Open Access Article
Open Access ArticleElectrostatic adsorptive loading of ciprofloxacin and metronidazole on chitosan nanoparticles to prolong the drug delivery process with preserved antibacterial activities: formulation and characterization
Fatema Tuj
Jahura
ab,
Farhana Khanam
Ferdousi
b,
Abu Hena
Mostofa Kamal
 c,
Anwar
Ul-Hamid
c,
Anwar
Ul-Hamid
 d,
Md. Qamrul
Ehsan
b and
Mohammad Abul
Hossain
d,
Md. Qamrul
Ehsan
b and
Mohammad Abul
Hossain
 *b
*b
aInstitute of Nuclear Science and Technology, Atomic Energy Research Establishment, Bangladesh Atomic Energy Commission, Dhaka-1349, Bangladesh
bDepartment of Chemistry, Faculty of Science, University of Dhaka, Dhaka-1000, Bangladesh. E-mail: hossainabul@yahoo.com; hossainabul@du.ac.bd
cInstitute of Food and Radiation Biology, Atomic Energy Research Establishment, Bangladesh Atomic Energy Commission, Dhaka-1349, Bangladesh
dCore Research Facilities, King Fahd University of Petroleum & Minerals, Dhahran 31261, Saudi Arabia
First published on 25th November 2024
Abstract
This study presents the formulation and evaluation of chitosan-based homogeneous nanoparticles of ciprofloxacin (CP) and metronidazole (MTZ) with improved loading efficiency to enhance the controlled release of drug within the human body as well as for the enhancement of the antibacterial activity. The drug-loaded chitosan nanoparticles (CSNPs) were prepared using deacetylated chitosan extracted from shrimp shells. The characterization of the drug-loaded CSNPs were performed by FTIR, XRD, SEM and TEM analyses. The association efficiencies of the drug-loaded CSNPs were found to be 93% ± 3% and 89% ± 3% for ciprofloxacin and metronidazole, respectively. TEM analysis confirmed the formation of homogeneous nano-sized particles of 0.1–1.0, 0.3–1.3, and 0.4–1.5 nm for CSNPs, CP-CSNPs and MTZ-CSNPs, respectively, which elucidated the adsorptive loading of the drug molecules on the surface of the chitosan nanoparticles. A comparison of the surface charges of the above nanoparticles suggested that the electrostatic adsorption dominated the drug loading process. Both of the drug-loaded CSNPs showed sustained release of drugs after an initial rapid release in the in vitro release kinetic studies. Thus, the adsorption of drugs on the surface of CSNPs resulted in the sustained release of the drugs from CSNPs without encapsulation. Positive results of the antibacterial activities of the CP-loaded CSNPs (CP-CSNPs) against both Gram-positive and Gram-negative bacteria were observed.
1. Introduction
The effectiveness of a large number of medicines is frequently limited by their ability to get to the target site for therapeutic intervention.1 In the case of a normal dosage volume, only a very small amount of the administered dose reaches the target site, while the maximum portion of the dose is dispersed in every part of the rest of the body according to its biochemical and physicochemical properties. Therefore, it is necessary to develop a drug administration system that optimizes the pharmaceutical performance of a drug, thereby reducing its toxic side effects.Controlled drug delivery increases the availability of the administered doses of drugs within the therapeutic window or at the target site for a sustained period of time. Nevertheless, this is difficult to achieve in the case of traditional drug delivery systems. In a controlled drug delivery system, drugs can be protected from degradation in vivo, which improves their half-life and also their therapeutic effects. To establish a controlled drug delivery system, the drug is incorporated into the carrier first. Then, after administration, the carrier system can release the drug over a sustained period of time, ranging from days to months.2
In the case of drug administration systems, nanoparticles have attracted much attention from pharmaceutical scientists since 1990 due to their various advantages, like controlled drug release, lesser frequency of dose administration and protection from in vivo chemical degradation of drugs.3,4 As a consequence, breathtaking improvements have been observed in patient compliance. Polymeric materials like chitosan have a long history of use in synthesizing biodegradable nanoparticles (CSNPs) for diverse applications (like biocatalyst encapsulation, wastewater treatment, food preservative and metal nanocomposites) due to their small size,5 high surface area,6 high stability and quantum size effect.7 In addition, the non-toxicity, non-antigenicity and good biocompatibility of chitosan nanoparticles (CSNPs) make them an ideal component for different biomedical and pharmaceutical applications, like molecular imprinting, pharmaceutical formulations and so on.8–13 In recent years, CSNPs have attracted particular attention for drug transportation because of their exotic ability to target specific tissues and enable controlled drug release. Many chitosan-controlled drug distribution systems have been developed in recent times through the chemical modification of functional groups in chitosan (chitosan derivatives), which ultimately upgrades the bioactivity and aqueous solubility of chitosan. However, such chemical modification does not affect the inherent properties of chitosan, but rather magnifies its scope of applications for a wide range of chemicals in multiple environments (acidic, alkaline, or neutral conditions). The administration system of many antibiotics (like ampicillin, doxycycline, and ceftriaxone) and anti-cancer drugs (such as 5-fluorouracil, paclitaxel, doxorubicin, letrozole, and saponin) have been extensively studied using CSNPs as a potential drug transporter. This research project was also focused on this field, with detailed investigations of the prolonged drug delivery process of two leading antibiotics, ciprofloxacin and metronidazole, loaded with chitosan nanoparticles. In a nutshell, the paper aimed to address the gap in the existing research on these two antibiotics.
Ciprofloxacin is a commonly used oral antimicrobial drug whose broad spectrum of activity means it has the potential to treat serious infections caused by Gram-negative and Gram-positive bacteria. However, it is a poorly cell-penetrating antibiotic since it is almost insoluble in aqueous medium and only slightly soluble in organic solvents, such as ethanol and methylene chloride.14 The problem of its lower drug solubility can be avoided and its bioavailability improved by formulating the drug in systems containing colloidal drug transporters, such as biodegradable polymeric nanoparticles. So far, various research groups have already reported ciprofloxacin-loaded different synthetic biodegradable polymeric nanoparticles for the sustained release of this antibiotic.15–17 Nevertheless, some of these biodegradable polymeric nanoparticles have concerns regarding toxicity.18 On the other hand, Jain and Banerjee19 showed that non-toxic natural polymer chitosan NPs can act as promising carriers for constant ciprofloxacin release in infective conditions in a controlled way. They prepared ciprofloxacin hydrochloride-loaded CNPs by the conventional ionotropic gelation method with tripolyphosphate anions to develop single-dose-delivery systems for the extended antibiotic release of this drug. Nevertheless, the major drawbacks of their study was the poor drug encapsulation of ciprofloxacin with CNPs (only 35.01% ± 2.66%; drug-to-carrier ratio of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; and average particle sizes of 247 ± 48 to 322 ± 52 nm for CSNPs). Later on, Ibrahim et al. (2015)20 further examined the ionic gelation of chitosan with tripolyphosphate (TPP) anions to optimize the particle size of CSNPs by varying the concentrations of chitosan and sodium (TPP) keeping the pH of the solution and ultrasonication time constant (at pH 5.5 and 45 min) for the intracellular delivery of ciprofloxacin. The results showed that the antibacterial activity of ciprofloxacin loaded with CSNPs increased with the increase in antibiotic concentration. However, the particle sizes were more favorable at the chitosan concentration of 0.2 g mL−1 (2–8 nm) at a constant concentration of TPP, while the size increased with the increase in TPP. However, the research lacked information regarding the encapsulation efficiency. Therefore, these gaps should be clarified and further detailed investigations in this area are desirable.
1; and average particle sizes of 247 ± 48 to 322 ± 52 nm for CSNPs). Later on, Ibrahim et al. (2015)20 further examined the ionic gelation of chitosan with tripolyphosphate (TPP) anions to optimize the particle size of CSNPs by varying the concentrations of chitosan and sodium (TPP) keeping the pH of the solution and ultrasonication time constant (at pH 5.5 and 45 min) for the intracellular delivery of ciprofloxacin. The results showed that the antibacterial activity of ciprofloxacin loaded with CSNPs increased with the increase in antibiotic concentration. However, the particle sizes were more favorable at the chitosan concentration of 0.2 g mL−1 (2–8 nm) at a constant concentration of TPP, while the size increased with the increase in TPP. However, the research lacked information regarding the encapsulation efficiency. Therefore, these gaps should be clarified and further detailed investigations in this area are desirable.
On the other hand, metronidazole is an amoebicide, antiprotozoan and antibiotic that is effective against anaerobic bowel amoebiasis, giardiasis, trichomoniasis, bacterial vaginosis, and surgical infections.21 The development of colon-targeted metronidazole-loaded nanoparticles has attracted attention in recent years. The prime goal of using this drug transporter is to deliver the medication exactly to the colon for achieving an efficient effect against E. histolytica. However, the administration of this drug as a classic tablet provides a minimum amount of metronidazole for local action in the colon.21,22 In fact, due to the presence of insoluble mucus hydrogel in our gastrointestinal tract, which is mainly glycoproteins made of ester sulfate and sialic acid groups, an overall strong net negative charge exists all over the gastrointestinal tract.23 Therefore, a mucoadhesive-featured chitosan may be a potential drug transporter to deliver any drug to mucosal tissues in the gastrointestinal tract. This adhesion mechanism involves hydration, hydrogen bonding, or ionic interactions.24–27 Elzatahry and Mohy Eldin (2008)28 formulated a mucoadhesive chitosan containing metronidazole (MTZ) for colon-specific delivery using the ionic gelation method and succeeded in controlling the release of MTZ over a 12 h period (CSNPs size: 280–300 nm; chitosan/TPP = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, drug-loaded nanoparticles sizes <300 nm). Nevertheless, a prominent linear relationship was observed between the chitosan/TPP weight ratio and the particle size, whereas, the mucoadhesive properties of chitosan were reduced with the increase in the concentration of antibiotic as well as the possible particle size. Therefore, further reduction of the size of the CSNPs through a modified process could be a new focus of much attention for scientists.
1, drug-loaded nanoparticles sizes <300 nm). Nevertheless, a prominent linear relationship was observed between the chitosan/TPP weight ratio and the particle size, whereas, the mucoadhesive properties of chitosan were reduced with the increase in the concentration of antibiotic as well as the possible particle size. Therefore, further reduction of the size of the CSNPs through a modified process could be a new focus of much attention for scientists.
In the present work, chitosan nanoparticles containing ciprofloxacin and metronidazole were prepared separately, using a modified ionic gelation method to reduce the homogeneous size of the CSNPs and chitosan-loaded drugs, which should ultimately enhance the efficacy of these two drugs. This new perspective enabled a more sustained release of these two antibiotics. The mechanisms of drug loading and release were established from TEM study and surface charge measurements. Accordingly, the antibacterial activity of the antibiotic drug-loaded chitosan nanoparticles were evaluated against Gram-positive and Gram-negative bacterial strains.
2. Experimental
2.1. Materials
Chitosan was extracted from waste shrimp shell using a traditional method described elsewhere in the literature.29 The physical characteristics of the obtained chitosan are summarized in Table 1. Sodium tripolyphosphate (Na5P3O10) (Na-TPP), dimethyl sulfoxide [(CH3)2SO], and di-sodium hydrogen phosphate, (Na2HPO4) were obtained from Merck Germany. Glacial acetic acid (CH3COOH) was purchased from Daejung Chemicals & Metals Co. Ltd, Korea. In order to load drugs within the chitosan nanoparticles, ciprofloxacin was obtained from Reneta Pharmaceuticals Ltd, Bangladesh. Metronidazole was provided by ACI Pharmaceuticals Ltd Bangladesh, with a potency of 99%. The bacterial strains for the antimicrobial study were collected from the Veterinary Drug Residue Analysis Division and Institute of Food and Radiation Biology, IFRB, AERE, Savar, Dhaka, Bangladesh.| Molecular formula | (C18H35N3O13)n |
|---|---|
| Molecular weight (g mol−1) | 2.3 × 105 |
| Solubility (%) | 82.85 (soluble in pH 3–4) |
| Degree of deacetylation (%) | 70 |
2.2. Preparation of the chitosan and drug-loaded chitosan nanoparticles
Chitosan nanoparticles (CSNPs) were prepared from extracted chitosan29 with minor modifications of the ionic gelation method.30 The characteristic features of the extracted chitosan are shown in Table 1. The parameters for the CSNPs preparation were optimized as described in a previous report.29 In brief, the ionic crosslinking of 2% chitosan (in 1% acetic acid) with 1% aqueous sodium tripolyphosphate (Na-TPP) was performed under constant magnetic stirring for 30 min at room temperature. According to TEM analysis,29 the prepared CSNPs with the smallest particle size (0.1–1.0 nm) were considered as the optimized formulation of CSNPs for the antibiotic drug loading.29For assessing the association of each drug with CSNPs in the optimized formulation, it was added to the chitosan solution (2%) before adding the TPP solution in it. Then, 1% Na-TPP solution was added to the mixture in a chitosan to TPP weight ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 under continuous magnetic stirring. The reaction was kept for 30 min at room temperature to form a suspension of the drug-loaded CSNPs. In this case, the effect of the concentration of ciprofloxacin in the chitosan solution on the ciprofloxacin-loaded chitosan nanoparticles (CP-CSNPs) preparation in terms of the loading or association efficiency was investigated by changing the ratio of ciprofloxacin to chitosan as 0.5
1 under continuous magnetic stirring. The reaction was kept for 30 min at room temperature to form a suspension of the drug-loaded CSNPs. In this case, the effect of the concentration of ciprofloxacin in the chitosan solution on the ciprofloxacin-loaded chitosan nanoparticles (CP-CSNPs) preparation in terms of the loading or association efficiency was investigated by changing the ratio of ciprofloxacin to chitosan as 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0, 0.65
1.0, 0.65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0, 0.8
1.0, 0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1.0
1, and 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0. In the case of MTZ loading, MTZ was added to chitosan solution in a MTZ to chitosan ratio of 0.5
1.0. In the case of MTZ loading, MTZ was added to chitosan solution in a MTZ to chitosan ratio of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 before adding Na-TPP. Higher ratios of MTZ
1.0 before adding Na-TPP. Higher ratios of MTZ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chitosan were not chosen because of the low solubility of MTZ in chitosan solution at higher concentration. Then the antibiotics-loaded CSNPs were separated from the suspension by centrifugation and were then freeze-dried at −50 °C using a freeze drier (LABCONCO, USA).
