Open Access Article
Open Access ArticleSubstrate softness increases magnetic microdiscs-induced cytotoxicity†
Andrea
Visonà
 ab,
Sébastien
Cavalaglio
ab,
Sébastien
Cavalaglio
 a,
Sébastien
Labau
a,
Sébastien
Soulan
a,
Hélène
Joisten
a,
Sébastien
Labau
a,
Sébastien
Soulan
a,
Hélène
Joisten
 b,
François
Berger
c,
Bernard
Dieny
b,
François
Berger
c,
Bernard
Dieny
 b,
Robert
Morel
b,
Robert
Morel
 b and
Alice
Nicolas
b and
Alice
Nicolas
 *a
*a
aUniv. Grenoble Alpes, CNRS, CEA/LETI-Minatec, Grenoble INP, LTM, Grenoble F-38000, France. E-mail: alice.nicolas@cea.fr
bUniv. Grenoble Alpes, CEA, CNRS, Spintec, Grenoble F-38000, France
cUniv. Grenoble Alpes, INSERM, CHU Grenoble, BrainTech Lab, Grenoble F-38000, France
First published on 12th November 2024
Abstract
Cytotoxicity of nanoparticles is primarily assessed on cells grown in plastic culture plates, a mechanical environment that is a million times stiffer than most of the human tissues. Here we question whether nanoparticles cytotoxicity is sensitive to the stiffness of the extracellular environment. To this end, we compare the metabolic activity, the proliferation and death rates, and the motility of a glioblastoma cancer cell line and a fibroblast cell line exposed to gold-coated Ni80Fe20 microdiscs when grown on a glass substrate or on a soft substrate whose mechanical properties are close to physiology. Our main result is that cells grown on soft substrates take up more microdiscs which results in greater toxic effects, but also that toxicity at similar particle load is more pronounced on soft substrates especially at large concentration of nanoparticles. These results suggest that both microdiscs uptake and their intracellular processing differ between soft and rigid substrates.
1 Introduction
Magnetic nanoparticles (NPs) are envisioned as a promising tool to locally alter cell behavior1,2 thanks to the ability of controlling them remotely via external magnetic field. Nanometric sized particles named SPIONs are already used in some clinical applications.3 The application of a high frequency magnetic field (≥100 kHz) results in magnetic heating, inducing an increase of the local temperature (hyperthermia) which in turn can alter cell fate, either leading to cell death or to cell reprogramming.4 In contrast to SPIONs (Superparamagnetic Iron Oxide Nanoparticles), vortex particles are much larger magnetic nanoparticles, which have recently attracted attention for their ability to exert mechanical forces in the nanoNewton range,5–8 comparable to the cellular forces.9 Their high shape anisotropy (micrometer sized disc with few tens of nanometers thickness, see Fig. 1c) results, in the absence of magnetic field, in an in-plane circular closed-flux vortex magnetization configuration.10 Under applied magnetic field (of the order of 10 to 100 mT) the particles polarize and, with low-frequency rotating or oscillating field, in the few Hertz range, they oscillate and give rise to a mechanical stress to the surrounding cells and tissues11–13 (Fig. S1†). In addition, calculations and observations showed that these particles have a low enough magnetic susceptibility to avoid self-polarization effects within chains of particles, allowing the particles to redisperse after the magnetic field has been removed.13 For these reasons, vortex microdiscs show great promise for future clinical applications based on mechanical stimulation. It has for instance been observed that the magneto-mechanical stress provided by these particles can induce cancer cell death11,12,14 or promote insulin secretion of pancreatic cells.15In what follows, we focus on these highly anisotropic microdiscs, which we refer to interchangeably as microdiscs or nanoparticles, as defined by the European Commission Recommendation 2022/C 229/01.16 To reach clinical applications, these composite NPs should better show the lowest possible toxicity in the absence of magnetic field. In this context, the gold coating aims to limit the release of metal ions, especially nickel ions, which were shown to be highly toxic.17 These particles have already been used in several studies for their capability to exert mechanical forces in the nN range.11,12,14,15,18 Up to now, they were used at very low concentration, below 50 NPs per cell.11,14,15 In these conditions, they have been reported to have no or a very limited toxicity, assessed by metabolic assays. This evaluation was performed on cells grown in vitro, in standard culture plates made of plastic or glass, thus in a mechanical environment that is a million times stiffer than living tissues.19 However, it is now well established that many cellular responses are sensitive to the mechanical properties of the extracellular environment. For instance, endocytosis was shown to be either enhanced, unaltered or limited when comparing cells grown on substrates of varying stiffness, depending on the precise mechanism of endocytosis.20 Since vortex particles were shown to be engulfed by cells,14,15 we questioned whether their cytotoxicity depends on the mechanical properties of the culture plate. To this end, we performed cytotoxic analysis with cells grown on a 2D soft matrix made of polyacrylamide hydrogel. Growing cells on these supports was recently shown to bring cells in a state of high physiological relevance for characterizing molecular events.21 Here we focused on two cell types, a human cancer cell line of glioblastoma, a disease for which these particles could show promise,22 the U87-MG cells, and a murine fibroblastic cell line that is often used as a reference for toxicity assays, the NIH 3T3 cells. The stiffness of the soft substrates were chosen according to the mechanical properties of their tissues of origin. For instance, we chose a value of 4 kPa to mimic the core of a glioblastoma tumor.23,24 The same value was first used for the NIH 3T3 cells, as a value of 5 kPa had been reported to allow dermal fibroblasts activation with a profile that resemble human biology.25 A second value, of 12 kPa, was also assayed that mimics stiffened regions associated with initial fibrosis and fibroblast activation.26
The cytotoxicity of the vortex microdiscs in cells grown on the soft substrates was compared to that measured in glass culture plates. In order to single out the role of the stiffness of the extracellular matrix alone, the soft and glass substrates were coated with identical surface density of fibronectin, an adhesion protein that enables the growth of the two cell types. Using this experimental approach, our main result is that the cytotoxicity of the vortex microdiscs is larger for cells grown on a soft substrate. In these conditions, the cells capture more particles, with the greater particle load leading to a slowdown of the proliferation rate, to an increased death rate, and to a reduced proportion of cells progressing in cell cycle. Nevertheless, we observed that these cellular responses do not result in a decreased metabolic activity, suggesting that when grown on a soft substrate, cells engage other energy producing mechanisms that compensate the adverse toxic effects.
2 Experimental section
2.1 Nanoparticles fabrication
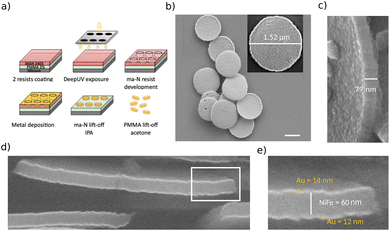
The vortex micro-discs particles were fabricated by a top-down approach as described previously27 (Fig. 1a). A double layer of, first, PMMA 2% AR-P 679.04 positive resist (Allresist, Strausberg, Germany) and, second, ma-N 2403 negative resist (Micro resist technology, Berlin, Germany) was spun onto 100 mm bare silicon wafers according to manufacturer's recommendations. DUV optical lithography (240 nm) with mask aligner (Süss MicroTec MJB4, Garching, Germany) allowed defining an array of circular wells of diameter 1.3 μm in the ma-N top resist. After ma-N resist development with AZ 326 MIF (MicroChemicals, Ulm, Germany), metal deposition (Au 10 nm/Ni80Fe20 60 nm/Au 10 nm) was made using a Plassys MEB550 HV e-beam evaporation system (Marolles-en-Hurepoix, France). A first ethanol lift-off allowed removing the ma-N resist, leaving the particles standing on the PMMA resist layer. A second acetone lift-off allowed to free the particles from the silicon substrate (Fig. 1b). Particles were washed three times in hot acetone (50 °C) in an ultrasonic tank, to allow for the removal of the PMMA monomers. They were then washed and sonicated three times with ethanol. A sample was taken from each batch for magnetization measurement in order to estimate the amount of produced vortex discs. They were then stored in ethanol, in sealed tubes, and kept in the refrigerator until use. This protocol produced disc-shaped flakes with a nominal diameter of 1.3 μm and a thickness of around 80 nm, consisting of a permalloy core (Ni80Fe20) with a nominal thickness of 60 nm covered on its upper and lower faces with a layer of gold with a nominal thickness of 10 nm (Fig. 1c and e). Before use in cell culture, the ethanol was substituted with culture medium sequentially to ensure cells are not exposed to harmful doses of ethanol. Dispersion of the particles was achieved by sonication and extensive flushing with pipette before adding the medium and NPs in culture.2.2 Electron microscopy
Few particles suspended in ethanol were deposited on a 10 × 10 mm2 silicon wafer and ethanol was let to evaporate. SEM imaging was performed on ZEISS Ultra Plus Scanning Electron Microscope equipped with an in-lens detector. Accelerating voltage was set to 20 kV. Cross-section views of the NPs were obtained using a Helios 450S Focused Ion Beam equiped with Scanning Transmission Electron Microscope FIB-STEM from FEI (Thermo Fisher Scientific Inc.), featuring a gallium FIB gun at 30 kV and a 30 kV SEM column. Prior to imaging, the sample was encapsulated in SU8-2002 resist (Microchemicals, Ulm, Germany) to prevent the detachment of the NPs. The resist was diluted in PGMEA (Sigma Aldrich, Saint Quentin Falavier, France) in ratio 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, spin coated at 2000 rpm for 30 s on the silicon piece according to manufacturer's recommendations in order to produce a layer with a thickness of less than one micron. This was followed by two anneals, one at 70 °C for drying and the second one at 95 °C for polymer cross-linking. 40 nm of platinum was deposited on the sample using a Bio-Rad SC500 sputter to avoid charging effects under electron beam conditions. The first treatments applied to the sample, carried out in the FIB-STEM chamber, involved a focused platinum deposit enhanced by an ion beam to obtain a protective mask over an area of 4 μm × 15 μm and a thickness of 1 μm. A series of focused cuts around the mask was used to extract a small volume of the wafer surface to a depth of 3 to 4 μm below the protection. The prism was then attached to a TEM grid to be eroded by the gallium beam. SEM images were captured at various points during this progression through the volume using the in-lens detector located in the pole piece of the electron emission column. This detector is capable of reconstructing the image of secondary electrons for topographical information, as well as BSE backscattered electrons giving good chemical contrasts. High acceleration of 29 kV and a low current of 6.3 pA were used for better resolution.
50, spin coated at 2000 rpm for 30 s on the silicon piece according to manufacturer's recommendations in order to produce a layer with a thickness of less than one micron. This was followed by two anneals, one at 70 °C for drying and the second one at 95 °C for polymer cross-linking. 40 nm of platinum was deposited on the sample using a Bio-Rad SC500 sputter to avoid charging effects under electron beam conditions. The first treatments applied to the sample, carried out in the FIB-STEM chamber, involved a focused platinum deposit enhanced by an ion beam to obtain a protective mask over an area of 4 μm × 15 μm and a thickness of 1 μm. A series of focused cuts around the mask was used to extract a small volume of the wafer surface to a depth of 3 to 4 μm below the protection. The prism was then attached to a TEM grid to be eroded by the gallium beam. SEM images were captured at various points during this progression through the volume using the in-lens detector located in the pole piece of the electron emission column. This detector is capable of reconstructing the image of secondary electrons for topographical information, as well as BSE backscattered electrons giving good chemical contrasts. High acceleration of 29 kV and a low current of 6.3 pA were used for better resolution.
2.3 Cell culture
GFP-tagged U87-MG and NIH 3T3 cells were purchased from ATCC. The cells were grown in Dulbecco's Modified Eagle's medium (DMEM, Gibco, ref. 31966047) completed with 10% fetal bovin serum (FBS, Gibco, ref. 10270106) and 1% ATAM (Gibco, ref. 15240062). The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2. For our experiments, the cells were grown either on glass culture plates coated with 1 μg cm−2 fibronectin (stiff substrate) or on plates having a layer of polyacrylamide hydrogel coated with 1 μg cm−2 fibronectin (soft substrate). The plates were purchased from Cell&Soft (Grenoble, France) and used following the manufacturer's instructions. We chose Young's moduli of the soft bottoms of 4 kPa for the study with U87-MG cells and both 4 kPa and 12 kPa for the NIH 3T3 cells. The choice of these values is explained in the Results section.2.4 Characterization of the coating of the culture plates
We checked that the fibronectin coating was identical between all the conditions by performing a comparative immunostaining assay. In the absence of cells, the wells were labeled with anti-fibronectin antibody (Sigma-Aldrich, F3648) diluted at 1/400 and conjugated with an Alexafluor 488 secondary antibody (Molecular Probes, A21206) diluted at 1/2000. Confocal imaging (LSM 880, Zeiss) was performed to compare the level of intensity in the 3 conditions (4 kPa, 12 kPa, glass). A pile of images was acquired at 20× magnification (NA 0.8) for 3 positions in each well with identical illumination and capture parameters. Images stacks were analyzed with Fiji.28 The pile of images was summed and the resulting image was blurred with a Gaussian filter of 10 pixels radius to remove the pixelisation coming from the confocal. The mean intensity and the standard deviations were then measured in every condition.2.5 Metabolic assay
A colorimetric assay (CCK-8, Sigma-Aldrich, ref. 96992) was used to assess cell metabolism. In this test, the highly water-soluble tetrazolium salt, WST-8, is reduced by dehydrogenase in living cells to give a yellow-coloured dye (formazan). The amount of the formazan dye generated by the activity of mitochondria is proportional to the energy production of cells. We compared the amount of formazan between cells that were grown in the presence or in the absence of NPs. Cells were seeded at three cell densities, either 4000 cells per cm2 (hereafter D1), 8000 cells per cm2 (hereafter D2) or 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 (hereafter D3), in 96 wells plates with either a stiff glass bottom or a soft one (4 kPa or 12 kPa). Two concentrations of NPs, of 5 and 50 μg cm−2, were compared to a control condition, defined by the absence of NPs. The NPs were introduced 6 h post seeding and incubated for 16 hours. Additional control wells containing the four concentrations of NPs (5, 10, 50 and 100 μg cm−2) but no cells were prepared to check whether the absorbance is altered by the particles. CCK-8 product was then added and incubated for 2 hours, after which the optical density (OD) was measured with a FLUOstar Omega reader (BMG Labtech) at 450 nm (formazan absorbance) and at 600 nm (control). Three independent experiments were performed using cell populations that had been separated two passages prior to the metabolic assay.
000 cells per cm2 (hereafter D3), in 96 wells plates with either a stiff glass bottom or a soft one (4 kPa or 12 kPa). Two concentrations of NPs, of 5 and 50 μg cm−2, were compared to a control condition, defined by the absence of NPs. The NPs were introduced 6 h post seeding and incubated for 16 hours. Additional control wells containing the four concentrations of NPs (5, 10, 50 and 100 μg cm−2) but no cells were prepared to check whether the absorbance is altered by the particles. CCK-8 product was then added and incubated for 2 hours, after which the optical density (OD) was measured with a FLUOstar Omega reader (BMG Labtech) at 450 nm (formazan absorbance) and at 600 nm (control). Three independent experiments were performed using cell populations that had been separated two passages prior to the metabolic assay.
2.6 Cell growth assays
The number of cells was monitored over time after the addition of NPs using a DNA stain (Incucyte Nuclight Rapid Red Dye, Sartorius, ref. 4717). Three populations of cells were seeded in stiff and soft 96 wells plates at two cell densities, D1 and D3. The DNA stain was added 5 h post seeding at a concentration of 1/2000. One hour later, the NPs were introduced in some wells at concentrations 5 or 50 μg cm−2, while the control wells were kept free of NP. The number of nuclei was monitored every hour for 48 h in fluorescence and phase contrast channels at 10× magnification using an inverted microscope equipped with a motorized stage (IX83 Olympus, Olympus France, Rungis, France) and an incubation chamber (Okolab, Rovereto, Italy).To determined the ratio of cells that had entered into cell cycle, we measured the number of cells expressing the Ki-67 protein. This protein is expressed into the nucleus during all phases of the cell cycle but the G0 phase, which is a phase of quiescence. Three independent populations of cells were seeded in stiff and soft 96 wells plates, and the NPs were added 6 h post seeding. The cells were then fixed 16 h post exposure to the NPs and cell nuclei were fluorescently labelled with the anti-Ki-67 conjugated antibody (eFluor 570, eBioscience, ref. 41-5698-82, dilution 1/200) and Hoechst stain (dilution 1/1000). One image per well of the two fluorescent channels were acquired at 10× magnification and analysed.
Cell death was measured by quantifying the proportion of cells positive to propidium iodide (PI, Molecular Probes, ref. P3566). This molecule stains nuclei only in presence of holes in the lipid membrane which usually anticipates cell death, both in case of necrosis or apoptosis. Cells were seeded at D1 in stiff and soft 24 wells plates. The NPs were added 6 h post seeding at the two concentrations, 5 and 50 μg cm−2 alongside with PI diluted at 1/2000 and Nuclight Rapid Red dye diluted at 1/2000. Images were taken 16, 24 and 48 h post exposure to the NPs. Three images per well were captured in phase contrast, CY3 and CY5 channels.
2.7 Quantification of the number of particles per cell
The amount of particles per cell was determined by measuring the magnetic moment of pellets of cells grown with NPs for 16 h. Three independent populations of cells were seeded in stiff and soft 24 wells plates. NPs were added 6 h post seeding and let incubate for 16 h. Each well was then treated separately. The cells were detached with 0.05% trypsin–EDTA (Gibco, ref. 25300062) and centrifuged at 1250 rpm for 5 min. The pellet was then immersed in a solution of PBS+/+ containing 1% Triton X-100 (Bio-Rad, ref. 1610407) to destroy the cell body. The Triton X-100 solution was substituted with ethanol and the pellet, with NPs and cell debris, was dried on a 5 × 5 mm silicon wafer positioned above a permanent magnet to avoid spillage of particles. A second piece of silicon wafer was glued on top of the first to prevent the pellet from detaching. Control samples were prepared with dilutions of the starting solutions of NPs used with the cells. In addition to detaching cells from the fibronectin coating, trypsin also removed some of the NPs lying at the bottom of the well, which were not bound to cells, and which should be discarded from the count. Therefore, additional wells were seeded with only particles in order to quantify the number of detached NPs, nw. We observed that nw is a constant proportion α of the initial number of NPs deposited in the well, n0:| nw = αn0 | (1) |
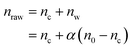 | (2) |
Eqn (2) takes into account the fact that, as nc NPs are attached to cells, the number of free NPs lying on the surface is no more n0 but n0 − nc. The number of NPs per cell is then, for a given cell number Nc:
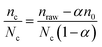 | (3) |
2.8 Immunofluorescence and confocal imaging
Cells were stained to visualize Ki-67 protein and the plasmic membrane. Ki-67 was immunostained with a eFluor 570-conjugated Ki-67 monoclonal antibody (eBioscience, ref. 41-5698-82, dilution 1/200) after cell permeabilization in a solution of 4% PFA, 0.5% Triton 100× in PBS+/+ for 15 min and fixation in a solution of 4% PFA in PBS+/+ for 45 min. The plasmic membrane was stained with DiL (Molecular Probes, ref. V-22885) diluted at 1/400 with cells being fixed but not permeabilized. In both case, the nuclei were stained with Hoechst dye at 1/1000. Fluorescently labeled samples were imaged with confocal microscopy (LSM 880, Zeiss) with a 63× magnification oil immersion objective (NA 1.4). A pile of images with spacing 0.25 μm was acquired. NPs were imaged with the same parameters using the reflected light at 488 nm.2.9 Image analysis
Nuclei were segmented with StarDist Fiji plugin29 or with Cellpose30 and the masks were post-processed with an in-house Python3 code to count the nuclei. Using Cellpose, a neural network was trained starting from cyto2 pre-trained model. The training method employed was the so called “Human in the loop”, where some exemplary images of the whole data set are sequentially used to refine the pre-trained network. This allowed to obtain greater precision without the need of a large data set with many ROIs per image, which would otherwise be required with a training performed from scratch. Five images were selected to span the different scenarios met in experiments: high and low cellular density, low or medium amount of NPs, loaded preferentially in the cell body or laid in majority on the surface. An example of the improvement in the segmentation, from the Cellpose pre-trained model to the refined one, is available in the ESI (Fig. S3).† However, it was not possible to automatically analyse images with high concentration of particles, especially at longer time, due to the excessive cells particles-loading. In this case the nuclei were counted manually. This was for instance the case with Ki-67 staining: the heterogeneity of the staining together with the presence of the NPs prevented automatic counting. Indeed, not only the level of Ki-67 expression depends on the specific phase and is maximum in the G2 phase, but it can either be localised into the chromosomes appearing like small dots, or be homogeneously distributed in the nucleus.31 Images were thresholded to only count cells with more than few small bright spots. The threshold was kept identical in all the conditions.Cell motility was quantified based on the images captured during the proliferation assay, with a lapse of 1 h between each image. Five images, between 14 h and 18 h post exposure to the NPs were segmented with Cellpose. Cell nuclei were tracked using the TrackMate Fiji plugin.32 The median velocity was calculated from the distribution of velocities at every time step. Mean of the medians was calculated for every condition. This methodology was evaluated by checking that the velocity does not significantly vary during the four hours lapse (Table S1†).
2.10 Statistical analysis
Each experiment was performed in triplicate with three independent populations of cells that were splitted from a shared population two passages prior to the experiment. Significance was assessed by performing bilateral Student's t-test when comparing two conditions. One sample t-test was used to evaluate significant variations in metabolic assays. Statistical differences between three or more samples were assessed by one-way ANOVA test.3 Results
3.1 The lateral faces of the vortex NPs are partly covered with gold
Vortex NPs are gold-covered permalloy (Ni80Fe20) thin discs. The gold coverage allows for the functionalization of the NPs, but also acts as a barrier to protect the iron and nickel alloy. Indeed, exposed permalloy have been shown to oxidize, giving rise to a few angström thick layer of nickel and iron oxides.33 Moreover, the toxicity of nickel oxide on cells grown in vitro has been reported.34 By performing SEM and FIB-SEM imaging (Fig. 1b–e), we observed with overall views and cross sections that the top and bottom surface gold coverage is continuous, with no exposed permalloy. Here, gold appears brighter due to its higher atomic number. On the other hand, we observed that the lateral faces of the NPs were covered with a non uniform thickness of gold. Some uncovered areas were visible. Even though the exposed surface amounts to only a few percent of the NP total surface, it opens up the risk of a residual toxicity dependent on the cell type, as described with nickel oxides.3.2 Vortex NPs are internalized by the cells
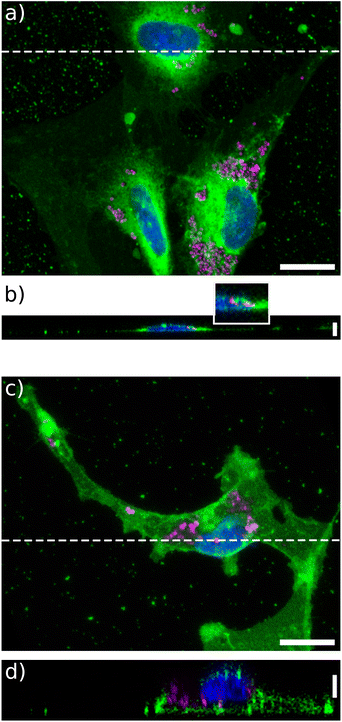
Then we wondered whether these particles would be internalized by U87-MG or NIH 3T3 cells, when grown on a soft extracellular environment. To this end, confocal imaging was performed. Some particles were clearly observed inside the cells grown on a soft matrix in both cell lines (Fig. 2b, d and S4†). Their precise location in cells grown on glass was more difficult to assess, as the cells were more spread and much thinner (Fig. 2b). Nonetheless, some particles were clearly seen embedded into the thin lamellipodia. In addition, particles were observed to concentrate around the nuclei for all stiffness, suggesting that they are trapped by the endosomal or lysosomal machinery35 (Movies S1–S4†).3.3 Vortex NPs do not affect U87-MG nor NIH 3T3 metabolic activity when the cells are grown on glass or on 4 kPa substrates
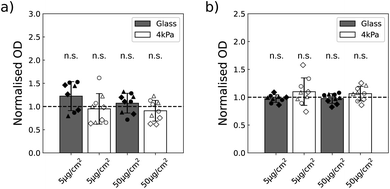
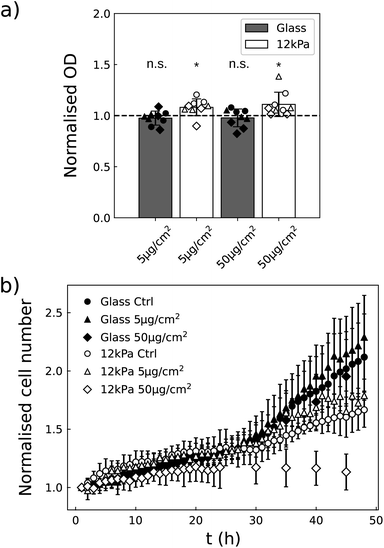
In order to assess their toxicity, we first quantified the metabolic activity of the two cell lines exposed to the NPs, when they are grown on glass (stiff substrate) or on 4 kPa polyacrylamide hydrogel (soft substrate) with a colorimetric assay based on the production of formazan dye. This assay is in general used to get information on cell viability and proliferation. And indeed when the cells are grown on a plastic or glass plate, the optical density resulting from formazan production often correlates with the cell number.36 The stiffness of the soft plate was chosen in the range of the stiffness met in glioblastoma tissues24 and in skin.25 The two substrates were identically coated with fibronectin to ensure U87-MG and NIH 3T3 cells adhesion (Fig. S5†). The metabolic activity of the two cell lines was quantified 16 h after exposing the cells to a low (5 μg cm−2) and a higher (50 μg cm−2) dose of NPs.A first study was performed to insure that the absorbance of the NPs is negligible at the wavelength of absorption of formazan dye, of 450 nm, so that no correction of the data are necessary. We indeed observed non-significant variations in the absorbance at 450 nm when varying the concentration of NPs, while at 600 nm the optical density was increased in proportion to the NPs concentration (Fig. S6†). This validated that the measure of absorbance at the wavelength of interest for formazan is not biased by the presence of the NPs. Using this methodology, we compared the metabolic activity of the two cell lines as a function of the concentration of NPs and of the cell density. Of note, the cells never reached confluency during the experiment time with the chosen densities. Since in most cases we did not observe any statistical difference as a function of cell densities (Table S2†), we pooled the data by density and a one sample Student's t-test was performed (Fig. 3). This approach enabled to increase the sensitivity of the statistical test. No alteration in the metabolic activity could nevertheless be detected at the two concentrations of NPs assayed.
3.4 NPs induce a dose effect on growth rate, sensitive to matrix stiffness
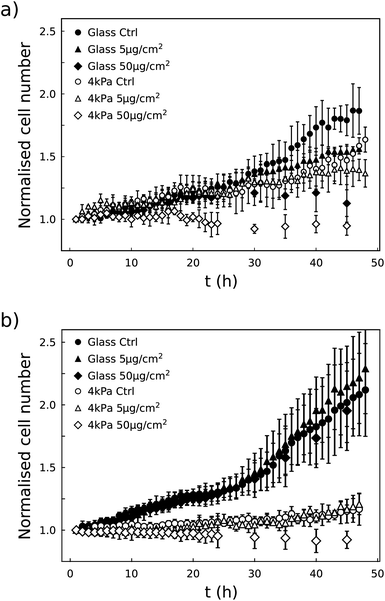
Although the metabolic activity of the cells was not affected by the presence of the particles, we observed that their growth dynamics was altered when exposed to 50 μg per cm2 NPs. This dose effect was observed in the two cell lines, although differently: U87-MG cells were affected whatever the stiffness of the substrate whereas the growth rate of the NIH 3T3 cells was only slowed down on the 4 kPa substrate (Fig. 4). As a whole, we observed that the dose effect of the vortex NPs on cell proliferation was markedly enhanced by the softness of the matrix.3.5 NPs alter the metabolic activity and growth rate of NIH 3T3 cells grown on a 12 kPa matrix
Fig. 4b shows that NIH 3T3 cells hardly proliferate on the 4 kPa substrate, even in the absence of NPs. This is not surprising as these cells do not proliferate much under physiological conditions, in which the stiffness of the tissue is close to 4 kPa,25 in contrast to stiff environments that induce altered proliferative behaviour. Although in this condition the decrease in cell number is clearly visible 24 h post exposure to NPs at a concentration of 50 μg cm−2, the absence of proliferation appears as an obstacle to finely analyse whether the NPs impede proliferation or induce death. To solve this issue, NIH 3T3 cells were also cultured on 12 kPa soft plates (Fig. 5). This stiffness restored cell proliferation. But their behavior contrasted with U87-MG cell line. While both are in a proliferative state on their respective soft support, the metabolic activity of the NIH 3T3 cells grown on 12 kPa substrate was enhanced by the NPs 16 h post exposure, while the proliferation was either fastened at the lowest concentration of NPs, of 5 μg cm−2, or slowed down at the highest concentration, of 50 μg cm−2. Here again, the dose-induced slowdown of the proliferation is more marked on the soft matrix than on the glass support (Fig. 5b).3.6 Cells capture more NPs when grown on a soft matrix
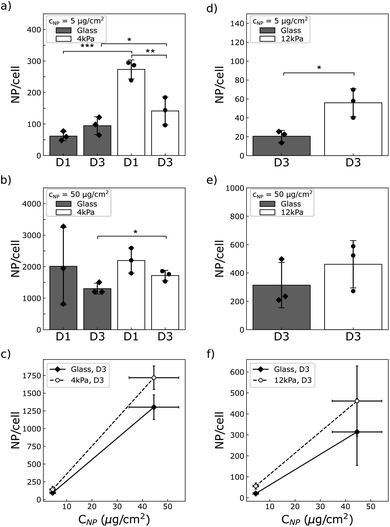
We wondered whether the stiffness-sensitive dose effect we report could be related, among other factors, to a different number of particles per cell depending on the stiffness of the substrate or the cell type. Qualitative observation of the cells by phase contrast microscopy suggests that cells grown on the soft substrates grab more particles than those grown on glass (compare Movies S1 and S3 to S2 and S4†). To quantify this we measured the amount of particles per cell using magnetometry, as described above.As a result, we observed that both U87-MG and NIH 3T3 cells were loaded with more NPs when grown on a soft support compared to glass (Fig. 6). The difference was more pronounced at low concentration of particles, where the cells grown on the soft supports were loaded with many more particles than those grown on glass (approximately 3 times more for the NIH 3T3 cells, and respectively 5 or 1.5 times more for the U87-MG cells seeded at D1 or D3). This difference was reduced at high particle concentration. For instance, for U87-MG cells, this difference dropped to a ratio of few tens of percent.
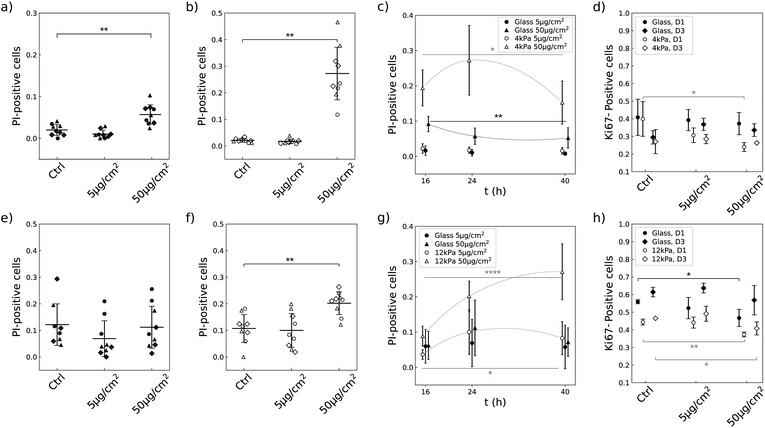
3.7 Soft substrates enhance the propensity of NPs to induce cell death and to decrease the proportion of cycling cells in a lineage-specific manner
The limited growth rate in presence of NPs that we observe on soft substrates could either come from dying cells or from the slowdown of the proliferation rate. We investigated the two hypothesis by measuring the number of dying cells using propidium iodide (PI) and the number of cells engaged in cell cycle using Ki-67 antibody. U87-MG and NIH 3T3 cells have a different sensitivity to the NPs when grown on glass (Compare Fig. 7a, e, d and h). Nevertheless, the two cell lines showed similar qualitative responses when grown on a soft substrate and exposed to NPs at a concentration of 50 μg cm−2: the proportion of dying cells was significantly increased (Fig. 7b and f) and the proportion of cells engaged in the replicative cycle was significantly reduced (Fig. 7d and h). But the amplitude of cell reactions was specific to each lineage as well as their dependence on cell density. For instance, NIH 3T3 cells showed a delayed reaction to the NPs compared to U87-MG cells: the proportion of dying NIH 3T3 cells increased with time and became significantly greater than the control more than 16 h post exposure to the NPs, while the proportion of dying U87-MG cells was already significantly greater than the control 16 h post exposure to the NPs and slightly decreased over time (Fig. 7c and g). Analysis of the number of cells engaged in the replicative cycle showed that indeed the number of proliferating U87-MG cells 16 h post exposure to the NPs is only significantly altered on the soft matrix, while NIH 3T3 cell proliferation is altered on the two substrates, with a more pronounced effect at low cell density (Fig. 7d and h).In contrast, none of the two cell types exposed to a concentration of 5 μg per cm2 NPs showed a significant increase in mortality or reduction in the number of cells engaged in cell cycle on either of the two substrates (Fig. 7c, g, d and h). In this latter case, the cells are loaded with less than 300 NPs per cell. This range of concentration of captured NPs thus does not seem to alter cell viability and cycling.
3.8 Alteration in cell motility does not correlate with cell metabolic activity
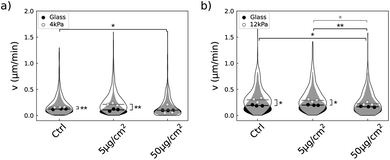
Quantification of variation in the metabolic activity of the two cell lines exposed to the NPs had shown that the U87-MG cells metabolic activity was unaltered at any of the two concentrations of the NPs while it was enhanced in NIH 3T3 cell line grown on 12 kPa (Fig. 3a and 5a). The results obtained with NIH 3T3 cells cannot be explained by the increased growth rate, at least for the higher concentration of NPs, since in this condition the growth rate is lower than in the absence of NPs. As cells look active even when the proliferation is slowed down (Movies S2 and S4†), we wondered whether cell motility could be at least partly responsible for the increased metabolic activity. Quantification of cell speed by tracking nuclei discarded this hypothesis (Fig. 8). Cell motility was quantified from 14 h to 18 h post exposure to the NPs, at the rate of one image every hour. During this lapse, the median cell velocity remained constant which enabled to pool the data from 14 h to 18 h (Table S1†). Comparison of the mean median values showed that increasing the NP concentration resulted in a reduction in NIH 3T3 velocity independent of substrate compliance (Fig. 8b). This effect is less marked for U87-MG cells, where only cells grown on glass show such a reduction in their speed (Fig. 8a). Furthermore, we observed that while both cell types moved faster on soft substrates than on glass, the difference in speed was reduced when the cells were exposed to a concentration of 50 μg per cm2 NP, suggesting that a high dose of NP inhibits the mechanosensitivity of cell migration. Overall, we found that the reduction in cell velocity in both cell lines is not consistent with the lack of variation in metabolic activity we report in U87-MG cells exposed to NPs nor can it explain the increase in metabolic activity in NIH 3T3 cells grown on the 12 kPa soft substrate in the presence of NPs.4 Discussion
It has been known for several years that in vitro assessment of the cytotoxicity of pharmacological compounds depends on the mechanical environment in which the cells are grown.37–40 This effect has been associated to the mechanosensitivity of endocytotic processes in relation to the intracellular tension and to the stiffness of the plasma membrane.20,41–43 However, no unified description valid for all types of cells or drugs could be identified. A reason could be that no universal relation has neither been found between membrane tension, involved in drug internalisation, and cytoskeletal tension, which cells vary to adapt to their mechanical environment.44 And in addition, cell phenotypic state has recently been shown to adapt to the mechanical environment, which can lead to over or under expression of signalling pathways targeted by the pharmacological compounds.21,45Despite the proximity of the issues met in NPs and drug delivery communities, the assessment of the NP cytotoxicity in relation to the stiffness of the extracellular environment has, to our best knowledge, not been addressed. Several studies have focused on the influence of the stiffness of the extracellular matrix on nanoparticle uptake.46–48 Controversial results were reported, some showing reduced endocytotic capabilities on soft substrates46,47 while others reporting increased internalisation of NPs for cells grown on a soft matrix.48 While these studies addressed particles of similar sizes, from 100 to 500 nm, and shape, close to spherical, the composition and surface properties of the NPs under study were different. In addition, the material properties of the soft environment (surface chemistry and rheology) and cell densities also differed from one study to another. The differences in the reported observations then suggest that all these parameters may indeed be of importance and have a strong impact on cell/NPs interaction. In this context, our study brings additional data to this topic. We show that U87-MG glioblastoma cells and NIH 3T3 fibroblast cells exhibit a higher uptake of gold coated-Ni80Fe20 microdiscs when grown on soft substrates made of polyacrylamide hydrogel coated with fibronectin (Fig. 6), at all concentrations assayed in this study (Table 1). In complement to the previous studies, we show that this increased uptake depends on cell density and NPs concentration in a complex manner. For instance, U87-MG cells exhibit a larger relative uptake on soft substrate when they are exposed to a low concentration of NPs and grown at low cell density. Similar trend are observed for the NIH 3T3 cell line. The difference in the amount of NP taken up per cell is reduced when the cells are exposed to the highest concentration of NPs probed here, about 50 μg cm−2. The variation of NP content in relation to cell density was not possible to quantify in NIH 3T3 cells due to the low amount of particles these cells capture when they are grown at low density, and the resulting low signal-to-noise ratio coming from the collection of NPs that were not interacting with cells (see Materials and methods).
| Glass | Soft | ||||
|---|---|---|---|---|---|
| 5 μg cm−2 | 50 μg cm−2 | 5 μg cm−2 | 50 μg cm−2 | ||
| a The number of captured NPs and of KI-67 positive cells are measured in cells seeded at D3 density. Significance compared to the control (absence of NPs) is shown in parenthesis: n.s.,* and ** denote respectively non significant, p < 0.05 and p < 0.01. | |||||
| U87-MG | # NPs | 95 ± 28 | 1231 ± 174 | 142 ± 44 | 1718 ± 166 |
| PI | 1% (n.s.) | 6% (**) | 2% (n.s.) | 27% (**) | |
| Ki-67 | 37% (n.s.) | 34% (n.s.) | 29% (n.s.) | 26% (n.s.) | |
| NIH 3T3 | # NPs | 21 ± 6 | 314 ± 160 | 56 ± 15 | 461 ± 167 |
| PI | 7% (n.s.) | 11% (n.s.) | 10% (n.s.) | 20% (**) | |
| Ki-67 | 64% (n.s.) | 57% (*) | 49% (n.s.) | 41% (*) | |
In order to investigate whether the toxicity of NPs could vary with the mechanical properties of the extracellular environment, we used flake-shaped NPs composed of a Ni80Fe20 core coated with gold (Fig. 1). These particles, also known as vortex microdiscs because of their magnetic properties, have been used in vitro and in vivo to either induce cell apoptosis5,11,12,14 or increase insulin production in pancreatic cells.15 Most of these studies used a very low number of NPs, below 50 NPs per cells and showed either no or a limited toxicity.14,15 Our results are in line with these earlier observations, since toxic effects are here seen for cells exposed to highest concentration of particles (50 μg cm−2), corresponding to several hundreds of NPs per cell, but not for the lowest concentration of particles (5 μg cm−2) similar to the one previously used in experiments of magneto-mechanical stimulation of cells. A dose assay conducted with renal carcinoma cells grown in plastic plates12 nevertheless showed a reduced and fairly stable low toxicity over several decades of NPs concentration, a result that differs from our study where we report a decrease in cell viability when increasing the concentration of NPs (Fig. 7).
A reason is that all these studies used the production of formazan dye to assess cell viability. Here we show that this approach does not give any direct information on cell viability when the cells are grown on a soft substrate. Indeed, although we observe an increased uptake of NPs accompanied with an increased death rate and a decreased proliferation rate in cells grown on a soft substrate (Fig. 7), we do not observe any decrease of the metabolic activity (Fig. 3). In contrast, the NIH 3T3 cells grown on 12 kPa substrate even exhibit an increased metabolic activity in parallel with an increased death rate (Fig. 5 and 7). Indeed, we observed that the cells remain very active under this condition (Movies S1–S4†). But the increased metabolic activity of the NIH 3T3 cells grown on 12 kPa support could not be explained by such enhanced motility (Fig. 8). These intriguing results highlight the complexity of the interaction between cells and NPs and lead to conclude that for cells grown on a soft substrate, metabolic assays are not predictive of cell viability: functions other than cell proliferation or migration, which we were unable to identify, make a dominant contribution to ATP production and compensate for toxicity-triggered deficits.
One plausible origin for the toxicity of these NPs could be the uneven gold coating on the lateral faces of the particles (Fig. 1), exposing some of the core nickel-iron alloy. Various experiments with gold, iron oxide, as well as with various types of NPs, indicate that in most cases the main toxicity effect arises from the release of metal ions, which is exacerbated when the particles are located in acidic lysosomes.49–52 And indeed, in consistence with previous studies,15 we observed that the particles either interact with the membrane or enter the cell body and concentrate around the nucleus being possibly trapped in endosomes or lysosomes (Fig. 2, S4 and Movies S1–S4†). Here we expect that the cytotoxicity mainly comes from the release of nickel ions as among the constituents of these magnetic microdiscs, nickel is potentially the most toxic. Nickel nanoparticles, in the form of nickel oxide or Ni-based compounds were shown to cause significant oxidative stress and cell alteration even at pharmacological dose,53 with differences in cytotoxicity and ROS production depending on particle size.54 It can be expected that, due to size-dependence in the endocytose mechanisms55 and endosomal/lysosomal pathways56 as well as the dependence of these pathways on the mechanical properties of the extracellular environment,20,57,58 some differences in these behaviors might be observed.
More precisely, in the present study, the cytotoxicity results in a reduction of the proliferation rate together with an alteration of the cell cycle as well as the increase of the death rate (Fig. 7). Compilation of all the results brings to light that besides the fact that the number of NPs captured by the cells differs on soft and stiff substrates, the dose effect also differs quantitatively between the two cell types (Table 1). For instance, while NIH 3T3 cells exposed to a NP concentration of 50 μg cm−2 take up approximately the same number of NPs when grown on any of the substrates when seeded at D3 (Fig. 6e), the outcome for the cells is much more severe when grown on the soft substrate: the proliferation is virtually stopped on the soft substrate unlike to glass (Fig. 5b), the death rate is drastically increased (Fig. 7e and f) and a significant proportion of the cells are put out of the replicative cycle (Fig. 7h). This effect is also visible in U87-MG cells which interact with a greater number of NPs, although it cannot easily be dissociated from the difference in particle uptake observed between the two substrates. As previously mentioned, many cellular pathways involved in the intracellular trafficking of the nanoparticles, including endocytosis or endosomal/lysosomal pathways, are differentially activated when varying the mechanical properties of the extracellular environment. However, our intriguing result that NIH 3T3 cells have a higher metabolic activity on 12 kPa compared to glass (Fig. 5a) while their proliferation is reduced (Fig. 5b) and their migration unaffected (Fig. 8b) suggests that the softness of the matrix could introduce differences in the integrated stress response associated to the oxidative stress. And indeed, variation in the activation of the stress response was shown to be responsible for the difference in toxicity of gold nanorods between normal and cancer cell lines.59,60 This hypothesis, that deserves further studies, is in line with the recent observation that the stress response is also differentially activated in cells grown on soft versus stiff supports.21
Of note, the comparison of the two cell types is performed 16 h post exposure to the NPs. As Fig. 7c and g show, the kinetics of cell/NPs interaction indeed differs between the two cell types. While the death rate of the U87-MG cells decreases in time, the death rate of the NIH 3T3 cells grown on the soft substrate significantly increases during the same time lapse. In addition, the visual observation of the cells over 24 h shows that NIH 3T3 cells take a longer time to eventually capture NPs than U87-MG cells (compare Movies S1, S2 and S3, S4†). These observations therefore suggest that U87-MG cells may indeed have grabbed most of the particles in a delay shorter than our first observation time point, and then share the particles with daughter cells following cell division as already reported.14 Subsequently, the toxicity would decrease in time because the number of NPs/cell also decreases. Consistently, alteration of the proliferation rate can be seen from the first hour following exposure to NPs (Fig. 4a). In contrast, NIH 3T3 cells need more time to take up the NPs, leading to an enhancement of the cytotoxic effects during the first 40 h. And consistently, the proliferation curve at the highest concentration of NPs shows a significant slowdown compared to the control only after a delay, of approximately 10 h (Fig. 5b), in contrast to U87-MG where the slowdown of proliferation is already observable 6 h post exposure to the NPs.
All these observations were made with cells grown on substrates of different stiffnesses but coated with an identical surface density of fibronectin (Fig. S5†), a protein that is both found in the skin61 and in the brain.62 It has been shown that growing cells on such mechanomimetic supports brings them into a state closer to human physiology than when they are grown on plastic plates.21 However, the stiffness of the extracellular environment is not the only factor influencing cellular responses; the surface coating may also play an important role.63,64 For instance metabolic activity in cells in relation to drug screening was shown to depend on the protein coatings.65,66 Therefore, to achieve a high level of prediction in in vitro cytotoxicity assays, the influence of the culture plate surface coating should also be assessed. Unlike mechanical properties, the latter evolves over time with cell secretions. It is therefore expected that varying the composition of the extracellular matrix will have a greater effect than simply adjusting its surface density.
5 Conclusions
Overall, just as the toxic effects of NPs depend on the type of cells or the material coating, we show that it also depends on the mechanical environment. Our findings are that cells grown on the soft matrix grab more particles than those grown on glass, and that cell reaction to the toxicity of the particles is enhanced on the soft substrate. This observation should be compared with the large impact the stiffness of the extracellular matrix has on basic cellular responses such as cell metabolism.57 It thus suggests that in vitro assays to assess the cytotoxicity of nanomaterials may gain relevance by considering growing the cells on substrates whose stiffness is representative of the tissue or the pathology of interest. Interestingly, our study also shows that the evaluation of the sensitivity of NP cytotoxicity to the compliance of the extracellular environment cannot be apprehended by solely measuring cell metabolic activity. Variations in the stiffness of the matrix impact many cellular responses, including cell proliferation, death, migration and endocytosis, which all contribute to the metabolic activity. And we could see that some of them contribute in opposite direction, for instance cell proliferation being slowed down and particle grabbing or cell migration being enhanced by the softness of the substrate.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
AV performed the experiments under the supervision of RM and AN. AV and SS implemented AI to analyze the data. SC and SL produced FIBSEM and SEM images. AV, RM and AN analyzed and curated the data. AV, FB, RM and AN designed the experiments. HJ, BD, RM and AN acquired the project funding, conceptualized the study and developed the methodology. AV, SC, RM and AN wrote the initial draft which was reviewed and edited by all authors.Conflicts of interest
AN is co-founder, shareholder and scientific advisor of Cell&Soft company that provided the pre-coated glass and soft plates for cell culture.Acknowledgements
This work is supported by the French National Research Agency in the framework of the “Investissements d'avenir” program (ANR-15-IDEX-02) and by the french Renatech network. This project received help from MuLife imaging facility, which is funded by GRAL, a programme from the Chemistry Biology Health Graduate School of University Grenoble Alpes (ANR-17-EURE-0003). S. C. thanks H. Dansas for very fruitful discussions.Notes and references
- D. Kilinc, C. L. Dennis and G. U. Lee, Adv. Mater., 2016, 28, 5672–5680 CrossRef CAS.
- A. Van de Walle, J. E. Perez, A. Abou-Hassan, M. Hemadi, N. Luciani and C. Wilhelm, Mater. Today Nano, 2020, 100084 CrossRef.
- A. V. Samrot, C. S. Sahithya, J. A. Selvarani, S. K. Purayil and P. Ponnaiah, Curr. Opin. Green Sustainable Chem., 2021, 4, 100042 CrossRef CAS.
- C. Pucci, A. Degl'Innocenti, M. Belenli Gümüş and G. Ciofani, Biomater. Sci., 2022, 10, 2103–2121 RSC.
- C. Naud, C. Thébault, M. Carrière, Y. Hou, R. Morel, F. Berger, B. Diény and H. Joisten, Nanoscale Adv., 2020, 2, 3632–3655 RSC.
- H. Brueckl, A. Shoshi, S. Schrittwieser, B. Schmid, P. Schneeweiss, T. Mitteramskogler, M. J. Haslinger, M. Muehlberger and J. Schotter, Sci. Rep., 2021, 11, 6039 CrossRef CAS.
- M. Goiriena-Goikoetxea, D. Muñoz, I. Orue, M. L. Fernández-Gubieda, J. Bokor, A. Muela and A. García-Arribas, Appl. Phys. Rev., 2020, 7, 011306 CAS.
- L. Peixoto, R. Magalhães, D. Navas, S. Moraes, C. Redondo, R. Morales, J. P. Araújo and C. T. Sousa, Appl. Phys. Rev., 2020, 7, 011310 CAS.
- U. Schwarz, Soft Matter, 2007, 3, 263–266 RSC.
- V. Novosad and E. A. Rozhkova, in Ferromagnets-Based Multifunctional Nanoplatform for Targeted Cancer Therapy, InTech, 2011, ch. 18, pp. 425–444 Search PubMed.
- D.-H. Kim, E. A. Rozhkova, I. V. Ulasov, S. D. Bader, T. Rajh, M. S. Lesniak and V. Novosad, Nat. Mater., 2010, 9, 165–171 CrossRef CAS.
- S. Leulmi, X. Chauchet, M. Morcrette, G. Ortiz, H. Joisten, P. Sabon, T. Livache, Y. Hou, M. Carrière, S. Lequien and B. Dieny, Nanoscale, 2015, 7, 15904–15914 RSC.
- S. Leulmi, H. Joisten, T. Dietsch, C. Iss, M. Morcrette, S. Auffret, P. Sabon and B. Dieny, Appl. Phys. Lett., 2013, 103, 132412 CrossRef.
- Y. Cheng, M. E. Muroski, D. C. Petit, R. Mansell, T. Vemulkar, R. A. Morshed, Y. Han, I. V. Balyasnikova, C. M. Horbinski, X. Huang, L. Zhang, R. P. Cowburn and M. S. Lesniak, J. Controlled Release, 2016, 223, 75–84 CrossRef CAS PubMed.
- S. Ponomareva, H. Joisten, T. François, C. Naud, R. Morel, Y. Hou, T. Myers, I. Joumard, B. Dieny and M. Carriere, Nanoscale, 2022, 14, 13274–13283 RSC.
- Official Journal of the European Union, https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32022H0614(01).
- G. Genchi, A. Carocci, G. Lauria, M. S. Sinicropi and A. Catalano, Int. J. Environ. Res. Public Health, 2020, 17, 679 CrossRef CAS.
- S. Ponomareva, M. Carriere, Y. Hou, R. Morel, B. Dieny and H. Joisten, Adv. Intell. Syst., 2023, 5, 2300022 CrossRef.
- I. Levental, P. C. Georges and P. A. Janmey, Soft Matter, 2007, 3, 299–306 RSC.
- D. Missirlis, PLoS One, 2014, 9, e96548 CrossRef.
- P.-A. Laval, M. Piecyk, P. Le Guen, M.-D. Ilie, J. Fauvre, I. Coste, T. Renno, N. Aznar, C. Hadji, C. Migdal, C. Duret, P. Bertolino, C. Ferraro-Peyret, A. Nicolas and C. Chaveroux, Acta Biomater., 2024, 182, 93–110 CrossRef CAS PubMed.
- B. Dieny, A. Visona, C. Naud, H. Joisten, R. Morel, A. Nicolas, M. Carriere and F. Berger, Int. J. Magn. Part. Imaging, 2024, 10, 787 Search PubMed.
- A. Sohrabi, A. E. Lefebvre, M. J. Harrison, M. C. Condro, T. M. Sanazzaro, G. Safarians, I. Solomon, S. Bastola, S. Kordbacheh, N. Toh, H. I. Kornblum, M. A. Digman and S. K. Seidlits, Cell Rep., 2023, 42, 113175 CrossRef CAS PubMed.
- G. Ciasca, T. E. Sassun, E. Minelli, M. Antonelli, M. Papi, A. Santoro, F. Giangaspero, R. Delfini and M. De Spirito, Nanoscale, 2016, 8, 19629–19643 RSC.
- V. F. Achterberg, L. Buscemi, H. Diekmann, J. Smith-Clerc, H. Schwengler, J.-J. Meister, H. Wenck, S. Gallinat and B. Hinz, J. Invest. Dermatol., 2014, 134, 1862–1872 CrossRef CAS.
- F. S. Younesi, A. E. Miller, T. H. Barker, F. M. V. Rossi and B. Hinz, Nat. Rev. Mol. Cell Biol., 2024, 25, 617–638 CrossRef CAS PubMed.
- H. Joisten, T. Courcier, P. Balint, P. Sabon, J. Faure-Vincent, S. Auffret and B. Dieny, Appl. Phys. Lett., 2010, 97, 253112 CrossRef.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS.
- U. Schmidt, M. Weigert, C. Broaddus and G. Myers, in Cell Detection with Star-Convex Polygons, Springer International Publishing, 2018, pp. 265–273 Search PubMed.
- C. Stringer, T. Wang, M. Michaelos and M. Pachitariu, Nat. Methods, 2020, 18, 100–106 CrossRef PubMed.
- X. Sun and P. D. Kaufman, Chromosoma, 2018, 127, 175–186 CrossRef CAS.
- J.-Y. Tinevez, N. Perry, J. Schindelin, G. M. Hoopes, G. D. Reynolds, E. Laplantine, S. Y. Bednarek, S. L. Shorte and K. W. Eliceiri, Methods, 2017, 115, 80–90 CrossRef CAS PubMed.
- M. Salou, B. Lescop, S. Rioual, A. Lebon, J. B. Youssef and B. Rouvellou, Surf. Sci., 2008, 602, 2901–2906 CrossRef CAS.
- M. H. Cambre, N. J. Holl, B. Wang, L. Harper, H.-J. Lee, C. C. Chusuei, F. Y. Hou, E. T. Williams, J. D. Argo, R. R. Pandey and Y.-W. Huang, Int. J. Mol. Sci., 2020, 21, 2355 CrossRef CAS.
- N. Gal, S. Massalha, O. Samuelly-Nafta and D. Weihs, Med. Eng. Phys., 2015, 37, 478–483 CrossRef CAS PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS.
- J. D. Mih, A. S. Sharif, F. Liu, A. Marinkovic, M. M. Symer and D. J. Tschumperlin, PLoS One, 2011, 6, e19929 CrossRef CAS PubMed.
- S. Zustiak, R. Nossal and D. L. Sackett, Biotechnol. Bioeng., 2014, 111, 396–403 CrossRef CAS.
- R. Krishnan, J.-A. Park, C. Y. Seow, P. V.-S. Lee and A. G. Stewart, Trends Pharmacol. Sci., 2016, 37, 87–100 CrossRef CAS PubMed.
- S. H. Medina, B. Bush, M. Cam, E. Sevcik, F. W. DelRio, K. Nandy and J. P. Schneider, Biomaterials, 2019, 202, 1–11 CrossRef CAS.
- S. Boulant, C. Kural, J.-C. Zeeh, F. Ubelmann and T. Kirchhausen, Nat. Cell Biol., 2011, 13, 1124–1131 CrossRef CAS PubMed.
- T. M. Kruger, K. J. Bell, T. I. Lansakara, A. V. Tivanski, J. A. Doorn and L. L. Stevens, ACS Chem. Neurosci., 2018, 10, 1284–1293 CrossRef.
- J. Planade, R. Belbahri, M. Boiero Sanders, A. Guillotin, O. du Roure, A. Michelot and J. Heuvingh, PLoS Biol., 2019, 17, e3000500 CrossRef CAS.
- P. A. Janmey, D. A. Fletcher and C. A. Reinhart-King, Physiol. Rev., 2020, 100, 695–724 CrossRef CAS PubMed.
- P. Romani, L. Valcarcel-Jimenez, C. Frezza and S. Dupont, Nat. Rev. Mol. Cell Biol., 2020, 1–17 Search PubMed.
- C. Huang, P. J. Butler, S. Tong, H. S. Muddana, G. Bao and S. Zhang, Nano Lett., 2013, 13, 1611–1615 CrossRef CAS.
- X. Wei, R. Wei, G. Jiang, Y. Jia, H. Lou, Z. Yang, D. Luo, Q. Huang, S. Xu, X. Yang, Y. Zhou, X. Li, T. Ji, J. Hu, L. Xi, D. Ma, F. Ye and Q. Gao, Nanomedicine, 2019, 14, 613–626 CrossRef CAS.
- A. Lee, M. Sousa de Almeida, D. Milinkovic, D. Septiadi, P. Taladriz-Blanco, C. Loussert-Fonta, S. Balog, A. Bazzoni, B. Rothen-Rutishauser and A. Petri-Fink, Nanoscale, 2022, 14, 15141–15155 RSC.
- P. J. Barnard and S. J. Berners-Price, Coord. Chem. Rev., 2007, 251, 1889–1902 CrossRef CAS.
- S. Sabella, R. P. Carney, V. Brunetti, M. A. Malvindi, N. Al-Juffali, G. Vecchio, S. M. Janes, O. M. Bakr, R. Cingolani, F. Stellacci and P. P. Pompa, Nanoscale, 2014, 6, 7052 RSC.
- A. Van de Walle, J. Kolosnjaj-Tabi, Y. Lalatonne and C. Wilhelm, Acc. Chem. Res., 2020, 53, 2212–2224 CrossRef CAS.
- A. Fromain, J. E. Perez, A. Van de Walle, Y. Lalatonne and C. Wilhelm, Nat. Commun., 2023, 14, 4637 CrossRef CAS PubMed.
- K. Peters, R. Unger, A. Gatti, E. Sabbioni, R. Tsaryk and C. Kirkpatrick, Int. J. Immunopathol. Pharmacol., 2007, 20, 685–695 CrossRef CAS.
- S. Latvala, J. Hedberg, S. Di Bucchianico, L. Möller, I. Odnevall Wallinder, K. Elihn and H. L. Karlsson, PLoS One, 2016, 11, e0159684 CrossRef PubMed.
- S. E. A. Gratton, P. A. Ropp, P. D. Pohlhaus, J. C. Luft, V. J. Madden, M. E. Napier and J. M. DeSimone, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 11613–11618 CrossRef CAS.
- J. Rejman, V. Oberle, I. S. Zuhorn and D. Hoekstra, Biochem. J., 2004, 377, 159–169 CrossRef CAS PubMed.
- K. M. Tharp, R. Higuchi-Sanabria, G. A. Timblin, B. Ford, C. Garzon-Coral, C. Schneider, J. M. Muncie, C. Stashko, J. R. Daniele, A. S. Moore, P. A. Frankino, S. Homentcovschi, S. S. Manoli, H. Shao, A. L. Richards, K.-H. Chen, J. t. Hoeve, G. M. Ku, M. Hellerstein, D. K. Nomura, K. Saijo, J. Gestwicki, A. R. Dunn, N. J. Krogan, D. L. Swaney, A. Dillin and V. M. Weaver, Cell Metab., 2021, 33, 1322–1341 CrossRef CAS PubMed.
- P. V. Taufalele and C. A. Reinhart-King, Trends Cancer, 2021, 7, 883–885 CrossRef CAS PubMed.
- L. Zhang, L. Wang, Y. Hu, Z. Liu, Y. Tian, X. Wu, Y. Zhao, H. Tang, C. Chen and Y. Wang, Biomaterials, 2013, 34, 7117–7126 CrossRef CAS PubMed.
- M. Awashra and P. Młynarz, Nanoscale Adv., 2023, 5, 2674–2723 RSC.
- K. Pfisterer, L. E. Shaw, D. Symmank and W. Weninger, Front. Cell Dev. Biol., 2021, 9, 682414 CrossRef PubMed.
- L. W. Lau, R. Cua, M. B. Keough, S. Haylock-Jacobs and V. W. Yong, Nat. Rev. Neurosci., 2013, 14, 722–729 CrossRef CAS PubMed.
- A. P. Kourouklis, K. B. Kaylan and G. H. Underhill, Biomaterials, 2016, 99, 82–94 CrossRef CAS PubMed.
- A. E. Stanton, X. Tong and F. Yang, Acta Biomater., 2019, 96, 310–320 Search PubMed.
- S. Forciniti, E. Dalla Pozza, M. R. Greco, T. M. Amaral Carvalho, B. Rolando, G. Ambrosini, C. A. Carmona-Carmona, R. Pacchiana, D. Di Molfetta, M. Donadelli, S. Arpicco, M. Palmieri, S. J. Reshkin, I. Dando and R. A. Cardone, Int. J. Mol. Sci., 2020, 22, 29 CrossRef PubMed.
- D. Di Molfetta, S. Cannone, M. R. Greco, R. Caroppo, F. Piccapane, T. M. A. Carvalho, C. Altamura, I. Saltarella, D. Tavares Valente, J. F. Desaphy, S. J. Reshkin and R. A. Cardone, Int. J. Mol. Sci., 2023, 24, 10632 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00704b |
| This journal is © The Royal Society of Chemistry 2025 |