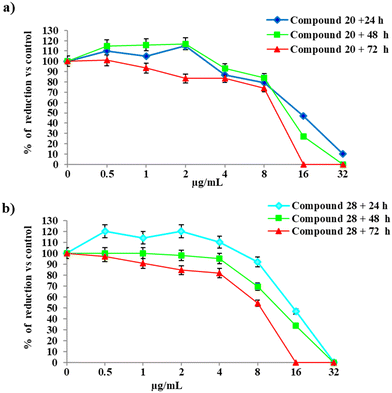
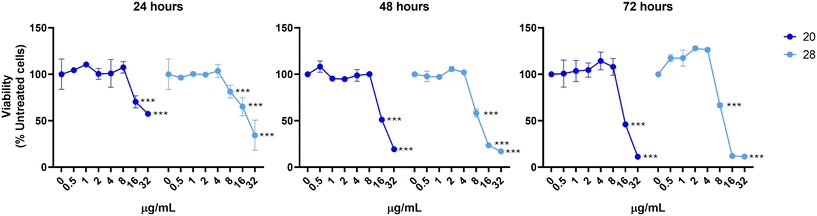
To evaluate the biological effects in vitro of 20 and 28, increasing concentrations of these compounds were administered to human gastric epithelial SV40-immortalized non-tumorigenic, GES-1,49 and normal epithelial cells from the small intestine, HIEC-6, up to 72 h of exposure (Fig. 6 and 7). These cell lines were chosen to overcome the limitations of the use of cancerous AGS, Caco-2, and HT-29 cell lines as in vitro models. In fact, GES-1 cells mimic the site of H. pylori infection and HIEC-6 polarized columnar cells represent a satisfactory alternative displaying human crypt-like features with dense microvilli and non-organized tight junctions.50At 24 h, both compounds are well tolerated in the low–medium concentration range, with the IC50 assessed at 14.99 μg mL−1 and 12.09 μg mL−1 for 20 and 28, respectively, in GES-1, and at 15.06 μg mL−1 and 13.60 μg mL−1, respectively, on HIEC-6 cells. Notably, the IC50 values obtained were significantly higher than the concentrations obtained as MICs for both compounds. At 48 h and 72 h, compound 28 was characterized by a major toxicity on HIEC-6 cells, with the IC50 assessed at 7.61 μg mL−1. On GES-1 cells the toxicity increased only at 72 h with the IC50 of 8.44 μg mL−1. In parallel, the cell viability trend in the presence of 20 is comparable with the one registered at 24 h on both cell lines. In particular, on GES-1 the IC50 at 48 h and 72 h was 12.09 μg mL−1 and 12.02 μg mL−1, respectively; at 48 h and 72 h, HIEC-6 showed an IC50 of 13.38 μg mL−1 and 13.26 μg mL−1, respectively. In conclusion, the discrete selective toxicity of these compounds, calculated as the IC50/MIC50 ratio, corroborates their further refinement for the design of new and selective anti-H. pylori agents.
3. Conclusions
In the frame of increasing antimicrobial resistance phenomena and the correlated decrease in efficacy of the current drugs, the search for new compounds with innovative mechanisms of action is recommended. For a long time, we have put great effort into the development of potent and broad-spectrum antibiotics,51–54 especially towards Hp,55 a pathogen threatening public health due to the associated severe diseases. Herein, we reported a series of 36 compounds resulting from the classical diazotization reaction of such phenols with structurally diverse aromatic amines. Interestingly, for Car and Thy, apart from the expected para-azo derivatives, being the sterically preferred isomers, disubstituted compounds with an ortho- and para-substitution pattern were also obtained, and in just one case, we succeeded to isolate the Carortho-azo derivative (20). Notably, the O-modified compounds 29–36 showed a complete loss of activity with respect to the corresponding free phenols 26 and 28, being among the most active and selective compounds in the series, thereby confirming the importance of the free phenolic group for the antibacterial activity. However, the mono-azo compounds resulted in exerting an anti-Hp activity, with Eu derivatives exerting the weakest activity or being totally inactive, whereas compounds generated from Van, Car, and Thy showed different bioactivity based on the aromatic amine-derived substituents. Likely due to the highest steric hindrance, the bis-azo derivatives resulted in not inhibiting the Hp growth.To search a putative target for this class of compounds, we investigated Hp urease, FabZ, G6PD, and IMPDH, being the most extensively studied Hp targets through reverse docking calculations by using homology models when required. Promising results were obtained with compounds 20 and 28 in the IMPDH active site acting as competitive or non-competitive ligand. Interestingly, the IMPDH enzyme is involved in microbial proliferation; thereby its inhibition could interfere with bacterial growth. In the end, a new series of 36 compounds derived from diazotization of phenolic monoterpenes has been designed and synthesized as selective and quite safe antibacterial agents active against Hp, and a pharmacological target has been suggested by in silico studies. Further investigations are necessary to confirm it and address the synthesis toward more potent antibacterial agents.
4. Materials and methods
4.1. Chemistry
(E)-4-Allyl-2-methoxy-6-(o-tolyldiazenyl)phenol (1). Brown solid, m.p. 82–85 °C, 69% yield. 1H NMR (600 MHz, CDCl3) δ 2.62 (s, 3H, Ar–CH3) 3.43 (d, J = 6.0 Hz, 2H, Ar–CH2), 3.94 (s, 3H, OCH3), 5.11–5.17 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.98–6.05 (m, 1H,
CH2), 5.98–6.05 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.81 (d, J = 6.0 Hz, 1H, Ar), 7.33–7.35 (m, 1H, Ar), 7.35–7.38 (m, 2H, Ar), 7.43 (d, J = 3.0 Hz, 1H, Ar), 7.83 (d, J = 6.0 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 18.1, 39.7, 56.5, 115.3, 115.7, 116.3, 124.1, 127.0, 130.7, 131.3, 131.5, 136.4, 137.3, 137.4, 142.0, 148.5, 148.9. Anal. calcd for C17H18N2O2: C, 72.32; H, 6.43; N, 9.92. Found: C, 72.29; H, 6.41; N, 9.95.
CH), 6.81 (d, J = 6.0 Hz, 1H, Ar), 7.33–7.35 (m, 1H, Ar), 7.35–7.38 (m, 2H, Ar), 7.43 (d, J = 3.0 Hz, 1H, Ar), 7.83 (d, J = 6.0 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 18.1, 39.7, 56.5, 115.3, 115.7, 116.3, 124.1, 127.0, 130.7, 131.3, 131.5, 136.4, 137.3, 137.4, 142.0, 148.5, 148.9. Anal. calcd for C17H18N2O2: C, 72.32; H, 6.43; N, 9.92. Found: C, 72.29; H, 6.41; N, 9.95.
(E)-4-Allyl-2-methoxy-6-((3-methoxyphenyl)diazenyl)phenol (2). Brown solid, m.p. 105–107 °C, 71% yield. 1H NMR (600 MHz, CDCl3) δ 3.42 (d, J = 6.0 Hz, 2H, Ar–CH2), 3.89 (s, 3H, OCH3), 3.93 (s, 3H, OCH3), 5.11–5.16 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.99–6.03 (m, 1H,
CH2), 5.99–6.03 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.81 (d, J = 6.0 Hz, 1H, Ar), 7.04 (dq, 1H, Ar), 7.39–7.41 (m, 1H, Ar), 7.41–7.43 (m, 1H, Ar), 7.44 (d, J = 6.0 Hz, 1H, Ar), 7.45–7.47 (m, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.7, 55.6, 56.6, 104.9, 115.7, 116.3, 116.8, 118.0, 124.0, 130.2, 130.8, 137.0, 137.2, 141.9, 148.8, 151.7, 160.7. Anal. calcd for C17H18N2O3: C, 68.44; H, 6.08; N, 9.39. Found: C, 68.47; H, 6.09; N, 9.41.
CH), 6.81 (d, J = 6.0 Hz, 1H, Ar), 7.04 (dq, 1H, Ar), 7.39–7.41 (m, 1H, Ar), 7.41–7.43 (m, 1H, Ar), 7.44 (d, J = 6.0 Hz, 1H, Ar), 7.45–7.47 (m, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.7, 55.6, 56.6, 104.9, 115.7, 116.3, 116.8, 118.0, 124.0, 130.2, 130.8, 137.0, 137.2, 141.9, 148.8, 151.7, 160.7. Anal. calcd for C17H18N2O3: C, 68.44; H, 6.08; N, 9.39. Found: C, 68.47; H, 6.09; N, 9.41.
(E)-4-Allyl-2-methoxy-6-((3-nitrophenyl)diazenyl)phenol (3). Red solid, m.p. 112–113 °C, 68% yield.1H NMR (600 MHz, CDCl3) δ 3.43 (dtd, J = 6.7, 1.5, 0.7 Hz, 2H, Ar–CH2), 3.94 (s, 3H, OCH3), 5.09–5.22 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.96–6.06 (m, 1H, CH
CH2), 5.96–6.06 (m, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.86 (d, J = 2.0 Hz, 1H, Ar), 7.44 (dd, J = 1.9, 0.9 Hz, 1H, Ar), 7.71 (t, J = 8.0 Hz, 1H, Ar), 8.18 (ddd, J = 7.9, 1.9, 1.0 Hz, 1H, Ar), 8.32 (ddd, J = 8.2, 2.3, 1.0 Hz, 1H, Ar), 8.69 (t, J = 2.1 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.6, 56.6, 116.0, 116.6, 116.8, 124.2, 125.0, 129.0, 130.4, 131.4, 136.9, 137.2, 141.9, 148.9, 149.3, 151.3. Anal. calcd for C16H15N3O4: C, 61.34; H, 4.83; N, 13.41. Found: C, 61.38; H, 4.85; N, 13.44.
), 6.86 (d, J = 2.0 Hz, 1H, Ar), 7.44 (dd, J = 1.9, 0.9 Hz, 1H, Ar), 7.71 (t, J = 8.0 Hz, 1H, Ar), 8.18 (ddd, J = 7.9, 1.9, 1.0 Hz, 1H, Ar), 8.32 (ddd, J = 8.2, 2.3, 1.0 Hz, 1H, Ar), 8.69 (t, J = 2.1 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.6, 56.6, 116.0, 116.6, 116.8, 124.2, 125.0, 129.0, 130.4, 131.4, 136.9, 137.2, 141.9, 148.9, 149.3, 151.3. Anal. calcd for C16H15N3O4: C, 61.34; H, 4.83; N, 13.41. Found: C, 61.38; H, 4.85; N, 13.44.
(E)-4-Allyl-2-methoxy-6-((4-nitrophenyl)diazenyl)phenol (4). Red-brown solid, m.p. 193–194 °C, 53% yield. 1H NMR (600 MHz, CDCl3) δ 3.43 (ddq J = 6.7, 1.5, 0.7 Hz, 2H, Ar–CH2), 3.94 (s, 3H, OCH3), 5.12–5.16 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.96–6.06 (m, 1H,
CH2), 5.96–6.06 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.86 (d, J = 2.0 Hz, 1H, Ar), 7.43–7.47 (m, 1H, Ar), 7.70 (d, J = 8.0 Hz, 1H, Ar), 8.18 (ddd, J = 7.9, 2.0, 1.0 Hz, 1H, Ar), 8.32 (ddd, J = 8.2, 2.3, 1.0 Hz, 1H, Ar), 8.69 (t, J = 2.1 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.6, 56.6, 116.0, 116.6, 124.2, 125.0, 128.9, 130.4, 131.4, 136.9, 137.2, 142.0, 148.9, 149.3, 151.3. Anal. calcd for C16H15N3O4: C, 61.34; H, 4.83; N, 13.41. Found: C, 61.30; H, 4.82; N, 13.43.
CH), 6.86 (d, J = 2.0 Hz, 1H, Ar), 7.43–7.47 (m, 1H, Ar), 7.70 (d, J = 8.0 Hz, 1H, Ar), 8.18 (ddd, J = 7.9, 2.0, 1.0 Hz, 1H, Ar), 8.32 (ddd, J = 8.2, 2.3, 1.0 Hz, 1H, Ar), 8.69 (t, J = 2.1 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.6, 56.6, 116.0, 116.6, 124.2, 125.0, 128.9, 130.4, 131.4, 136.9, 137.2, 142.0, 148.9, 149.3, 151.3. Anal. calcd for C16H15N3O4: C, 61.34; H, 4.83; N, 13.41. Found: C, 61.30; H, 4.82; N, 13.43.
(E)-4-Allyl-2-((2,4-dichlorophenyl)diazenyl)-6-methoxyphenol (5). Red solid, m.p. 137–139 °C, 54% yield. 1H NMR (300 MHz, CDCl3) δ 3.42 (d, J = 6.6 Hz, 2H, Ar–CH2), 3.94 (s, 3H, OCH3), 5.12–5.18 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.95–6.08 (m, 1H, CH
CH2), 5.95–6.08 (m, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.84 (d, J = 1.2 Hz, 1H, Ar), 7.33–7.40 (m, 2H, Ar), 7.58 (d, J = 1.8 Hz, 1H, Ar), 7.87 (d, J = 9.0 Hz, 1H, Ar), 13.18 (s, 1H, OH). 13C NMR (151 MHz, CDCl3) δ 39.5, 56.5, 116.2, 116.3, 118.0, 124.3, 128.1, 130.3, 130.9, 134.6, 136.9, 137.3, 137.6, 141.9, 149.0. Anal. calcd for C16H14Cl2N2O2: C, 56.99; H, 4.18; N, 8.31. Found: C, 56.94; H, 4.15; N, 8.28.
), 6.84 (d, J = 1.2 Hz, 1H, Ar), 7.33–7.40 (m, 2H, Ar), 7.58 (d, J = 1.8 Hz, 1H, Ar), 7.87 (d, J = 9.0 Hz, 1H, Ar), 13.18 (s, 1H, OH). 13C NMR (151 MHz, CDCl3) δ 39.5, 56.5, 116.2, 116.3, 118.0, 124.3, 128.1, 130.3, 130.9, 134.6, 136.9, 137.3, 137.6, 141.9, 149.0. Anal. calcd for C16H14Cl2N2O2: C, 56.99; H, 4.18; N, 8.31. Found: C, 56.94; H, 4.15; N, 8.28.
(E)-4-Allyl-2-(benzo[d][1,3]dioxol-5-yldiazenyl)-6-methoxyphenol (6). Brown solid, m.p. 142–144 °C, 73% yield. 1H NMR (600 MHz, CDCl3) δ 3.42 (d, J = 6.0 Hz, 2H, Ar–CH2), 3.93 (s, 3H, OCH3), 5.10–5.17 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.97–6.06 (m, 1H, CH
CH2), 5.97–6.06 (m, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.07 (s, 2H, OCH2O), 6.79 (s, 1H, Ar), 6.93 (d, J = 9.0 Hz, 1H, Ar), 7.35 (s, 1H, Ar), 7.42 (s, 1H, Ar), 7.43 (s, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.7, 56.5, 98.6, 102.2, 108.3, 115.1, 116.2, 122.6, 123.6, 130.7, 136.9, 137.3, 141.2, 146.4, 148.7, 149.2, 150.6. Anal. calcd for C17H16N2O4: C, 65.38; H, 5.15; N, 8.97. Found: C, 65.35; H, 5.17; N, 9.00.
), 6.07 (s, 2H, OCH2O), 6.79 (s, 1H, Ar), 6.93 (d, J = 9.0 Hz, 1H, Ar), 7.35 (s, 1H, Ar), 7.42 (s, 1H, Ar), 7.43 (s, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.7, 56.5, 98.6, 102.2, 108.3, 115.1, 116.2, 122.6, 123.6, 130.7, 136.9, 137.3, 141.2, 146.4, 148.7, 149.2, 150.6. Anal. calcd for C17H16N2O4: C, 65.38; H, 5.15; N, 8.97. Found: C, 65.35; H, 5.17; N, 9.00.
(E)-4-Allyl-2-methoxy-6-(naphthalen-1-yldiazenyl)phenol (7). Brown solid, m.p. 113–115 °C, 68% yield. 1H NMR (600 MHz, CDCl3) δ 3.45 (dtd, J = 6.7, 1.4, 0.7 Hz, 2H, Ar–CH2), 3.97 (s, 3H, OCH3), 5.10–5.21 (m, 2H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 6.03 (ddt, J = 16.8, 10.0, 6.7 Hz, 1H, CH
CH2), 6.03 (ddt, J = 16.8, 10.0, 6.7 Hz, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.84 (d, J = 2.0 Hz, 1H, Ar), 7.49 (dd, J = 1.8, 0.9 Hz, 1H, Ar), 7.54–7.64 (m, 2H, Ar), 7.63 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H, Ar), 7.94 (ddt, J = 8.1, 1.3, 0.6 Hz, 1H, Ar), 7.98 (ddd, J = 10.6, 7.9, 1.1 Hz, 2H, Ar), 8.50 (dq, J = 8.6, 0.9 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.8, 56.6, 113.3, 115.6, 116.4, 122.2, 124.0, 125.9, 126.8, 127.7, 128.5, 129.6, 131.0, 131.7, 134.4, 137.2, 137.9, 142.1, 145.9, 148.9. Anal. calcd for C20H18N2O2: C, 75.45; H, 5.70; N, 8.90. Found: C, 75.49; H, 5.73; N, 8.93.
), 6.84 (d, J = 2.0 Hz, 1H, Ar), 7.49 (dd, J = 1.8, 0.9 Hz, 1H, Ar), 7.54–7.64 (m, 2H, Ar), 7.63 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H, Ar), 7.94 (ddt, J = 8.1, 1.3, 0.6 Hz, 1H, Ar), 7.98 (ddd, J = 10.6, 7.9, 1.1 Hz, 2H, Ar), 8.50 (dq, J = 8.6, 0.9 Hz, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 39.8, 56.6, 113.3, 115.6, 116.4, 122.2, 124.0, 125.9, 126.8, 127.7, 128.5, 129.6, 131.0, 131.7, 134.4, 137.2, 137.9, 142.1, 145.9, 148.9. Anal. calcd for C20H18N2O2: C, 75.45; H, 5.70; N, 8.90. Found: C, 75.49; H, 5.73; N, 8.93.
(E)-3-((2-Chlorophenyl)diazenyl)-4-hydroxy-5-methoxybenzaldehyde (8). Brown solid, m.p. 200–202 °C, 56% yield. Characterization data are in agreement with previous literature.56 Anal. calcd for C14H11ClN2O3: C, 57.84; H, 3.81; N, 9.64. Found: C, 57.88; H, 3.83; N, 9.66.
(E)-3-((3-Chlorophenyl)diazenyl)-4-hydroxy-5-methoxybenzaldehyde (9). Red-brown solid, m.p. 180–182 °C, 72% yield. Characterization data are in agreement with previous literature.56 Anal. calcd for C14H11ClN2O3: C, 57.84; H, 3.81; N, 9.64. Found: C, 57.81; H, 3.79; N, 9.67.
(E)-4-Hydroxy-3-methoxy-5-((3-nitrophenyl)diazenyl)benzaldehyde (10). Red-brown solid, m.p. 112–113 °C, 76% yield. Characterization data are in agreement with previous literature.57 Anal. calcd for C14H11N3O5: C, 55.82; H, 3.68; N, 13.95. Found: C, 55.85; H, 3.70; N, 13.98.
(E)-4-Hydroxy-3-methoxy-5-(p-tolyldiazenyl)benzaldehyde (11). Red solid, m.p. 82–84 °C, 90% yield. Characterization data are in agreement with previous literature.57 Anal. calcd for C15H14N2O3: C, 66.66; H, 5.22; N, 10.36. Found: C, 66.70; H, 5.24; N, 10.39.
(E)-4-Hydroxy-3-methoxy-5-((4-methoxyphenyl)diazenyl)benzaldehyde (12). Orange solid, m.p. 82–83 °C, 66% yield. Characterization data are in agreement with previous literature.58 Anal. calcd for C15H14N2O4: C, 62.93; H, 4.93; N, 9.79. Found: C, 62.96; H, 4.95; N, 9.77.
(E)-3-((4-Bromophenyl)diazenyl)-4-hydroxy-5-methoxybenzaldehyde (13). Red solid, m.p. 176–178 °C, 76% yield. 1H NMR (600 MHz, CDCl3) δ 4.03 (s, 3H, OCH3), 7.53 (d, J = 2.4 Hz, 1H, Ar), 7.72 (dt, J = 13.5, 3.3 Hz, 2H, Ar), 7.79 (dt, J = 9.9, 3.6 Hz, 2H, Ar), 8.10 (d, J = 3.0 Hz, 1H, Ar), 9.97 (s, 1H, CHO). 13C NMR (151 MHz, CDCl3) δ 56.6, 111.0, 123.8, 126.5, 128.7, 130.3, 132.9, 149.7. Anal. calcd for C14H11BrN2O3: C, 50.17; H, 3.31; N, 8.36. Found: C, 50.20; H, 3.33; N, 8.39.
(E)-4-((5-Formyl-2-hydroxy-3-methoxyphenyl)diazenyl)benzonitrile (14). Brown solid, m.p. 203–204 °C, 90% yield. 1H NMR (300 MHz, CDCl3) δ 4.01 (s, 3H, OCH3), 7.53 (s, 1H, Ar), 7.86 (d, J = 8.7 Hz, 2H, Ar), 7.99 (d, J = 8.1 Hz, 2H, Ar), 8.01 (d, J = 1.8 Hz, 1H, Ar), 9.96 (s, 1H, CHO), 13.69 (s, 1H, OH). 13C NMR (75 MHz, CDCl3) δ 56.6, 111.7, 114.8, 118.0, 122.8, 129.0, 130.5, 133.6, 136.4, 150.2, 151.7, 189.8. Anal. calcd for C15H11N3O3: C, 64.05; H, 3.94; N, 14.94. Found: C, 64.09; H, 3.96; N, 14.91.
(E)-3-((3,4-Dichlorophenyl)diazenyl)-4-hydroxy-5-methoxybenzaldehyde (15). Red-brown solid, m.p. 188–190 °C, 81% yield. 1H NMR (300 MHz, CDCl3) δ 4.01 (s, 3H, OCH3), 7.52 (s, 1H, Ar), 7.63 (d, J = 8.7 Hz, 1H, Ar), 7.76 (dd, J = 8.7, 2.0 Hz, 1H, Ar), 8.08 (dd, J = 22.8, 1.8 Hz, 2H, Ar), 9.95 (s, 1H, CHO), 13.48 (s, 1H, OH). 13C NMR (75 MHz, CDCl3) δ 56.6, 111.4, 122.3, 123.2, 128.8, 130.2, 131.3, 134.2, 136.0, 136.1, 148.8, 149.3, 150.0, 189.9. Anal. calcd for C14H10Cl2N3O3: C, 51.72; H, 3.10; N, 8.62. Found: C, 51.75; H, 3.12; N, 8.65.
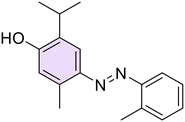
(E)-4-((2-Chlorophenyl)diazenyl)-5-isopropyl-2-methylphenol (16). Orange solid, m.p. 162–163 °C, 70% yield. 1H NMR (400 MHz, DMSO) δ 1.27 (d, J = 6.8 Hz, 6H, 2 iPr CH3), 2.15 (s, 3H, Ar–CH3), 4.02–4.08 (m, 1H, iPr CH), 6.90 (s, 1H, Ar), 7.44–7.49 (m, 2H, Ar), 7.51 (s, 1H, Ar), 7.58–7.60 (m, 1H, Ar), 7.65–7.67 (m, 1H, Ar), 10.28 (s, 1H, OH). 13C NMR (100 MHz, CDCl3) δ 17.2, 22.4, 27.2, 115.2, 117.2, 117.9, 127.2, 130.5, 130.6, 133.0, 134.6, 138.7, 145.5, 149.3, 156.3. Anal. calcd for C16H17ClN2O: C, 66.55; H, 5.93; N, 9.70. Found: C, 66.59; H, 5.91; N, 9.67.
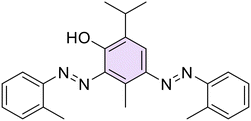
2,4-Bis((E)-(2-chlorophenyl)diazenyl)-3-isopropyl-6-methylphenol (17). Brown solid, m.p. 109–111 °C, 15% yield. 1H NMR (500 MHz, CDCl3) δ 1.66 (d, J = 8.5 Hz, 6H, 2 iPr CH3), 2.37 (d, J = 0.5 Hz, 3H, Ar–CH3), 4.88–4.94 (m, 1H, iPr CH), 7.38–7.41 (m, 2H, Ar), 7.45–7.49 (m, 2H, Ar), 7.59–7.65 (m, 2H, Ar), 7.71–7.74 (m, 1H, Ar), 7.95 (s, 1H, Ar), 7.98–8.01 (m, 1H, Ar), 15.31 (s, 1H, OH). 13C NMR (125 MHz, CDCl3) δ 15.6, 24.4, 26.9, 117.7, 118.0, 127.3, 127.9, 130.8, 130.8, 131.0, 131.6, 133.4, 135.1, 146.0, 149.0, 150.0. Anal. calcd for C22H20Cl2N4O: C, 61.83; H, 4.72; N, 13.11. Found: C, 61.86; H, 4.75; N, 13.13.
(E)-5-Isopropyl-4-((4-methoxyphenyl)diazenyl)-2-methylphenol (18). Orange solid, m.p. 110–112 °C, 60% yield. 1H NMR (400 MHz, DMSO) δ 1.26 (d, J = 6.8 Hz, 6H, 2 iPr CH3), 2.14 (s, 3H, Ar–CH3), 3.85 (s, 3H, OCH3), 3.98–4.03 (m, 1H, iPr CH), 6.86 (s, 1H, Ar), 7.10 (d, J = 9.2 Hz, 2H, Ar), 7.46 (s, 1H, Ar), 7.80 (d, J = 8.8 Hz, 2H, Ar). 13C NMR (100 MHz, DMSO) δ 16.1, 24.3, 27.3, 56.0, 112.0, 115.0, 117.6, 122.9, 124.3, 142.0, 147.4, 147.9, 159.4, 161.4. Anal. calcd for C17H20N2O2: C, 71.81; H, 7.09; N, 9.85. Found: C, 71.78; H, 7.06; N, 9.81.
3-Isopropyl-2,4-bis((E)-(4-methoxyphenyl)diazenyl)-6-methylphenol (19). Orange solid, m.p. 206–208 °C, 19% yield. 1H NMR (600 MHz, CDCl3) δ 1.66 (d, J = 10.8 Hz, 6H, 2× iPr CH3), 2.34 (d, J = 1.2 Hz, 3H, Ar–CH3), 3.89 (s, 3H, OCH3), 3.90 (s, 3H, OCH3), 4.83–4.94 (m, 1H, iPr CH), 7.03–7.07 (m, 4H, Ar), 7.77 (s, 1H, Ar), 7.86–7.90 (m, 2H, Ar), 7.93–7.96 (m, 2H, Ar). 13C NMR (151 MHz, CDCl3) δ 15.4, 24.2, 26.8, 55.6, 55.6, 114.2, 114.7, 122.3, 124.0, 124.6, 125.6, 134.2, 143.2, can144.4, 147.5, 147.6, 155.3, 161.4, 162.0. Anal. calcd for C24H26N4O3: C, 68.88; H, 6.26; N, 13.39. Found: C, 68.90; H, 6.28; N, 13.42.
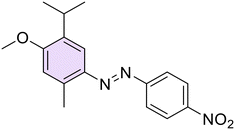
(E)-3-Isopropyl-6-methyl-2-((4-nitrophenyl)diazenyl)phenol (20). Orange solid, m.p. 103–105 °C, 64% yield. 1H NMR (300 MHz, CDCl3) δ 1.36 (d, J = 6.6 Hz, 6H, 2 iPr CH3), 2.22 (s, 3H, Ar–CH3), 3.96–4.06 (m, 1H, iPr CH), 6.88 (d, J = 7.2 Hz, 1H, Ar), 7.27 (d, J = 7.2 Hz, 1H, Ar), 7.94 (d, J = 8.1 Hz, 2H, Ar), 8.39 (d, J = 8.1 Hz, Ar). 13C NMR (151 MHz, CDCl3) δ 15.0, 24.0, 28.0, 116.5, 122.4, 125.1, 137.2, 150.4, 154.2. Anal. calcd for C16H17N3O3: C, 64.20; H, 5.72; N, 14.04. Found: C, 64.24; H, 5.73; N, 14.05.
2,4-Bis((E)-(4-bromophenyl)diazenyl)-3-isopropyl-6-methylphenol (21). Brown solid, m.p. 218–220 °C, 18% yield. 1H NMR (500 MHz, CDCl3) δ 1.62 (d, J = 9.0 Hz, 6H, 2 iPr CH3), 2.33 (s, 3H, Ar–CH3), 4.82–4.86 (m, 1H, iPr CH), 7.67–7.73 (m, 4H, Ar), 7.78–7.85 (m, 5H, Ar), 14.99 (s, 1H, OH). 13C NMR (125 MHz, CDCl3) δ 15.3, 24.3, 26.8, 123.5, 124.3, 124.6, 125.3, 126.3, 132.9, 148.8, 149.4, 151.6. Anal. calcd for C22H20Br2N4O: C, 51.19; H, 3.91; N, 10.85. Found: C, 51.15; H, 3.90; N, 10.87.
(E)-2-Isopropyl-5-methyl-4-(o-tolyldiazenyl)phenol (22). Orange solid, m.p. 110–111 °C, 60% yield. Characterization data are in agreement with previous literature.59 Anal. calcd for C17H20N2O: C, 76.09; H, 7.51; N, 10.44. Found: C, 76.13; H, 7.53; N, 10.42.
6-Isopropyl-3-methyl-2,4-bis((E)-o-tolyldiazenyl)phenol (23). Brown solid, m.p. 143–145 °C, 20% yield. 1H NMR (500 MHz, CDCl3) δ 1.23 (d, J = 8.5 Hz, 6H, 2 iPr CH3), 2.59 (s, 3H, Ar–CH3), 2.67 (s, 3H, Ar–CH3), 3.06 (s, 3H, Ar–CH3), 3.33–3.36 (m, 1H, iPr CH), 7.17–7.29 (m, 6H, Ar), 7.59 (d, J = 9.5 Hz, 1H, Ar), 7.81–7.84 (m, 2H, Ar), 15.30 (s, 1H, OH). 13C NMR (125 MHz, CDCl3) δ 11.6, 17.7, 18.3, 22.4, 26.5, 115.9, 116.3, 119.2, 126.4, 127.1, 130.1, 130.7, 131.2, 131.5, 135.4, 136.2, 137.5, 140.8, 143.8, 151.1, 156.5. Anal. calcd for C24H26N4O: C, 74.58; H, 6.78; N, 14.50. Found: C, 74.61; H, 6.80; N, 14.52.
(E)-4-((2-Chlorophenyl)diazenyl)-2-isopropyl-5-methylphenol (24). Orange solid, m.p. 182–183 °C, 70% yield. 1H NMR (600 MHz, CDCl3) δ 1.32 (d, J = 10.8 Hz, 6H, 2 iPr CH3), 2.69 (s, 3H, Ar–CH3), 3.17–3.24 (m, 1H, iPr CH), 5.25 (s, 1H, OH), 7.33–7.36 (m, 2H, Ar), 7.55–7.57 (m, 1H, Ar), 7.65–7.66 (m, 1H, Ar), 7.77 (s, 1H, Ar). 13C NMR (151 MHz, CDCl3) δ 17.2, 22.4, 27.2, 115.2, 117.2, 117.9, 127.2, 130.5, 130.6, 133.0, 134.6, 138.7, 145.5, 149.3, 156.3. Anal. calcd for C16H17ClN2O: C, 66.55; H, 5.93; N, 9.70. Found: C, 66.51; H, 5.91; N, 9.73.
2,4-Bis((E)-(2-chlorophenyl)diazenyl)-6-isopropyl-3-methylphenol (25). Brown solid, m.p. 188–190 °C, 16% yield. 1H NMR (500 MHz, CDCl3) δ 1.35 (d, J = 8.5 Hz, 6H, 2 iPr CH3), 3.14 (s, 3H, Ar–CH3), 3.45–3.47 (m, 1H, iPr CH), 7.36–7.38 (m, 2H, Ar), 7.41–7.44 (m, 2H, Ar), 7.56–7.59 (m, 2H, Ar), 7.73–7.75 (m, 1H, Ar), 8.00–8.03 (m, 2H, Ar), 15.19 (s, 1H, OH). 13C NMR (125 MHz, CDCl3) δ 11.6, 22.2, 26.5, 117.7, 117.9, 120.4, 127.2, 127.7, 130.5, 130.6, 130.9, 131.4, 133.0, 134.9, 135.4, 136.8, 141.8, 143.7, 145.8, 149.0, 157.2. Anal. calcd for C22H20Cl2N4O: C, 61.83; H, 4.72; N, 13.11. Found: C, 61.80; H, 4.70; N, 13.14.
(E)-2-Isopropyl-4-((4-methoxyphenyl)diazenyl)-5-methylphenol (26). Yellow solid, m.p. 88–90 °C, 58% yield. Characterization data are in agreement with previous literature.57 Anal. calcd for C17H20N2O2: C, 71.81; H, 7.09; N, 9.85. Found: C, 71.78; H, 7.07; N, 9.82.
6-Isopropyl-2,4-bis((E)-(4-methoxyphenyl)diazenyl)-3-methylphenol (27). Brown solid, m.p. 186–187 °C, 30% yield. 1H NMR (600 MHz, CDCl3) δ 1.35 (d, J = 10.8 Hz, 6H, 2 iPr CH3), 3.10 (s, 3H, Ar–CH3), 3.41–3.45 (m, 1H, iPr CH), 3.92 (s, 3H, OCH3), 3.92 (s, 3H, OCH3), 7.03–7.06 (m, 4H, Ar), 7.85 (s, 1H, Ar), 7.91 (dd, J = 10.2, 1.8 Hz, 2H, Ar), 7.97 (dd, J = 10.2, 1.5 Hz, 2H, Ar), 14.92 (s, 1H, OH). 13C NMR (151 MHz, CDCl3) δ 11.6, 22.4, 26.7, 55.6, 55.7, 114.1, 114.7, 117.8, 123.9, 124.5, 134.7, 135.5, 139.7, 143.4, 144.5, 147.5, 154.3, 161.4, 162.0. Anal. calcd for C24H26N4O3: C, 68.88; H, 6.26; N, 13.39. Found: C, 68.91; H, 6.28; N, 13.41.
(E)-2-Isopropyl-5-methyl-4-((4-nitrophenyl)diazenyl)phenol (28). Orange solid, m.p.: 176–177 °C, 74% yield. Characterization data are in agreement with previous literature.59 Anal. calcd for C16H17N3O3: C, 64.20; H, 5.72; N, 14.04. Found: C, 64.24; H, 5.70; N, 14.02.
(E)-1-(5-Isopropyl-4-methoxy-2-methylphenyl)-2-(4-methoxyphenyl)diazene (29). Orange solid, m.p.: 64–66 °C, 76% yield. 1H NMR (300 MHz, CDCl3) δ 1.22 (d, J = 6.6 Hz, 6H, 2 iPr CH3), 2.71 (s, 3H, Ar–CH3), 3.26–3.31 (m, 1H, iPr CH), 3.89 (s, 3H, OCH3), 3.90 (s, 3H, OCH3), 6.76 (s, 1H, Ar), 7.00 (dt, J = 9.5, 2.6 Hz, 2H, Ar), 7.65 (s, 1H, Ar), 7.90 (dt, J = 9.6, 2.7 Hz, 2H, Ar). 13C NMR (75 MHz, CDCl3) δ 17.4, 22.6, 26.9, 55.5, 110.0, 112.06, 113.4, 114.1, 124.3, 135.4, 137.5, 144.5, 147.7, 159.1, 161.2. Anal. calcd for C18H22N2O2: C, 72.46; H, 7.43; N, 9.39. Found: C, 72.49; H, 7.45; N, 9.41.
(E)-1-(5-Isopropyl-4-methoxy-2-methylphenyl)-2-(4-nitrophenyl)diazene (33). Red solid, m.p.: 122–123 °C, 79% yield. 1H NMR (300 MHz, CDCl3) δ 1.25 (d, J = 6.6 Hz, 6H, 2 iPr CH3), 2.75 (s, 3H, Ar–CH3), 3.26–3.31 (m, 1H, iPr CH), 3.93 (s, 3H, OCH3), 6.80 (s, 1H, Ar), 7.72 (s, 1H, Ar), 7.98 (dt, J = 9.3, 2.4 Hz, 2H, Ar), 8.36 (dt, J = 9.3, 2.4 Hz, 2H, Ar). 13C NMR (75 MHz, CDCl3) δ 17.5, 22.5, 26.8, 55.6, 112.1, 113.6, 123.1, 124.7, 135.9, 140.4, 144.6, 147.9, 156.6, 161.0. Anal. calcd for C18H22N2O2: C, 72.46; H, 7.43; N, 9.39. Found: C, 72.49; H, 7.41; N, 9.37.
(E)-2-Isopropyl-4-((4-methoxyphenyl)diazenyl)-5-methylphenyl acetate (30). Red solid, m.p.: 102–104 °C, 64% yield. Characterization data were in agreement with previous literature.26 Anal. calcd for C19H22N2O3: C, 69.92; H, 6.79; N, 8.58. Found: C, 69.95; H, 6.77; N, 8.60.
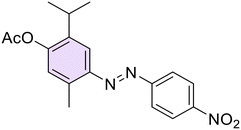
(E)-2-Isopropyl-5-methyl-4-((4-nitrophenyl)diazenyl)phenyl acetate (34). Red solid, m.p.: 137–138 °C, 78% yield. Characterization data were in agreement with previous literature.26 Anal. calcd for C19H22N2O3: C, 69.92; H, 6.79; N, 8.58. Found: C, 69.96; H, 6.81; N, 8.55.
(E)-2-Isopropyl-4-((4-methoxyphenyl)diazenyl)-5-methylphenyl dimethylcarbamate (31). Orange solid, m.p.: 90–92 °C. 64% yield. 1H NMR (300 MHz, CDCl3) δ 1.26 (d, J = 6.6 Hz, 6H, 2 iPr CH3), 2.65 (s, 3H, Ar–CH3), 3.05 (s, 3H, NCH3), 3.07–3.11 (m, 1H, iPr CH), 3.15 (s, 3H, NCH3), 3.89 (s, 3H, OCH3), 7.03 (m, 3H, Ar), 7.60 (s, 1H, Ar), 7.91 (d, J = 9.0 Hz, 2H, Ar). 13C NMR (75 MHz, CDCl3) δ 17.1, 22.9, 27.7, 36.5, 36.8, 55.6, 110.0, 113.5, 114.1, 124.7, 124.8, 136.4, 138.5, 147.5, 148.2, 150.5, 154.7, 161.7. Anal. calcd for C19H22N2O3: C, 67.58; H, 7.09; N, 11.82. Found: C, 67.62; H, 7.07; N, 11.80.
(E)-O-(2-Isopropyl-4-((4-methoxyphenyl)diazenyl)-5-methylphenyl)dimethylcarbamothioate (32). Orange solid, m.p.: 123–124 °C, 60% yield. 1H NMR (300 MHz, CDCl3) δ 1.26 (d, J = 7.2 Hz, 6H, 2 iPr CH3), 2.62 (s, 3H, Ar–CH3), 2.95–3.04 (m, 1H, iPr CH), 3.37 (s, 3H, NCH3), 3.48 (s, 3H, NCH3), 3.89 (s, 3H, OCH3), 6.96 (s, 1H, Ar), 6.99 (d, J = 9.0 Hz, 2H, Ar), 7.64 (s, 1H, Ar), 7.91 (d, J = 9.0 Hz, 2H, Ar). 13C NMR (75 MHz, CDCl3) δ 17.2, 23.0, 27.5, 38.7, 43.3, 55.6, 113.7, 114.2, 124.7, 125.5, 136.3, 139.1, 147.4, 148.6, 152.9, 161.8, 187.5. Anal. calcd for C20H25N3O2S: C, 64.66; H, 6.78; N, 11.31. Found: C, 64.70; H, 6.80; N, 11.28.
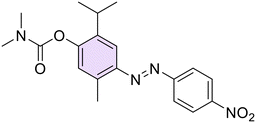
(E)-2-Isopropyl-5-methyl-4-((4-nitrophenyl)diazenyl)phenyl dimethylcarbamate (35). Red solid, m.p.: 129–130 °C, 92% yield. 1H NMR (300 MHz, CDCl3) δ 1.27 (d, J = 6.9 Hz, 6H, 2 iPr CH3), 2.71 (s, 3H, Ar–CH3), 3.06 (s, 3H, NCH3), 3.09–3.13 (m, 1H, iPr CH), 3.16 (s, 3H, NCH3), 7.12 (s, 1H, Ar), 7.68 (s, 1H, Ar), 8.01 (dt, J = 11.6, 2.3 Hz, 2H, Ar), 8.40 (dt, J = 9.5, 2.3 Hz, 2H, Ar). 13C NMR (75 MHz, CDCl3) δ 17.1, 22.8, 27.57, 36.5, 36.8, 113.6, 123.4, 124.7, 125.2, 138.7, 139.0, 148.0, 148.4, 152.4, 154.4, 156.2. Anal. calcd for C19H22N2O3: C, 67.58; H, 7.09; N, 11.82. Found: C, 67.62; H, 7.11; N, 11.85.
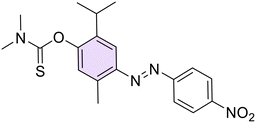
(E)-O-(2-Isopropyl-5-methyl-4-((4-nitrophenyl)diazenyl)phenyl) dimethylcarbamothioate (36). Red solid, m.p.: 166–170 °C, 86% yield. 1H NMR (300 MHz, CDCl3) δ 1.27 (d, J = 6.9 Hz, 6H, 2 iPr CH3), 2.72 (s, 3H, Ar–CH3), 2.97–3.06 (m, 1H, iPr CH), 3.93 (s, 3H, NCH3), 3.49 (s, 3H, NCH3), 7.02 (s, 1H, Ar), 7.71 (s, 1H, Ar), 8.04 (d, J = 9.6 Hz, 2H, Ar), 8.37 (d, J = 9.6 Hz, 2H, Ar). 13C NMR (75 MHz, CDCl3) δ 17.2, 23.0, 23.6, 27.4, 37.1, 38.7, 43.3, 112.7, 113.8, 123.4, 123.6, 124.7, 126.1, 138.4, 139.6, 148.4, 148.4, 150.9, 154.6, 156.2, 187.1. Anal. calcd for C20H25N3O2S: C, 64.66; H, 6.78; N, 11.31. Found: C, 64.62; H, 6.76; N, 11.29.
4.2. Antibacterial activity evaluation
H. pylori strains were NCTC 11637 and clinical isolates F1, 23, and F4. The MIC determination was performed using a modified broth microdilution assay as previously described.60 For MBC, 10 μL of suspensions without visible growth were spotted on the surface of Skirrow agar plates and incubated for 72 h at 37 °C under microaerophilic conditions and 100% humidity. The MBC was defined as the concentration that killed 99.9% of the initial inoculum. The MIC and MBC assessment was performed in triplicate, at least on three different days.Escherichia coli ATCC 25922 was cultured on MacConkey agar (Oxoid) for 18 h at 37 °C, Pseudomonas aeruginosa ATCC 27853 on agar cetrimide for 18 h at 37 °C, and Staphylococcus aureus on mannitol salt agar (MSA) (Oxoid) for 24 h at 37 °C. MIC determination was performed on 96-well microtiter plates using a microdilution method in cation-adjusted Mueller Hinton broth (CAMHB).61 All the plates were incubated at 37 °C for 24 h. After incubation, the lowest concentration of the compounds inhibiting visible growth was considered to be the MIC. For MBC, aliquots (10 μL) of suspensions without visible growth were plated on Mueller Hinton agar and incubated at 37 °C for 24 h. The MBC was defined as the lowest concentration of compounds that killed ≥99.9% of the initial inoculum.
4.3. In silico studies
4.4. Cell-based assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with fresh growth medium for 4 h at 37 °C and 5% CO2. Then, the MTT solution was removed to be replaced with 100 μL per well DMSO. Cells were further incubated for 20 min in the same conditions and gently swirled for 10 min at r.t. Absorbances were registered by means of a multiscan GO microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
10 with fresh growth medium for 4 h at 37 °C and 5% CO2. Then, the MTT solution was removed to be replaced with 100 μL per well DMSO. Cells were further incubated for 20 min in the same conditions and gently swirled for 10 min at r.t. Absorbances were registered by means of a multiscan GO microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Data availability
This published article and its ESI† include all data generated or analyzed during this study.Author contributions
Conceptualization, review and editing: S. C., I. D. A., A. A., F. S., and A. A.-S. Methodology and formal analysis: F. M., I. D. A., M. F., M. A., N. M., M. G., M. S., P. G., S. H. A, F. S., and S.C. Writing-original draft preparation: F. M., S. C., I. D. A., and P. G. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by FAR funding (Italian Ministry for Instruction, University, and Research) assigned to S. C. This article is based upon work from COST Action EURESTOP, CA21145, supported by COST (European Cooperation in Science and Technology) to S. C. and I. D. A. This publication was produced while attending the PhD programme in Biomolecular and Pharmaceutical Sciences at the University of Chieti-Pescara, Cycle XXXIX, with the support of a scholarship co-financed by the Ministerial Decree no. 117 of 02.03.2023, based on the NRRP - funded by the European Union - NextGenerationEU - Mission 4 “Education and Research”, Component 2 “From Research to Business”, Investment 3.3, and by the company IDI s.r.l. (N. M.).References
- A. Ali and K. I. AlHussaini, Helicobacter pylori: A Contemporary Perspective on Pathogenesis, Diagnosis and Treatment Strategies, Microorganisms, 2024, 12(1), 222 CAS.
- C. Lu, Y. Yu, L. Li, C. Yu and P. Xu, Systematic review of the relationship of Helicobacter pylori infection with geographical latitude, average annual temperature and average daily sunshine, BMC Gastroenterol., 2018, 18(1), 50 CrossRef PubMed.
- Y. C. Chen, P. Malfertheiner, H. T. Yu, C. L. Kuo, Y. Y. Chang and F. T. Meng, et al., Global Prevalence of Helicobacter pylori Infection and Incidence of Gastric Cancer Between 1980 and 2022, Gastroenterology, 2024, 166(4), 605–619 CrossRef.
- P. Malfertheiner, M. C. Camargo, E. El-Omar, J. M. Liou, R. Peek and C. Schulz, et al., Helicobacter pylori infection, Nat. Rev. Dis. Primers, 2023, 9(1), 1–24 CrossRef.
- Schistosomes, liver flukes and Helicobacter pylori, IARC Monogr Eval Carcinog Risks Hum., 1994, vol. 61, pp. 1–241 Search PubMed.
- T. Furuta, M. Yamade, T. Kagami, T. Uotani, T. Suzuki and T. Higuchi, et al., Dual Therapy with Vonoprazan and Amoxicillin Is as Effective as Triple Therapy with Vonoprazan, Amoxicillin and Clarithromycin for Eradication of Helicobacter pylori, Digestion, 2020, 101(6), 743–751 CAS.
- L. Boyanova, P. Hadzhiyski, R. Gergova and R. Markovska, Evolution of Helicobacter pylori Resistance to Antibiotics: A Topic of Increasing Concern, Antibiotics, 2023, 12(2), 332 CrossRef CAS.
- J. J. Lima, C. D. Thomas, J. Barbarino, Z. Desta, S. L. Van Driest and N. El Rouby, et al., Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing, Clin. Pharmacol. Ther., 2021, 109(6), 1417–1423 CrossRef PubMed.
- G. Benito, I. D'Agostino, S. Carradori, M. Fantacuzzi, M. Agamennone and V. Puca, et al., Erlotinib-containing benzenesulfonamides as anti-Helicobacter pylori agents through carbonic anhydrase inhibition, Future Med. Chem., 2023, 15(20), 1865–1883 CrossRef CAS PubMed.
- I. D'Agostino, C. Ardino, G. Poli, F. Sannio, M. Lucidi and F. Poggialini, et al., Antibacterial alkylguanidino ureas: Molecular simplification approach, searching for membrane-based MoA, Eur. J. Med. Chem., 2022, 231, 114158 CrossRef.
- C. Ardino, F. Sannio, G. Poli, S. Galati, E. Dreassi and L. Botta, et al., An update on antibacterial AlkylGuanidino ureas: Design of new derivatives, synergism with colistin and data analysis of the whole library, Eur. J. Med. Chem., 2024, 116362 CrossRef CAS.
- FDA, FDA Approves New Antibiotic for Three Different Uses [Internet], available from: https://www.fda.gov/news-events/press-announcements/fda-approves-new-antibiotic-three-different-uses.
- D. M. Klug, F. I. M. Idiris, M. A. T. Blaskovich, F. von Delft, C. G. Dowson and C. Kirchhelle, et al., There is no market for new antibiotics: this allows an open approach to research and development, Wellcome Open Res., 2021, 6, 146 Search PubMed.
- M. Di Martino, L. Sessa, M. Di Matteo, B. Panunzi, S. Piotto and S. Concilio, Azobenzene as Antimicrobial Molecules, Molecules, 2022, 27(17), 5643 CrossRef CAS.
- T. Tahir, M. I. Shahzad, R. Tabassum, M. Rafiq, M. Ashfaq and M. Hassan, et al., Diaryl azo derivatives as anti-diabetic and antimicrobial agents: synthesis, in vitro, kinetic and docking studies, J. Enzyme Inhib. Med. Chem., 2021, 36(1), 1508–1519 CrossRef CAS PubMed.
- A. M. Khedr, M. Gaber and E. H. Abd El-Zaher, Synthesis, Structural Characterization, and Antimicrobial Activities of Mn(II), Co(II), Ni(II), Cu(II) and Zn(II) Complexes of Triazole-based Azodyes, Chin. J. Chem., 2011, 29(6), 1124–1132 CrossRef CAS.
- S. Jangra, G. Purushothaman, K. Juvale, S. Ravi, A. Menon and V. Thiruvenkatam, et al., Synthesis and In Vitro Enzymatic Studies of New 3-Aryldiazenyl Indoles as Promising Helicobacter pylori IMPDH Inhibitors, Curr. Top. Med. Chem., 2019, 19(5), 376–382 CrossRef CAS.
- K. T. Chung, G. E. Fulk and M. Egan, Reduction of azo dyes by intestinal anaerobes, Appl. Environ. Microbiol., 1978, 35(3), 558–562 CrossRef CAS PubMed.
- F. Sisto, S. Carradori, P. Guglielmi, C. B. Traversi, M. Spano and A. P. Sobolev, et al., Synthesis and Biological Evaluation of Carvacrol-Based Derivatives as Dual Inhibitors of H. pylori Strains and AGS Cell Proliferation, Pharmaceuticals, 2020, 13(11), 405 CrossRef CAS PubMed.
- F. Sisto, S. Carradori, P. Guglielmi, M. Spano, D. Secci and A. Granese, et al., Synthesis and Evaluation of Thymol-Based Synthetic Derivatives as Dual-Action Inhibitors against Different Strains of H. pylori and AGS Cell Line, Molecules, 2021, 26(7), 1829 CrossRef CAS PubMed.
- S. Carradori, A. Ammazzalorso, S. Niccolai, D. Tanini, I. D'Agostino and F. Melfi, et al., Nature-Inspired Compounds: Synthesis and Antibacterial Susceptibility Testing of Eugenol Derivatives against H. pylori Strains, Pharmaceuticals, 2023, 16(9), 1317 CrossRef CAS.
- C. Kantar and N. Baltaş, Azo dyes containing eugenol and guaiacol, synthesis, antioxidant and enzyme inhibitory, 2016 Search PubMed.
- C. Kantar, N. Baltaş and Ş. Alpay Karaoğlu, Some azo dyes containing eugenol and guaiacol, synthesis, antioxidant capacity, urease inhibitory properties and anti-helicobacter pylori activity, Rev. Roum. Chim., 2018, 63, 189–197 Search PubMed.
- C. Kantar, N. Baltaş, Ş. A. Karaoğlu and S. Şaşmaz, New Potential Monotherapeutic Candidates for Helicobacter pylori: Some Pyridinazo Compounds Having Both Urease Enzyme Inhibition and Anti-Helicobacter pylori Effectiveness, Pharm. Chem. J., 2021, 55(3), 246–252 CrossRef CAS.
- C. Kantar, N. Baltaş, Ş. A. Karaoğlu and S. Şaşmaz, Some Schiff Bases Containing Eugenol and Guaiacol: Comparison of Urease Inhibition and Anti-Helicobacter pylori Activities with Its Azo Analogs, Pharm. Chem. J., 2024, 57(11), 1738–1744 CrossRef CAS.
- S. Moffa, S. Carradori, F. Melfi, A. Fontana, M. Ciulla and P. Di Profio, et al., Fine-tuning of membrane permeability by reversible photoisomerization of aryl-azo derivatives of thymol embedded in lipid nanoparticles, Colloids Surf., B, 2024, 241, 114043 CrossRef CAS PubMed.
- S. Galati, M. Di Stefano, E. Martinelli, G. Poli and T. Tuccinardi, Recent Advances in In Silico Target Fishing, Molecules, 2021, 26(17), 5124 CrossRef CAS PubMed.
- R. Amoroso, L. De Lellis, R. Florio, N. Moreno, M. Agamennone and B. De Filippis, et al., Benzothiazole Derivatives Endowed with Antiproliferative Activity in Paraganglioma and Pancreatic Cancer Cells: Structure-Activity Relationship Studies and Target Prediction Analysis, Pharmaceuticals, 2022, 15(8), 937 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules, Nucleic Acids Res., 2019, 47(W1), W357–W364 CrossRef CAS.
- F. Ciriaco, N. Gambacorta, D. Trisciuzzi and O. Nicolotti, PLATO: A Predictive Drug Discovery Web Platform for Efficient Target Fishing and Bioactivity Profiling of Small Molecules, Int. J. Mol. Sci., 2022, 23(9), 5245 CrossRef CAS PubMed.
- J. Nickel, B. O. Gohlke, J. Erehman, P. Banerjee, W. W. Rong and A. Goede, et al., SuperPred: update on drug classification and target prediction, Nucleic Acids Res., 2014, 42(Web Server issue), W26–W31 CrossRef CAS PubMed.
- M. Awale and J. L. Reymond, Polypharmacology Browser PPB2: Target Prediction Combining Nearest Neighbors with Machine Learning, J. Chem. Inf. Model., 2019, 59(1), 10–17 CrossRef CAS.
- M. J. Keiser, B. L. Roth, B. N. Armbruster, P. Ernsberger, J. J. Irwin and B. K. Shoichet, Relating protein pharmacology by ligand chemistry, Nat. Biotechnol., 2007, 25(2), 197–206 CrossRef CAS PubMed.
- I. D'Agostino and C. S. Urease, in Metalloenzymes: From Bench to Bedside, ed. C. T. Supuran and W. A. Donald, 2023, pp. 393–410 Search PubMed.
- C. Li, P. Huang, K. Wong, Y. Xu, L. Tan, H. Chen, Q. Lu, C. Luo, C. Tam, L. Zhu, Z. Su and J. Xie, Coptisine-induced inhibition of Helicobacter pylori: elucidation of specific mechanisms by probing urease active site and its maturation process, J. Enzyme Inhib. Med. Chem., 2018, 33(1), 1362–1375 CrossRef CAS PubMed.
- E. S. Cunha, X. Chen, M. Sanz-Gaitero, D. J. Mills and H. Luecke, Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 Å resolution, Nat. Commun., 2021, 12(1), 230 CrossRef CAS PubMed.
- W. Liu, C. Luo, C. Han, S. Peng, Y. Yang and J. Yue, et al., A new beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Helicobacter pylori: Molecular cloning, enzymatic characterization, and structural modeling, Biochem. Biophys. Res. Commun., 2005, 333(4), 1078–1086 CrossRef CAS.
- L. He, L. Zhang, X. Liu, X. Li, M. Zheng and H. Li, et al., Discovering Potent Inhibitors Against the β-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) of Helicobacter pylori: Structure-Based Design, Synthesis, Bioassay, and Crystal Structure Determination, J. Med. Chem., 2009, 52(8), 2465–2481 CrossRef CAS.
- B. Hernández-Ochoa, G. Navarrete-Vázquez, R. Aguayo-Ortiz, P. Ortiz-Ramírez, L. Morales-Luna, V. Martínez-Rosas, A. González-Valdez, F. Gómez-Chávez, S. Enríquez-Flores, C. Wong-Baeza, I. Baeza-Ramírez, V. Pérez de la Cruz and S. Gómez-Manzo, Identification and In Silico Characterization of Novel Helicobacter pylori Glucose-6-Phosphate Dehydrogenase Inhibitors, Molecules, 2021, 26(16), 4955 CrossRef PubMed.
- Schrödinger Release 2024-1, Maestro, Glide, Protein Preparation Wizard, Epik, MacroModel, Prime, Schrödinger, LLC, New York (NY), 2024 Search PubMed.
- H. M. Berman, T. Battistuz, T. N. Bhat, W. F. Bluhm, P. E. Bourne and K. Burkhardt, et al., The Protein Data Bank, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58(Pt 6 1), 899–907 CrossRef.
- C. Camacho, G. Coulouris, V. Avagyan, N. Ma, J. Papadopoulos and K. Bealer, et al., BLAST+: architecture and applications, BMC Bioinf., 2009, 10(1), 421 CrossRef PubMed.
- I. Berneburg, S. Rahlfs, K. Becker and K. Fritz-Wolf, Crystal structure of Leishmania donovani glucose 6-phosphate dehydrogenase reveals a unique N-terminal domain, Commun. Biol., 2022, 5(1), 1–14 CrossRef.
- R. Zhang, G. Evans, F. Rotella, E. Westbrook, E. Huberman and A. Joachimiak, et al., Differential signatures of bacterial and mammalian IMP dehydrogenase enzymes, Curr. Med. Chem., 1999, 6(7), 537–543 CrossRef CAS.
- RCSB Protein Data Bank, RCSB PDB - 4MZ1: Crystal Structure of the Inosine 5′-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 [Internet], available from: https://www.rcsb.org/structure/4mz1.
- K. Juvale, G. Purushothaman, V. Singh, A. Shaik, S. Ravi and V. Thiruvenkatam, et al., Identification of selective inhibitors of Helicobacter pylori IMPDH as a targeted therapy for the infection, Sci. Rep., 2019, 9(1), 190 CrossRef PubMed.
- D. E. V. Pires, T. L. Blundell and D. B. Ascher, pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures, J. Med. Chem., 2015, 58(9), 4066–4072 CrossRef CAS.
- B. H. Jin, B. W. Yoo, J. Park, J. H. Kim, J. Y. Lee and J. S. Shin, et al., Pharmacokinetic drug interaction and safety after coadministration of clarithromycin, amoxicillin, and ilaprazole: a randomised, open-label, one-way crossover, two parallel sequences study, Eur. J. Clin. Pharmacol., 2018, 74(9), 1149–1157 CrossRef CAS.
- L. Chen, Y. Gao, L. Zhu, H. Song, L. Zhao and A. Liu, et al., Establishment and characterization of a GES-1 human gastric epithelial cell line stably expressing miR-23a, Oncol. Lett., 2018, 16(1), 977–983 Search PubMed.
- A. Fedi, C. Vitale, G. Ponschin, S. Ayehunie, M. Fato and S. Scaglione, In vitro models replicating the human intestinal epithelium for absorption and metabolism studies: A systematic review, J. Control. Release, 2021, 335, 247–268 CrossRef CAS PubMed.
- C. Ardino, F. Sannio, C. Pasero, L. Botta, E. Dreassi and J. D. Docquier, et al., The impact of counterions in biological activity: case study of antibacterial alkylguanidino ureas, Mol. Diversity, 2023, 27(3), 1489–1499 CrossRef CAS PubMed.
- I. D'Agostino, G. E. Mathew, P. Angelini, R. Venanzoni, G. Angeles Flores and A. Angeli, et al., Biological investigation of N-methyl thiosemicarbazones as antimicrobial agents and bacterial carbonic anhydrases inhibitors, J. Enzyme Inhib. Med. Chem., 2022, 37(1), 986–993 CrossRef PubMed.
- C. Zamperini, G. Maccari, D. Deodato, C. Pasero, I. D'Agostino and F. Orofino, et al., Identification, synthesis and biological activity of alkyl-guanidine oligomers as potent antibacterial agents, Sci. Rep., 2017, 7(1), 8251 CrossRef CAS PubMed.
- C. Pasero, I. D'Agostino, F. De Luca, C. Zamperini, D. Deodato and G. I. Truglio, et al., Alkyl-guanidine Compounds as Potent Broad-Spectrum Antibacterial Agents: Chemical Library Extension and Biological Characterization, J. Med. Chem., 2018, 61(20), 9162–9176 CrossRef CAS.
- V. Puca, G. Turacchio, B. Marinacci, C. T. Supuran, C. Capasso and P. Di Giovanni, et al., Antimicrobial and Antibiofilm Activities of Carvacrol, Amoxicillin and Salicylhydroxamic Acid Alone and in Combination vs. Helicobacter pylori: Towards a New Multi-Targeted Therapy, Int. J. Mol. Sci., 2023, 24(5), 4455 CrossRef CAS.
- B. R. Pirgonde and M. A. Pujar, Reactions of dichlorobis(cyclopentadienyl)titanium(IV) with diazo compounds, Acta Chim. Hung., 1989, 126(2), 247–249 CAS.
- P. Sagar Vijay Kumar, L. Suresh, T. Vinodkumar, R. BenjaramM and G. V. P. Chandramouli, Zirconium Doped Ceria Nanoparticles: An Efficient and Reusable Catalyst for a Green Multicomponent Synthesis of Novel Phenyldiazenyl–Chromene Derivatives Using Aqueous Medium, ACS Sustainable Chem. Eng., 2016, 4(4), 2376–2386 CrossRef CAS.
- M. Amaravathi, S. Kanakaraju and G. V. P. Chandramouli, Synthesis and Characterization of Azobenzene–Porphyrins, J. Heterocyclic Chem., 2013, 50(2), 268–271 CrossRef CAS.
- S. M. Koshti, J. P. Sonar, A. E. Sonawane, Y. A. Pawar, P. S. Nagle, P. P. Mahulikar and D. H. More, Synthesis of Azo Compounds Containing Thymol Moiety, Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem., 2008, 47(2), 329–331 Search PubMed.
- F. Sisto, M. M. Scaltrito, G. Russello, A. Bonomi and F. Dubini, Antimicrobial susceptibility testing of Helicobacter pylori determined by microdilution method using a new medium, Curr. Microbiol., 2009, 58(6), 559–563 CrossRef CAS PubMed.
- Clinical and Laboratory Standard Institute (CLSI), Development of In Vitro Susceptibility Testing Criteria and Quality Control Parameters: Approved Guideline, Clinical and Laboratory Standard Institute, Wayne, PA, USA, 3rd edn, 2008, vol. 28 Search PubMed.
- J. C. Shelley, A. Cholleti, L. L. Frye, J. R. Greenwood, M. R. Timlin and M. Uchimaya, Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules, J. Comput.-Aided Mol. Des., 2007, 21(12), 681–691 CrossRef CAS.
- UniProt Consortium, UniProt: the Universal Protein Knowledgebase in 2023, Nucleic Acids Res., 2023, 51(D1), D523–D531 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The supplementary material file contains the NMR spectra of the compounds reported in this article. See DOI: https://doi.org/10.1039/d4md00511b |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2025 |