chitosan were not chosen because of the low solubility of MTZ in chitosan solution at higher concentration. Then the antibiotics-loaded CSNPs were separated from the suspension by centrifugation and were then freeze-dried at −50 °C using a freeze drier (LABCONCO, USA).
2.3. Measurement of the drug loading efficiency
The association efficiency (AE) or drug loading efficiency (LE) indicates the efficiency of the preparation method as it expresses the total amount of drug within the nanoparticles. After separating the antibiotic drug-loaded CSNPs from the suspension, the amount of free drug in the supernatant was measured using a UV-spectrophotometer (UV-1800 Spectrophotometer, Shimadzu, Japan) at the appropriate wavelength for the respective drugs. The amounts of drugs loaded or associated were evaluated from the difference between the total quantity of drug added to the loading solution and the quantity of non-entrapped drug remaining in the supernatant. Finally, the association efficiencies (AE) of the nanoparticles were calculated using eqn (1).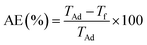 | (1) |
2.4. Characterization of the nanoparticles
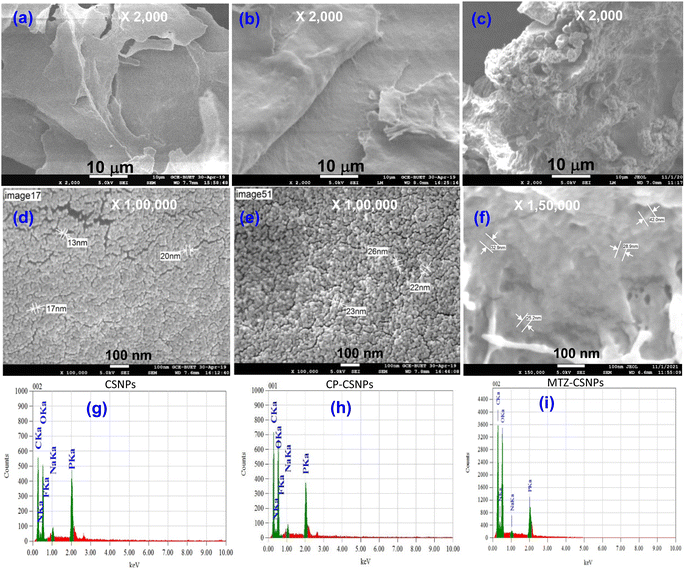
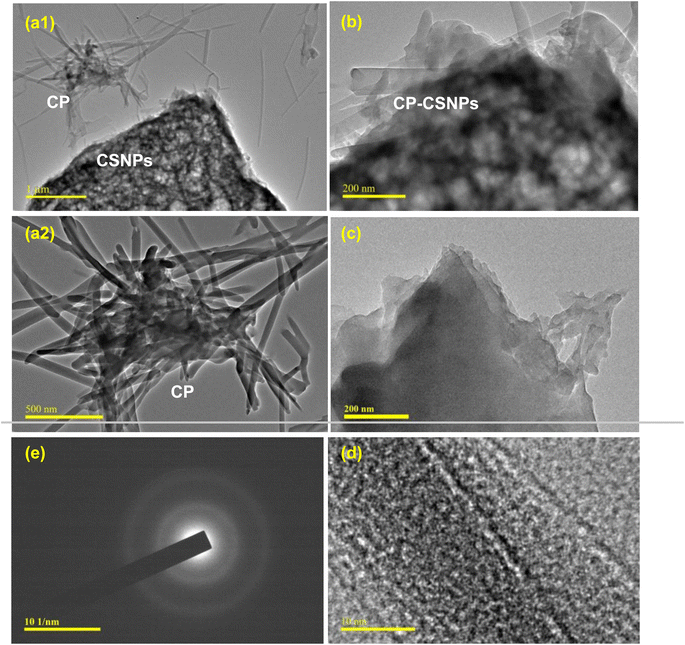
Morphological characterization, and qualitative and quantitative elemental analyses of the nanoparticles were done using a field emission scanning electron microscopy system (FESEM) (JEOL JSM-7600F), equipped with an energy dispersive X-ray (EDX) unit. The samples for FESEM analysis were prepared by completely drying the sample and then coating with platinum in vacuum. Then the micrographs of the particles were captured by FESEM. The morphology and size characteristics of the unmodified CSNPs loaded with different drugs were also observed by transmission electron microscopy (TEM) (JEOL, JEM 21000F). The TEM samples were obtained using a pastor pipette to place a drop of the chitosan nanoparticle suspension onto carbon-coated copper grids. The samples were dried at room temperature and then examined without any further modification or negative staining. Fourier transform infrared (FTIR) spectra of the materials were recorded in the range of 400–4000 cm−1 on an IR Prestige-21 system (Shimadzu, Japan) with 4 cm−1 resolution using the potassium bromide (KBr) disk method. The crystalline phase of the nanoparticles was analyzed by powder X-ray diffraction (XRD) patterns using an XRD 7000 diffractometer (Shimadzu, Japan) equipped with Ni-filtered Cu-Kα radiation (λ = 1.5406 Å) operated at 40 kV and 40 mA. Diffraction patterns were recorded within the 2θ range of 5°–70° in the continuous mode. The surface charge or zeta potential of the prepared chitosan nanoparticles, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles were measured as a function of pH by the acid–base titration method proposed by Parks et al.31 and Hossain.322.5. Biomedical application of drug-loaded chitosan nanoparticles
3. Results and discussion
3.1. Drug association efficiency
The association efficiencies (AEs) of CSNPs for CP and MTZ were determined using eqn (1). The average percentage association efficiencies of these formulations are shown in Table 2. From Table 2, it is seen that the loading process of ciprofloxacin was amount dependent, although the dependency was not very significant. An increase in ciprofloxacin concentration led to a slight increase in association efficiency. However, the ratios of 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.8 and 1
0.8 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for chitosan
1 for chitosan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CP in the loading procedure resulted in almost no change in the association efficiency of ciprofloxacin (varying from 83 ± 3% to 93 ± 3%). Finally, the association efficiency of the CS4 formulation containing the CS
CP in the loading procedure resulted in almost no change in the association efficiency of ciprofloxacin (varying from 83 ± 3% to 93 ± 3%). Finally, the association efficiency of the CS4 formulation containing the CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CP ratio of 1
CP ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was almost 93 ± 3%, which was the highest association efficiency and was due to electrostatic interaction between CS and CP, and thus, this formulation of CS4 was selected for further analysis.
1 was almost 93 ± 3%, which was the highest association efficiency and was due to electrostatic interaction between CS and CP, and thus, this formulation of CS4 was selected for further analysis.
| No. | Drug![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chitosan chitosan |
AE of CP-CSNPs (%) | AE of MTZ-CSNPs (%) |
|---|---|---|---|
| 1 | 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 1.0 |
83 ± 3 | 89 ± 3 |
| 2 | 0.65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 1.0 |
90 ± 3 | — |
| 3 | 0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 1.0 |
92 ± 3 | — |
| 4 | 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 1.0 |
93 ± 3 | — |
In the case of MTZ, the metronidazole![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chitosan ratio of 0.5
chitosan ratio of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 was selected for loading metronidazole within the chitosan nanoparticles because of the low solubility of metronidazole in chitosan solution at a higher concentration level. In addition, it was evident from the results of previous reports34 that increasing the metronidazole concentration leads to a decrease in metronidazole loading on CSNPs. Finally, the percentage of AE was found to be 89%, which is satisfactory and higher than that of previous studies.34
1.0 was selected for loading metronidazole within the chitosan nanoparticles because of the low solubility of metronidazole in chitosan solution at a higher concentration level. In addition, it was evident from the results of previous reports34 that increasing the metronidazole concentration leads to a decrease in metronidazole loading on CSNPs. Finally, the percentage of AE was found to be 89%, which is satisfactory and higher than that of previous studies.34
3.2. Characterization of the nanoparticles
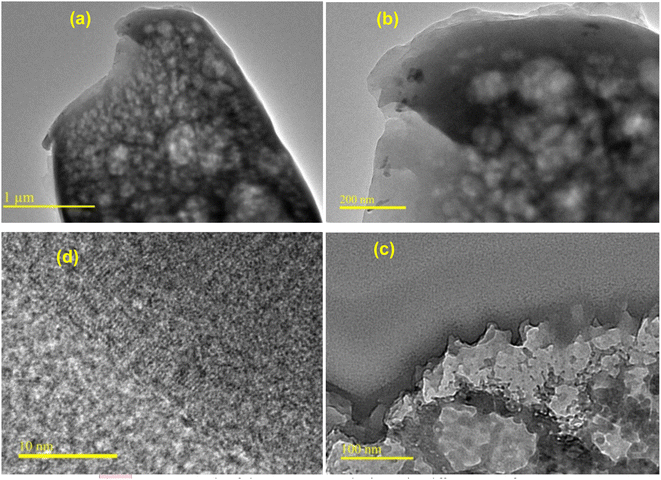 | ||
| Fig. 2 TEM micrographs of chitosan nanoparticles (CSNPs) at different magnifications: (a) ×2000, (b) ×50 000, (c) ×100 000 and (d) ×300 000. | ||
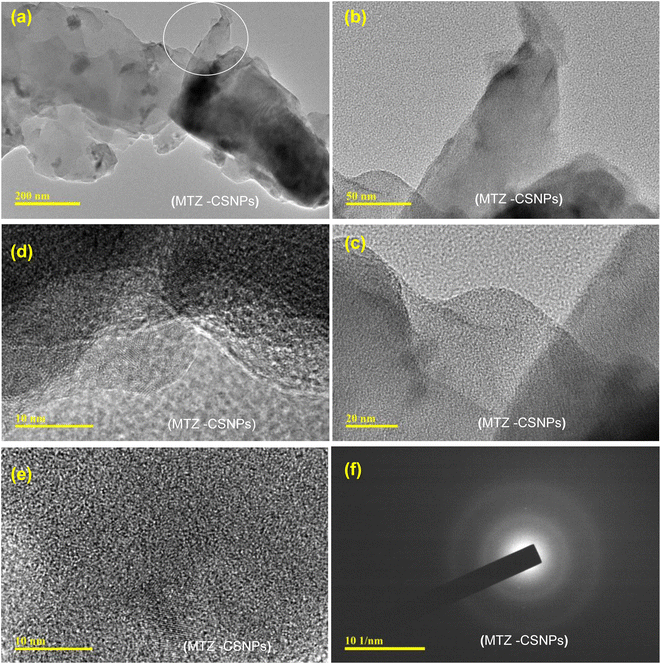 | ||
| Fig. 4 TEM micrographs of metronidazole-loaded chitosan nanoparticles (MTZ-CSNPs): (a–e) MTZ-CSNPs at different magnifications, and (f) image showing the non-crystalline nature of the MTZ-CSNPs. | ||
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration (amide I band) and the NH2 group bending vibration.35 The band at 894 cm−1 was related to the anhydro glucosidic ring. The bands at 2918 and 2878 cm−1 were related to C–H stretching, 2372 cm−1 was due to asymmetric C–N band stretching, 1379 cm−1 arose because of the asymmetric C–H bending of CH2 group, and 1079 cm−1 was for the skeletal vibration involving C–O stretching. In the case of CP-CSNPs, the peak at 3421 cm−1 corresponding to the hydroxyl groups in pure chitosan was shifted to 3428 cm−1, indicating that there could be some hydrogen bonding between the ciprofloxacin and chitosan nanoparticles. The bands at 1657 and 1595 cm−1 (amide I and amide II band), as clearly observed in pure chitosan, were decreased in the case of the CP-CSNPs, while two new absorption bands appeared at 1633 and 1536 cm−1 in the CP-CSNPs. This result indicated that the NH3+ groups of pure chitosan were crosslinked with the TPP groups of sodium polyphosphate, which helped to enhance both the inter- and intra-molecular interactions within the chitosan nanoparticles.36 Besides, the bands at 2945 cm−1 (2918 cm−1 in chitosan) and 2878 cm−1 in CP-CSNPs were related to symmetric and asymmetric CH3 stretching vibrations attributed to the pyranose ring of chitosan, while the band at 2358 cm−1 was due to asymmetric stretching of the C–N band, which was found at 2372 cm−1 in pure chitosan, while the band at 1299–1155 cm−1 was related to the C–F band stretching of ciprofloxacin.
O stretching vibration (amide I band) and the NH2 group bending vibration.35 The band at 894 cm−1 was related to the anhydro glucosidic ring. The bands at 2918 and 2878 cm−1 were related to C–H stretching, 2372 cm−1 was due to asymmetric C–N band stretching, 1379 cm−1 arose because of the asymmetric C–H bending of CH2 group, and 1079 cm−1 was for the skeletal vibration involving C–O stretching. In the case of CP-CSNPs, the peak at 3421 cm−1 corresponding to the hydroxyl groups in pure chitosan was shifted to 3428 cm−1, indicating that there could be some hydrogen bonding between the ciprofloxacin and chitosan nanoparticles. The bands at 1657 and 1595 cm−1 (amide I and amide II band), as clearly observed in pure chitosan, were decreased in the case of the CP-CSNPs, while two new absorption bands appeared at 1633 and 1536 cm−1 in the CP-CSNPs. This result indicated that the NH3+ groups of pure chitosan were crosslinked with the TPP groups of sodium polyphosphate, which helped to enhance both the inter- and intra-molecular interactions within the chitosan nanoparticles.36 Besides, the bands at 2945 cm−1 (2918 cm−1 in chitosan) and 2878 cm−1 in CP-CSNPs were related to symmetric and asymmetric CH3 stretching vibrations attributed to the pyranose ring of chitosan, while the band at 2358 cm−1 was due to asymmetric stretching of the C–N band, which was found at 2372 cm−1 in pure chitosan, while the band at 1299–1155 cm−1 was related to the C–F band stretching of ciprofloxacin.
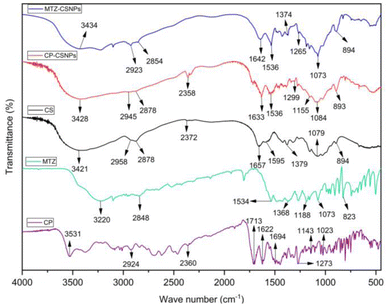 | ||
| Fig. 6 FTIR spectra of chitosan, ciprofloxacin-loaded chitosan nanoparticles (CP-CSNPs), and metronidazole-loaded chitosan nanoparticles (MTZ-CSNPs). | ||
In the case of the metronidazole-loaded chitosan nanoparticles, the peak for –OH stretching was slightly shifted to 3434 from 3421 cm−1, indicating the enhancement of hydrogen bonding in it. The peak for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching was found at 1642 cm−1 instead of 1657 cm−1. The peak at around 1595 cm−1 was assigned to the NH bending vibration of amide II groups, which could be observed clearly in the pure chitosan but was decreased dramatically, and a new absorption band appeared at 1536 cm−1, indicating the ammonium groups were crosslinked with tripolyphosphate molecules. The peak present at 1265 cm−1 corresponded to the N
O stretching was found at 1642 cm−1 instead of 1657 cm−1. The peak at around 1595 cm−1 was assigned to the NH bending vibration of amide II groups, which could be observed clearly in the pure chitosan but was decreased dramatically, and a new absorption band appeared at 1536 cm−1, indicating the ammonium groups were crosslinked with tripolyphosphate molecules. The peak present at 1265 cm−1 corresponded to the N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of metronidazole in the nanoparticles.
O stretching of metronidazole in the nanoparticles.
Finally, the FTIR spectra of these two drug-loaded CSNPs showed that some peaks observed for pure chitosan were shifted, but the shifting was not very significant. So, it could be concluded that the drugs were loaded on to the chitosan nanoparticles without functional group interaction but there was electrostatic interaction between the drugs and chitosan nanoparticles.
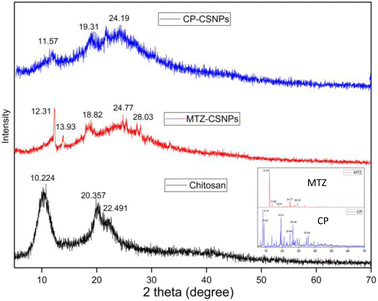 | ||
| Fig. 7 XRD patterns of chitosan, MTZ-CSNPs, and CP-CSNPs. Inset shows the XRD patterns of pure CP and MTZ. | ||
In the case of CP-CSNPs, the decrease in % relative intensity of the (110) and (020) planes compared to the pure chitosan showed that the native chitosan had been successfully transformed into nanoparticles. Besides, these relatively broad and weaker peaks were found at 2θ = 24.19. In the case of MTZ-CSNPs these diffraction peaks were decreased after metronidazole loading, which indicates that the crystallinity was completely destroyed in the nanoparticles. At the same time, the metronidazole powder showed several sharp and intense peaks, which indicated the crystalline pattern of the drug. The XRD pattern of the metronidazole-loaded chitosan nanoparticles presented some small diffraction peaks for metronidazole at 2θ near 12°, 13°, 18°, 24°, and 28° when compared to free metronidazole (inset). However, these peaks were very broad and weaker and the other peaks had disappeared in the MTZ-CSNPs. This might be due to the loading of metronidazole on the nanoparticles, while the modification procedure destroyed the crystal structure of the metronidazole.
3.3. Surface charge of the nanoparticles
The surface charge or zeta potential was determined from the measurement zero point of charge (ZPC or PZC), which is defined as the pH value where the surface positive and negative charges are equal.31,32 The surface charges of the chitosan, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles were determined from measurement of the pH as a function of acid–base titration.31,32Fig. 8 shows the variation of the surface charge density of chitosan, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles as a function of pH in aqueous solution of 0.001 M NaNO3. The point of zero charge (PZC) of chitosan, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles was obtained from the intersection of σovs. pH curves in Fig. 8. The values of pHPZC for the chitosan, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles were 3.15 ± 0.05, 3.95 ± 0.05, and 4.29 ± 0.05, respectively which indicate that at pH 3.15, 3.95, and 4.29, the surface charges of the chitosan, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles, respectively, were zero (σo), and at pH values above or below the mentioned values, the above surfaces became negative or positive, respectively. The higher values of pHZPC for the ciprofloxacin-loaded chitosan and metronidazole-loaded chitosan nanoparticles than that for the chitosan nanoparticles suggested that the negative species of ciprofloxacin and metronidazole were electrostatically adsorbed on the chitosan nanoparticles.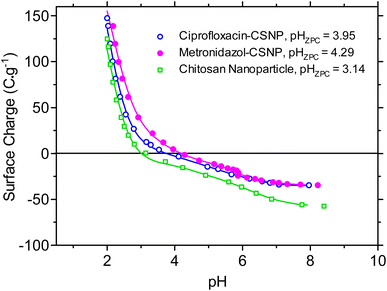 | ||
| Fig. 8 Surface charges pH (pHZPC) or zeta potentials of chitosan, ciprofloxacin-loaded chitosan, and metronidazole-loaded chitosan nanoparticles. | ||
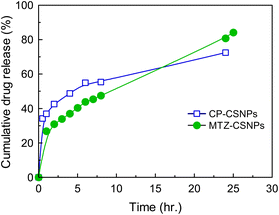
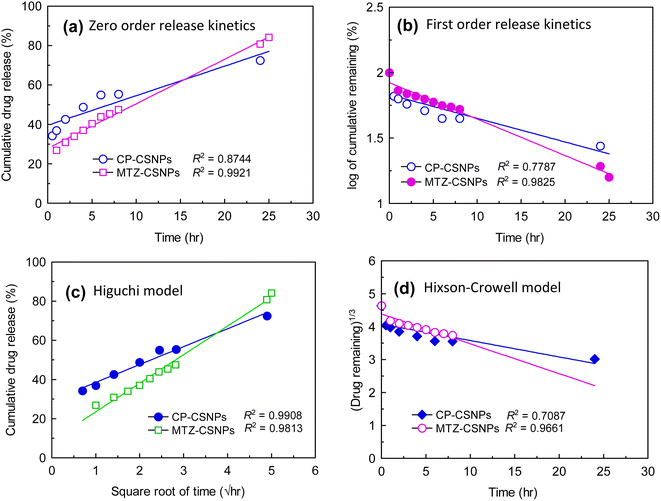
3.4. In vitro drug release kinetic study
Fig. 9 presents the average cumulative percentage release of drugs (ciprofloxacin and metronidazole) from chitosan nanoparticles at various time intervals. In both cases, the in vitro release profile showed a biphasic pattern. These two phases in the release profile occurred due to “sudden release” and “sustained release”. The sudden release or the initial rapid release can be characterized as a “burst effect”. From the release profile, it is very clear that the sudden release phase occurred in the first ½ h for CP and 1 h for MTZ. As the rapid or burst release period ended, the rate of release of the drugs fell and the second phase then proceeded, which was the “sustained release” phase, which was slower than previous. These slower and sustained releases occurred throughout the incubation period.Sustained drug release is the key strategy for reducing the systemic side effects of drugs in the human body associated with the frequency of drug administration and high drug concentrations. One of the most common methods used and studied to control drug release is encapsulation within a polymer matrix, which slows diffusion. However in the present study, the CSNPs were used for controlled release of the drug without encapsulation. Here, adsorption due to electrostatic interactions between the drugs and the CSNPs was the governing factor for drug release in this system.
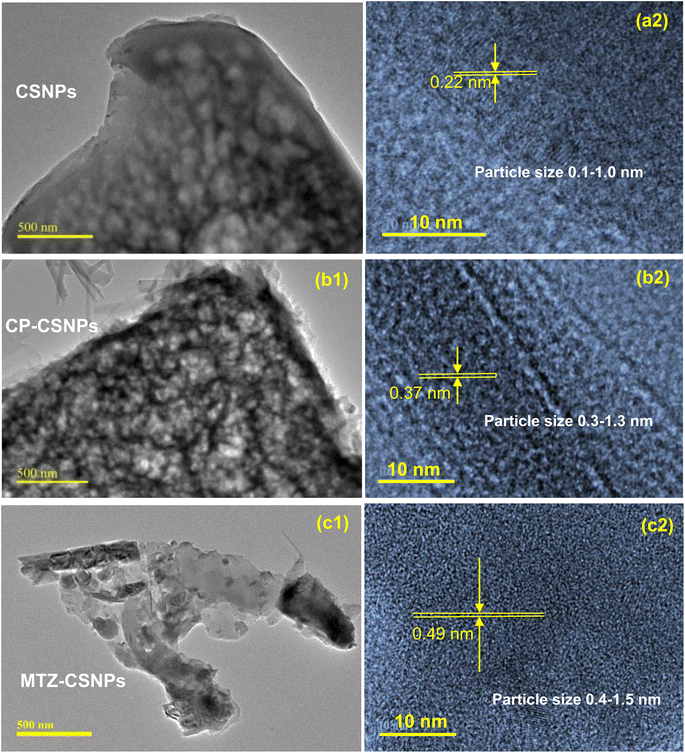
To understand the release mechanism, we first examined the physical characteristics of this release system. The morphology and sizes of the nanoparticles were characterized by SEM and TEM analysis. The CP-CSNPs were spherical with diameters ranging from 0.3–1.3 nm, which were higher than the blank chitosan nanoparticles (0.1–1.0 nm). This was due to the adsorption of drugs on the surfaces of the nanoparticles according to the TEM analysis (Fig. 5). The size of the unmodified chitosan nanoparticles was too small to encapsulate the drug molecules into the nanoparticles matrix. However, our results showed very similar release profiles as for drugs encapsulated in nanoparticles, though they were was not encapsulated in the chitosan nanoparticles, indicating that another mechanism must be involved. To describe the release mechanism, we postulated that the sustained release that we observed was primarily caused by adsorption due to electrostatic interactions between the drugs and CSNPs.39 We also postulated that initially the drugs are fully adsorbed on the nanoparticle surface. As the nanoparticle begins to degrade, the local pH of the system changes. At a certain threshold, the surface of the nanoparticles becomes neutral, weakening the electrostatic interactions and initiating release. Release can then be governed by nanoparticle degradation and drug diffusion or desorption, depending on the system. Finally, we can describe the mechanism of drug release from chitosan nanoparticles as usually involving polymer degradation, erosion, and diffusion of the drugs.40 From the above results, it is clear that the chitosan nanoparticles had the effect of prolonging the release of these two drugs in the body.
The dissolution or release profile of drugs can be described by several mathematical models representing several release kinetics, as represented in Fig. 10. Data obtained from the in vitro release studies of the drug-loaded CSNPs were fitted to zero-order, first-order, Higuchi model, and Hixson-Crowell models. The optimum model was chosen on the basis of the highest correlation coefficient (R2) value of these models.
Thus, the in vitro drug release profile applied in different mathematical models was evaluated by the correlation coefficient (R2), as presented in Table 3. The highest degree of correlation coefficient determines the most suitable mathematical model that follows the drug release kinetics. From the above comparison, it was found that the Higuchi square root model showed a higher degree of correlation coefficient (R2) than the other models.
| Samples | Saturation release (%) | R 2 | |||
|---|---|---|---|---|---|
| Zero order | First order | Higuchi square root | Hixson-Crowell | ||
| a C t is the amount of drug released at time t, Co is the initial concentration of drug, Ct is the percentage of drug remaining at time t, Q is the cumulative amount of drug released in time t, Wo is the initial amount of drug in the pharmaceutical dosage form, Wt is the remaining amount of drug in the pharmaceutical dosage form at time t, and K is the rate constant of the individual release kinetics. | |||||
| CP-CSNPs | 72 ± 3 | 0.874 | 0.779 | 0.991 | 0.709 |
| MTZ-CSNPs | 84 ± 3 | 0.992 | 0.983 | 0.981 | 0.966 |
| Equation | C t = Co + Kt | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C = log C = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Co − t/2.303 Co − t/2.303 |
Q = K × t1/2 | W o 1/3 − Wt1/3 = Kt | |
So drug release was found to be best fitted by the Higuchi square root model (R2 = 0.991 and 0.981 for ciprofloxacin and MTZ, respectively), which implies that the release of drug from matrix was a square root of the time dependent process and was diffusion controlled. Hence, the drug release profile of ciprofloxacin followed a diffusion mechanism.
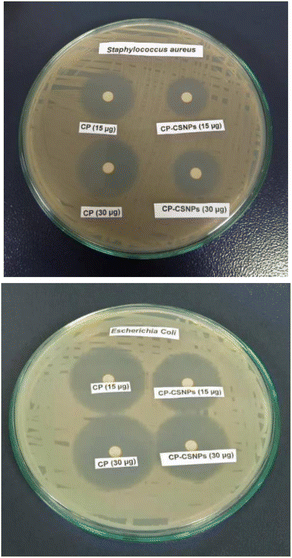
3.5. Antibacterial activity analysis
Microbiological studies were performed to observe the antibacterial activities of the ciprofloxacin-loaded chitosan nanoparticles (CP-CSNPs) against two types of bacteria, namely Escherichia coli (Gram negative) and Staphylococcus aureus (Gram positive).The results imply that both free CP and CP-CSNPs showed similar effects in liquid media. Sobhani reported that CP-CSNPs showed better antibacterial activity than free CP,42 whereas Jeong41 suggested the opposite, whereby the in vitro antimicrobial activity of CP-encapsulated PLGA nanoparticles was relatively lower than that of free CP against Escherichia coli. However, our study showed no difference between the free antibiotic and antibiotic-loaded nanoparticles in liquid media. So, we can say that the loading of ciprofloxacin on chitosan nanoparticles did not show any negative effect on the antibacterial activity, rather it could be highly active for inhibiting bacterial growth in vivo.
4. Conclusion
In the current study, two different antibiotic drugs, namely ciprofloxacin and metronidazole, were successfully loaded on to chitosan nanoparticles prepared from the crosslinking of chitosan and TPP to investigate potential biomedical application. The drugs were loaded with high association efficiencies of 93 ± 3% and 90 ± 3% for ciprofloxacin and metronidazole, respectively. The CSNPs and CSNPs-loaded antibiotics were characterized by SEM, EDX, TEM, FTIR, and XRD. The morphology analysis of all the drug-loaded nanoparticles observed by SEM suggested that the particles were spherical in shape with homogeneous surfaces. The TEM analysis showed that the average diameters were increased in the drug-loaded nanoparticles compared to the blank one (0.1–1 nm), such as 0.3–1.3 nm and 0.4–1.5 nm for the CP-CSNPs and MTZ-CSNPs, respectively. These indicate the drugs adsorption on the surface of the nanoparticles rather than encapsulation. Comparison of the surface charges of the prepared nanoparticles and the small shifting of some characteristic peaks in the FTIR spectra of both the drug-loaded nanoparticles from pure chitosan indicated the existence of electrostatic interactions between the drugs and chitosan nanoparticles. The in vitro release profile study of CP-CSNPs and MTZ-CSNPs showed initial burst releases at ½ h and 1 h followed by sustained release of 73 ± 3%, and 84 ± 3% of the drugs at 24 and 25 h, respectively, with the release profile governed by the Higuchi square root model for each case, which implies that the release of drugs was diffusion controlled. Due to the adsorption of drugs on the nanoparticles, the electrostatic interaction between the drugs and nanoparticles became the governing factor in sustaining the drug release effectively. Moreover, the antibacterial activity of CP-CSNPs observed by determination of the MIC and MBC showed similar results in the case of the loaded CP and free CP. This result implies that the loading of drugs on chitosan nanoparticles had no negative impact on the antibacterial activity. Furthermore, they may show better effectiveness to inhibit bacterial growth in vivo due to sustained release property according to several research studies. Finally, it can be concluded that the prepared nanoparticles have promising potential for use in drug delivery.Data availability
The data used to support the findings of this study are included within the article.Author contributions
Fatema Tuj Jahura: conceptualisation, methodology, investigation and writing. Farhana Khanam Ferdousi: formal analysis, resources and visualization. Abu Hena Mostofa Kamal: investigation of antibacterial activities and visualization. Anwar Ul-Hamid: TEM analysis and visualization. Md. Qamrul Ehsan: conceptualization, methodology, visualization and editing. Mohammad Abul Hossain: conceptualization, methodology, investigation, resources, writing, review editing, etc.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the Centennial Research Grant (CRG: 2020-2021) of Dhaka University. Fatema Tuj Jahura has given her thanks to the Bangabandhu Science and Technology Fellowship Trust, Bangladesh for the financial support.References
- M. P. Patel, R. R. Patel and J. K. Patel, J. Pharm. Sci., 2010, 13(4), 536–557 CAS.
- D. Bhowmik, H. Gopinath, B. Kumar, S. Duraivel and K. Kumar, Pharma Innov., 2012, 1(10), 24–32 CAS.
- T. Sagir, M. Huysal, M. Senel, S. Isık, N. Burgucu, O. Tabakoglu and M. Zaim, J. Colloid Interface Sci., 2022, 625, 711–721 CrossRef CAS.
- U. Garg, S. Chauhan, U. Nagaich and N. Jain, Adv. Pharm. Bull., 2019, 9(2), 195–204 CrossRef CAS.
- S. Laurent, D. Forge, M. Port, A. Roch, C. Robic, L. V. Elst and R. N. Muller, Chem. Rev., 2008, 108, 2064–2110 CrossRef CAS.
- A. Grenha, B. Seijo, C. Serra and C. R. Lopez, Biomacromolecules, 2007, 8, 2072–2078 CrossRef CAS PubMed.
- British Pharmacopia, The stationary office, London, 2009, pp. 493–498 Search PubMed.
- D. Bhowmik, H. Gopinath, B. Kumar, S. Duraivel and K. Kumar, Pharma Innov., 2012, 1(10), 24–32 CAS.
- E. Cevher, Z. Orhan, L. Mulazimoglu, D. Sensoy, M. Alper and A. Yildiz, et al , Int. J. Pharm., 2006, 317(2), 127–135 CrossRef CAS PubMed.
- M. Gomez-Burgaz, G. Torrado and S. Torrado, Eur. J. Pharm. Biopharm., 2009, 73(1), 130–139 CrossRef CAS PubMed.
- M. Kong, X. G. Chen, K. Xing and H. J. Park, Int. J. Food Microbiol., 2010, 144(1), 51–63 CrossRef CAS PubMed.
- K. S. V. K. Rao, P. R. Reddy, Y. I. Lee and C. Kim, Carbohydr. Polym., 2012, 87(1), 920–925 CrossRef.
- L. H. Li, J. C. Deng, H. R. Deng, Z. L. Liu and X. L. Li, Chem. Eng. J., 2010, 160(1), 378–382 CrossRef CAS.
- A. I. Caco, F. Varanda, M. J. Pratas de Melo, A. M. A. Dias, R. Dohrn and I. M. Marrucho, Ind. Eng. Chem. Res., 2008, 47, 8083–8089 CrossRef CAS.
- F. Fawaz, F. Bonini, J. Maugein and A. M. Lagueny, Int. J. Pharm., 1998, 168, 255–259 CrossRef CAS.
- M.-E. Page-Clisson, H. Pinto-Alphandary, M. Ourevitch, A. Andremont and P. Couvreur, J. Contr. Release, 1998, 56, 23–32 CrossRef CAS.
- K. Dillen, J. Vandervoort, G. Van den Mooter and A. Ludwig, Int. J. Pharm., 2006, 314, 72–82 CrossRef CAS PubMed.
- Y. C. Tseng, Y. Tabata, S. H. Hyon and Y. Ikada, J. Biomed. Mater. Res., 1990, 24, 1355–1367 CrossRef CAS.
- D. Jain and R. Banerjee, J. Biomed. Mater. Res., Part B, 2008, 86(1), 105–112 CrossRef.
- H. M. Ibrahim, M. K. El-Bisi, G. M. Taha and E. A. El-Alfy, J. Appl. Pharmaceut. Sci., 2015, 5(10), 85–90 CrossRef CAS.
- S. Latha, P. Selvamani, C. S. Kumar, P. Sharavanan, G. Suganya, V. S. Beniwal and P. R. Rao, J. Magn. Magn Mater., 2009, 32, 1580–1585 CrossRef.
- G. Patel, R. B. Patel and H. R. Patel, J. Sci. Technol., 2011, 6(2), 33–45 Search PubMed.
- A. Allen, G. Flemstrom, A. Garner and E. Kivilaakso, Physiol. Rev., 1993, 73, 823–857 CrossRef CAS PubMed.
- M. George and T. E. Abraham, J. Contr. Release, 2006, 114, 1–14 CrossRef CAS.
- J. D. Smart, I. W. Kellaway and H. E. Worthington, J. Pharm. Pharmacol., 1998, 36, 295–299 CrossRef.
- V. Gonzalez, C. Guerrero and U. Ortiz, J. Appl. Polym. Sci., 2000, 78, 850–857 CrossRef CAS.
- M. R. Kasaai and M. Malaekeh, Adv. Chitin Sci., 2003, 7, 178–180 CAS.
- A. A. Elzatahry and M. S. Mohy Eldin, Polym. Adv. Technol., 2008, 19(12), 1787–1791 CrossRef CAS.
- F. Tuj-Jahura, Department of Chemistry, PhD thesis, University of Dhaka, Bangladesh, 2022.
- P. Calvo, J. L. Remunan-Lopez and A. M. J. Vila-Jato, J. Appl. Polym. Sci., 1997, 63(1), 125–132 CrossRef CAS.
- S. G. A. Parks and P. L. de Bruyn, J. Phys. Chem., 1962, 66, 967–973 CrossRef.
- M. A. Hossain, PhD thesis, Kanazawa University, Japan, 2006.
- I. Wiegand, K. Hilpert and R. E. W. Hancock, Nat. Protoc., 2008, 3(2), 163–170 CrossRef CAS PubMed.
- H. Ruchika, Int. J. Sci. Res. Methodol., 2016, 4(4), 1–17 Search PubMed.
- R. C. Nagarwal, P. N. Singh, S. Kant, P. Maiti and J. K. Pandit, Chem. Pharm. Bull., 2011, 59, 272–278 CrossRef CAS.
- L. Qi, Z. Xu, X. Jiang, C. Hu and X. Zou, Carbohydr. Res., 2004, 339(16), 2693–2700 CrossRef CAS.
- J. R. Anusha, A. T. Fleming, A. M. Valan, K. B. Chul, N. A. Al-Dhabi, K. H. Yu and R. C. Justin, J. Adv. Res., 2016, 7, 863–871 CrossRef CAS.
- E. M. A. Hejjaji, A. M. Smith and G. A. Morris, Int. J. Biol. Macromol., 2017, 95, 564–573 CrossRef CAS.
- H. F. G. Barbosa, D. S. Francisco, A. P. G. Ferreira and E. T. G. Cavalheiro, Carbohydr. Polym., 2019, 225, 115232 CrossRef.
- M. M. Pakulska, I. E. Donaghue, J. M. Obermeyer, A. Tuladhar, C. K. McLaughlin, T. N. Shendruk and M. S. Shoichet, Sci. Adv., 2016, 2(5), e1600519 CrossRef.
- Y. I. Jeong, H. S. Na, D. H. Seo, D. G. Kim, H. C. Lee and M. K. Jang, Int. J. Pharm., 2008, 352(1–2), 317–323 CrossRef CAS PubMed.
- Z. Sobhani, S. M. Samani, H. Montaseri and E. Khezri, Adv. Pharm. Bull., 2017, 7(3), 427–432 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |