Open Access Article
Open Access ArticleExtracellular vesicles of different cellular origin feature distinct biomolecular corona dynamics†‡
Angelo
Musicò§
ab,
Andrea
Zendrini§
 ab,
Santiago Gimenez
Reyes
cd,
Valentina
Mangolini
ab,
Lucia
Paolini
bh,
Miriam
Romano
ab,
Andrea
Papait
ef,
Antonietta Rosa
Silini
i,
Paolo
Di Gianvincenzo
c,
Arabella
Neva
g,
Marina
Cretich¶
ab,
Santiago Gimenez
Reyes
cd,
Valentina
Mangolini
ab,
Lucia
Paolini
bh,
Miriam
Romano
ab,
Andrea
Papait
ef,
Antonietta Rosa
Silini
i,
Paolo
Di Gianvincenzo
c,
Arabella
Neva
g,
Marina
Cretich¶
 j,
Ornella
Parolini
ef,
Camillo
Almici
j,
Ornella
Parolini
ef,
Camillo
Almici
 g,
Sergio E.
Moya
g,
Sergio E.
Moya
 c,
Annalisa
Radeghieri§
c,
Annalisa
Radeghieri§
 ab and
Paolo
Bergese§
ab and
Paolo
Bergese§
 *abk
*abk
aDepartment of Molecular and Translational Medicine, University of Brescia, 25123 Brescia, Italy. E-mail: paolo.bergese@unibs.it
bCSGI, Center for Colloid and Surface Science, 50019 Florence, Italy
cSoft Matter Nanotechnology, Center for Cooperative Research in Biomaterials (CIC biomaGUNE), Basque Research and Technology Alliance (BRTA), Paseo de Miramon 194, Donostia-San Sebastián, Spain
dInstituto de Fisica del Sur (IFISUR-CONICET), Av. Alem, Bahia Blanca, Argentina
eDepartment of Life Science and Public Health, Università Cattolica del Sacro Cuore, 00168 Rome, Italy
fFondazione Policlinico Universitario “Agostino Gemelli” IRCCS, 00168 Rome, Italy
gLaboratory for Stem Cells Manipulation and Cryopreservation, Department of Transfusion Medicine, ASST Spedali Civili of Brescia, 25123, Brescia, Italy
hDepartment of Medical and Surgical Specialties, Radiological Sciences and Public Health (DSMC), University of Brescia, Brescia, Italy
iCentro di Ricerca Eugenia Menni, Fondazione Poliambulanza Istituto Ospedaliero, 25124, Brescia, Italy
jIstituto di Scienze e Tecnologie Chimiche “Giulio Natta” – National Research Council of Italy (SCITEC-CNR), 20131 Milan, Italy
kNational Inter-university Consortium of Materials Science and Technology (INSTM), Via Giuseppe Giusti 9, 50121 Firenze, Italy
First published on 7th November 2024
Abstract
Initially observed on synthetic nanoparticles, the existence of biomolecular corona and its role in determining nanoparticle identity and function are now beginning to be acknowledged in biogenic nanoparticles, particularly in extracellular vesicles – membrane-enclosed nanoparticle shuttling proteins, nucleic acids, and metabolites which are released by cells for physiological and pathological communication – we developed a methodology based on fluorescence correlation spectroscopy to track biomolecular corona formation on extracellular vesicles derived from human red blood cells and amniotic membrane mesenchymal stromal cells when these vesicles are dispersed in human plasma. The methodology allows for tracking corona dynamics in situ under physiological conditions. Results evidence that the two extracellular vesicle populations feature distinct corona dynamics. These findings indicate that the dynamics of the biomolecular corona may ultimately be linked to the cellular origin of the extracellular vesicles, revealing an additional level of heterogeneity, and possibly of bionanoscale identity, that characterizes circulating extracellular vesicles.
New conceptsThe EV biomolecular corona (BC) is a mesoscale contextual property emerging from the multiscale interactions occurring at the interface between an extracellular vesicle (EV) and the molecular and nanoscale components of the surrounding media. Therefore, the BC concurs in determining EV surface identity and in turn its very function, including biodistribution and cellular uptake.EV–BC investigation is in the very beginning and has mainly relied on “ex situ” measurements, complicated by the fact that the physicochemical characteristics of the EV–BC nanosystem make it very challenging to separate BC and EVs without “scrambling” them. We developed a new method for non-destructive, in situ tracking of EV–BC formation, based on fluorescence correlation spectroscopy (FCS), and implemented it to compare the formation of a BC from human plasma onto red blood cells (RBC) EVs and mesenchymal stromal cells (MSC) EVs under physiological conditions. These two EV types are vying for the leading role in translational medicine: RBC-EVs in RNA, drug and biological delivery, MSC-EVs in regenerative medicine. Our findings indicate that BC dynamics differ by EV origin, identifying BC–EV dynamics as a key parameter to be considered in both fundamental investigations in mesoscale extracellular biology as well as in designing EV systems for specific tasks. |
Introduction
The biomolecular corona (BC) refers to the dynamic coating that biomolecules form on the surface of nanoparticles (NPs) when these are immersed in biological fluids. BC redefines some of the native physicochemical properties of NPs (synthetic identity) – including surface composition, structure and energy, surface charge, NP hydrodynamic radius, and aggregation/stability – giving the NPs diverse biological identities at the nanoscale, and ultimately impacting their biological behavior.1–3The concept of BC was originally introduced in 2007, exclusively referring to synthetic NPs.4,5 Currently, within that context, it is a topic of great research vitality and far from being exhausted. For example, lively debates on the composition, structure, and kinetics of BC formation are still underway.6 On the other hand, the fact that the BC enriches specific biomolecular patterns “hidden” in the biological fluid represents a potential breakthrough for diagnostics.7,8
In recent years, it has been realized that biological fluids inherently display nanoscale features given that, besides soluble proteins, they contain several kinds of extracellular NPs, e.g. exomeres, lipoproteins, extracellular vesicles, midbody remnants, and many others.9 In turn, since 2018,10 few researchers have started to highlight the presence and biological significance of “BC-like” dynamic associations involving soluble proteins, lipoproteins, and extracellular vesicles (EVs).
Complexes formed by EVs and lipoproteins (EV–LP) have been shown to occur under physiological conditions11 and during nanoscale manipulation,12 exhibiting features like EV–LP fusion.13 EVs derived from brain metastases have been observed to induce the binding and aggregation of lipoproteins.14 It has also been reported that a BC forms around EVs in blood plasma,15 and that this BC has a functional impact,16,17 enhancing angiogenesis, skin regeneration, and immunomodulation.18,19 The EV BC also has potential in diagnostics8,20 and should be critically considered in EV surface engineering.21 The formation of the EV–BC is mainly driven by non-covalent interactions, probably based on van der Waals forces. Additionally, the formation of hydrogen bonds between the EV surface and the components of the biomolecular corona cannot be excluded, as well as pi interactions. Finally, hydrophobic interactions may play a role. For such interactions to occur, adsorbed proteins might need to expose their hydrophobic regions, facilitating interaction with the hydrophobic part of the EV membrane.
Both the specific cell type from which EVs originate and the EV intracellular biogenesis pathway shape the surface of each EV type. As a result, EVs derived from various cells can exhibit distinct surface properties. We speculate that this variability also influences, together with the molecular composition of the BC10,15 its dynamics, because changes in EV surface physicochemical properties are expected to be mirrored in time required for BC formation, BC evolution over time, and biomolecule exchange rate at the interface.
To investigate the dynamics of the EV BC, it is necessary to put in place new in situ methodologies that work under physiological conditions and in real-time, overcoming the limitations associated with static ex situ methods, which have been conventionally employed in EV–BC analysis. It is worth noting that measuring BC formation even with ex situ approaches previously developed for synthetic NPs has proven to be extremely challenging, due to the inherent physicochemical characteristics of EVs, e.g. difficulty of not altering or even breaking the EVs and/or distinguishing corona components from the internal EV cargo upon corona “peeling”.10,15,17,19,22 Furthermore, ex-situ methods, such as mass spectrometry23 and gel electrophoresis,20,21 cannot provide information about the BC dynamics as they require pre-analytical separation of the EV–BC complexes (i.e., by size exclusion chromatography, SEC, or ultracentrifugation) which may result in altering the complexes by the loss of weakly associated components.18
Fluorescence correlation spectroscopy (FCS) is an in situ spectroscopic technique that measures the fluctuations of fluorescence in a confocal volume, which can be related to the movement of the fluorescent species in and out of the volume.24 From the autocorrelation function describing the time evolution of fluorescence, it is possible to extrapolate the diffusion coefficient of the fluorescent species and relate it to their size and mass.25,26 FCS has been successfully used to characterize the colloidal stability of synthetic NPs in different biological fluids.27–29 FCS is an optimal technique for in situ measurements as it allows to trace diffusion of specific molecular components, provided that these are fluorescently labelled. Therefore, if we selectively label proteins present in media or the EVs, or even both with different fluorophores with non overlapping fluorescence, we will be capable of tracing the fate of proteins, EVs or both, respectively, without the need for separating EV–BC complexes from other proteins or biomolecules present in the media.30
In this work, we implement, for the first time, a dedicated FCS methodology based on competition experiments of physisorption to track the dynamic formation of plasma BC on two different model EV types: red blood cells EVs (REVs) and amniotic membrane mesenchymal stromal cells EVs (MEVs). Both EV types are now in the spotlight for their great potential in clinical translation. REVs for personalized medicine and drug delivery,31–33 MEVs for regenerative medicine and immunomodulation.34–37 We will show that our methodology does not require the separation of the EVs from the media, thus reducing potential artifacts due to the separation processes. Furthermore, we implemented stoichiometric and geometric models to use FCS results for estimating the architecture of the BC.
Results and discussion
Cetuximab modification and characterization
Cetuximab (CTX) was labelled with Sulfo Cyanine 5 NHS ester (Cy5) and characterized following the protocol previously described in Musicò et al.21 Sulfo Cyanine 5 NHS-ester was used instead of Sulfo Cyanine 7.5 NHS-ester, due to better suitability for FCS measurements. According to stoichiometric estimations, each fluorescently labelled CTX (mCTX) bears approximately 3.4 fluorophore molecules and 1.3 DBCO molecules. Detailed mCTX preparation protocol and characterization are reported in the ESI‡ and Fig. S1.Separation, biophysical, and biochemical characterization of REVs and MEVs
REVs and MEVs were separated and characterized according to the most updated international standards.38 Briefly (see the ESI‡ for full details), EVs were separated by differential centrifugation of, respectively, a suspension of red blood cells induced by calcium ionophore following protocols reported in ref. 39 and a mesenchymal stromal cell conditioned medium following protocols reported in ref. 40. Nanoparticle tracking analysis (NTA), dynamic light scattering (DLS), bicinchonicic acid assay (BCA), transmission electron microscopy (TEM), western blot, and the Colorimetric NANoplasmonic (CONAN) assay41 were used to determine EV particle number density, EV size distribution, (sample) protein concentration, EV morphology, EV protein markers, and the presence of exogenous single or aggregated soluble protein contaminants, respectively. Characterization data highlighted that REV and MEV preparations were similar in terms of size distribution (MEVs 78.88 ± 3.12 nm and REVs 108 ± 5.22 nm in hydrodynamic diameter (HD), Fig. S2A and C, ESI‡), protein content (MEVs 833.72 ± 257.02 μg mL−1 and REVs 942.99 ± 105.08 μg mL−1, Fig. S2B, ESI‡), and morphology (Fig. S2E, ESI‡), which appear to be cup-shaped.42 This is a typical TEM artifact caused by imaging dried EVs in a vacuum, which leads to EV deflation and collapse, resulting in altered diameter and morphology that hinder accurate EV sizing. REVs and MEVs have, as expected, different protein markers, due to different cell sources (Fig. S2G, ESI‡). Additionally, the minimal presence of soluble proteins43,44 in both EV types (Fig. S2F, ESI‡), which otherwise could generate artifacts during EV functionalization, makes them suitable for both the mCTX physisorption and the chemisorption processes.Preparation of REVs and MEVs with the (physisorbed) fluorescent cetuximab probe (mCTX)
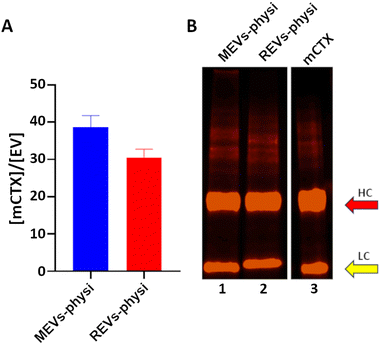
We previously demonstrated that functionalization of REVs with mCTX using physisorption or chemisorption strategies led to the formation of BCs with different stability in a biological environment.21 For this reason, we decided to use mCTX physisorbed on both REVs and MEVs as a probe for studying BC dynamics. As a control for the physisorption competition experiments REVs and MEVs were also chemisorbed through the covalent binding of mCTX via bioorthogonal click-chemistry as previously described.21 Full details on labelling are also given in the ESI.‡After REVs and MEVs functionalization we quantified mCTX per EV ex situ by combining fluorimetry and NTA (Fig. 1A). We concluded that by physisorption we obtained a consistent mCTX coverage of the same order of magnitude for the two EV types, with slightly higher coverage for MEVs. This preliminary ex situ data first points to the fact that different protein coronas may form on REVs and MEVs. In both cases, assuming an even distribution on all the EVs, the number of 30-40 mCTX per EV indicates a non-uniform EV surface coverage (between 3–5% of the total EV surface. For a uniform monolayer a minimum of about 800 mCTX can be estimated, by using the procedure reported in the ESI‡).
The SDS-PAGE densitometric profiles of mCTX obtained from the mCTX physisorbed EV samples and the stock solution of free mCTX (Fig. 1B), are experimentally identical, indicating no covalent interactions of the antibody with EV surface proteins. Differences in the densitometric profiles are visible in chemisorbed samples, as shown in Fig S3B (ESI‡).
Determination of plasma BC dynamics by fluorescence correlation spectroscopy (FCS)
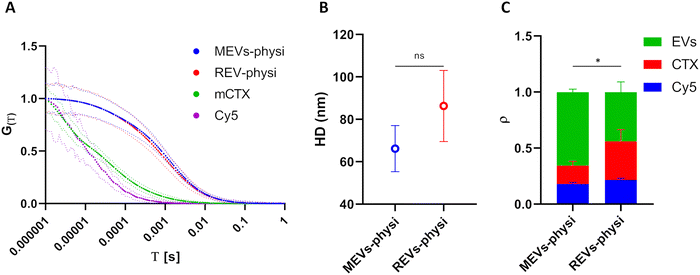
To set up FCS measurements (Fig. 2), we first measured by fluorimetry the fluorescence spectra of all the fluorescent molecules employed in this study, determining the dynamic range of measurement for the detector. As highlighted in ESI‡ Fig. S4A, no significant differences in fluorescence spectra were found between unbound Cy5, free mCTX, and mCTX physisorbed and chemisorbed onto EVs. This suggests that the association of the fluorophore to other macromolecules and nanostructures did not significantly alter its excitation and emission ranges.FCS experimental methodology is explained in the ESI.‡ The HD of unbound Cy5 and free mCTX were extrapolated from the respective FCS autocorrelation functions, resulting in 1.93 ± 0.06 nm and 20.82 ± 2.24 nm, respectively, showing good concordance with those reported in the literature.45 Moreover, the measurement also highlighted the presence of free (unbound) Cy5 in mCTX preparations (Fig. S4B, ESI‡), which could not be removed, regardless of the number of washing steps performed. Normalized autocorrelation functions of Cy5, mCTX, and mCTX physisorbed MEVs and REVs (hereafter referred to as MEVs-physi and REVs-physi) are reported in Fig. 3A (Cy5 in violet, mCTX in green, MEVs-physi in blue, and REVs-physi in red). The shift trend in the diffusion time of each fluorescent species is as expected since it depends on their HD. By extrapolating the HD from the autocorrelation functions of the EV samples, we found that MEVs-physi and REVs-physi have an HD equal to 66.18 ± 10.86 nm and 86.28 ± 16.76 nm, respectively, as reported in Fig. 3B. The size of pristine MEVs and REVs was also measured using NTA and DLS (Fig. S2A and C, ESI‡), which together provided an average EV size range of 70 to 160 nm. It is important to note that FCS, NTA, and DLS rely on different physical principles to size nano-objects in solution—fluorescence, Brownian motion, and light scattering, respectively. These differences can influence the final readout, leading to the reported discrepancies, which are therefore expected.46 Then, from the fitting of the autocorrelation functions of all samples (the method is reported in the ESI‡) we determined the fraction (ρ) of the three fluorescent entities tracked by FCS, namely unbound Cy5, free mCTX, and EVs. mCTX is either free or attached to the EVs. When mCTX attaches to the EVs, these become fluorescent objects. Each labeled EV, independent of the number of mCTX associated will count as one single fluorescent object. Therefore, from FCS we detect the relative number of free mCTX molecules and the number of labeled EVs in the confocal volume. Results are summarized in Fig. 3C, showing three main features. First, an equal fraction of unbound Cy5 (blue bars) is present in both MEV-physi and REV-physi samples and can therefore be considered as background noise. Second, MEV-physi samples have a significantly more abundant fraction of EVs (green bar) than REV-physi samples. In turn, the REV-physi sample displays a larger fraction of free mCTX (red bar) than the MEV-physi sample, indicating that mCTX is weakly associated with (has a lower affinity for) the REV surface than to the MEV surface. Or, in other words, the physisorption equilibrium (viz. the mCTX surface-to-solution partition) is less favored for REVs compared to MEVs.
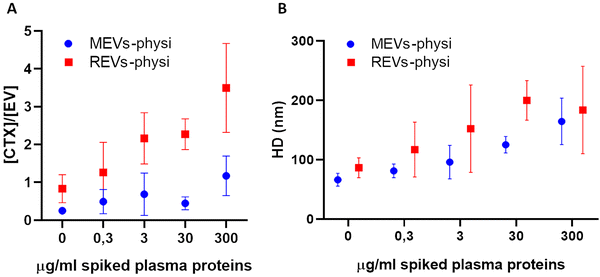
Titration experiments were performed by the addition of plasma to MEVs-physi and REV-Physi to gain information about (i) the tendency of the two EV populations to form the BC from plasma and (ii) the average architecture of such BC. Defined amounts (0.3, 3, 30, 300 μg mL−1 of proteins, corresponding to 0.001%, 0.01%, 0.1% and 1% human plasma, respectively) of EV-depleted plasma, were added to the EV samples. The system was incubated O/N to attain equilibrium and was subsequently measured by FCS to determine the competitive substitution of mCTX by plasma proteins, that is the formation of a plasma BC. Results are reported in Fig. 4. In Fig. 4A reports the variation of the molar concentration of mCTX in solution with respect to the molar concentration of EVs, which is related to the amount of mCTX competitively displaced by plasma proteins. In Fig. 4b the variations in the hydrodynamic diameter (HD) of the EVs, which is related to the amount of physisorbed plasma proteins, are reported.
Interestingly, REVs-physi and MEVs-physi displayed a significant detachment of mCTX (Fig. 4A), confirming the weak interaction between the antibody and the EV surface,21 which led to mCTX displacement due to the adsorption of plasma proteins. Moreover, despite both REVs-physi and MEVs-physi showing a consistent detachment of mCTX after the addition of plasma proteins, for REVs-physi the mCTX detachment is much more pronounced than for MEVs-physi (viz. REVs exchange rate of mCTX with plasma protein is higher), confirming that the two EV types have different affinities for mCTX and plasma proteins.
Indeed, the detachment of mCTX from the EV surface is probably dependent on the affinity competition between plasma proteins and mCTX. These data strongly support that physisorbed mCTX on MEVs-physi is more stably anchored than mCTX on REVs-physi, being also evident for not-spiked samples.
The analysis of the HD fluctuations on the physisorbed samples showed that plasma protein adsorption onto REVs-physi and MEVs-physi occurred following two different dynamics. In specific, REVs-physi HD grows rapidly, reaching a plateau at the concentration of 30 μg mL−1 of spiked plasma proteins (Fig. 4B, red squares). On the contrary, MEVs-physi HD increases slowly and does not reach a plateau, even at the higher concentration of spiked plasma proteins tested (Fig. 4B, blue dots). To our knowledge, no appropriate models can correctly fit our data providing apparent affinity constants and/or repartition coefficients. This is probably due to the complexity and heterogeneity of the interactions that can occur during the formation of the BC on the EV surface, which cannot be described by classic fitting models.
Chemisorbed REVs and MEVs were prepared to generate a negative control for mCTX displacement, as mCTX bound to the EV surface in this way is stably anchored and should not be exchanged with other proteins.21 Accordingly, these samples showed a minimal detachment of mCTX after the addition of plasma proteins (Fig. S6E, ESI‡), confirming that mCTX in the preparation is mostly covalently bound to the EV surface and not physisorbed (autocorrelation functions of physisorbed and chemisorbed MEVs and REVs after every spike of plasma proteins are reported in ESI‡ Fig. S6A–D).
Considerations on the BC architecture
The possible architecture of the formed plasma BC can be inferred by comparing the experimental and nominal HD, where the experimental HD is the one determined by FCS, and the nominal is calculated by the following stoichiometric and geometric arguments.The nominal HD is obtained by assuming that all the plasma proteins introduced in each spike are physisorbed onto each EV by an identical layer-by-layer process, where each monolayer beging to form only after the underlying layer has been completed. Pristine EVs are modeled as identical spheres (with the diameter determined by FCS, 66 ± 10.9 nm and 86 ± 16.8 nm for MEVs-physi and REVs-physi, respectively) and all the plasma proteins as identical spheres of 7 nm diameter adsorbing in monolayers with a packing factor of 0.63 (see the section “Stoichiometric evaluation of BC corona formation” in the ESI‡ for details). This results in a monolayer thickness of 7 nm and, in turn, a 14 nm increase in the EV HD. From this, the nominal HD of the EVs (hereafter referred HDnom) that builds from the number of the nominal monolayers for a given spike of plasma proteins can be calculated, and finally compared with the experimental HD (hereafter referred HDexp) determined by FCS.
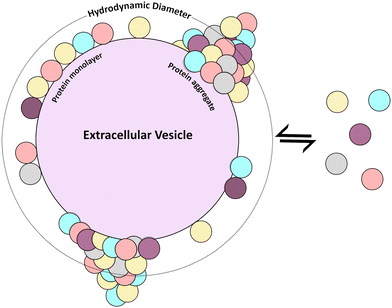
Results are summarized in Table 1. In the case of REVs-physi, for the first plasma spike, corresponding to 0.3 μg mL−1 of proteins, the number of proteins is far lower than that needed to complete a monolayer, as indicated by the nominal number of monolayers, which is 0.02. Considering that this tiny fraction of monolayer (corresponding to less than 10 proteins) cannot cause an increase in the HD of the pristine EV, one obtains HDnom = 86 ± 16.8 nm, which is lower than HDexp = (116 ± 36.2) nm. This discrepancy suggests that the BC accumulates in discrete regions forming protruding aggregates (Fig. 5), which cause an overall increase in the HDexp. The significant error on HDexp, about 30%, indicates a substantial particle-to-particle variability, suggesting that BC can also organize in non-uniform monolayers (Fig. 5), with negligible impact on HDexp. Analogous reasoning holds for REVs-physi when exposed to 3.0 μg mL−1 plasma protein spikes, as well as for MEVs-physi upon the 0.3 μg mL−1 and 3.0 μg mL−1 plasma protein spikes.
| Spiked plasma proteins (μg mL−1) | REVs-physi | MEVs-physi | ||||||
|---|---|---|---|---|---|---|---|---|
| HDexp (nm) | HDnom (nm) | Nominal number of monolayers | Probable BC architecture(s) | HDexp (nm) | HDnom (nm) | Nominal number of monolayers | Probable BC architecture(s) | |
| 0.3 | 116 ± 36.2 | 86 ± 16.8 | 0.02 | Non-uniform monolayer and aggregates | 81 ± 11.5 | 66 ± 10.9 | 0.03 | Non-uniform monolayer and aggregates |
| 3.0 | 152 ± 53.6 | 86 ± 16.8 | 0.17 | Non-uniform monolayer and aggregates | 95 ± 18.2 | 66 ± 10.9 | 0.27 | Non-uniform monolayer and aggregates |
| 30.0 | 199 ± 53.6 | 114 ± 16.8 | 1.5 | Monolayers and aggregates | 125 ± 13.9 | 94 ± 10.9 | 2 | Monolayers and aggregates |
| 300.0 | 183 ± 33.2 | 142 ± 16.8 | 4 | Monolayers and aggregates | 164 ± 39.1 | 136 ± 10.9 | 5 | Monolayers and aggregates |
On the other hand, when spiking REVs-physi and MEVs-physi with higher concentrations of plasma proteins (30.0 and 300.0 μg mL−1) we estimated an increase in the HD of the pristine EVs due to the formation of multiple monolayers of BC. Specifically, with a spike of 30.0 μg mL−1 of plasma proteins, HDnom for MEVs and REVs are respectively 94 ± 10.9 and 114 ± 16.8 nm, while with 300.0 μg mL−1 of plasma proteins, HDnom for the two EV subtypes are 136 ± 10.9 nm (MEVs) and 142 ± 16.8 nm (REVs). Again, the HDexp is higher than the HDnom in both cases and at every spike of plasma protein, specifically for MEVs-physi and REVs-physi, measured HDexp are respectively 125 ± 13.9 nm and 199 ± 53.6 nm at 30.0 μg mL−1, while, at 300.0 μg mL−1, 164 ± 39.1 nm and 183 ± 33.2 nm. These discrepancies suggest again that the BC accumulates in discrete regions of protruding aggregates, causing such an increase in the HDexp, however in this case we can speculate about the possible formation of complete layers of BC on the EV surface due to the high concentration of proteins. Notably, the difference between HDnom and HDexp in MEVs, spiked with 300.0 μg mL−1, is around 28 nm, while for REVs is 57 nm, which corresponds to 2 and 4 further BC layers respectively, thus indicating the formation of aggregates with a bigger size in REVs-physi samples compared to MEVs.
On a final note, we remark that the above variety in BC architectures, holds at the “collective level” (viz. the suspension level that is averaged over the whole population of EVs47), but is also well mirrored at “single-EV level” as previously reported.15
Conclusions
In this study, we developed an FCS-based methodology to monitor BC formation on REVs and MEVs in physiological conditions by an in situ protocol, unbiased from separation methods.By spiking our EVs with different concentrations of plasma proteins and using the fluctuation of the HD as a readout of the adsorption of such proteins at the EV surface we evaluated the dynamic formation of the BC.
Our findings reveal distinct BC dynamics between REVs and MEVs, with REVs featuring a higher exchange. By interpreting experimental observations with geometrical and stoichiometric considerations, it also turned out that in both the EV populations the BC does not simply (dynamically) self-organize in a layer-by-layer process, but instead may also pile up into discrete islands of protruding protein aggregates, with REVs showing a higher number of aggregates, or aggregates of a bigger size.
These findings provide new insight into the EV–BC. They underscore that BC formation, evolution, and architecture may be influenced by the EV's origin, and in turn surface properties, highlighting an additional layer of heterogeneity among circulating EVs. Future efforts shall be devoted to applying the strategy proposed in the manuscript to compare the influence of different EV separation protocols18 (i.e. tangential flow filtration, size exclusion chromatography, microfluidic techniques, etc.) on BC formation and stability. From an applicative perspective, our findings suggest that BC dynamics should be a key consideration when evaluating the use of EVs as therapeutics and in diagnostics.
Author contributions
Conceptualization: P. B, A. M., S. M., A. R., and A. Z.; data curation: A. M., S. G. R., and A. Z.; funding acquisition: P. B., A. M., S. M., and A. R.; investigation: M. C., A. M., L. P., A. P., M. R., S. G. R., and A. Z.; methodology: P. B., A. M., S. M., A. R., and A. Z.; project administration: P. B. and S. M.; resources: C. A., P. B., M. C., S. M., A. N., O. P., and A.R.; supervision: P. B., S. M., and A. R. Visualization: A. M. and A. Z.; writing – original draft: P. B. A. M., and A. Z.; writing – review & editing: all authors. Please turn to the CRediT taxonomy for the term explanation.Data availability
The data supporting this article have been included as part of the ESI.‡Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been supported by MIUR through PRIN 2017E3A2NR_004 project, by European Union's Horizon 2020 research and innovation program through the BOW project (H2020-EIC-FETPROACT-2019, ID: 952183); by the NFFA-Europe-Pilot Transnational Access Activity (ID 101007417), having benefited from the access provided by CIC biomaGUNE, proposal ID 362. S. E. M. thanks the PID2020-114356RB-I00 project from the Ministry of Science and Innovation of the Government of Spain; and by Consorzio Interuniversitario Nazionale per la Scienza e Tecnologia dei Materiali – INSTM. The authors would like to thank Prof. Fabio Corsi and Serena Mazzucchelli (University of Milan, Italy) for providing the Cetuximab antibody, and Dr Desirè De Silvio and Dr Irantzu Llarena for the support for FCS measurements.References
- K. A. Dawson and Y. Yan, Current understanding of biological identity at the nanoscale and future prospects, Nat. Nanotechnol., 2021, 16(3), 229 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7(12), 779 CrossRef CAS PubMed.
- J. Ren, N. Andrikopoulos, K. Velonia, H. Tang, R. Cai and F. Ding, et al., Chemical and Biophysical Signatures of the Protein Corona in Nanomedicine, J. Am. Chem. Soc., 2022, 144, 9184 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin and H. Nilsson, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. 2007. Available from: https://www.pnas.org/cgi/content/full/.
- M. Mahmoudi, M. P. Landry, A. Moore and R. Coreas, The protein corona from nanomedicine to environmental science, Nat. Rev. Mater., 2023, 8, 422 CrossRef PubMed.
- P. L. Latreille, M. Le Goas, S. Salimi, J. Robert, G. De Crescenzo and D. C. Boffito, et al., Scratching the Surface of the Protein Corona: Challenging Measurements and Controversies, ACS Nano, 2022, 16, 1689 CrossRef CAS.
- D. N. Trinh, R. A. Gardner, A. N. Franciosi, C. McCarthy, M. P. Keane and M. G. Soliman, et al., Nanoparticle Biomolecular Corona-Based Enrichment of Plasma Glycoproteins for N-Glycan Profiling and Application in Biomarker Discovery, ACS Nano, 2022, 16(4), 5463 CrossRef CAS.
- A. Radeghieri and P. Bergese, The biomolecular corona of extracellular nanoparticles holds new promises for advancing clinical molecular diagnostics, Expert Rev. Mol. Diagn., 2023, 23, 471 CrossRef CAS.
- S. Busatto, A. Zendrini, A. Radeghieri, L. Paolini, M. Romano and M. Presta, et al., The nanostructured secretome, Biomater. Sci., 2020, 8, 39–63 RSC.
- E. I. Buzás, E. Tóth, B. W. Sódar and K. Szabó-Taylor, Molecular interactions at the surface of extracellular vesicles, Semin. Immunopathol., 2018, 40, 453 CrossRef.
- E. Lozano-Andrés, A. Enciso-Martinez, A. Gijsbers, A. Ridolfi, G. Van Niel and S. F. W. M. Libregts, et al., Physical association of low density lipoprotein particles and extracellular vesicles unveiled by single particle analysis, J. Extracell. Vesicles, 2023, 12(11), e12376 CrossRef.
- A. Ridolfi, L. Conti, M. Brucale, R. Frigerio, J. Cardellini and A. Musicò, et al., Particle profiling of EV-lipoprotein mixtures by AFM nanomechanical imaging, J. Extracell. Vesicles, 2023, 12(10), 12349 CrossRef CAS PubMed.
- S. Busatto, Y. Yang, D. Iannotta, I. Davidovich, Y. Talmon and J. Wolfram, Considerations for extracellular vesicle and lipoprotein interactions in cell culture assays, J. Extracell. Vesicles, 2022, 11, e12202 CrossRef CAS PubMed.
- S. Busatto, Y. Yang, S. A. Walker, I. Davidovich, W. H. Lin and L. Lewis-Tuffin, et al., Brain metastases-derived extracellular vesicles induce binding and aggregation of low-density lipoprotein, J. Nanobiotechnol., 2020, 18(1), 162 CrossRef CAS PubMed.
- E. Tóth, L. Turiák, T. Visnovitz, C. Cserép, A. Mázló and B. W. Sódar, et al., Formation of a protein corona on the surface of extracellular vesicles in blood plasma, J. Extracell. Vesicles, 2021, 10(11), 12140 CrossRef.
- F. G. Gomes, A. C. Andrade, M. Wolf, S. Hochmann, L. Krisch and N. Maeding, et al., Synergy of Human Platelet-Derived Extracellular Vesicles with Secretome Proteins Promotes Regenerative Functions, Biomedicines, 2022, 10(2), 238 CrossRef CAS PubMed.
- R. Liam-Or, F. N. Faruqu, A. Walters, S. Han, L. Xu and J. T. W. Wang, et al., Cellular uptake and in vivo distribution of mesenchymal-stem-cell-derived extracellular vesicles are protein corona dependent, Nat. Nanotechnol., 2024, 846–855 CrossRef CAS.
- M. Wolf, R. W. Poupardin, P. Ebner-Peking, A. C. Andrade, C. Blöchl and A. Obermayer, et al., A functional corona around extracellular vesicles enhances angiogenesis, skin regeneration and immunomodulation, J. Extracell. Vesicles, 2022, 11(4), 12207 CrossRef PubMed.
- L. Dietz, J. Oberländer, A. Mateos-Maroto, J. Schunke, M. Fichter and E. M. Krämer-Albers, et al., Uptake of extracellular vesicles into immune cells is enhanced by the protein corona, J. Extracell. Vesicles, 2023, 12(12), 12399 CrossRef.
- A. Radeghieri, S. Alacqua, A. Zendrini, V. Previcini, F. Todaro and G. Martini, et al., Active antithrombin glycoforms are selectively physiosorbed on plasma extracellular vesicles, J. Extracell. Biol., 2022, 1(9), 57 CrossRef.
- A. Musicò, R. Zenatelli, M. Romano, A. Zendrini, S. Alacqua and S. Tassoni, et al., Surface functionalization of extracellular vesicle nanoparticles with antibodies: a first study on the protein corona variable, Nanoscale Adv., 2023, 5(18), 4703 RSC.
- E. I. Buzas, Opportunities and challenges in studying the extracellular vesicle corona, Nat. Cell Biol., 2022, 24, 1322 CrossRef CAS PubMed.
- D. Choi, G. Go, D. K. Kim, J. Lee, S. M. Park and D. Di Vizio, et al., Quantitative proteomic analysis of trypsin-treated extracellular vesicles to identify the real-vesicular proteins, J. Extracell. Vesicles, 2020, 9(1), 1757209 CrossRef CAS.
- D. Di Silvio, A. Silvestri, L. Lay, L. Polito and S. E. Moya, Impact of ConcanavalinA affinity in the intracellular fate of Protein Corona on Glucosamine Au nanoparticles, Sci. Rep., 2018, 8(1), 9046 CrossRef PubMed.
- T. Liedl, S. Keller, F. C. Simmel, J. O. Rädler and W. J. Parak, Fluorescent nanocrystals as colloidal probes in complex fluids measured by fluorescence correlation spectroscopy, Small, 2005, 1(10), 997–1003 CrossRef CAS PubMed.
- N. Joshi, S. Basak, S. Kundu, G. De, A. Mukhopadhyay and K. Chattopadhyay, Attenuation of the early events of α-synuclein aggregation: A fluorescence correlation spectroscopy and laser scanning microscopy study in the presence of surface-coated Fe3O4 nanoparticles, Langmuir, 2015, 31(4), 1469 CrossRef CAS.
- L. Shang and G. U. Nienhaus, In Situ Characterization of Protein Adsorption onto Nanoparticles by Fluorescence Correlation Spectroscopy, Acc. Chem. Res., 2017, 50(2), 387 CrossRef CAS PubMed.
- H. Wang, Y. Lin, K. Nienhaus and G. U. Nienhaus, The protein corona on nanoparticles as viewed from a nanoparticle-sizing perspective, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10(4), 1500 Search PubMed.
- G. U. Nienhaus, P. Maffre and K. Nienhaus, in Studying the protein corona on nanoparticles by FCS, Methods in Enzymology, Academic Press Inc, 2013. p. 115 Search PubMed.
- M. Martinez-Moro, D. Di Silvio and S. E. Moya, Fluorescence correlation spectroscopy as a tool for the study of the intracellular dynamics and biological fate of protein corona, Biophys. Chem., 2019, 253 Search PubMed.
- M. K. Jayasinghe, M. Pirisinu, Y. Yang, B. Peng, T. T. Pham and C. Yu Lee, et al., Surface-engineered extracellular vesicles for targeted delivery of therapeutic RNAs and peptides for cancer therapy, Theranostics, 2022, 12(8), 3288 CrossRef CAS PubMed.
- B. Peng, Y. Yang, Z. Wu, R. Tan, T. T. Pham and E. Y. M. Yeo, et al., Red blood cell extracellular vesicles deliver therapeutic siRNAs to skeletal muscles for treatment of cancer cachexia, Mol. Ther., 2023, 31(5), 1418 CrossRef CAS PubMed.
- P. H. D. Nguyen, M. K. Jayasinghe, A. H. Le, B. Peng and M. T. N. Le, Advances in Drug Delivery Systems Based on Red Blood Cells and Their Membrane-Derived Nanoparticles, ACS Nano, 2023, 17, 5187 CrossRef CAS.
- C. Gissi, A. Radeghieri, C. A. Lamorgese Passeri, M. Gallorini, L. Calciano and F. Oliva, et al., Extracellular vesicles from rat-bone-marrow mesenchymal stromal/stem cells improve tendon repair in rat Achilles tendon injury model in dose-dependent manner: A pilot study, PLoS One, 2020, 15(3), e0229914 CrossRef CAS.
- V. Marassi, G. La Rocca, A. Placci, A. Muntiu, F. Vincenzoni and A. Vitali, et al., Native characterization and QC profiling of human amniotic mesenchymal stromal cell vesicular fractions for secretome-based therapy, Talanta, 2024, 276 Search PubMed.
- P. Valiukevičius, J. Mačiulaitis, D. Pangonytė, V. Siratavičiūtė, K. Kluszczyńska and U. Kuzaitytė, et al., Human Placental Mesenchymal Stem Cells and Derived Extracellular Vesicles Ameliorate Lung Injury in Acute Respiratory Distress Syndrome Murine Model, Cells, 2023, 12(23), 2729 CrossRef.
- M. Romano, A. Zendrini, L. Paolini, S. Busatto, A. C. Berardi and P. Bergese, et al., Extracellular vesicles in regenerative medicine, Nanomater. Theranostics Tissue Eng., 2020, 29–58 CAS.
- J. A. Welsh, D. C. I. Goberdhan, L. O’Driscoll, E. I. Buzas, C. Blenkiron and B. Bussolati, et al., Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches, J. Extracell. Vesicles, 2024, 13(2), e12404 CrossRef.
- W. M. Usman, T. C. Pham, Y. Y. Kwok, L. T. Vu, V. Ma and B. Peng, et al., Efficient RNA drug delivery using red blood cell extracellular vesicles, Nat. Commun., 2018, 9, 2359 CrossRef PubMed.
- A. R. Silini, A. Papait, A. Cargnoni, E. Vertua, P. Romele and P. Bonassi Signoroni, et al., CM from intact hAM: an easily obtained product with relevant implications for translation in regenerative medicine, Stem Cell Res. Ther., 2021, 12, 540 CrossRef CAS PubMed.
- A. Zendrini, L. Paolini, S. Busatto, A. Radeghieri, M. Romano and M. H. M. Wauben, et al., Augmented COlorimetric NANoplasmonic (CONAN) Method for Grading Purity and Determine Concentration of EV Microliter Volume Solutions, Front. Bioeng. Biotechnol., 2020, 7 Search PubMed.
- L. G. Rikkert, R. Nieuwland, L. W. M. M. Terstappen and F. A. W. Coumans, Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent, J. Extracell. Vesicles, 2019, 8, 1555419 CrossRef CAS.
- A. Zendrini, G. Guerra, K. Sagini, T. Vagner, D. Di Vizio and P. Bergese, On the surface-to-bulk partition of proteins in extracellular vesicles, Colloids Surf., B, 2022, 218 Search PubMed.
- E. D. Sverdlov, Amedeo Avogadro's cry: What is 1 μg of exosomes?, BioEssays, 2012, 34(10), 873 CrossRef CAS PubMed.
- H. Imamura, A. Sasaki and S. Honda, Fate of a Stressed Therapeutic Antibody Tracked by Fluorescence Correlation Spectroscopy: Folded Monomers Survive Aggregation, J. Phys. Chem. B, 2017, 121(34), 8085 CrossRef CAS.
- C. Montis, A. Zendrini, F. Valle, S. Busatto, L. Paolini, A. Radeghieri, A. Salvatore, D. Berti and P. Bergese, Size distribution of extracellular vesicles by optical correlation techniques, Colloids Surf., B, 2017, 158, 331–338 CrossRef CAS.
- J. M. Rabanel, V. Adibnia, S. F. Tehrani, S. Sanche, P. Hildgen and X. Banquy, et al., Nanoparticle heterogeneity: an emerging structural parameter influencing particle fate in biological media?, Nanoscale, 2019, 11, 383–406 RSC.
Footnotes |
| † We dedicate this work to Marina Cretich, who left us during the final stages of its preparation. Our deep yet light and delightful discussions, your incisive yet kind comments, your unique intelligence and creativity, which you truly shared with everyone, your generosity will be irreplaceable. |
| ‡ Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nh00320a |
| § These authors equally contributed to the work. |
| ¶ This author passed away. |
| This journal is © The Royal Society of Chemistry 2025 |