Functional antimicrobial peptide-loaded 3D scaffolds for infected bone defect treatment with AI and multidimensional printing
Mengmeng
Li
ab,
Peizhang
Zhao
ab,
Jingwen
Wang
ab,
Xincai
Zhang
 *c and
Jun
Li
*c and
Jun
Li
 *ab
*ab
aOrthopedic Research Institute, Department of Orthopedics, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, PR China. E-mail: mrorthopedics@sina.com
bTrauma Center, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, PR China
cDepartment of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA. E-mail: xingcai@stanford.edu
First published on 22nd October 2024
Abstract
Infection is the most prevalent complication of fractures, particularly in open fractures, and often leads to severe consequences. The emergence of bacterial resistance has significantly exacerbated the burden of infection in clinical practice, making infection control a significant treatment challenge for infectious bone defects. The implantation of a structural stent is necessary to treat large bone defects despite the increased risk of infection. Therefore, there is a need for the development of novel antibacterial therapies. The advancement in antibacterial biomaterials and new antimicrobial drugs offers fresh perspectives on antibacterial treatment. Although antimicrobial 3D scaffolds are currently under intense research focus, relying solely on material properties or antibiotic action remains insufficient. Antimicrobial peptides (AMPs) are one of the most promising new antibacterial therapy approaches. This review discusses the underlying mechanisms behind infectious bone defects and presents research findings on antimicrobial peptides, specifically emphasizing their mechanisms and optimization strategies. We also explore the potential prospects of utilizing antimicrobial peptides in treating infectious bone defects. Furthermore, we propose that artificial intelligence (AI) algorithms can be utilized for predicting the pharmacokinetic properties of AMPs, including absorption, distribution, metabolism, and excretion, and by combining information from genomics, proteomics, metabolomics, and clinical studies with computational models driven by machine learning algorithms, scientists can gain a comprehensive understanding of AMPs' mechanisms of action, therapeutic potential, and optimizing treatment strategies tailored to individual patients, and through interdisciplinary collaborations between computer scientists, biologists, and clinicians, the full potential of AI in accelerating the discovery and development of novel AMPs will be realized. Besides, with the continuous advancements in 3D/4D/5D/6D technology and its integration into bone scaffold materials, we anticipate remarkable progress in the field of regenerative medicine. This review summarizes relevant research on the optimal future for the treatment of infectious bone defects, provides guidance for future novel treatment strategies combining multi-dimensional printing with new antimicrobial agents, and provides a novel and effective solution to the current challenges in the field of bone regeneration.
Wider impactThis comprehensive review provides a concise overview of recent research on the multifaceted factors influencing bone regeneration in large bone defects, with a specific focus on open fractures and the intricate pathways of infection that hinder successful healing. Additionally, we offer an insightful summary of remarkable advancements in novel 3D materials and antimicrobial peptides (AMPs), an innovative class of antibacterial agents, for effectively repairing infected bone defects while addressing challenges posed by bacterial resistance skillfully. For the repair and regeneration of these complex bone defects, ingenious 3D scaffolds incorporating AI and multidimensional printing not only provide essential structural support for optimal cell proliferation but also create a favorable microenvironment for efficient osteogenesis through controlled release mechanisms involving pivotal factors such as BMP-2 and various essential metal ions. This review also highlights groundbreaking research findings from recent years that have significantly contributed to our current understanding in this field. The synergistic combination of antimicrobial peptides with state-of-the-art 3D materials, AI, and multidimensional printing holds immense potential to revolutionize the treatment landscape for challenging bone defects. Our findings may facilitate the development of more efficient strategies by providing insights into infected bone defects and cutting-edge approaches for effective therapeutic interventions. |
1. Introduction
Fracture is one of the most common orthopedic conditions, and especially with the aging of the population, bone regeneration is an important clinical problem. High-energy or refractory fractures can result in large bone loss and minor bone defects that do not “spontaneously” heal.1 Therefore, a stable support structure and exogenous “healing stimulation” are essential to promote bone defect regeneration.1 Appropriate mechanical strength is crucial for bone tissue engineering materials to provide sufficient support and avoid stress-shielding effects in bone repair.2 Currently, there are three methods of bone grafting: autologous transplantation, allograft transplantation, and artificial scaffolds.3 Although there are many studies on biomaterials for bone defect repair, the limited promotion of cell proliferation and osteogenesis cannot meet the physiological structure requirements.4 On the other hand, studies have shown that surface topographical cues are also essential for bone repair, which may be related to the polarization of macrophages induced by them.5 Also, MSCs exhibited different osteogenic abilities on materials that mimic natural surfaces made by plants or animals, which may be related to changes in the differentiation potential of MSCs. Similar effects also have been shown for bacterial colonization, such as for Pseudomonas aeruginosa.6In recent years, significant advancements have been made in 3D printing technology, enabling the production of biomimetic scaffolds with precise geometric shapes and structures for joint regeneration.4 Moreover, biological 3D printing integrates bioactive factors, mesenchymal stem cells, and other active components to create a highly biologically interactive microenvironment that facilitates extensive new bone formation.7 In addition to bacterial colonization after direct wound contamination, infections associated with orthopedic biomaterials are expected, with Staphylococcus aureus being the most common pathogen.8 The risk of bacterial infection is especially high in implants that enhance cell adhesion and proliferation, and even a minor bacterial infection can result in the development of biofilms with significant implications.9 Non-traditional antibacterial agents, such as antimicrobial peptides, offer new opportunities for treating infections with different mechanisms of action and have been increasingly researched over the past three decades.10
AMPs, also known as defensins, are present in almost all life forms, from bacteria to vertebrates, and are an important part of innate defense.11 AMPs play an important role in many fields due to their antibacterial, anticancer, anti-inflammatory, and autoimmune modulatory abilities.12 In addition to their clinical use as anti-infectives, antimicrobial peptides are used in other industries, such as microecological modulators.13 AMPs are typically 12–50 residues long and contain large amounts of cationic (arginine and lysine) and hydrophobic (leucine, leucine, valine, phenylalanine, and tryptophan) amino acids.14 This structure means that AMPs are amphoteric molecules, both hydrophilic and lipophilic, and mostly positively charged.14,15 One classification method divides AMPs into four categories according to their secondary structure. Class α antimicrobial peptides only have an α-helix, class β antimicrobial peptides only have a β-sheet, class αβ antimicrobial peptides have both structures and non-αβ antimicrobial peptides have neither structure.16 Disulfide bonds can extend or stabilize the α-helical, β-lamellar, and β-hairpin structures of AMPs, enhancing the resistance of AMPs to proteolytic hydrolysis.10 In this review, we discuss the infection mechanism of infective bone defects, current research on repair materials for bone defects, and biomaterials related to antibacterial agents. In particular, we discuss the antibacterial mechanism of novel antibacterial drugs called antimicrobial peptides and current research on their improvement. In addition, we have synthesized current cutting-edge research on the application of antimicrobial peptides in infectious bone defects, with a particular focus on studies exploring the integration of AMPs with 3D bioprinting scaffolds. We aspire for this article to highlight the distinctive advantages and significant developmental potential inherent in combining AMPs with 3D scaffolds for addressing infectious bone defects. We hope that in the future, antimicrobial peptides can be applied to treat infective bone defects to better control infection and promote defect repair.
2. Infected bone defects
2.1 Mechanism and method of bone repair
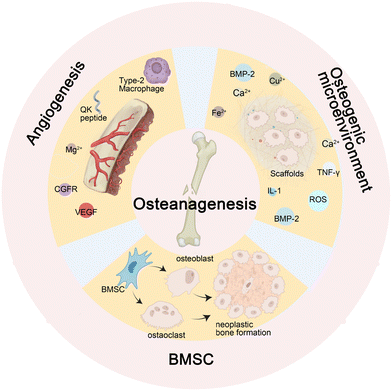
During the normal process of bone defect repair, numerous influencing factors are involved. Given a sufficient blood supply, mesenchymal stem cells (MSCs) differentiate into osteoblasts and osteoclasts under the influence of osteogenic factors. The functional coordination between the two cell types promotes callus formation and remodeling. Nevertheless, infectious bone defects, due to the stimulation of pathogens, result in local inflammation, disrupting the balance between the functions of osteoblasts and osteoclasts. Furthermore, long-term inflammatory stimulation can aggravate local tissue damage, interfere with normal signaling pathways, affect cell differentiation and proliferation, cause inflammatory bone hyperplasia and avascular necrosis,17 and eventually present poor bone formation and affect the repair of bone defects.The activation of β-catenin and its target gene activating transcription factor 4 (ATF4) can significantly promote the proliferation and osteogenic differentiation of Gli1+ BMSCs. This process is influenced by Piezo1, a major mechanosensitive ion channel involved in bone homeostasis, which can activate the β-catenin pathway.24 Reversely, microRNA-138a is a negative regulator of β-catenin.25 Besides, the Wnt protein family promotes the self-renewal of human skeletal stem cells (hSSCs) by activating the Wnt/β-catenin pathway.26 For instance, the Wnt3a bandage reported by Okuchi et al. has a good effect on promoting osteogenesis. Research shows that the activation of the ERK1/2 and Wnt/β-catenin signaling pathways are the main factors that initiate the osteogenic function of osteoblasts27 (Fig. 2A). Another target of BMSCs is ubiquitin-specific peptidase 53 (USP53). Research in vitro denotes that USP53 can promote the osteogenic differentiation of human BMSCs.28 USP53 interacts with F-box only protein 31 to facilitate the proteasomal degradation of β-catenin, thereby augmenting the level of active β-catenin. In animal experiments, overexpression of USP3 in BMSCs significantly enhanced bone regeneration in mice with cranial defects28 (Fig. 2B).
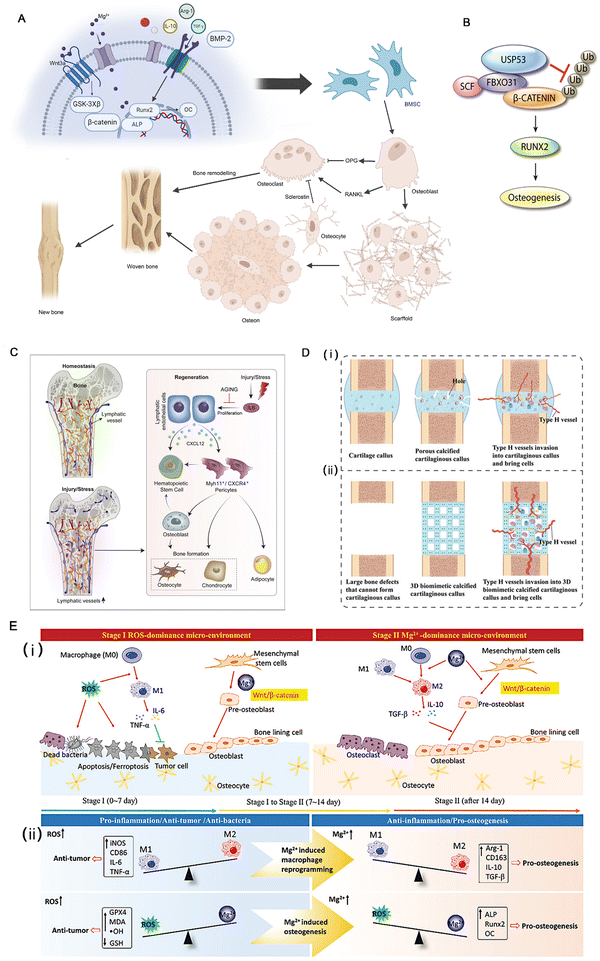 | ||
| Fig. 2 (A) A brief description of the process of bone regeneration and the Wnt3a signaling pathway. (B) Diagram showing the molecular mechanism through which FBXO31 and USP53 regulate the Wnt signaling mediator, β-catenin, during the osteogenic differentiation of hBMSCs. Reproduced with permission.28 Copyright 2021. (C) Lymphatic vessels not only connect secondary lymphoid organs with tissue sites throughout the body but are also now shown directly irrigating bones and participating in their regeneration. Reproduced with permission.29 Copyright 2023, Elsevier Inc. (D) Construction of a 3D biomimetic cartilaginous callus and its mechanism of promoting the regeneration of large-segment bone defects. (i) Normal biological process of bone regeneration. (ii) Large-segment bone defects cannot form calcified cartilaginous calluses or type H vessels. A 3D biomimetic calcified cartilaginous callus with coupled type H vessels formation was constructed to promote the regeneration of large-segment bone defects. Reproduced with permission.22 Copyright 2023, Wiley-VCH GmbH. (E) The time-sequential functional mechanism of the MgO2/PLGA scaffold for repairing the bone defect, the red arrows for promotion, and green “T” for inhibition. Reproduced with permission.30 Copyright 2023, Wiley-VCH GmbH. | ||
A concise UTR sequence incorporating TISU and 5-iodopyrimidine nucleotides can significantly enhance transcriptional efficiency while mitigating mRNA-induced inflammation. During BMP-2 encoding, this modified cmRNA exhibits a clear dose-dependent correlation and demonstrates the potential to facilitate osteogenesis.35 The hypertrophic cartilage material generated by gene-edited human mesenchymal cells expressing nerve growth factor receptor and a high level of BMP-2 was subjected to inactivation and implanted into the body, resulting in significant induction of subchondral ossification. However, this process was found to rely more on the properties of cartilage than the dosage of BMP-2.31
Type M1 macrophages play an important role in inhibiting inflammation and promoting angiogenesis and tissue repair37 by inhibiting the generation of reactive oxygen (ROS).38 The calcitonin gene-related peptide (CGFR)-focal adhesion kinase (FAK)-vascular endothelial growth factor (VEGF) signaling axis is one of the main signaling pathways of angiogenesis.18 VEGF can promote angiogenesis and induce osteoblasts, and it has been used in some research to improve the osteogenic effect of a scaffold.39 VEGF analog QK peptide has the effect of inducing angiogenesis.33 In Dominic et al.'s study, QK peptide showed good promoting effects on angiogenesis,40 and the 3D printing material engineered by their team, which integrates QK peptide and collagen cross-linking to form a scaffold while incorporating diverse catalytic enzymes, represents a highly versatile substance that can substantially enhance angiogenesis. It has been used in research on osteogenic scaffolds,33 but few studies have demonstrated that QK peptide has osteogenic activity similar to VEGF.39 Sensory nerves play an important role in bone regeneration. The concentration of Tublin, a sensory neuropeptide involved in bone defect healing, was significantly increased in the bone defect area, and it played an important role in the bone defect repair promoted by Mg2+ through the CGRP-FAK-VEGF signaling axis.18
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 have the effect of promoting angiogenesis related to VEGF. For these metal ions, the utilization of Layered Double Hydroxides (LDHs) with a positive charge as a carrier demonstrates remarkable potential in facilitating drug delivery within cells and bacteria, thereby augmenting therapeutic efficacy. In environments characterized by acidity or an abundance of H2O2, such as bone infections, bone tumors, osteoarthritis, and similar conditions, alkaline LDHs exhibit accelerated degradation rates and enhanced drug release capabilities.2 The metal frame can also promote the generation of OH− ions to form an alkaline microenvironment, which is conducive to bone tissue regeneration.42
30 have the effect of promoting angiogenesis related to VEGF. For these metal ions, the utilization of Layered Double Hydroxides (LDHs) with a positive charge as a carrier demonstrates remarkable potential in facilitating drug delivery within cells and bacteria, thereby augmenting therapeutic efficacy. In environments characterized by acidity or an abundance of H2O2, such as bone infections, bone tumors, osteoarthritis, and similar conditions, alkaline LDHs exhibit accelerated degradation rates and enhanced drug release capabilities.2 The metal frame can also promote the generation of OH− ions to form an alkaline microenvironment, which is conducive to bone tissue regeneration.42
External physical stimulation has clear benefits for enhancing bone regeneration. Appropriate stimulation, such as light and electrical stimulation, can enhance the adhesion, proliferation, and differentiation of osteoblasts in the scaffold, which is related to changes in intracellular behavior, stimulation of PI3K/Akt signaling, pathway, inner electronic signals, increased ATP production, and activation of Ca channels.47 External electrical stimulation can stimulate the endogenous electric field of bone tissue to promote bone regeneration, which has the potential to treat nonunion.36,48 Combined with light-activated nanoparticles, it can clear bacterial infections.38 However, the limited penetration distance of light limits the application of this method in deep tissues.38 The trace element silicon, a non-metal, is essential for bone regeneration. Research has demonstrated that biomaterials incorporating silicon can induce the differentiation and activation of BMSCs, facilitate cell-to-cell interactions, and promote angiogenesis. Consequently, scholars have proposed incorporating silane coupling agents in preparing biological scaffolds to augment their efficacy.49
2.2 Application of 3D printing technology in bone defect repair
Using implanted fillers to create an osteogenic microenvironment is a promising research direction for bone defect repair.36 Tissue engineering materials used for bone defect repair should be biocompatible and not harmful to system function.50 Commonly used materials are bioceramics, organic polymers, biodegradable metals, and carbon-based nanomaterials.3,50 Meanwhile, 3D printing technology can produce structural elements with ideal geometric shapes and structures for bone and joint regeneration.7 For example, sponge collagen protein,51 calcium sulfate/hydroxyapatite (CaS/HA) biomaterial,1 and biodegradable metal scaffolds,52 are 3D materials that have been proposed or have potential. HA is one of the most commonly used stent materials. However, many 3D materials cannot continuously induce cell migration and proliferation, and induction factors often need to be added.1,51 Considering the limitations, biological 3D printing has emerged as a promising area of research.7 Biological 3D printing employs techniques such as jetting or extrusion to deliver growth factors and live cells into hydrogels and other materials, creating intricate functional living tissues. It offers enhanced cell integration strength, improved distribution of diverse cells and growth factors, and the ability to establish a biologically active microenvironment within the scaffold. One of the most commonly used bioactive molecules is BMP-2.BMP-2 and negatively charged polydeoxyribonucleotide (PDRN) can synergistically affect bone regeneration and angiogenesis. The poly(lactic-co-glycolic)acid/nanocomplex scaffold developed based on this shows excellent potential in the repair of bone defects in rats.3 The ability of sponge collagen nanofibers to promote cell migration was significantly improved after cross-linking with BMP-2 analog KP peptide.51 In addition to autologous bone grafting, collagen sponge carriers impregnated with high-dose BMP-2 have emerged as a promising approach for repairing extensive bone defects.53 The 3D scaffolds modified with the combination of QK peptide and KP peptide were significantly better than the collagen sponge scaffolds in promoting osteogenesis and angiogenesis.33
In addition to BMP-2-based biological scaffolds, there are also some studies on bioactive substances of other signaling pathways in osteogenesis, such as hedgehog signaling and the smoothened agonist sterosome-immobilized 3D hybrid scaffold based on it54 and hydrogels with Mg2+ added to them.30 Also, in rats, the transverse and longitudinal arrangement of polycaprolactone (PCL) nanofibers, coated gradually, could guide cell migration and induce the expression of bone repair-related genes.4 The collagen/PCL composite mineralized by HA nanocrystals forms a porous matrix. At the same time, patterned monocrystalline Si films are embedded into the mineralized collagen/PCL scaffold to provide optoelectronic stimulation for bone regeneration.48 Besides, the calcium phosphate composite scaffold modified with graphene oxide also uses the photoelectric effect to promote the repair of bone defects.55
Compared to inorganic materials, biomaterial 3D scaffolds have unique advantages. They have good biocompatibility and tend to have a weaker stimulation on the body. Li et al. has reported that 3D-printed hydrogel scaffolds, loaded with exosomes, effectively inhibited inflammation and facilitated bone and cartilage regeneration without evident systemic toxicity in rats.23 These scaffolds comprised decellularized components derived from porcine cartilage and cancellous bone, hyaluronic acid, dopamine, and oxyhalogenated hyaluronic acid. They were crosslinked using methacrylated gelatin to enhance rigidity and brittleness.
2.3 Bone infections can severely impair bone regeneration
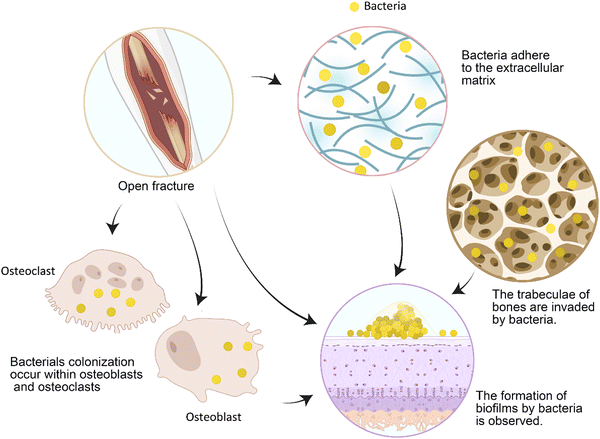
Bone cell infection induces the secretion of osteoclast cytokines, resulting in pathological bone loss, which directly affects the process of bone repair.56 The local excessive inflammation caused by pathogen-associated molecular patterns is an important reason for the poor healing of bone defects.57 Infection causes an increase in the generation of reactive oxygen species (ROS) in mitochondria, exacerbating local cell and tissue damage.57 However, some scholars believe that infection does not cause nonunion but leads to the formation of bone sockets and implant loosening, which causes nonunion.19 As the most common infectious pathogen, Staphylococcus aureus can colonize osteoblasts and osteoclasts (Fig. 3), and it will not be eradicated by vancomycin treatment.56 Animal experiments show that long-term asymptomatic local infection can inhibit the immune function of the whole body, which is manifested by the decrease in the number of circulating immune cells, such as B cells, and the incoordination of cytokines and chemokines. This is associated with an increase in the number of immunosuppressive myeloid-derived suppressor cells (MDSCs).8Bloodless materials, in particular, easily become objects of bacterial adhesion. After a large number of bacteria adhere and form a biofilm (Fig. 3), the infection is difficult to cure. Proteins secreted by bacteria in biofilms, such as S. aureus, also enhance immune evasion, facilitating bacterial invasion of the bone canal system.8 Patients' immune functions are decreased after surgery, so bacteria are more likely to develop drug resistance.2 Human serum can also induce the development of bacterial resistance, such as S. aureus resistance to tobramycin.58 As bacterial drug resistance becomes a massive burden in the biomedical field, it is very important to prevent infection during bone repair.
2.4 Antibiotic-releasing scaffolds in the treatment of infected bone defects
To solve the problems related to bone infection, more and more scholars have focused on integrating antibacterial effects into bone repair scaffolds. However, the impact of materials on osteogenesis should also be considered to achieve the antibacterial purpose of finding a balance between the two. For example, the appropriate level of reactive oxygen species can achieve an antibacterial effect without damaging new bone tissue.59 Metal ions such as Zn2+, Mg2+, Cu2+, etc., can be adsorbed and destroyed in bacterial cell membranes and are used as coating materials for scaffolds.2 FeSAC-BG is a scaffold made by dispersing highly catalytically active single-atom iron catalysts (FeSAC) into bioactive glass (BG). The roughness of FeSAC-BG allows BMSCs to adhere better and proliferate, and its amorphous carbon substrate also has the effect of inducing osteogenesis. Moreover, the Fe catalyzes H2O2 to produce ·OH through the Fenton reaction, which has a strong bactericidal effect.60 The MgO/FeOx nanoflake films can create an alkaline microenvironment abundant in OH− ions, which effectively deplete protons from the external surface of bacteria to weaken bacterial ATP synthesis42 (Fig. 4A). Combining the antibacterial properties of the alkaline environment enhances bacteria's thermosensitivity, lowers the required killing temperature, and minimizes tissue damage. Additionally, the photothermal effect induced by metal ions can deactivate bacterial proteins, DNA, etc., resulting in rapid bactericidal action. Unfortunately, the efficacy of antimicrobial implants is still uncertain, as antimicrobial agents on the surface of implants can reduce histocompatibility and inhibit angiogenesis and osteogenesis, which in turn delays bone healing and increases the risk of infection in vivo.39 Furthermore, antibiotic coatings may have some antibacterial effects, but there is also a risk of inducing bacterial resistance.61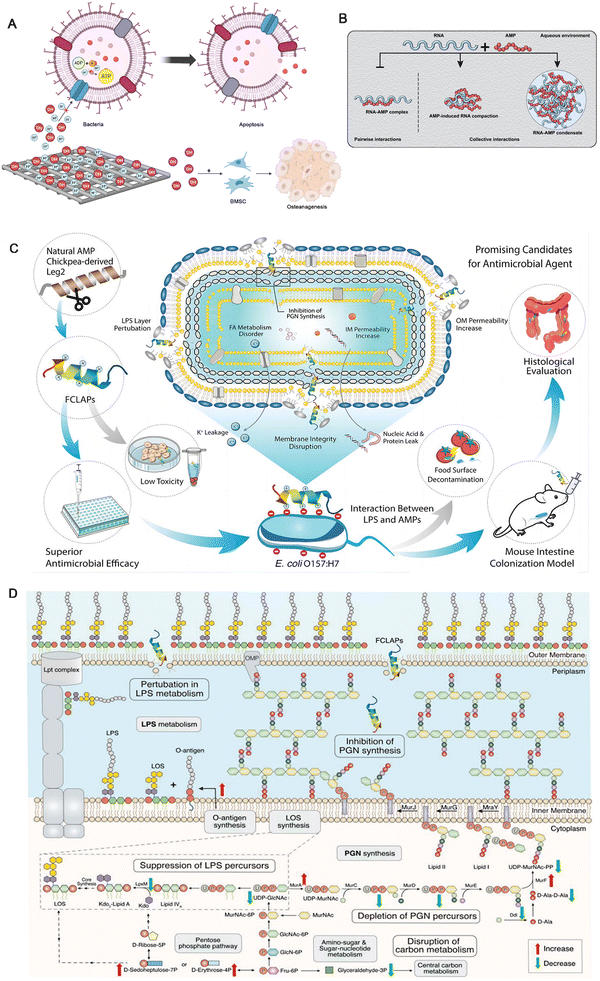 | ||
| Fig. 4 (A) A schematic diagram illustrating the electrochemical generation of H+ for bactericidal activity and OH− for osteogenic promotion by metal stents. (B) Schematic illustration depicting the formation of distinct AMP-RNA assemblies that are held together via interactions of different strengths and nature. Source data are provided as a Source data file. Reproduced with permission.62 Copyright 2023, © The Author(s) 2020. (C) Schematic illustration of FCLAPs combating E. coli O157:H7 both in vitro and in vivo. FCLAPs kill bacteria through a dual-targeting mechanism: (1) interaction with the LPS leaflet of the outer membrane, causing areas of destabilization and facilitating the entry of FCLAPs; (2) FCLAPs further insert into the PGN layer, blocking the PGN synthesis, resulting in a disrupted cell wall architecture. The rupture of the cell envelope further induces metabolic disorder of membrane components, leakage of cytoplasmic content, and eventually causes bacterial death. Reproduced with permission.63 Copyright 2022, Wiley-VCH GmbH. (D) Schematic diagram summarizing the main antibacterial mechanisms of KTA in combating E. coli O157:H7. The most significant findings on the inactivation mechanism of KTA include inhibition of LPS and PGN metabolism, depletion of key precursors for LPS and PGN biosynthesis, and perturbation of central carbon metabolism and energy metabolism. Reproduced with permission.63 Copyright 2022, Wiley-VCH GmbH. | ||
3. Antibacterial peptides
3.1 Mechanism of antimicrobial peptides
The target sites of antimicrobial peptides range from extracellular to intracellular (Table 1).64 The review by Portelinha et al. describes several models of AMPs' action on cell membranes and transmembranes, such as the toroidal pore model and transmembrane models, and related mechanisms in detail.11 The most in-depth research has been conducted on the mechanism by which AMP damages cell membranes and causes cell lysis.| AMPsa | Resource | Target points | Pathogen | Time | Ref. |
|---|---|---|---|---|---|
| a All the names of the AMPs come from the original papers. | |||||
| Ω76 | Ω-family peptides | Bacterial membranes | Carbapenem- and tigecycline-resistant A. baumannii | 2019 | 65 |
| Functionalized chickpea-derived Leg2 antimicrobial peptides | Natural chickpea-derived AMP-Leg2 | Peptidoglycan; cell membranes | Escherichia coli O157:H7 | 2022 | 63 |
| VTK-LL37 | Heptapeptide VTKLGSL (VTK) fusion with LL-37 | Production of HMGB1 | Escherichia coli MSI001 | 2022 | 66 |
| Macrocyclic peptide | Tranzyme pharma | The LptB2 FGC complex (related to the transport of lipopolysaccharides) | Carbapenem-resistant Acinetobacter baumannii (CRAB) | 2024 | 67 |
| AMP1, AMP2 and AMP3 | PAM1 transgenic N. benthamiana plants | Bacterial membrane | Antibiotic resistant ESKAPE pathogens | 2023 | 68 |
| Drosocin | Fruit flies | Ribosome | E. coli BL21 | 2023 | 69 |
| Epifadin | Nasal Staphylococcus epidermidis IVK83 | Bacterial membrane | S. aureus | 2023 | 70 |
| Peptide YY (PYY1–36) | Mammalian gut epithelial paneth cells | Hyphal membranes | Virulent Candida albicans hyphae | 2023 | 71 |
There is also a synergistic effect between different antimicrobial peptides in permeating cell membranes.75 For example, magainin 2 and PGLa are complementary in the rate and stability of channel formation.73 The above research findings clearly indicate that the insertion of AMP into bacterial cell membranes is primarily influenced by its physical properties and structural features, exhibiting low specificity. And it is imperative to acknowledge the potential toxicity to human cells.
3.2 LL-37
LL-37 is an alpha-helical peptide, one of several cleaved products of 23hcap18, a product of the human version of the only antimicrobial peptide gene CAMP26.15 LL-37 is the only cathelicidin family antimicrobial peptide found in the human body, which can protect the body through multiple mechanisms.15 Current research has found that LL-37 can be found in various cell types, tissues, and body fluids. The present study demonstrated that the activation of p38 and JNK is required to induce the transcriptional activation of LL-37. This activation involves downstream activation of MSK-1 and NF-κB, and NF-κB protein is essential for the signaling.61 LL-37 has significant potential in preventing and controlling drug-resistant bacterial infections and inhibiting biofilm formation. The amino acid residues 17 to 19 of LL-37 form very stable fibrils rich in arginine residues in vivo that bind to and disrupt lipid bilayers, such as bacterial cell membranes.77 In addition to electrostatic effects affecting LL-37 interaction with the membrane, hydrophobicity also has an impact.12 The binding of LL-37 and LPS has been proposed by some scholars to induce the dissociation of LPS aggregates and inhibit the proinflammatory cascade.78 This may be due to the enhanced stability of the α-helical structure of LL-37 induced by LPS.78Different antimicrobial peptides can also produce synergistic antimicrobial effects when used in combination.79 Nevertheless, some scholars have proposed that the current studies are insufficient to prove the synergistic cytotoxicity between antibiotics and antimicrobial peptides.64 Also, LL-37 assists histone H2A in piercing the cell membranes of E. coli and S. aureus and produces synergistic effects in membrane penetration and destruction.80 Except for its direct antibacterial activity, LL-37 modulates host immune function and is an important component of innate immunity.81 Based on the above characteristics, LL-37 is often used as a template or molecular skeleton for the development of new antibacterial drugs.66 Jin et al. further demonstrated the osteogenic potential of LL-37 in various aspects following biomaterial implantation, thereby enhancing bone regeneration82 (Fig. 5A).
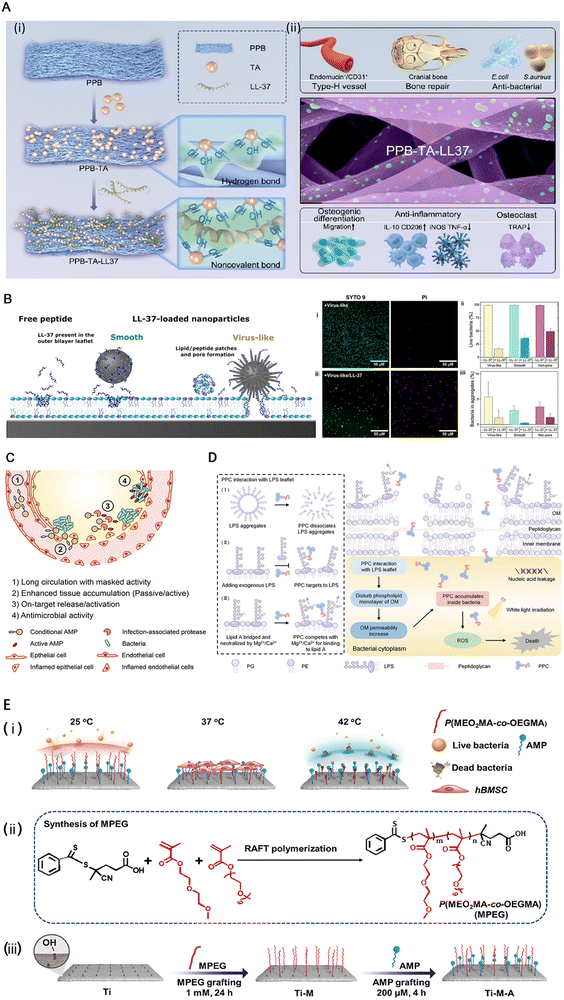 | ||
| Fig. 5 (A) Schematic of (i) the PPB (a multifunctional core–shell structured electrospun fiber scaffold composed of polycaprolactone loaded with baicalin as the core, and poly(lactic-co-glycolic acid) combined with fish collagen as the shell) fibrous scaffold modified using tannic acid (TA) and LL-37 peptide and (ii) the LL-37 peptide-grafted fibrous scaffold significantly enhanced angiogenesis, anti-inflammatory effects, and type-H vascularized bone regeneration. Reproduced with permission.82 Copyright 2023, © 2023 American Chemical Society. (B) (Left) Schematic illustration of the structural effects observed by neutron reflectivity after interaction of POPC/POPG bilayers with virus-like and smooth mesoporous silica nanoparticles loaded with LL-37, as well as with free LL-37. As shown, structural effects induced by the peptide-loaded smooth nanoparticles are modest, and confined to the outer bilayer leaflet. In contrast, LL-37-loaded virus-like nanoparticles induce drastic structural changes, spanning the entire bilayer and more pronounced than the effects of free LL-37. Reproduced with permission.12 Copyright 2021, American Chemical Society. (C) Desirable features of conditional AMP therapeutics include (1) long circulation with masked activity, (2) accumulation at infection site either by passive or active targeting, (3) activation by an infection microenvironmental trigger, and (4) exhibition of on-target antimicrobial activity. Reproduced with permission.83 Copyright 2022, American Chemical Society. (D) Schematic illustration of the antibacterial mechanism of a peptide–photosensitizer conjugate toward Gram-negative bacteria. Reproduced with permission.84 Copyright 2024, © 2023 American Chemical Society. (E) The preparation of the functional titanium surface. (i) The functional surface could adapt throughout the service life to exhibit specific biofunctions. (ii) and (iii) Schematic illustration of the preparation process of the functional titanium implant. Reproduced with permission.9 Copyright 2023, Elsevier Ltd. | ||
In addition to antibacterial activity, LL-37 can inhibit the self-assembly of islet amyloid protein, thereby alleviating the degeneration of islet β cells and improving the pathogenesis of type II diabetes.81 Moreover, LL-37 also inhibits αS-amyloid protein aggregation and mitigates αS-oligomers-induced cell damage, attributed to the α-helical structure, amphiphilic, and positively charged properties of LL-37.85 LL-37 can activate the formyl peptide receptor-like 1 receptor, thereby promoting angiogenesis. So, it also plays a role in wound healing.86
3.3 The advantages of AMPs
Antimicrobial peptides come from various sources and have evolved along with the evolution of bacteria,87 as shown in Table 1. The resident microbiome of the human body is an important producer of AMPs, and the AMPs produced by it can effectively inhibit their competitors, including drug-resistant bacteria, playing an important role in maintaining the microbial ecological balance.88 The current consensus is that traditional antibiotic resistance mechanisms do not impact the activity of antimicrobial peptides.14 Bacteria develop resistance to antimicrobial peptides much more slowly than other antibiotics.723.4 The challenges in the application of AMPs
The main limitations in the application of antimicrobial peptides are high production cost, acid resistance, easy degradation by protease in vivo, short half-life in blood volume and instability,68 and hemolytic toxicity.89 Due to their special folding patterns and bactericidal properties, antimicrobial peptides are often challenging to produce in large quantities by chemical or recombinant methods.74 Host factors such as saline concentration, apolipoproteins, DNA, host proteins, glycosaminoglycans,15 and lipid droplets90 affect the bactericidal activity of antimicrobial peptides, including LL-37.15 In addition, one of the main reasons AMP is less effective in vivo than in vitro is its poor absorption.14 In addition to paying attention to its biological activity, we still need to note that about 40% of AMPs in clinical trials are from humans. But once the human body develops resistance to these peptides, it may induce immune disorders, so we should be cautious when using AMPs.75Ngambenjawong et al. designed a special albumin binding domain (ABD)–AMP conjugate, which consists of four essential structures, namely ABD, anion block, protease cleavage linker, and AMP. It not only prolongs the circulation time of AMP (the conjugate's size ensures that the kidneys cannot filter it out) but also activates AMP by proteolytic cleavage of the ABD linker chain in infected organs.83 The hydrophilic cationic AMP-grafted polymer (PAMP) proposed by Kang et al., along with the hydrophobic reactive oxygen species (ROS) sensitive phenylboronic ester-grafted polymer (PPBE), presents a well-designed approach for cladding AMP, effectively preventing damage to cell membranes during circulation and enabling degradation and exposure of AMP in ROS-rich environments, thereby enhancing its therapeutic efficacy.89 Conversely, using cell-penetrating peptides (CPPs) as carriers for compounds across cell membranes can deliver a variety of antibacterial compounds, such as phosphorodiamidate morpholino oligomers (PMOs), to increase drug absorption91 (Fig. 5D).
3.5 Development and application of AI-enabled AMPs
The identification of antibacterial peptides with antimicrobial activity from organisms remains a significant challenge in the production and application of such peptides. AI technology has very considerable potential for application in screening potential antimicrobial peptides. It is possible to discover potential antimicrobial peptides through AI, thereby significantly reducing the production costs associated with their development.95 Furthermore, this approach eliminates the need for traditional methods of obtaining new antimicrobial peptides through amino acid insertion or modification based on existing AMPs, thus enhancing the diversity within this class of peptides.72At present, big data analysis is widely used in antimicrobial peptide mining,64 such as Conditional Latent (attribute) Space Sampling (CLaSS),96 sequential model ensemble pipeline (SMEP),97 combination of deep learning models and cell–free protein synthesis (CFPS),95 HydrAMP.72 The main process involves selecting a gene cluster from the Human Microbiome Project database, reconstructing it, and finally expressing it in E. coli and screening the expression product activity using human commensal bacteria and pathogens.88
4. Application of AMPs in the treatment of infected bone defects
In the study of Zhou et al., a novel thermoresponsive peptide implant, developed by combining AMP HHC36 with polyethylene glycol (PEG)–based thermoresponsive polymer (MPEG) and titanium, demonstrates responsiveness to different microenvironments. At 25 °C, the implant exhibits an anti-contamination function by relaxing the MPEG chain and inhibiting biofilm formation while maintaining AMP coverage. At 37 °C, the implant's anti-infection activity decreases due to the partial collapse of the MPEG chain; however, it represents good biocompatibility and osteogenic activity. In an infected environment where the local temperature rises to 42 °C, further collapse of MPEG occurs, exposing AMP and restoring its antibacterial activity (Fig. 5E). Furthermore, this change in biological activity is reversible and allows for good adaptation to changes in the microenvironment.9Another promising strategy entails the synthesis of mesoporous silica nanoparticles (MSNs) characterized by a diselenide-bridged architecture, which serve as carriers for the subsequent loading of AMPs.98 The resulting nanomaterial can be utilized as a coating on titanium alloy implants, effectively synergizing the osteoinductive properties of selenium embedded within the porous nanoceramic matrix with the antibacterial potency of AMPs.98 This innovative combination is designed to inhibit bacterial adhesion while fostering normal tissue proliferation and differentiation, thereby reducing bacterial competitiveness relative to normal tissue cells in adhering to the implant surface and promoting rapid osseointegration alongside mitigating implant-associated infections.
5. Conclusion and perspectives
This paper provides a comprehensive review of the infection and repair mechanisms of infective bone defects, along with various treatment methods, focusing mainly on the current research advancements in antibacterial 3D scaffolds and novel antibacterial therapy AMPs. By combining the action of osteogenic factors, the 3D structure offers optimal support for the skeleton structure and microenvironment for cell adhesion and proliferation, thereby enhancing bone repair efficacy. This unique structural advantage has made 3D printing a hot topic for research and has the potential to expand to more dimensions. The introduction of antibacterial biomaterials presents a novel approach to infection control and prevention of chronic infections. Currently, metal ion-based 3D scaffolds are widely utilized as antibacterial implantable scaffolds due to their inherent ability to promote osteogenic differentiation and create an alkaline microenvironment that inhibits bacterial growth. However, it should be noted that metal ions exhibit relatively weak antibacterial effects. In contrast, AMPs have emerged as promising therapeutic agents for combating bacterial infections. Notably, AMPs possess diverse sources, and their unique mechanism of action results in slower development of resistance than antibiotics. They also indirectly regulate host immune function against bacteria.In contrast to the conventional vancomycin treatment regimens currently utilized in clinical settings, the antibiotic-loaded AMP anti-infection stent presents a more targeted therapeutic approach, effectively minimizing both the volume of medication and dosage required. This strategy mitigates adverse effects associated with prolonged or high-dose antibiotic administration, including disruption of human microecology and compromised fracture healing. Moreover, the modulation of host immune system functionality, combined with the intrinsic biological properties of AMP, demonstrates particular efficacy against complex infections induced by drug-resistant bacteria. Furthermore, this innovative stent significantly curtails infections arising from exogenous non-vascular materials while effectively inhibiting biofilm formation, thereby providing a new strategy for dealing with chronic infections.
In recent years, significant progress has been made by researchers in developing diverse strategies to address the hemolytic toxicity associated with AMPs and enhance their pharmacokinetics. One such strategy involves the utilization of nanocarriers, which are minute particles capable of encapsulating AMPs and delivering them specifically to targeted sites within the body. This approach not only safeguards AMPs from degradation but also enables controlled release, ensuring optimal therapeutic efficacy. Another promising technique is amino acid modification, wherein specific amino acids within AMPs are altered or substituted to improve their stability and reduce toxicity. By carefully selecting and modifying these amino acids, scientists have successfully enhanced the overall safety profile of AMPs while retaining their potent antimicrobial activity. These advancements in mitigating hemolytic toxicity and improving pharmacokinetics have significantly expanded the clinical potential of AMPs. With reduced side effects and enhanced drug delivery capabilities, these modified peptides hold great promise as a new generation of antibiotics. They offer a potential solution for combating antibiotic resistance by targeting bacterial membranes through multiple mechanisms of action. Furthermore, the development of novel formulations incorporating nanotechnology has opened up possibilities for targeted therapy against specific pathogens or infections. By engineering nanocarriers with surface modifications that enable selective binding to bacteria or infected cells, researchers aim to maximize treatment efficacy while minimizing off-target effects on healthy tissues.
In addition to its role in screening and discovering new AMP candidates, artificial intelligence (AI) technology has revolutionized the drug development process. With its ability to rapidly and accurately analyze vast amounts of data, AI has significantly expedited the identification of potential drug targets and enhanced lead optimization efficiency. Furthermore, AI algorithms can be utilized for predicting the pharmacokinetic properties of AMPs, including absorption, distribution, metabolism, and excretion. This predictive capability enables researchers to prioritize promising candidates for further investigation while minimizing costly experimental failures. Additionally, AI-powered platforms have facilitated the integration of diverse data sources from various disciplines. By combining information from genomics, proteomics, metabolomics, and clinical studies with computational models driven by machine learning algorithms, scientists can gain a comprehensive understanding of AMPs' mechanisms of action and therapeutic potential. As research progresses and databases continue to expand with new experimental results and clinical data on AMPs' efficacy and safety profiles across different populations or disease conditions, AI will play an increasingly vital role in optimizing treatment strategies tailored to individual patients. The ability to personalize medicine based on patient-specific characteristics will not only enhance therapeutic outcomes but also reduce adverse effects associated with traditional broad-spectrum antibiotics. Overall, the application of AI technology in screening new AMP candidates represents just one aspect of its transformative impact on antimicrobial research. As we delve deeper into this field's complexities through interdisciplinary collaborations between computer scientists, biologists, and clinicians, the full potential of AI in accelerating the discovery and development of novel AMPs will be realized.
In the future, there is a potential trend towards systemic targeted delivery of AMPs, combined with AI and 3D biomaterial scaffolds for localized application. This emerging approach holds great promise in addressing the challenges associated with treating infectious bone defects. 3D printing technology can create scaffolds with more intricate microstructures, which can change the microstructure not only to optimize the physical and chemical properties of the material itself, such as making the scaffold more wear-resistant, but also to alter the delivery for AMPs and make AMP controlled release possible. By utilizing systemic targeted delivery, AMPs can be specifically directed to the site of infection within the bone, maximizing their effectiveness while minimizing potential side effects. The use of AI further enhances this precision by analyzing patient-specific data and optimizing treatment strategies based on individual characteristics such as infection severity, immune response, and drug resistance patterns. Moreover, incorporating 3D biomaterial scaffolds into this therapeutic approach offers several advantages. These scaffolds provide a supportive structure that mimics the natural environment of bone tissue, promoting cell growth and regeneration. They also allow for controlled release of AMPs over an extended period, ensuring sustained antimicrobial activity at the site of infection. The combination of these advanced technologies has the potential to revolutionize the treatment of infectious bone defects. By tailoring therapy to each patient's unique needs through targeted delivery and personalized AI algorithms, healthcare professionals can optimize outcomes while minimizing adverse effects. Furthermore, this approach may have broader implications beyond infectious bone defects. In particular, the advent of 3D printing technology has enabled contemporary medical practices to fabricate complex structures specifically tailored to meet the individualized needs of patients. This methodology effectively addresses the distinct characteristics inherent in each injury or defect, thereby unveiling novel avenues for medical treatment. Notably, it provides more efficient, customizable, and effective solutions to tackle the unique challenges posed by various tissue repair and regeneration protocols. The integration of AI and 3D biomaterial scaffolds could potentially be applied to other areas in regenerative medicine or tissue engineering where precise drug delivery is crucial for successful treatment outcomes.
In the future, with the development and advancement of multidimensional 3D/4D/5D/6D printing technology, the integration of temporal dimensions into bone scaffold materials will enable dynamic adjustments to external conditions and specific functionalities, thereby enhancing adaptability to changes in the microenvironment.99 Expanding upon this foundation, 5D/6D technology significantly enhances product precision, facilitating the production of more intricate structures, functions, and personalized products. For instance, flexible materials exhibit superior flexibility and malleability while providing stable support that seamlessly conforms to the shape and movement functions of human bones. If combined with wearable technology for real-time monitoring of the skeletal system, it would greatly benefit diagnosis and treatment procedures for patients. With the continuous advancements in 4D/5D/6D technology and its integration into bone scaffold materials, we anticipate remarkable progress in the field of regenerative medicine. These technological developments have opened up new possibilities for enhancing tissue regeneration and promoting healing processes. One potential application lies in the fabrication of smart scaffolds that can dynamically respond to physiological cues within the body. By incorporating 4D printing techniques, these scaffolds can be designed to change their shape or release specific growth factors at different stages of tissue regeneration. This personalized approach has the potential to significantly improve patient outcomes by providing tailored support for each individual's unique healing process. Furthermore, with the integration of 5D/6D technology, it becomes possible to incorporate additional functionalities into bone scaffold materials. For instance, sensors embedded within the scaffolds could monitor key parameters such as pH levels or oxygen concentration, allowing real-time feedback on tissue health and guiding treatment decisions. Additionally, advanced imaging techniques enabled by these technologies could provide detailed insights into tissue development and help researchers better understand how regenerative processes occur at a cellular level. Moreover, advancements in 4D/5D/6D technology offer exciting prospects for biofabrication methods used in regenerative medicine. The ability to precisely control material properties at multiple dimensions allows for more accurate replication of native tissues' complex structures and functions. This opens up avenues for creating biomimetic scaffolds that closely mimic natural tissues' mechanical properties and biochemical signaling cues necessary for successful regeneration.
Data availability
No primary research results, software, or code have been included, and no new data were generated or analyzed as part of this review.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by the National Natural Science Fund of China (no. 82202726); the National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Z20192013); Key research and development project of Sichuan Science and Technology Department (2023YFG0219); “Zero to One” Innovation Research Project of Sichuan University (2022SCUH0014); Frontiers Medical Center, Tianfu Jincheng Laboratory Foundation (TFJC2023010001).References
- D. B. Raina, L. M. Matuszewski, C. Vater, J. Bolte, H. Isaksson, L. Lidgren, M. Tägil and S. Zwingenberger, Sci. Adv., 2020, 6, eabc1779 CrossRef CAS.
- Y. Bian, X. Cai, Z. Lv, Y. Xu, H. Wang, C. Tan, R. Liang and X. Weng, Adv. Sci., 2023, 10, e2301806 CrossRef.
- D. S. Kim, J. K. Lee, J. H. Kim, J. Lee, D. S. Kim, S. An, S. B. Park, T. H. Kim, J. S. Rim, S. Lee and D. K. Han, Sci. Adv., 2021, 7, eabj1083 CrossRef CAS PubMed.
- S. Chen, H. Wang, V. L. Mainardi, G. Talò, A. McCarthy, J. V. John, M. J. Teusink, L. Hong and J. Xie, Sci. Adv., 2021, 7, eabg3089 CrossRef CAS PubMed.
- Y. Zhu, H. Liang, X. Liu, J. Wu, C. Yang, T. M. Wong, K. Y. H. Kwan, K. M. C. Cheung, S. Wu and K. W. K. Yeung, Sci. Adv., 2021, 7, eabf6654 CrossRef CAS.
- S. Vermeulen, F. Honig, A. Vasilevich, N. Roumans, M. Romero, A. Dede Eren, U. Tuvshindorj, M. Alexander, A. Carlier, P. Williams, J. Uquillas and J. de Boer, Adv. Mater., 2021, 33, e2102084 CrossRef.
- T. Li, Z. Ma, Y. Zhang, Z. Yang, W. Li, D. Lu, Y. Liu, L. Qiang, T. Wang, Y. Ren, W. Wang, H. He, X. Zhou, Y. Mao, J. Zhu, J. Wang, X. Chen and K. Dai, Adv. Sci., 2023, 10, e2205059 CrossRef.
- C. E. Vantucci, H. Ahn, T. Fulton, M. L. Schenker, P. Pradhan, L. B. Wood, R. E. Guldberg, K. Roy and N. J. Willett, Biomaterials, 2021, 264, 120405 CrossRef CAS.
- H. Zhou, S. Ye, M. Xu, L. Hao, J. Chen, Z. Fang, K. Guo, Y. Chen and L. Wang, Biomaterials, 2023, 301, 122200 CrossRef CAS.
- W. Li, F. Separovic, N. M. O'Brien Simpson and J. D. Wade, Chem. Soc. Rev., 2021, 50, 4932–4973 RSC.
- J. Portelinha, S. S. Duay, S. I. Yu, K. Heilemann, M. D. J. Libardo, S. A. Juliano, J. L. Klassen and A. M. Angeles Boza, Chem. Rev., 2021, 121, 2648–2712 CrossRef CAS PubMed.
- S. M. Häffner, E. Parra Ortiz, K. L. Browning, E. Jørgensen, M. W. A. Skoda, C. Montis, X. Li, D. Berti, D. Zhao and M. Malmsten, ACS Nano, 2021, 15, 6787–6800 CrossRef PubMed.
- I. Sugrue, R. P. Ross and C. Hill, Nat. Rev. Microbiol., 2024, 22, 556–571 CrossRef CAS.
- R. E. W. Hancock, M. A. Alford and E. F. Haney, Nat. Rev. Microbiol., 2021, 19, 786–797 CrossRef CAS PubMed.
- N. Mookherjee, M. A. Anderson, H. P. Haagsman and D. J. Davidson, Nat. Rev. Drug Discovery, 2020, 19, 311–332 CrossRef CAS PubMed.
- B. H. Gan, J. Gaynord, S. M. Rowe, T. Deingruber and D. R. Spring, Chem. Soc. Rev., 2021, 50, 7820–7880 RSC.
- Z. Li, D. He, B. Guo, Z. Wang, H. Yu, Y. Wang, S. Jin, M. Yu, L. Zhu, L. Chen, C. Ding, X. Wu, T. Wu, S. Gong, J. Mao, Y. Zhou, D. Luo and Y. Liu, Nat. Commun., 2023, 14, 6963 CrossRef CAS PubMed.
- L. Ye, J. Xu, J. Mi, X. He, Q. Pan, L. Zheng, H. Zu, Z. Chen, B. Dai, X. Li, Q. Pang, L. Zou, L. Zhou, L. Huang, W. Tong, G. Li and L. Qin, Biomaterials, 2021, 275, 120984 CrossRef.
- B. Wildemann, A. Ignatius, F. Leung, L. A. Taitsman, R. M. Smith, R. Pesántez, M. J. Stoddart, R. G. Richards and J. B. Jupiter, Nat. Rev. Dis. Primers, 2021, 7, 57 CrossRef.
- S. Hochmann, K. Ou, R. Poupardin, M. Mittermeir, M. Textor, S. Ali, M. Wolf, A. Ellinghaus, D. Jacobi, J. A. J. Elmiger, S. Donsante, M. Riminucci, R. Schäfer, U. Kornak, O. Klein, K. Schallmoser, K. Schmidt Bleek, G. N. Duda, J. K. Polansky, S. Geissler and D. Strunk, Sci. Transl. Med., 2023, 15, eabm7477 CrossRef CAS PubMed.
- H. S. Shu, Y. L. Liu, X. T. Tang, X. S. Zhang, B. Zhou, W. Zou and B. O. Zhou, Cell Stem Cell, 2021, 28, 2122–2136.e2123 CrossRef CAS.
- M. Qiu, C. Li, Z. Cai, C. Li, K. Yang, N. Tulufu, B. Chen, L. Cheng, C. Zhuang, Z. Liu, J. Qi, W. Cui and L. Deng, Adv. Sci., 2023, 10, e2303650 CrossRef PubMed.
- Q. Li, H. Yu, F. Zhao, C. Cao, T. Wu, Y. Fan, Y. Ao and X. Hu, Adv. Sci., 2023, 10, e2303650 CrossRef.
- Y. Hu, H. Tian, W. Chen, Y. Liu, Y. Cao, H. Pei, C. Ming, C. Shan, X. Chen, Z. Dai, S. Yang, Z. Shao, S. Lan, Y. Liu and W. Tong, Adv. Sci., 2023, 10, e2303375 CrossRef.
- J. M. Sadowska, R. N. Power, K. J. Genoud, A. Matheson, A. González Vázquez, L. Costard, K. Eichholz, P. Pitacco, T. Hallegouet, G. Chen, C. M. Curtin, C. M. Murphy, B. Cavanagh, H. Zhang, D. J. Kelly, A. R. Boccaccini and F. J. O'Brien, Adv. Mater., 2024, 36, e2307639 CrossRef.
- Y. Okuchi, J. Reeves, S. S. Ng, D. H. Doro, S. Junyent, K. J. Liu, A. J. El Haj and S. J. Habib, Nat. Mater., 2021, 20, 108–118 CrossRef CAS.
- L. Zheng, D. Zhou, F. Ju, Z. Liu, C. Yan, Z. Dong, S. Chen, L. Deng, S. Chan, J. Deng and X. Zhang, Adv. Sci., 2023, 10, e2204592 CrossRef.
- D. Baek, K. H. Park, K. M. Lee, S. Jung, S. Joung, J. Kim and J. W. Lee, Cell Death Dis., 2021, 12, 238 CrossRef CAS.
- L. Biswas, J. Chen, J. De Angelis, A. Singh, C. Owen Woods, Z. Ding, J. M. Pujol, N. Kumar, F. Zeng, S. K. Ramasamy and A. P. Kusumbe, Cell, 2023, 186, 382–397.e324 CrossRef CAS.
- C. Li, W. Zhang, Y. Nie, X. Du, C. Huang, L. Li, J. Long, X. Wang, W. Tong, L. Qin and Y. Lai, Adv. Mater., 2024, 36, e2308875 CrossRef.
- S. Pigeot, T. Klein, F. Gullotta, S. J. Dupard, A. Garcia Garcia, A. García García, S. Prithiviraj, P. Lorenzo, M. Filippi, C. Jaquiery, L. Kouba, M. A. Asnaghi, D. B. Raina, B. Dasen, H. Isaksson, P. Önnerfjord, M. Tägil, A. Bondanza, I. Martin and P. E. Bourgine, Adv. Mater., 2021, 33, e2103737 CrossRef.
- S. Lin, H. Maekawa, S. Moeinzadeh, E. Lui, H. V. Alizadeh, J. Li, S. Kim, M. Poland, B. C. Gadomski, J. T. Easley, J. Young, M. Gardner, D. Mohler, W. J. Maloney and Y. P. Yang, Nat. Commun., 2023, 14, 4455 CrossRef CAS PubMed.
- Y. Wei, H. Pan, J. Yang, C. Zeng, W. Wan and S. Chen, Sci. Adv., 2024, 10, eadk6722 CrossRef CAS PubMed.
- C. T. Johnson, M. C. P. Sok, K. E. Martin, P. P. Kalelkar, J. D. Caplin, E. A. Botchwey and A. J. García, Sci. Adv., 2019, 5, eaaw1228 CrossRef CAS.
- R. E. De La Vega, M. van Griensven, W. Zhang, M. J. Coenen, C. V. Nagelli, J. A. Panos, C. J. Peniche Silva, J. Geiger, C. Plank, C. H. Evans and E. R. Balmayor, Sci. Adv., 2022, 8, eabl6242 CrossRef CAS PubMed.
- T. Wang, H. Ouyang, Y. Luo, J. Xue, E. Wang, L. Zhang, Z. Zhou, Z. Liu, X. Li, S. Tan, Y. Chen, L. Nan, W. Cao, Z. Li, F. Chen and L. Zheng, Sci. Adv., 2024, 10, eadi6799 CrossRef CAS.
- D. S. Sparks, F. M. Savi, C. E. Dlaska, S. Saifzadeh, G. Brierly, E. Ren, A. Cipitria, J. C. Reichert, M. L. Wille, M. A. Schuetz, N. Ward, M. Wagels and D. W. Hutmacher, Sci. Adv., 2023, 9, eadd6071 CrossRef CAS.
- J. Fu, Y. Li, Y. Zhang, Y. Liang, Y. Zheng, Z. Li, S. Zhu, C. Li, Z. Cui and S. Wu, Adv. Mater., 2021, 33, e2102926 CrossRef PubMed.
- J. Chen, G. Hu, T. Li, Y. Chen, M. Gao, Q. Li, L. Hao, Y. Jia, L. Wang and Y. Wang, Biomaterials, 2021, 264, 120446 CrossRef CAS PubMed.
- D. Rütsche, M. Nanni, S. Rüdisser, T. Biedermann and M. Zenobi Wong, Adv. Mater., 2023, 35, e2209476 CrossRef PubMed.
- R. Yang, G. Li, C. Zhuang, P. Yu, T. Ye, Y. Zhang, P. Shang, J. Huang, M. Cai, L. Wang, W. Cui and L. Deng, Sci. Adv., 2021, 7, eabg3816 CrossRef CAS PubMed.
- D. Zhang, J. Tan, R. Xu, H. Du, J. Xie, F. Peng and X. Liu, Small, 2023, 19, e2204852 CrossRef.
- Z. Yang, X. W. Zhang, F. F. Zhuo, T. T. Liu, Q. W. Luo, Y. Z. Zheng, L. Li, H. Yang, Y. C. Zhang, Y. H. Wang, D. Liu, P. F. Tu and K. W. Zeng, Adv. Sci., 2023, 10, e2206533 CrossRef PubMed.
- J. Suh, N. K. Kim, W. Shim, S. H. Lee, H. J. Kim, E. Moon, H. Sesaki, J. H. Jang, J. E. Kim and Y. S. Lee, Cell Metab., 2023, 35, 345–360.e347 CrossRef CAS PubMed.
- J. Xu, Z. Li, R. J. Tower, S. Negri, Y. Wang, C. A. Meyers, T. Sono, Q. Qin, A. Lu, X. Xing, E. F. McCarthy, T. L. Clemens and A. W. James, Sci. Adv., 2022, 8, eabl5716 CrossRef CAS PubMed.
- J. Long, Z. Yao, W. Zhang, B. Liu, K. Chen, L. Li, B. Teng, X. F. Du, C. Li, X. F. Yu, L. Qin and Y. Lai, Adv. Sci., 2023, 10, e2302539 CrossRef PubMed.
- H. Wei, J. Cui, K. Lin, J. Xie and X. Wang, Bone Res., 2022, 10, 17 CrossRef CAS.
- H. Wang, J. Tian, Y. Jiang, S. Liu, J. Zheng, N. Li, G. Wang, F. Dong, J. Chen, Y. Xie, Y. Huang, X. Cai, X. Wang, W. Xiong, H. Qi, L. Yin, Y. Wang and X. Sheng, Sci. Adv., 2023, 9, eabq7750 CrossRef CAS.
- L. Tong, X. Pu, Q. Liu, X. Li, M. Chen, P. Wang, Y. Zou, G. Lu, J. Liang, Y. Fan, X. Zhang and Y. Sun, Adv. Sci., 2023, 10, e2300038 CrossRef.
- G. L. Koons, M. Diba and A. G. Mikos, Nat. Rev. Mater., 2020, 5, 584–603 CrossRef CAS.
- S. E. Eldridge, A. Barawi, H. Wang, A. J. Roelofs, M. Kaneva, Z. Guan, H. Lydon, B. L. Thomas, A. S. Thorup, B. F. Fernandez, S. Caxaria, D. Strachan, A. Ali, K. Shanmuganathan, C. Pitzalis, J. R. Whiteford, F. Henson, A. W. McCaskie, C. De Bari and F. Dell’Accio, Sci. Transl. Med., 2020, 12, eaax9086 CrossRef CAS.
- X. Wang, A. Liu, Z. Zhang, D. Hao, Y. Liang, J. Dai, X. Jin, H. Deng, Y. Zhao, P. Wen and Y. Li, Adv. Sci., 2024, 11, e2307329 CrossRef.
- S. Herberg, A. M. McDermott, P. N. Dang, D. S. Alt, R. Tang, J. H. Dawahare, D. Varghai, J. Y. Shin, A. McMillan, A. D. Dikina, F. He, Y. B. Lee, Y. Cheng, K. Umemori, P. C. Wong, H. Park, J. D. Boerckel and E. Alsberg, Sci. Adv., 2019, 5, eaax2476 CrossRef CAS PubMed.
- C. S. Lee, S. Kim, J. Fan, H. S. Hwang, T. Aghaloo and M. Lee, Sci. Adv., 2020, 6, eaaz7822 CrossRef CAS.
- K. Zhang, Y. Zhou, C. Xiao, W. Zhao, H. Wu, J. Tang, Z. Li, S. Yu, X. Li, L. Min, Z. Yu, G. Wang, L. Wang, K. Zhang, X. Yang, X. Zhu, C. Tu and X. Zhang, Sci. Adv., 2019, 5, eaax6946 CrossRef CAS PubMed.
- E. A. Masters, B. F. Ricciardi, K. L. D. M. Bentley, T. F. Moriarty, E. M. Schwarz and G. Muthukrishnan, Nat. Rev. Microbiol., 2022, 20, 385–400 CrossRef CAS PubMed.
- X. Chen, Q. He, Q. Zhai, H. Tang, D. Li, X. Zhu, X. Zheng, G. Jian, R. D. Cannon, L. Mei, S. Wang, P. Ji, J. Song and T. Chen, ACS Nano, 2023, 17, 22960–22978 CrossRef CAS PubMed.
- E. V. K. Ledger, S. Mesnage and A. M. Edwards, Nat. Commun., 2022, 13, 2041 CrossRef CAS.
- J. Li, J. Ma, H. Sun, M. Yu, H. Wang, Q. Meng, Z. Li, D. Liu, J. Bai, G. Liu, X. Xing, F. Han and B. Li, Sci. Adv., 2023, 9, eadf8645 CrossRef CAS PubMed.
- L. Wang, Q. Yang, M. Huo, D. Lu, Y. Gao, Y. Chen and H. Xu, Adv. Mater., 2021, 33, e2100150 CrossRef.
- C. Pineda Molina, G. S. Hussey, A. Liu, J. Eriksson, W. A. D'Angelo and S. F. Badylak, Biomaterials, 2021, 267, 120493 CrossRef CAS.
- T. Sneideris, N. A. Erkamp, H. Ausserwöger, K. L. Saar, T. J. Welsh, D. Qian, K. Katsuya Gaviria, M. L. L. Y. Johncock, G. Krainer, A. Borodavka and T. P. J. Knowles, Nat. Commun., 2023, 14, 7170 CrossRef CAS PubMed.
- Q. He, Z. Yang, Z. Zou, M. Qian, X. Wang, X. Zhang, Z. Yin, J. Wang, X. Ye, D. Liu and M. Guo, Adv. Sci., 2023, 10, e2205301 Search PubMed.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez, A. Ioannidis, M. A. Giulianotti, Y. Apidianakis, S. Bradfute, A. L. Ferguson, A. Cherkasov, M. N. Seleem, C. Pinilla, C. de la Fuente Nunez, T. Lazaridis, T. Dai, R. A. Houghten, R. E. W. Hancock and G. P. Tegos, Lancet Infect. Dis., 2020, 20, e216–e230 CrossRef CAS PubMed.
- D. Nagarajan, N. Roy, O. Kulkarni, N. Nanajkar, A. Datey, S. Ravichandran, C. Thakur, T. Sandeep, I. V. Aprameya, S. P. Sarma, D. Chakravortty and N. Chandra, Sci. Adv., 2019, 5, eaax1946 CAS.
- X. Zhang, S. Li, H. Luo, S. He, H. Yang, L. Li, T. Tian, Q. Han, J. Ye, C. Huang, A. Liu and Y. Jiang, Signal Transduction Targeted Ther., 2022, 7, 245 CrossRef CAS PubMed.
- C. Zampaloni, P. Mattei, K. Bleicher, L. Winther, C. Thäte, C. Bucher, J. M. Adam, A. Alanine, K. E. Amrein, V. Baidin, C. Bieniossek, C. Bissantz, F. Boess, C. Cantrill, T. Clairfeuille, F. Dey, P. Di Giorgio, P. du Castel, D. Dylus, P. Dzygiel, A. Felici, F. García Alcalde, A. Haldimann, M. Leipner, S. Leyn, S. Louvel, P. Misson, A. Osterman, K. Pahil, S. Rigo, A. Schäublin, S. Scharf, P. Schmitz, T. Stoll, A. Trauner, S. Zoffmann, D. Kahne, J. A. T. Young, M. A. Lobritz and K. A. Bradley, Nature, 2024, 625, 566–571 CAS.
- S. Chaudhary, Z. Ali, M. Tehseen, E. F. Haney, A. Pantoja Angles, S. Alshehri, T. Wang, G. J. Clancy, M. Ayach, C. Hauser, P. Y. Hong, S. M. Hamdan, R. E. W. Hancock and M. Mahfouz, Nat. Commun., 2023, 14, 1464 CrossRef CAS PubMed.
- K. Mangano, D. Klepacki, I. Ohanmu, C. Baliga, W. Huang, A. Brakel, A. Krizsan, Y. S. Polikanov, R. Hoffmann, N. Vázquez Laslop and A. S. Mankin, Nat. Chem. Biol., 2023, 19, 1082–1090 CrossRef CAS PubMed.
- B. O. Torres Salazar, T. Dema, N. A. Schilling, D. Janek, J. Bornikoel, A. Berscheid, A. M. A. Elsherbini, S. Krauss, S. J. Jaag, M. Lämmerhofer, M. Li, N. Alqahtani, M. J. Horsburgh, T. Weber, J. M. Beltrán Beleña, H. Brötz Oesterhelt, S. Grond, B. Krismer and A. Peschel, Nat. Microbiol., 2024, 9, 200–213 CrossRef CAS PubMed.
- J. Pierre, B. Peters, D. La Torre, A. Sidebottom, Y. Tao, X. Zhu, C. Cham, L. Wang, A. Kambal, K. Harris, J. Silva, O. Zaborina, J. Alverdy, H. Herzog, J. Witchley, S. Noble, V. Leone and E. J. S. Chang, Science, 2023, 381, 502–508 CrossRef CAS PubMed.
- P. Szymczak, M. Możejko, T. Grzegorzek, R. Jurczak, M. Bauer, D. Neubauer, K. Sikora, M. Michalski, J. Sroka, P. Setny, W. Kamysz and E. Szczurek, Nat. Commun., 2023, 14, 1453 CrossRef CAS PubMed.
- J. T. Mhlongo, A. Y. Waddad, F. Albericio and B. G. de la Torre, Adv. Sci., 2023, 10, e2300472 CrossRef.
- X. Gao, J. Ding, C. Liao, J. Xu, X. Liu and W. Lu, Adv. Drug Delivery Rev., 2021, 179, 114008 CrossRef CAS.
- B. P. Lazzaro, M. Zasloff and J. Rolff, Science, 2020, 368, eaau5480 CrossRef CAS.
- D. Minns, K. J. Smith, V. Alessandrini, G. Hardisty, L. Melrose, L. Jackson Jones, A. S. MacDonald, D. J. Davidson and E. Gwyer Findlay, Nat. Commun., 2021, 12, 1285 CrossRef CAS PubMed.
- Y. Engelberg and M. Landau, Nat. Commun., 2020, 11, 3894 CrossRef CAS PubMed.
- L. Hong, M. Gontsarik, H. Amenitsch and S. Salentinig, Small, 2021, 18, e2104211 CrossRef.
- M. P. Hoelscher, J. Forner, S. Calderone, C. Krämer, Z. Taylor, F. V. Loiacono, S. Agrawal, D. Karcher, F. Moratti, X. Kroop and R. Bock, Nat. Commun., 2022, 13, 5856 CrossRef CAS.
- T. Doolin, H. M. Amir, L. Duong, R. Rosenzweig, L. A. Urban, M. Bosch, A. Pol, S. P. Gross and A. Siryaporn, Nat. Commun., 2020, 11, 3888 CrossRef CAS.
- V. Armiento, K. Hille, D. Naltsas, J. S. Lin, A. E. Barron and A. Kapurniotu, Angew. Chem., Int. Ed., 2020, 59, 12837–12841 CrossRef CAS PubMed.
- S. Jin, R. Yang, C. Hu, S. Xiao, Y. Zuo, Y. Man, Y. Li and J. Li, ACS Appl. Mater. Interfaces, 2023, 15, 7804–7820 CrossRef CAS PubMed.
- C. Ngambenjawong, L. W. Chan, H. E. Fleming and S. N. Bhatia, ACS Nano, 2022, 16, 15779–15791 CrossRef CAS PubMed.
- W. Zhou, L. Chen, H. Li, M. Wu, M. Liang, Q. Liu, W. Wu, X. Jiang and X. Zhen, ACS Nano, 2024, 18(30), 19771–19782, DOI:10.1021/acsnano.4c05443.
- J. Santos, P. Gracia, S. Navarro, S. Peña Díaz, J. Pujols, N. Cremades, I. Pallarès and S. Ventura, Nat. Commun., 2021, 12, 3752 CrossRef CAS.
- J. V. John, N. S. Sharma, G. Tang, Z. Luo, Y. Su, S. Weihs, S. M. S. Shahriar, G. Wang, A. McCarthy, J. Dyke, Y. S. Zhang, A. Khademhosseini and J. Xie, Adv. Funct. Mater., 2022, 33, 2206936 CrossRef.
- M. A. Hanson, L. Grollmus and B. Lemaitre, Science, 2023, 381, eadg5725 CrossRef CAS.
- A. M. King, Z. Zhang, E. Glassey, P. Siuti, J. Clardy and C. A. Voigt, Nat. Microbiol., 2023, 8, 2420–2434 CrossRef CAS PubMed.
- Z. Kang, C. Wang, Z. Zhang, Q. Liu, Y. Zheng, Y. Zhao, Z. Pan, Q. Li, L. Shi and Y. Liu, Adv. Mater., 2022, 34, e2201945 CrossRef.
- M. Bosch, M. Sánchez Álvarez, A. Fajardo, R. Kapetanovic, B. Steiner, F. Dutra, L. Moreira, J. A. López, R. Campo, M. Marí, F. Morales Paytuví, O. Tort, A. Gubern, R. M. Templin, J. E. B. Curson, N. Martel, C. Català, F. Lozano, F. Tebar, C. Enrich, J. Vázquez, M. A. Del Pozo, M. J. Sweet, P. T. Bozza, S. P. Gross, R. G. Parton and A. Pol, Science, 2020, 370, eaay8085 CrossRef CAS.
- M. E. Schafer, H. Browne, J. B. Goldberg and D. E. Greenberg, Acc. Chem. Res., 2021, 54, 2377–2385 CrossRef CAS PubMed.
- S. Shabani, S. Hadjigol, W. Li, Z. Si, D. Pranantyo, M. B. Chan Park, N. M. O’Brien-Simpson and G. G. Qiao, Nat. Rev. Bioeng., 2024, 2, 343–361 CrossRef CAS.
- G. Li, Z. Lai and A. Shan, Adv. Sci., 2023, 10, e2206602 CrossRef.
- C. Wang, T. Hong, P. Cui, J. Wang and J. Xia, Adv. Drug Delivery Rev., 2021, 175, 113818 CrossRef CAS PubMed.
- A. Pandi, D. Adam, A. Zare, V. T. Trinh, S. L. Schaefer, M. Burt, B. Klabunde, E. Bobkova, M. Kushwaha, Y. Foroughijabbari, P. Braun, C. Spahn, C. Preußer, E. Pogge von Strandmann, H. B. Bode, H. von Buttlar, W. Bertrams, A. L. Jung, F. Abendroth, B. Schmeck, G. Hummer, O. Vázquez and T. J. Erb, Nat. Commun., 2023, 14, 7197 CrossRef CAS.
- P. Das, T. Sercu, K. Wadhawan, I. Padhi, S. Gehrmann, F. Cipcigan, V. Chenthamarakshan, H. Strobelt, C. dos Santos, P. Y. Chen, Y. Y. Yang, J. P. K. Tan, J. Hedrick, J. Crain and A. Mojsilovic, Nat. Biomed. Eng., 2021, 5, 613–623 CrossRef CAS.
- J. Huang, Y. Xu, Y. Xue, Y. Huang, X. Li, X. Chen, Y. Xu, D. Zhang, P. Zhang, J. Zhao and J. Ji, Nat. Biomed. Eng., 2023, 7, 797–810 CrossRef CAS.
- J. Dong, F. Chen, Y. Yao, C. Wu, S. Ye, Z. Ma, H. Yuan, D. Shao, L. Wang and Y. Wang, Biomaterials, 2024, 305, 122465 CrossRef CAS PubMed.
- X. Han, Q. Saiding, X. Cai, Y. Xiao, P. Wang, Z. Cai, X. Gong, W. Gong, X. Zhang and W. Cui, Nanomicro Lett., 2023, 15, 239 CAS.
| This journal is © The Royal Society of Chemistry 2025 |