Open Access Article
Open Access ArticleNear-infrared DNA biosensors based on polysulfonate coatings for the sensitive detection of microRNAs
Xianghang
Lin†
ab,
Yang
Yang†
b,
Wenzhang
Zhu†
c,
Xiaorong
He
*d and
Yunliang
Liu
 *ab
*ab
aDepartment of Otolaryngology, Fujian Children's Hospital (Fujian Branch of Shanghai Children's Medical Center), College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, 350014, China. E-mail: liuyunliang2024@yeah.net
bDepartment of Otolaryngology, Fujian Maternity and Child Health Hospital, College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, 350001, China
cCollege of Materials Science and Engineering, Fuzhou University, Fuzhou, 350108, China
dDermatology Institute of Fuzhou, Dermatology Hospital of Fuzhou, Fuzhou, 350001, China. E-mail: xiaorong.he93@gmail.com
First published on 26th November 2024
Abstract
MicroRNAs (miRNAs) play crucial roles in the regulation of immune cell differentiation and the immune response during allergic rhinitis (AR). Studies have shown that miRNA-155 is significantly upregulated in AR pathogenesis. Therefore, miRNA-155 can be used as a biomarker for AR diagnosis. Although fluorescent biosensors based on upconversion nanoparticles (UCNPs) have made significant advances in the detection of miRNAs, developing UCNPs with polymer coatings, efficient surface passivation, and DNA functionalization for hybrid sensing in biological media remains challenging. Herein, hairpin DNA1 (H1) is modified into a thin polysulfonic acid layer on UCNPs by sulfonamide bonds, and the fluorescence of the UCNPs is quenched by the fluorescence resonance energy transfer (FRET) process of BHQ3 carried by H1. When the target miRNA-155 is present, the hairpin structure of H1 is opened, allowing BHQ3 to move away from the UCNP surface, and the fluorescence of UCNP is restored. At the same time, hairpin DNA1 (H2) can combine with H1 to replace the miRNA-155 that is bound to H1 with the help of the opening stem ring structure of H1, and the replaced miRNA-155 can continue to react with H1 to amplify the fluorescence signal. Under the optimal experimental conditions, the linear range of miRNA-155 is 0.01–3 nM, with a detection limit of 1.14 pM. Furthermore, the constructed biosensor has been applied to determine miRNA-155 in serum samples, and the spiked recoveries range from 99.8% to 104.8%, which indicates that the developed assay has potential applications in monitoring allergic rhinitis or other miRNA related diseases.
1 Introduction
Allergic rhinitis (AR), also known as hay fever, is a nasal mucosal allergic disease caused by the release of histamine and other mediators mediated by IgE in atopic individuals after exposure to allergens.1 Multiple immune-active cells and cytokines are involved in pathogenesis. Previous studies have shown that there is differential expression of miRNAs during the pathogenesis of AR, such as significant upregulation or downregulation of miR-155.2 Therefore, the development of microRNA (miRNA) detection methods aims to provide a new perspective for the effective treatment of AR. Traditional methods for detecting miRNAs, such as polymerase chain reaction (PCR),3 northern blotting,4 and microarrays,5 require expensive and demanding nucleases,6 resulting in a complex and costly detection process. In addition, considering the characteristics of miRNAs, such as long chain length, high sequence likeness, and low abundance in biological fluids,7 it is necessary to develop sensitive, specific, and easy-to-use detection methods.At present, many methods have been used to detect miRNA-155, including electrochemical methods,8–10 colorimetric methods11 and fluorescence methods.12 The above methods often need to combine oligonucleotide signal amplification methods to achieve sensitive detection,13 such as hybridization chain reaction (HCR),14 strand displacement reaction (SDR)15 and catalytic hairpin assembly (CHA).16 In addition, the fluorescence method is considered an effective method for detecting miRNAs because of its high sensitivity, low cost, easy operation, and ease of reading.17 At present, the luminescent materials used in fluorescence methods include mainly fluorescent dyes,18 nanoclusters,19 quantum dots (QDs),20 and upconversion nanoparticles (UCNPs).21 Compared with other fluorescent material, UCNPs have many unique merits, such as greater tissue penetration depth, excellent photochemical stability and weak autofluorescence, because they can assimilate near-infrared (NIR) radiation and then emit radiation from ultraviolet or visible light.22 Therefore, upconversion luminescence (UCL) has the ability to effectively ameliorate the behavior of bioanalytical sensors and approaches.23 In addition, the use of fluorescence resonance energy transfer (FRET) to determine biological analytes has the merits of a simple step, good stability, sensitive measurement, and easy loading on various nanomaterial-based platforms.24 UCNP-based FRET assays have been widely used for miRNA detection.25 However, the diameters of UCNPs are usually greater than 20 nm, causing UCL to be quenched in aqueous media, so their surfaces are generally coated.26 Furthermore, proper surface adjustment with a functional identification group is necessary for efficacious biosensing.27 Given the distance between the donor and acceptor for FRET and the specific combination of another biomolecule labelled with a FRET acceptor, it is still challenging to use surface-modified UCNPs to construct a simple, sensitive, and specific UCNP-based FRET biosensor for the detection of miRNAs.
Common surface modification techniques for UCNPs include ligand exchange,28 ligand removal,29 silica shell capping,30 and polymer coating.31 Among them, polymer shell capping can prevent interactions between the UCNP surface and the phosphate skeleton of DNA as well as other biomolecules, and the large number of functional groups carried can achieve covalent crosslinking with DNA. Tian et al.32 coated N-hydroxysuccinimide ester polyethylene glycol maleimide onto the surface of UCNPs to obtain amino-functionalized UCNPs, which were used for the intracellular detection of miRNAs. However, existing polymer encapsulation often leads to strong electrostatic interactions between modified UCNPs and DNA. This nonspecific influence can cause UCNP accumulation and/or inhibit DNA binding with complementary strands, hindering miRNA sensing.33 Polysulfonates can effectively prevent the annihilation of UCNPs, thereby preventing adverse cell toxicity and ensuring long-term stability. In addition, studies have shown that compared with other polymers such as polyethylene glycol, given the large number of sulfonate groups, a thin and adjustable polymer shell can be grown on the outside of UCNPs, which is essential for UCNPs to act as FRET donors. However, there is currently limited research on the use of polysulfonate-coated UCNPs for detecting miRNAs.
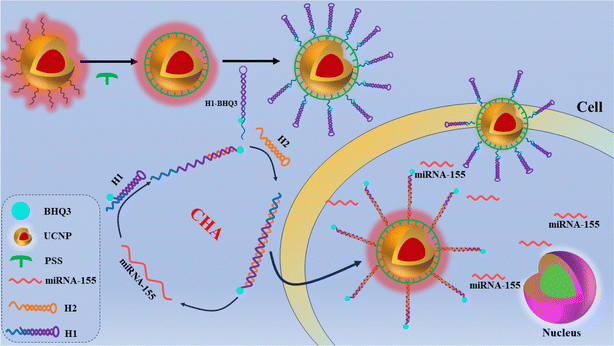
In this study, considering the potential advantages of polysulfonates, we designed UCNP-DNA-based FRET biosensors. Fig. 1 illustrates the detection principle of the constructed sensor for miRNA-155. First, poly(4-styrenesulfonic acid sodium) (PSS) was modified onto the surface of the UCNPs, and the sulfonate groups on the surface of the UCNPs were combined with hairpin DNA (H1) modified with amino functional groups at the 5′ end through a sulfonamide reaction. The other end of H1 is modified with a black hole quencher (BHQ3). The hairpin structure of H1 can cause the distance between BHQ3 and UCNP@PSS to be less than 10 nm, and the FRET process occurs between them, quenching the fluorescence of the UCNPs. When the target miRNA-155 is present, it reacts with H1 to open the stem loop structure of H1, forming a heteroduplex structure of H1-miRNA, causing the quenching group at the 3′ end to move away from the UCNP, terminating the FRET process, and restoring the red fluorescence of the UCNP. At the same time, the previously locked toe portion is exposed, and H2 can react with H1. Owing to the stronger binding force between H1 and H2 than between H1 and miRNA-155, miRNA-155 is replaced from H1 and reacts with a new H1 again, triggering catalytic hairpin amplification. This work provides an efficient, sensitive, and specific biosensor for detecting miRNAs.
2 Experimental
2.1 Materials
ErCl3·6H2O, TmCl3·6H2O, YCl3·6H2O, GdCl3·6H2O, YbCl3·6H2O, oleic acid (OA), octadecene (ODE), ammonium fluoride (NH4F), sodium hydroxide (NaOH), nitrosyl tetrafluoroborate (NOBF4), cyanuric chloride (TCT), poly(sodium 4-styrenesulfonate) (PSS), magnesium chloride (MgCl2), sodium chloride (NaCl) and tris(hydroxymethyl)aminomethane (Tris) were obtained from Aladdin Reagent Co., Ltd (Shanghai, China). Methanol (CH3OH), ethanol (C2H5OH), cyclohexane (C6H12), N,N-dimethylformamide (DMF), dichloromethane (CH2Cl2), and hydrochloric acid (HCl) were obtained from Shanghai Chemical Reagent Company (Shanghai, China). Tris + EDTA buffer (TE Buffer) and phosphate-buffered saline (PBS) (0.01 M, pH 7.2–7.4) were purchased from Sangon, Inc. (Shanghai, China). The oligonucleotides were synthesized and purified via HPLC by Sangon Biotech Co. The base sequences from left to right (from 5′ to 3′) of the miRNAs are listed below:H1: NH2GGGGGGGGGGGTTTGTTGAATCGTGATAGGGGTCCATGTGTAGAACCCCTATCACGATTAGCATTAA-BHQ3
H2: CCATGTGTAGAAATCGTGATAGGGGTTCTACACATGGACCCCTAT
miRNA-155: UUAAUGCUAAUCGUGAUAGGGGU
miRNA-222: AGCUACAUCUGGCUACUGGGU
miRNA-214: UGCCUGUCUACACUUGCUGCUGC
miRNA-21: UAGCUUAUCAGACUGAUGUUGA
2.2 Instruments
UV-vis absorption spectra were acquired with a LAMBDA 950-visible spectrophotometer (PerkinElmer, USA). The fluorescence spectra were recorded using a FluoroMax-4 (Horiba Jobin Yvon, France). Fourier-transform infrared (FTIR) analysis was performed with a Nicolet 5700 instrument (Thermal Power Company, USA). X-ray diffraction (XRD) patterns were recorded using a Miniflex600 instrument (Rigaku Corporation, Japan). Zeta potential and dynamic light scattering (DLS) experiments were conducted on a Nano-ZS90 instrument (Malvern Instruments, UK). Transmission electron microscopy (TEM) was performed using a Talos F200i electron microscope (FEI, America).2.3 Preparation of NaErF4: Tm@NaGdF4:Yb (UCNP) core–shell nanoparticles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) was produced. The mixture was subsequently heated to 160 °C in an argon atmosphere and kept warm for 30 min to ensure that the raw material was completely dissolved, after which the mixture was naturally cooled to 40 °C. Soon afterwards 10 mL of methanol solution containing 296.4 mg of NH4F and 200 mg of NaOH was added to the above mixed liquid and nucleated at 40 °C for 30 min. The mixed solution was then heated to 110 °C and held for 20 min to remove the water and methanol. The mixed liquid was then heated to 300 °C and reacted for 1.5 h to form nanocrystals. After the reaction was complete, the mixture was cooled naturally, centrifugally collected, and washed with ultrapure water and anhydrous ethanol 3 times. Finally, NaErF4:0.5%Tm was obtained by centrifugation and dispersed in 5.0 mL of cyclohexane for use.
5) was produced. The mixture was subsequently heated to 160 °C in an argon atmosphere and kept warm for 30 min to ensure that the raw material was completely dissolved, after which the mixture was naturally cooled to 40 °C. Soon afterwards 10 mL of methanol solution containing 296.4 mg of NH4F and 200 mg of NaOH was added to the above mixed liquid and nucleated at 40 °C for 30 min. The mixed solution was then heated to 110 °C and held for 20 min to remove the water and methanol. The mixed liquid was then heated to 300 °C and reacted for 1.5 h to form nanocrystals. After the reaction was complete, the mixture was cooled naturally, centrifugally collected, and washed with ultrapure water and anhydrous ethanol 3 times. Finally, NaErF4:0.5%Tm was obtained by centrifugation and dispersed in 5.0 mL of cyclohexane for use.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio) was added, and the mixture was agitated continuously. The mixture was then heated to 160 °C in an argon atmosphere for 30 min and then cooled naturally to 80 °C. Subsequently, 5.0 mL of nuclear NaErF4:0.5%Tm dispersed in cyclohexane was added dropwise to the mixture, which was immediately heated to 110 °C and held for 10 min to dislodge the solvent cyclohexane. At the end of the experiment, the mixture was heated to 40 °C. Then, 10 mL of methanol solution containing 296.4 mg of NH4F and 200 mg of NaOH was added, and the mixture was kept warm for 30 min, heated to 110 °C and maintained for 40 min to dislodge the water and methanol. Thereafter, the temperature was increased to 300 °C for 1.5 h to obtain core–shell nanocrystals. Ultimately, the mixture was cooled naturally at room temperature with heating, centrifuged, collected, and washed with ultrapure water and anhydrous ethanol 3 times. Finally, the obtained oleic acid-coated core–shell UCNPs were centrifuged and dispersed in 5 mL of cyclohexane. For simplicity, all subsequent NaErF4:0.5%Tm nuclei were referred to as nuclear UCNPs, and NaErF4:0.5%Tm@NaGdF4:40%Yb was referred to as core–shell UCNPs.
1 volume ratio) was added, and the mixture was agitated continuously. The mixture was then heated to 160 °C in an argon atmosphere for 30 min and then cooled naturally to 80 °C. Subsequently, 5.0 mL of nuclear NaErF4:0.5%Tm dispersed in cyclohexane was added dropwise to the mixture, which was immediately heated to 110 °C and held for 10 min to dislodge the solvent cyclohexane. At the end of the experiment, the mixture was heated to 40 °C. Then, 10 mL of methanol solution containing 296.4 mg of NH4F and 200 mg of NaOH was added, and the mixture was kept warm for 30 min, heated to 110 °C and maintained for 40 min to dislodge the water and methanol. Thereafter, the temperature was increased to 300 °C for 1.5 h to obtain core–shell nanocrystals. Ultimately, the mixture was cooled naturally at room temperature with heating, centrifuged, collected, and washed with ultrapure water and anhydrous ethanol 3 times. Finally, the obtained oleic acid-coated core–shell UCNPs were centrifuged and dispersed in 5 mL of cyclohexane. For simplicity, all subsequent NaErF4:0.5%Tm nuclei were referred to as nuclear UCNPs, and NaErF4:0.5%Tm@NaGdF4:40%Yb was referred to as core–shell UCNPs.
2.4 Preparation of UCNPs coated with poly(sodium 4-styrene sulfonate) (PSS) (UCNP@PSS)
2.5 Construction of the UCNP@PSS@H1 biosensor
The traditional sulfonamide synthesis method was used to synthesize UCNP@PSS@H1.37 Briefly, 5 mg of TCT was stirred at room temperature in 500 μL of DMF for 1 h to form the TCT–DMF complex. Then, 100 μL of the mixture was added, 250 μL of UCNP@PSS (30 mg mL−1) and 2 mL of CH2Cl2 were added, and the mixture was agitated for 30 min. Subsequently, 100 μL of amino-grafted H1 (10 μM) was added, and the mixture was agitated overnight. Then, the nanoparticles were centrifuged, precipitated and washed twice with DMF and ultrapure water. Finally, UCNP@PSS@H1 was dispersed in 1 mL of ultrapure water.2.6 Fluorescence detection of miRNA-155
Before the fluorescence test, to ensure the formation of the H2 hairpin structure, H2 was annealed. First, 10 μL of UCNP@PSS@H1 was added to 70 μL of 20 mM Tris–HCl (pH 7.4, 5 mM MgCl2, 50 mM NaCl). Different concentrations of miRNA-155 (0.01, 0.2, 0.5, 1, 1.5, 2.5 and 3) were also added, followed by the addition of 110 nM H2. Finally, the total volume was diluted to 250 μL with water, and the reaction mixture was reacted at 37 °C for 90 min. Under an excitation light source of 980 nm, the fluorescence spectra of the reaction mixture in the wavelength range of 550–780 nm were measured and recorded. Each experiment was repeated three times.2.7 Fluorescence measurement of miRNA-155 in actual samples
Human serum samples were obtained from Fujian Children's Hospital (Fuzhou, China). All experiments on the sample were conducted in accordance with the guidelines of the corresponding author's institutional ethics committee, which is consistent with all experiments with the consent of randomized individuals. Before detection of miRNA-155 at UCNP@PSS@H1, human serum samples were diluted 20 times in 20 mM Tris–HCl (pH 7.4) and miRNA-155 in human serum samples was detected via the standard addition approach. The design method was subsequently used for analysis and the specific steps were the same as those for miRNA-155 detection described in Section 2.6. Finally, recovery experiments were carried out by adding various amounts of miRNA-155. The labelled solution was tested separately with commercial detection kits according to the above methods.3 Results and discussion
3.1 Characterization of the prepared nanoparticles
The morphology and structure of the prepared naked core, UCNPs, and UCNP@PSS were observed via TEM. Fig. 2(A and B) show that both the bare core and UCNP crystals exhibit hexagonal structures and the nanoparticle size increases from 22 to 27 nm, indicating the successful preparation of core–shell UCNP crystals. Fig. 2(C) shows that the morphology of the obtained UCNP@PSS was spherical, with a mean size of 29 nm. Compared with the core–shell UCNPs, the average size increased by approximately 2 nm, indicating that the thickness of the PSS shell was about 2 nm. Therefore, the ultrathin PSS shell effectively passivated the surface of the UCNPs while not affecting the FRET process, preventing their disintegration and enhancing the potential application of UCNPs in the field of biosensing. XRD patterns were also utilized to explore the crystal phase of the resulting nanoparticles. Fig. 2(D) shows that the crystal structures of the naked core, core–shell UCNP and UCNP@PSS coincide with the diffraction peak positions (2θ) at 17.1°, 30.0°, 30.8°, 43.5°, 53.2° and 53.7° of the NaErF4 standard card (JCPDs No. 270689), indicating that the resultant nanoparticles are all standard pure hexagonal phases with high crystallinity and that the coating layer does not affect the crystal structure of the UCNPs.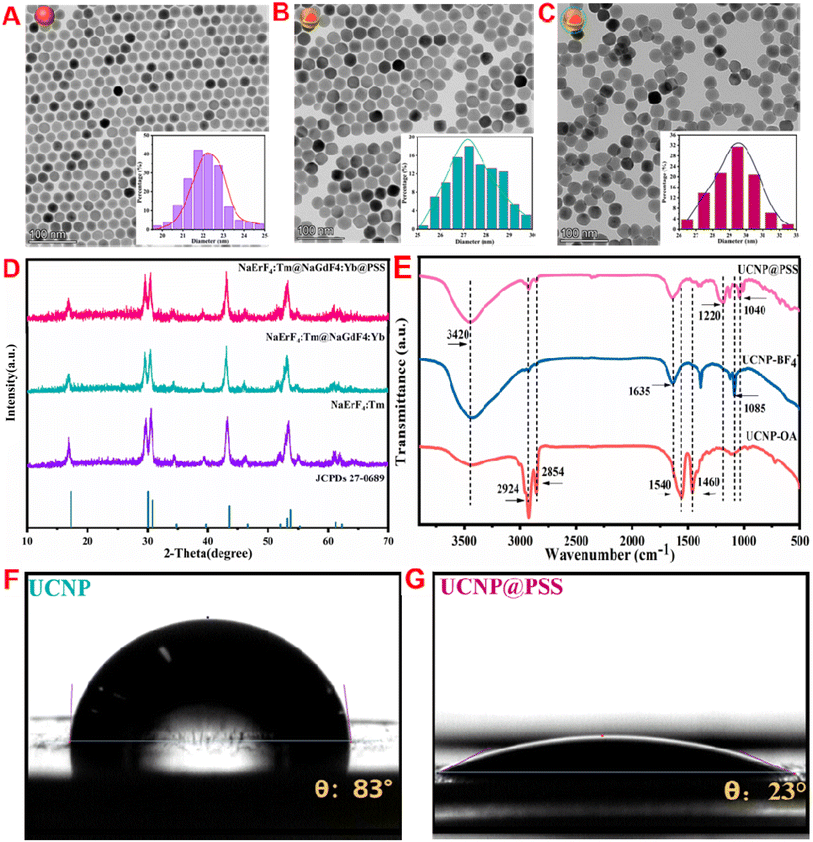 | ||
| Fig. 2 (A–C) TEM, (D) XRD, (E) FT-IR, and (F and G) water contact angles of the prepared nanoparticles. | ||
In addition, to verify ligand exchange and encapsulation of the PSS shell, the molecular structure and functional groups of UCNP, UCNP@NOBF4 and UCNP@PSS were further investigated by FT-IR analysis. Fig. 2(E), orange line, shows that the characteristic peaks of the UCNPs were located at 2924, 2854, 1540 and 1460 cm−1, representing the extended oscillations of the –CH2 and –COO− long alkyl chains of the OA molecules, respectively. After ligand exchange with NOBF4, as shown in Fig. 2(E), blue line, the absorption peak of the –CH2 stretching vibration caused by OA molecules disappears at 2800–3000 cm−1, the vibration peak intensity caused by –COO− decreases at 1600–1400 cm−1, and a new characteristic peak belonging to carbonyl groups and BF4− appears at 1635 cm−1 and 1085 cm−1, respectively, all of which are consistent with previous reports.38Fig. 2(E), pink line, shows that after the PSS shell was coated, two characteristic peaks appeared at 1220 and 1040 cm−1, originating from the extended oscillations of the S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in the sulfonic acid polymer, indicating the successful formation of the PSS shell on the surface of the UCNPs. The change in water contact between the UCNPs and UCNP@PSS verified that the water solubility of the UCNPs greatly improved after PSS coating. As shown in Fig. 2(F and G), after PSS coating, the water contact angle of the UCNPs was 83°, and the water contact angle decreased to 23°. The results show that UCNP@PSS has good water solubility.
O group in the sulfonic acid polymer, indicating the successful formation of the PSS shell on the surface of the UCNPs. The change in water contact between the UCNPs and UCNP@PSS verified that the water solubility of the UCNPs greatly improved after PSS coating. As shown in Fig. 2(F and G), after PSS coating, the water contact angle of the UCNPs was 83°, and the water contact angle decreased to 23°. The results show that UCNP@PSS has good water solubility.
3.2 Luminescent properties of the prepared nanoparticles
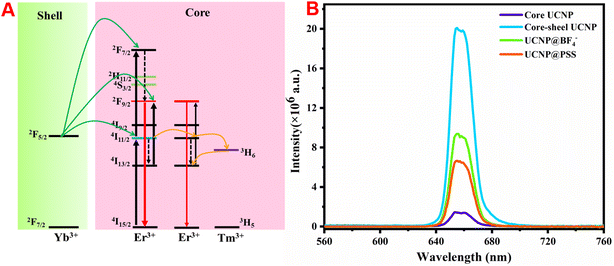
The fluorescence spectra of the prepared naked core and core–shell UCNPs under 980 nm excitation were measured. Fig. 3(A) shows that both display a single red emission peak at approximately 656 nm. However, the fluorescence intensity of the core–shell UCNPs is evidently greater than that of the bare core, which is attributed to the energy capture center Tm3+ and the active shell layer of NaGdF4:Yb growing in the core region. This not only achieves efficient energy transfer of near-infrared photons to the upconversion area, but also suppresses the luminescence quenching caused by surface defects and surface-related ligands, resulting in high-purity bright red light emission. The energy transfer mechanism of the core–shell structure is shown in Fig. 3(B). The red emission of Er3+ mainly comes from the transition between the 4F9/2 and 4I15/2 energy levels and this excited state can be filled with nonradiative relaxation of the 4I7/2 excited state, or can be directly transitioned to the 4I13/2 excited state by energy transfer from the excited Yb3+. Here, Er3+ has dual functions of energy collection and energy transfer. As the concentration of Er3+ increases, the distance between its particles decreases, which intensifies the energy transfer between Er3+. Additionally, the introduction of Tm3+ captures the energy in the 4I11/2 state of Er3+via the 3H5 level of Tm3+, which then undergoes a reverse energy transfer towards the 4I13/2 level. As a result, the red emission at 656 nm dominates the upconversion process. Notably, the UCL intensity of UCNP@PSS was much lower than that of the preassembled UCNPs; however, its fluorescence intensity was still as high as 4 million. Furthermore, in this work, the detection process is based on the amount of target added and the UCL recovery efficiency of the sensing system. Therefore, although the UCL intensity of UCNP@PSS has decreased, it still meets the determination requirements.3.3 Construction of the UCNP@PSS@H1 biosensor
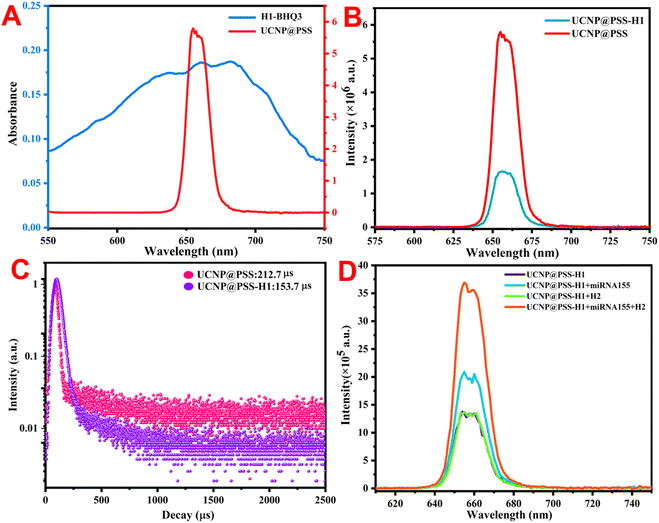
The UCNP@PSS@H1 biosensor was constructed through a sulfonamide reaction between the sulfonic acid group on the periphery of UCNP@PSS and the 5′-terminal amino-functionalized H1 to form sulfonamide. To verify the occurrence of this process, the UV-vis absorption spectra of UCNP@PSS and UCNP@PSS@H1 were first obtained. As shown in Fig. 4(A), compared with that of UCNP@PSS, the UV-vis absorption spectrum of UCNP@PSS@H1 has a distinct characteristic peak at 260 nm, which is attributed to the characteristic absorption of purine and pyrimidine rings in DNA oligonucleotides, indicating that H1 has successfully coupled to the surface of UCNP@PSS. Concerning the zeta potential measurements, negative values of UCNP@PSS and UCNP@PSS@H1 were observed (−27.9 mV and −36 mV) in Fig. 4(B), as expected, because of the large number of negative sulfonate and phosphate groups. Finally, Fig. 4(C) shows that the average sizes of UCNP@PSS and UCNP@PSS@H1 determined via DLS were 142.9 nm and 191.2 nm, respectively, further confirming the successful coupling of DNA strands on the UCNPs.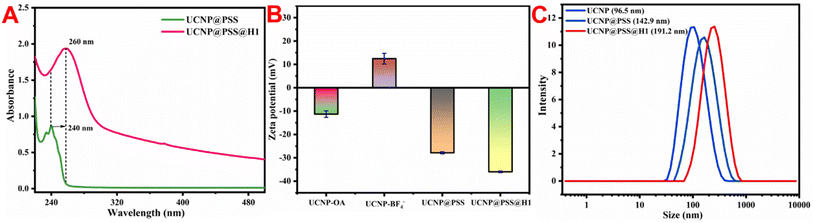 | ||
| Fig. 4 (A) UV-vis absorption spectra, (B) zeta potential and (C) DLS before and after UCNP modification. | ||
3.4 Feasibility of constructing sensors for detecting miRNAs
Fig. 1 illustrates the detection principle of the constructed sensor for miRNA-155. To verify the feasibility of constructing sensors for detecting miRNA, fluorescence spectra and UV-vis absorption spectra of various detection systems were measured. As shown in Fig. 5(A), compared with conventional upconversion nanoparticles, which use Er3+ as a sensitizer, owing to the introduction of an appropriate amount of Tm3+, which has a 3H5 energy level that can act as a reverse energy transfer, UCNP@PSS has bright red fluorescence. BHQ3 exhibited a distinct UV-vis absorption peak between 550 and 750 nm that overlapped with the fluorescence emission spectrum of UCNP@PSS located at 655 nm, indicating the possibility of FRET between the two, quenching the red fluorescence of UCNP@PSS. Fig. 5(B) also confirms that the fluorescence of UCNP@PSS is quenched, with a quenching efficiency of 72%. To confirm the occurrence of the FRET process between UCNP@PSS and H1-BHQ3, the fluorescence lifetimes were also measured. Fig. 5(C) shows that the fluorescence lifetimes of UCNP@PSS before and after coupling with H1-BHQ3 are 212.7 μs and 153.7 μs, respectively. The decrease in fluorescence lifetime after assembly confirmed the occurrence of FRET between UCNP@PSS and H1-BHQ3.Finally, to validate the sensitive detection of miRNA through the catalytic hairpin amplification strategy, the fluorescence spectra of different detection systems, including UCNP@PSS@H1, UCNP@PSS@H1 + miRNA-155, UCNP@PSS@H1 + H2 and UCNP@PSS@H1 + miRNA-155 + H2, were measured. Fig. 5(D) shows that in the presence of miRNA-155, the upconversion fluorescence emission of UCNP@PSS-H1 was restored by approximately 50%. With the help of H2, the fluorescence intensity of UCNP@PSS-H1 significantly increased and was restored by about 174%, indicating that the catalytic hairpin amplification strategy can significantly increase the determination sensitivity of miRNA. Moreover, the fluorescence of the system did not recover after the addition of H2 alone, indicating that the catalytic hairpin amplification process can only be initiated in the presence of miRNA.
3.5 Optimization of detection conditions
To obtain accurate detection results, specific parameters were optimized on the basis of the fluorescence quenching process of UCNP@PSS (i.e., H1-BHQ3 reacts with UCNP@PSS through sulfonamide), the fluorescence recovery process (i.e., BHQ3-H1 opens its hairpin structure in the presence of the analyte substance) and the luminescence signal amplification process (catalytic hairpin amplification is activated with the assistance of H2) to obtain accurate experimental results. Optimization was carried out for several key experimental conditions, including the amount of H1, concentration of H2, and reaction time. The optimization criteria depend on the fluorescence quenching (F0 − F)/F0 and recovery efficiency (F1 − F)/F of the system, where F0 and F represent the fluorescence intensity before and after the reaction between UCNP@PSS and H1, respectively. F1 represents the luminescence intensity of the system after the addition of the target substance. H1-BHQ3 not only acts as a quencher in the detection system, but also plays a role in the specific recognition of target substances. Therefore, the concentration of H1 was first optimized. Fig. 6(A) shows that as the concentration of H1 increased, the quenching efficiency increased. When the concentration of H1 was 300 nM, the quenching efficiency reached a maximum. With increasing H1 concentration, the quenching efficiency of the system remains stable. Therefore, 300 nM was set as the optimal concentration for H1. The reaction time (Fig. 6(B)) and H2 content (Fig. 6(C)) were also studied and determined to be 90 min and 110 nM, respectively.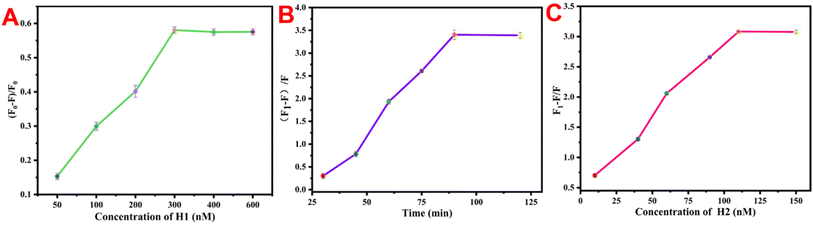 | ||
| Fig. 6 Effects of (A) H1 concentration, (B) reaction time and (C) H2 concentration on the detection system. | ||
3.6 The detection performance of the proposed sensors
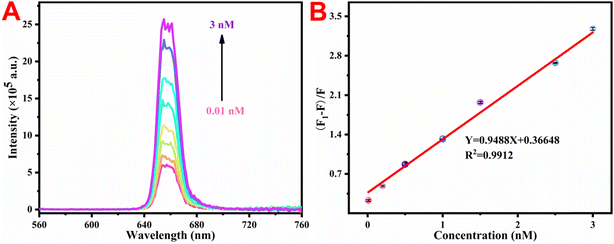
Under the best experimental conditions mentioned above, the upconversion luminescence response of the developed UCNP@PSS@H1 sensor was tested with various amounts of miRNA-155. Fig. 7(A) shows that as the content of miRNA-155 increased, the fluorescence intensity of the system increased step by step. Fig. 7(B) shows a linear relationship between the upconversion luminescence intensity at 655 nm and the content of miRNA-155 within the range of 0.01–3 nM. The fitted linear data can be expressed as Y = 0.9488X + 0.3665 (R2 = 0.9912). A higher R2 reflects the excellent linear response of the DNA-functionalized biosensor to miRNA-155. The LOD was calculated according to the following formula: LOD = 3 × δ/S, where δ expresses the noise level, which is the standard deviation from 10 blank trials, and S represents the slope of the linear regression equation in sensitivity testing. Therefore, the calculated LOD was 1.14 pM, which is superior to those of the other methods reported in Table 1 for determining miRNA-155.| Methods | Linear range (nM) | Limit of detection (pM) | References |
|---|---|---|---|
| AuNPs/3D DNA walker | 0.2–10 nM | 130 | 39 |
| DNA-AgNCs | 0.2–30 | 100 | 40 |
| DNA tetrahedron | 15–40 | 85 | 41 |
| DNA nanowheel | 0.1–50 | 82.3 | 42 |
| AgNCs | 0.25–1000 | 67 | 43 |
| Polystyrene beads | 0.01–1 | 30 | 44 |
| CuNC | 0.05–10 | 11 | 45 |
| DNA micelles | 0.05–4 | 3.1 | 46 |
| UCNP@PSS@H1 | 0.01–3 | 1.14 | This work |
3.7 Specific performance and stability of the proposed sensors
Because the miRNA family consists of a series of members with similar sequences, investigating the specific ability of biosensors to recognize miRNA sequences to better understand the physiological behavior and functions of specific miRNAs in major diseases is important. On this basis, three other miRNAs (miRNA-21, miRNA-214 and miRNA-222) were treated as interference sequences, and the concentration of the interfering miRNAs was increased to four times (6 nM) that of the target miRNA-155 to better validate the specificity of the UCNP@PSS@H1 biosensors. Moreover, a mixture of three interfering miRNAs and a specific recognition target, miRNA-155, was added to simulate the specificity of the fluorescent sensor in the presence of multiple interfering factors. As depicted in Fig. 8(A), the luminescence intensity of the experimental group with miRNA-21, miRNA-214 and miRNA-222 added slightly increased compared with that of the blank group, but the recovery of fluorescence intensity was negligible compared with that of the experimental group with miRNA-155 (1.5 nM) added. Moreover, the fluorescence intensity of the mixed group also greatly improved. These results suggest that the constructed biosensor has good specificity for the target miRNA-155. In addition, the detection performance of the developed method within 15 days was also investigated. Fig. 8(B) shows that the luminescence response of the obtained fluorescent biosensor to miRNA-155 remained almost unchanged. The above results indicate that the constructed UCNP@PSS@H1 biosensor has good specificity and stability and can realize the specific recognition and detection of miRNA-155 in a complex physiological environment.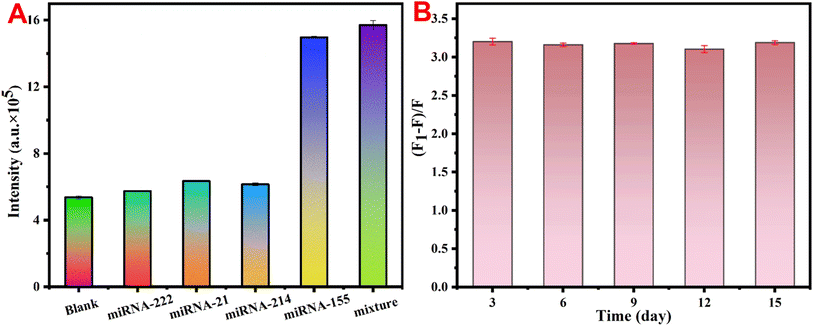 | ||
| Fig. 8 (A) Effects of different miRNAs on the fluorescence intensity and (B) stability of the detection system. | ||
3.8 Actual sample testing
To evaluate the applicability and reliability of the proposed method, miRNA-155 in serum samples was measured using UCNP@PSS@H1 biosensors. Table 2 reveals that the spiked restoration efficiency in actual samples ranged from 99.8% to 104.8%, with a relative standard deviation (RSD, n = 3) of less than 4.7%. These results indicate that the constructed UCNP@PSS@H1 sensor system has good detection ability for miRNA-155 in complex biological samples.| Samples | Added (nM) | Found (nM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Serum 1 | 0.04 | 0.041 ± 0.003 | 103.2 | 4.18 |
| 0.75 | 0.770 ± 0.035 | 102.8 | 4.70 | |
| 1.5 | 1.572 ± 0.017 | 104.8 | 1.10 | |
| Serum 2 | 0.04 | 0.04 ± 0.003 | 99.8 | 2.32 |
| 0.75 | 0.752 ± 0.016 | 100.7 | 2.10 | |
| 1.5 | 1.532 ± 0.19 | 102.1 | 1.20 |
4 Conclusion
In this work, a biosensor based on sulfonic acid polymer-coated UCNPs for sensitive detection of miRNAs was constructed. The proposed method has the following advantages: (1) compared with other fluorescent agents, UCNPs have many unique merits, such as greater tissue penetration depth, excellent photostability and weak autofluorescence, resulting in higher sensitivity and accuracy. (2) Polysulfonic acid salts can effectively prevent the annihilation of UCNPs, thereby ensuring long-term stability. In addition, compared with other polymers such as polyethylene glycol, owing to the presence of many sulfonic acid groups, a thin and adjustable polymer shell can be grown on the outside of UCNPs, which is beneficial for the use of UCNPs as FRET donors. (3) Compared with enzyme-assisted amplification, the catalytic hairpin assembly used in this work has the advantages of high stability, simplicity, rapid reaction and low cost. (4) This work develops a simple, rapid, sensitive and specific method for detecting miRNA-155, providing a new perspective for monitoring the progression of allergic rhinitis or other miRNA-related diseases. However, the proposed method also has several limitations, such as the loss of fluorescence performance due to the improved biocompatibility of UCNPs. Therefore, we will further improve the luminescence performance by in situ deposition of metal nanoparticles in the next step.Data availability
The authors confirm that the data supporting the findings of this study are available within the article.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Startup Fund for scientific research, Fujian Medical University (2020QH1209); Joint Funds for the Innovation of Science and Technology, Fujian province (2023Y9382); Fujian Provincial Natural Science Foundation of China (2023J011304); and Fujian provincial health technology project (2023GGA060, 2021GGA004).References
- J. A. Bernstein, J. S. Bernstein, R. Makol and S. Ward, Allergic Rhinitis: A Review, JAMA, 2024, 331(10), 866–877 CrossRef CAS PubMed.
- Q. X. Zeng, W. L. Liu, R. Z. Luo and G. Lu, MicroRNA-181a and microRNA-155 are involved in the regulation of the differentiation and function of regulatory T cells in allergic rhinitis children, Pediatr. Allergy Immunol., 2019, 30(4), 434–442 CrossRef PubMed.
- Y. P. Gao, K. J. Huang, B. Y. Wang, Q. Xu, H. Shuai and G. Li, Constructed a self-powered sensing platform based on nitrogen-doped hollow carbon nanospheres for ultra-sensitive detection and real-time tracking of double markers, Anal. Chim. Acta, 2023, 1267, 341333 CrossRef CAS PubMed.
- A. Válóczi, C. Hornyik, N. Varga, J. Burgyán, S. Kauppinen and Z. Havelda, Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes, Nucleic Acids Res., 2004, 32(22), e175 CrossRef PubMed.
- Y. Kitano, K. Aoki, F. Ohka, S. Yamazaki, K. Motomura, K. Tanahashi and A. Natsume, Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds, ACS Appl. Mater. Interfaces, 2021, 13(15), 17316–17329 CrossRef CAS PubMed.
- Y. P. Gao, K. J. Huang, F. T. Wang, Y. Y. Hou, J. Xu and G. Li, Recent advances in biological detection with rolling circle amplification: design strategy, biosensing mechanism, and practical applications, Analyst, 2022, 147, 3396–3441 RSC.
- O. A. Goryacheva, A. M. Vostrikova, A. A. Kokorina, E. A. Mordovina, D. V. Tsyupka, A. A. Bakal and I. Y. Goryacheva, Luminescent carbon nanostructures for microRNA detection, Trends Anal. Chem., 2019, 119, 115613 CrossRef CAS.
- Y. P. Gao, K. J. Huang, Y. Y. Hou, B. Y. Wang, Q. Y. Xu, H. L. Shuai, J. Xu and G. Q. Li, Nitrogen-doped dodecahedral hollow carbon combined with amplification strategy to construct ultra-sensitive self-powered biosensor for highly stable and real-time monitoring microRNA, Sens. Actuators, B, 2023, 387, 133816 CrossRef CAS.
- Y. P. Gao, K. J. Huang, B. Y. Wang, Q. Y. Xu, H. L. Shuai and G. Q. Li, Constructed a self-powered sensing platform based on nitrogen-doped hollow carbon nanospheres for ultra-sensitive detection and real-time tracking of double markers, Anal. Chim. Acta, 2023, 1267, 341333 CrossRef CAS PubMed.
- H. L. Shuai, K. J. Huang, Y. X. Chen, L. X. Fang and M. P. Jia, Au nanoparticles/hollow molybdenum disulfide microcubes based biosensor for microRNA-21 detection coupled with duplex-specific nuclease and enzyme signal amplification, Biosens. Bioelectron., 2017, 89, 989–997 CrossRef CAS PubMed.
- J. Y. Shi, S. Y. Liu, P. Y. Li, Y. Lin, H. Luo, Y. Wu, J. Yan, K. J. Huang and X. C. Tan, Self-powered dual-mode sensing strategy based on graphdiyne and DNA nanoring for sensitive detection of tumor biomarker, Biosens. Bioelectron., 2023, 237, 115557 CrossRef CAS PubMed.
- J. M. Zhang, Y. C. Jiang, Y. B. Wang and Z. S. Zhang, DNA-Functionalized Gold Nanorods for One-Step Simultaneous Determination of Multiple microRNAs Based upon Plasma-Enhanced Fluorescence, Anal. Lett., 2024, 1–13 Search PubMed.
- Y. Y. Hou, J. Xu, F. T. Wang, Z. Dong, X. C. Tan, K. J. Huang, J. Q. Li, C. Y. Zuo and S. Q. Zhang, Construction of an integrated device of a self-powered biosensor and matching capacitor based on graphdiyne and multiple signal amplification: ultrasensitive method for microRNA detection, Anal. Chem., 2021, 93, 15225–15230 CrossRef CAS PubMed.
- F. T. Wang, Y. Y. Hou, X. C. Tan, K. J. Huang, J. Xu and R. Cai, Real-time multiple signal amplification self-powered biosensing platform for ultrasensitive detection of microRNA, Biosens. Bioelectron., 2023, 222, 114933 CrossRef CAS PubMed.
- Y. C. Deng, P. Li, J. T. Li, D. L. Sun and H. R. Li, Color-Tunable Aqueous Room-Temperature Phosphorescence Supramolecular Assembly, ACS Appl. Mater. Interfaces, 2021, 13, 14407–14416 CrossRef CAS PubMed.
- D. Y. Tang, J. Y. Shi, Y. Wu, H. Luo, J. Yan, K. J. Huang and X. C. Tan, Flexible Self-Powered Sensing System Based on Novel DNA Circuit Strategy and Graphdiyne for Thalassemia Gene by Rapid Naked-Eye Tracking and Open-Circuit Voltage, Anal. Chem., 2023, 95, 16374–16382 CrossRef CAS PubMed.
- M. Norouzi, S. Yasamineh, M. Montazeri, M. Dadashpour, R. Sheervalilou, M. Abasi and Y. Pilehvar-Soltanahmadi, Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection, Mater. Sci. Eng. C., 2019, 104, 110007 CrossRef CAS PubMed.
- Y. Han, J. Zhou, F. Liu, Y. Ouyang, R. Yuan and Y. Q. Chai, pH-Stimulated Self-Locked DNA Nanostructure for the Effective Discrimination of Cancer Cells and Simultaneous Detection and Imaging of Endogenous Dual-MicroRNAs, Anal. Chem., 2023, 95(34), 12754–12760 CrossRef CAS PubMed.
- E. A. Sangachin, M. Hosseini and J. Mohammadnejad, MicroRNA sensing using integrating DNA-functionalized hydrogels with aggregation-induced emission of silver nanoclusters, Sens. Actuators, B, 2023, 390, 134012 CrossRef.
- P. Wang, X. W. Wei, L. M. Shen, K. X. Xu, Z. T. Wen, N. J. Gao and Y. F. Zhu, Amplification-Free Analysis of Bladder Cancer MicroRNAs on Wrinkled Silica Nanoparticles with DNA-Functionalized Quantum Dots, Anal. Chem., 2024, 96(12), 4860–4867 CrossRef CAS PubMed.
- X. Wu, Y. Li, M. Y. Yang and C. B. Mao, Simultaneous ultrasensitive detection of two breast cancer microRNA biomarkers by using a dual nanoparticle/nanosheet fluorescence resonance energy transfer sensor, Mater. Today Adv., 2021, 12, 100163 CrossRef CAS.
- X. M. Zhou, H. Y. Geng, P. F. Shi, H. J. Wang, G. F. Zhang, Z. M. Cui and S. Bi, NIR-driven photoelectrochemical-fluorescent dual-mode biosensor based on bipedal DNA walker for ultrasensitive detection of microRNA, Biosens. Bioelectron., 2024, 247, 115916 CrossRef CAS PubMed.
- K. Zhang, N. Zhang, Y. Wu, W. Zhang, F. J. Li and Y. Z. Shen, Upconversion nanoamplicon with confined emitters for precise reporting of microRNA-21 levels originated from cancer cells, Sens. Actuators, B, 2021, 342, 130062 CrossRef CAS.
- S. Bhuckory, S. Lahtinen, N. Höysniemi, J. Guo, X. Qiu, T. Soukka and N. Hildebrandt, Understanding FRET in Upconversion Nanoparticle Nucleic Acid Biosensors, Nano Lett., 2023, 23(6), 2253–2261 CrossRef CAS PubMed.
- H. Ren, Z. Long, X. T. Shen, Y. Zhang, J. H. Sun, J. Ouyang and N. Na, Sandwich DNA Hybridization Fluorescence Resonance Energy-Transfer Strategy for miR-122 Detection by Core–Shell Upconversion Nanoparticles, ACS Appl. Mater. Interfaces, 2021, 10(30), 25621–25628 CrossRef PubMed.
- U. Resch-Genger and H. H. Gorris, Perspectives and challenges of photon-upconversion nanoparticles - Part I: routes to brighter particles and quantitative spectroscopic studies, Anal. Bioanal. Chem., 2017, 409(25), 5855–5874 CrossRef CAS PubMed.
- D. L. Zhang, R. Z. Peng, W. F. Liu, M. J. Donovan, L. L. Wang, I. Ismail and W. H. Tan, Engineering DNA on the Surface of Upconversion Nanoparticles for Bioanalysis and Therapeutics, ACS Nano, 2021, 15(11), 17257–17274 CrossRef CAS PubMed.
- J. N. Liu, W. B. Bu, L. M. Pan, S. J. Zhang, F. Chen, L. P. Zhou and J. L. Shi, Simultaneous nuclear imaging and intranuclear drug delivery by nuclear-targeted multifunctional upconversion nanoprobes, Biomaterials, 2012, 33(29), 7282–7290 CrossRef CAS PubMed.
- J. Zhou, Q. Liu, W. Feng, Y. Sun and F. Y. Li, Upconversion Luminescent Materials: Advances and Applications, Chem. Rev., 2015, 115(1), 395–465 CrossRef CAS PubMed.
- N. J. J. Johnson, N. M. Sangeetha, J. C. Boyer and F. C. J. M. Veggel, Facile ligand-exchange with polyvinylpyrrolidone and subsequent silica coating of hydrophobic upconverting β-NaYF4:Yb3+/Er3+nanoparticles, Nanoscale, 2010, 2(5), 771–777 RSC.
- D. Mendez-Gonzalez, S. Lahtinen, M. Laurenti, E. López-Cabarcos, J. Rubio-Retama and T. Soukka, Photochemical Ligation to Ultrasensitive DNA Detection with Upconverting Nanoparticles, Anal. Chem., 2018, 90(22), 13385–13392 CrossRef CAS PubMed.
- F. Tian, Q. Wang, C. F. Jiang, M. M. Li, Y. Q. Zhang, J. Yu and X. Y. Song, Construction of an Endogenously Activated Signal Amplification Assay for In Situ Imaging of MicroRNA and Guided Precise Photodynamic Therapy for Cancer Cells, Anal. Chem., 2023, 95(13), 5601–5609 CrossRef CAS PubMed.
- W. Kong, T. Y. Sun, B. Chen, X. Chen, F. J. Ai, X. Y. Zhu and F. Wang, A General Strategy for Ligand Exchange on Upconversion Nanoparticles, Inorg. Chem., 2017, 56(2), 872–877 CrossRef CAS PubMed.
- G. Y. Hu, D. P. Yue, W. H. Chen, Q. Q. Lin and H. X. Lyu, Dual-mode upconversion sensors for detecting differently charged biotargets based on the oxidase-mimicking activity of Ce4+and electrostatic control, Talanta, 2024, 277, 126392 CrossRef CAS PubMed.
- H. Q. Chen, W. Wang, C. C. Ji and L. Wang, Dye-sensitized core–shell NaGdF4: Yb, Er@NaGdF4: Yb, Nd upconversion nanoprobe for determination of H2S, Spectrochim. Acta, Part A, 2021, 248, 119281 CrossRef CAS PubMed.
- N. Estebanez, M. González-Béjar and J. Pérez-Prieto, Polysulfonate Cappings on Upconversion Nanoparticles Prevent Their Disintegration in Water and Provide Superior Stability in a Highly Acidic Medium, ACS Omega, 2019, 4(2), 3012–3019 CrossRef CAS PubMed.
- L. Frances-Soriano, N. Estebanez, J. Perez-Prieto and N. Hildebrandt, DNA-Coated Upconversion Nanoparticles for Sensitive Nucleic Acid FRET Biosensing, Adv. Funct. Mater., 2022, 32, 2201541 CrossRef CAS.
- T. H. Wan, W. Song, H. L. Wen, Q. Xue, Q. Q. Zhan, W. Chen, H. J. Yu, Yu Lin and R. A. Abdur, The exploration of upconversion luminescence nanoprobes for tobramycin detection based on Förster resonance energy transfer, Mater. Today Adv., 2023, 19, 100409 CrossRef CAS.
- W. K. Zhong, J. Y. Wu, Y. Q. Huang, C. Xing and C. H. Lu, Target-Activated, Light-Actuated Three-Dimensional DNA Walker Nanomachine for Amplified miRNA Detection, Langmuir, 2022, 38(3), 1151–1157 CrossRef CAS PubMed.
- M. Hosseini, A. Akbari, M. R. Ganjali, M. Dadmehr and A. H. Rezayan, A Novel Label-Free microRNA-155 Detection on the Basis of Fluorescent Silver Nanoclusters, J. Fluoresc., 2015, 25(4), 925–929 CrossRef CAS PubMed.
- C. H. Li, W. Y. Lv, F. F. Yang, S. J. Zhen and C. Z. Huang, Simultaneous Imaging of Dual microRNAs in Cancer Cells through Catalytic Hairpin Assembly on a DNA Tetrahedron, ACS Appl. Mater. Interfaces, 2016, 14(10), 12059–12067 CrossRef PubMed.
- F. Yang, Y. Cheng, Y. Cao, Y. Y. Zhang, H. F. Dong, H. T. Lu and X. J. Zhang, MicroRNA Triggered DNA “Nano Wheel” for Visualizing Intracellular microRNA via Localized DNA Cascade Reaction, Anal. Chem., 2019, 91(15), 9828–9835 CrossRef CAS PubMed.
- C. Li, Z. H. Chen, Y. X. Zhang, J. Y. He, R. Yuan and W. J. Xu, Guanine-Lighting-Up Fluorescence Biosensing of Silver Nanoclusters Populated in Functional DNA Constructs by a pH-Triggered Switch, Anal. Chem., 2020, 92(19), 13369–13377 CrossRef CAS PubMed.
- N. N. Wang, L. R. Song, T. Deng and J. S. Li, Microsphere-based suspension array for simultaneous recognition and quantification of multiple cancer-associated miRNA via DNAzyme-Mediated signal amplification, Anal. Chim. Acta, 2020, 1140, 69–77 CrossRef CAS PubMed.
- Y. S. Borghei, M. Hosseini and M. R. Ganjali, Fluorescence based turn-on strategy for determination of microRNA-155 using DNA-templated copper nanoclusters, Microchim. Acta, 2017, 184(8), 2671–2677 CrossRef CAS.
- L. P. Cao, C. M. Li, S. J. Zhen and C. Z. Huang, A General Signal Amplifier of Self-Assembled DNA Micelles for Sensitive Quantification of Biomarkers, Anal. Chem., 2023, 95(3), 1794–1800 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |