Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
G-quadruplex-driven molecular disassembly and type I-to-type II photophysical conversion of a heavy-atom-free photosensitizer for site-specific oxidative damage†
Karolina
Saczuk
 a,
Maria V.
Cottini
b,
Marta
Dudek
a,
Maria V.
Cottini
b,
Marta
Dudek
 a,
Leszek M.
Mazur
a,
Leszek M.
Mazur
 a,
Dario Puchán
Sánchez
c,
Lucía
López-Pacios
d,
Ahmad
Kassem
c,
Katarzyna
Matczyszyn
a,
Dario Puchán
Sánchez
c,
Lucía
López-Pacios
d,
Ahmad
Kassem
c,
Katarzyna
Matczyszyn
 a,
Juan J.
Nogueira
a,
Juan J.
Nogueira
 de,
Cyrille
Monnereau
de,
Cyrille
Monnereau
 f,
Lara
Martínez-Fernández
f,
Lara
Martínez-Fernández
 *g,
Jan
Jamroskovic
*g,
Jan
Jamroskovic
 b,
Clément
Cabanetos
b,
Clément
Cabanetos
 *c and
Marco
Deiana
*c and
Marco
Deiana
 *a
*a
aInstitute of Advanced Materials, Faculty of Chemistry, Wrocław University of Science and Technology, Wyb. Wyspiańskiego 27, 50-370 Wrocław, Poland. E-mail: m.deiana@pwr.edu.pl
bDepartment of Microbial Genetics, Slovak Academy of Sciences, Institute of Molecular Biology, Dubravska cesta 21, 845 51 Bratislava, Slovakia
cUniv Angers, CNRS MOLTECH-ANJOU, SFR MATRIX, F-49000 Angers, France. E-mail: clement.cabanetos@univ-angers.fr
dDepartment of Chemistry, Universidad Autónoma de Madrid, Calle Francisco Tomás y Valiente, 7, 28049 Madrid, Spain
eIADCHEM, Institute for Advanced Research in Chemistry, Universidad Autónoma de Madrid, Calle Francisco Tomás y Valiente, 7, 28049 Madrid, Spain
fENS de Lyon, CNRS, Laboratoire de Chimie, UMR 5182, 46 allée d’Italie, F-69342 Lyon, France
gDepartamento de Química Física de Materiales, Instituto de Química Física Blas Cabrera, CSIC 28006, Madrid, Spain. E-mail: lmartinez@iqf.csic.es
First published on 18th June 2025
Abstract
G-quadruplex (G4)-targeted photosensitizers (PSs) are advancing precision oncology by confining DNA damage to malignant cells while sparing healthy tissue. Yet, molecular-level studies on the mechanisms and dynamics of G4 structure damage under PSs light-activation are limited. Here, we introduce DBI-POE, an activatable, heavy-atom-free PS derived from the G4-specific sulfur-substituted dibenzothioxanthene imide (S-DBI) and modified with a hydrophilic, bio-compatible polyoxyethylene (POE) side chain. In aqueous solution, owing to its amphiphilic character, DBI-POE self-assembles into nanoaggregates that disassemble upon binding to G4 DNA. This disassembly switches its photophysical behavior “turning on” its fluorescence while enabling two-photon near-infrared (NIR) excitation. Moreover, while DBI-POE follows a type I pathway in the aggregated state, producing superoxide anion (O2˙−) and hydroxyl (OH˙) radicals, it shifts to a type II mechanism that predominantly generates singlet oxygen (1O2) upon G4 binding. The generated 1O2 selectively oxidizes guanine residues, triggering G4 unfolding, a mechanism validated through biophysical experiments, dot blot assay and molecular dynamics (MD) simulations. Furthermore, biochemical experiments at single-base resolution reveal that photoactivated DBI-POE induces site-specific oxidative lesions at G4 sites, stalling DNA polymerase, while non-G4 regions remain unaffected. This combination of supramolecular disassembly, photophysical pathway switching, and G4-selective oxidative damage underscores the high specificity of DBI-POE, opening new avenues for the design of next-generation G4-targeted PSs for photodynamic cancer therapies.
New conceptsThis pioneering study explores, for the first time, the mechanistic and dynamic basis of photodamage in G-quadruplex (G4) DNA structures, which are emerging as promising anticancer targets in photodynamic therapy. To this end, we designed DBI-POE, a hydrophilic, sulfur-substituted dibenzothioxanthene imide derivative derived from the most potent G4-specific photosensitizer reported, strategically modified with a neutral polyoxyethylene side chain to overcome the hydrophobic limitations of its precursor. In aqueous solution, DBI-POE self-assembles into aggregation-quenched nanostructures, with fluorescence restored upon selective binding and monomerization to G4 structures, a phenomenon termed disaggregation-induced emission. This transformation not only amplifies fluorescence intensity and extends excited-state lifetimes but also enables efficient two-photon excitation in the near-infrared region. Notably, the G4-induced disassembly shifts its photochemical pathway from a type I mechanism, characterized by superoxide and hydroxyl radical generation, to a guanine-selective type II mechanism that yields singlet oxygen exclusively. This precise switch triggers site-specific oxidative guanine lesions and complete G4 unfolding, as mapped by high-resolution DNA polymerase stop assays with single-base precision. Our multidisciplinary approach, integrating photochemistry, biophysics, biochemistry, quantum mechanical calculations, and molecular dynamics simulations, provides unprecedented mechanistic insights that pave the way for designing a brand-new generation of structure-specific, G4-targeted photosensitizers in anticancer phototherapy. |
Introduction
Light-activated therapies have rapidly emerged as a transformative approach in oncology, offering the unprecedented ability to confine cytotoxic events with exquisite spatiotemporal control.1–3 This precision minimizes systemic toxicity while maximizing therapeutic efficacy.4 Among these approaches, photodynamic therapy (PDT) harnesses photosensitizers (PSs) that, upon illumination, generate reactive oxygen species (ROS) to induce localized oxidative stress and cell death.2 Despite their promise, next-generation PSs face significant challenges, including aggregation in physiological media, suboptimal optical properties for deep-tissue penetration, and undesirable off-target interactions.5,6 In particular, the development of heavy-atom-free PSs with superior photostability, selective accumulation, and robust ROS generation, preferentially in the near-infrared (NIR) therapeutic window, remains a critical objective.7,8A compelling strategy to overcome these challenges involves the use of supramolecularly responsive systems.9–13 While aggregation-induced emission (AIE) strategies have enhanced the photophysical performance of certain luminophores,14–18 many conventional PSs suffer from aggregation-caused quenching (ACQ),18–24 which diminishes their optical output and therapeutic efficiency. Recently, disaggregation-induced emission (DIE) approaches have been advanced to reverse ACQ effects in biological settings, thereby simultaneously amplifying fluorescence, improving bioavailability, and increasing therapeutic potency.18,25
Within this framework, G-quadruplex (G4) DNA has emerged as a potent trigger for controlled disassembly.26–32 Imaging studies using G4-specific antibodies such as BG4,33,34 alongside fluorescent reporters,35,36 have provided direct evidence of G4 DNA formation in living cells. These structures are particularly enriched during the S phase of the cell cycle, suggesting a functional role in DNA replication.33 Their formation is thought to be facilitated by duplex unwinding, which helps relieve thermodynamic strain.37
Computational analyses have predicted over 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 potential G4-forming sequences in the human genome,38 while experimental approaches such as G4-seq have identified more than 700
000 potential G4-forming sequences in the human genome,38 while experimental approaches such as G4-seq have identified more than 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 G4 DNA sites,39 including many that are not computationally predictable. Chromatin immunoprecipitation with BG4 followed by high-throughput sequencing (G4 ChIP-seq) has mapped G4 structures in vivo, revealing strong concordance with G4-seq data.40
000 G4 DNA sites,39 including many that are not computationally predictable. Chromatin immunoprecipitation with BG4 followed by high-throughput sequencing (G4 ChIP-seq) has mapped G4 structures in vivo, revealing strong concordance with G4-seq data.40
Studies in immortalized human epidermal keratinocyte HaCaT cells have shown increased G4 levels in certain tissues compared to healthy controls.40 Furthermore, G4 abundance has been reported to be significantly higher in cancer cells with impaired DNA repair mechanisms, such as triple-negative breast cancer (TNBC) cells, highlighting a potential link between genomic instability and G4 accumulation.33 Overall, these findings indicate that G4s are predominantly located in regulatory regions, nucleosome-depleted zones, and promoters of actively transcribed genes in cancer cells, highlighting their key roles in gene regulation and genome organization.40–42
The physiological relevance of G4s is further supported by G4-interacting proteins, such as helicases.43 These helicases unwind G4 structures to ensure proper DNA replication and transcription.44,45 Mutations or loss of function in these helicases disrupt G4 resolution, leading to transcriptional dysregulation and genomic instability, hallmarks of cancer.42
The evidence that G4 DNA folds in a temporally regulated manner in vivo, and that these structures are concentrated in oncogenes and cancer-related genes, supports their relevance as targets for anticancer therapeutics.46–49
Beyond their regulatory roles in oncogene regulation,41,42,50–52 G4s are also inherently prone to oxidation.53–56 Since, among all nucleobases, singlet oxygen (1O2) preferentially oxidizes guanine residues,57,58 G4 motifs are ideal targets for selective oxidative disruption.53 However, despite this promise, only a handful of G4 targeting-PS structures have been developed, primarily operating via a type I mechanism.59–64 In those rare instances, the molecular details of G4 oxidation remain unclear, as it is well established that type I radicals such as hydroxyl (OH˙), generally react non-selectively with nucleobases.58,65 For example, Chen et al. employed an acridinium derivative to target RNA G4s while generating superoxide anion (O2˙−) and OH˙ radicals.59 Similarly, Holden et al. used a Ru(II) complex to induce photodamage in mitochondrial G4s, although this compound failed to distinguish between G4 and duplex DNA.63 In studies with supercoiled pUC19 plasmid DNA, light-induced cleavage persisted even in the presence of the 1O2 trap sodium azide (NaN3), suggesting involvement of multiple ROS.63 Additionally, Zhang et al. reported that a triazole-attached dibenzoquinoxaline targeting G4 motifs produces O2˙− and OH˙ radicals, but not 1O2.60
Clearly, although previous studies have advanced our understanding of PS-mediated phototherapy at both cellular and in vivo levels, they have not yet clarified the molecular mechanisms underlying selective G4 damage.
In our earlier work, we demonstrated that heavy-atom-free mono-(BTI) and dibenzothioxanthene (DBI) PSs exhibited potent phototherapeutic activity with negligible dark toxicity.66–71 Notably, sulfur-substituted S-DBI showed exceptional G4-binding affinity and nearly unitary 1O2 generation quantum efficiency.67 However, its intrinsic hydrophobicity resulted in significant aggregation in aqueous solution, complicating detailed mechanistic studies of its G4-mediated phototherapeutic action.67
To address these challenges, gain deeper insights into selective G4 targeting, and ultimately facilitate future clinical translation, we developed DBI-POE, an amphiphilic, heavy-atom-free PS derived from S-DBI and modified with a hydrophilic polyoxyethylene (POE) side chain (Scheme 1). In aqueous media, DBI-POE spontaneously self-assembles into well-defined nanoaggregates that disassemble upon selective binding to G4 DNA. This disassembly not only activates the PS's emissive properties and two-photon absorption but also reprograms its photophysical behavior from a type I pathway, which produces O2˙− and OH˙ radicals, to a type II pathway that predominantly generates 1O2.
Comprehensive biophysical and theoretical studies, including circular dichroism (CD), nuclear magnetic resonance (NMR), molecular dynamics (MD) simulations and quantum mechanical (QM) calculations, confirmed that the photoinduced 1O2 selectively oxidized guanine residues, triggering precise unfolding of the G4 structure. Moreover, primer extension assays at single-base pair resolution revealed that photoactivated DBI-POE induced site-specific oxidative lesions that stalled DNA polymerase exclusively at damaged G4 regions, while, critically, non-G4 sequences remained unaffected.
Results and discussion
Photosensitizer design and characterization
Previously, we demonstrated that S-DBI exhibits exceptional G4-binding affinity and potent photodynamic therapeutic effects across 2D and 3D cancer cell models, as well as in a zebrafish rhabdomyosarcoma model.67,72 However, the intrinsic propensity of this compound to form highly stable nanoaggregates has impeded detailed mechanistic investigations into its phototriggered oxidative DNA damage at G4 sites (Scheme 1).To overcome this limitation and, for the first time, elucidate the molecular basis of G4-mediated photodynamic performance, we designed and synthesized DBI-POE, a hydrophilic variant of S-DBI modified with a polyoxyethylene (POE) side chain. DBI-POE was synthesized via a straightforward route starting from the previously prepared compound DBA (Scheme 1 and Fig. S1–S3, ESI†).67 As expected, the introduction of the POE substituent significantly enhances solubility by approximately 4.0-fold and reduces aggregation under biologically relevant conditions (vide infra).
In organic solvents such as methanol (MeOH)22,31 and dimethyl sulfoxide (DMSO),26,27 both recognized for their strong solubilizing and disaggregating properties, DBI-POE exhibited a sharp absorption band at approximately 485 nm (Fig. S4 and S5, ESI†). This observation aligns with computational predictions (S1(ππ*) state of DBI-POE, oscillator strength of 0.59, at 2.99 eV [413 nm], Table S4, ESI†) and closely mirrors the spectral characteristics of its sulfur-substituted parent structure in organic solvents, clearly indicating that the compound is well dissolved in these high polarity solvents.67 Upon incremental water addition, this absorption band gradually decreased in intensity, broadened, and underwent a bathochromic shift (S1(ππ*) state of the dimer DBI-POE2, oscillator strength of 0.03, at 2.89 eV [429 nm], Table S4 (ESI†), being the spectroscopic state S2), indicative of a self-assembly process driven by interchromophoric interactions reminiscent of slip-stacked arrangement (Fig. S4 and S5, ESI†).30,73 Concomitantly, fluorescence studies in pure MeOH revealed an emission band centered at 551 nm that experienced approximately a five-fold quenching and a shift to 564 nm with increasing water content, pointing to the existence of competitive less emissive or non-radiative decay pathways (Table S5 and Fig. S4B, ESI†).22
The UV/vis and emission spectra of DBI-POE, recorded in MeOH or in cell culture medium supplemented with 10% fetal calf serum after exposure to 485 ± 20 nm light for 1 hour at an irradiance of 10 mW cm−2, demonstrated excellent photostability, an essential prerequisite for effective long-term photosensitization (Fig. S6, ESI†).
Temperature-dependent UV/vis and emission experiments further underscored the reversible nature of DBI-POE self-assembly in aqueous media: at 25 °C, the dye exhibited broad absorption and quenched emission, whereas at 95 °C, it reverted to a well-defined absorption profile with progressively enhanced emission (Fig. S7, ESI†).28 Moreover, the addition of the anionic surfactant sodium dodecyl sulfate (SDS), widely used as a disaggregating agent to facilitate the dispersion and solubilization of aggregated compounds, induced pronounced changes in both the absorption and emission spectra, consistent with the disassembly of DBI-POE aggregates into monomeric species (Fig. 1A).26–29 Dynamic light scattering (DLS) measurements corroborated these spectroscopic findings by revealing the formation of uniform DBI-POE nanoaggregates with an average diameter of 190 nm in aqueous media (Fig. 1B). Calculations predicted the formation of the most stable dimeric species (DBI-POE2), with an aggregation free energy of −4.6 kcal mol−1 (Fig. 1C, Fig. S22 and Table S3, ESI†).
Collectively, these results confirm the reversible aggregation behavior of DBI-POE and highlight its potential for further sensing applications through modulation of its photophysical properties.
G4-mediated molecular disassembly and enhanced optical outputs
Encouraged by the activatable assembly–disassembly behavior of DBI-POE, we investigated its molecular recognition capacity toward G4 structures under physiologically relevant ionic conditions (100 mM K+). In aqueous solution, DBI-POE exhibited its characteristic broad absorption band and low-intensity emission, with a lowered fluorescence quantum yield (ΦF) of 4% (∼10% in organic solvents),67 which is indicative of dye self-assembly (Fig. 2A, Table 1 and Table S1, ESI†). Upon incremental addition of the c-MYC Pu22 sequence,27,48,51 a well-established intramolecular parallel G4 structure (Fig. S25, ESI†), located in the MYC promoter and commonly used in structural studies to investigate ligand-binding interactions,27,48,74 we observed a gradual increase in the absorption (S1(ππ*) state of DBI-POE:c-MYC Pu22, oscillator strength of 0.41, at 2.89 eV, Table S4, ESI†) accompanied by the emergence of two well-resolved isosbestic points (Fig. 2A). These spectral transitions signified the formation of a well-defined DBI-POE:c-MYC Pu22 complex that mirrored the optical behavior of DBI-POE observed in organic solvents (Fig. S4 and S5, ESI†), in the presence of SDS (Fig. 1A), or at elevated temperatures (Fig. S7, ESI†), thereby corroborating G4-mediated disaggregation.25–29 These results are consistent with computational docking simulations, where the stacked dimer disaggregates upon G4 intercalation of one of the monomers, while the other monomer binds externally (see ESI† for details and Fig. S27). Concurrent fluorescence measurements revealed a progressive “turn-on” of emission, reaching a ΦF of 15% at 5.0 equiv., which confirmed the conversion of weakly emissive nanoaggregates into emissive monomeric species (Fig. 2A and Table S1, ESI†). Indeed, the QM results for DBI-POE within the c-MYC Pu22 structure revealed the existence of just one emissive minimum, similar to that of the monomeric DBI-POE species (Table S5, ESI†).| System | Topologya | K a /M−1 | λ em/nm | Φ F /% | 〈τ〉/ns | k r/106 s−1 | k nr/106 s−1 | σ 2 /GM | Φ Δ/% |
|---|---|---|---|---|---|---|---|---|---|
a Topological structure, P = parallel, H = hybrid, AP = antiparallel, and NG4 = non-G4 structure.
b Data fitting to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model on the fluorimetric titration data was performed using Bindfit, which employed multiple global fitting methods (Nelder–Mead method).
c Not determined (ND) due to the absence of a saturation binding profile.
d Fluorescein is used as the reference standard (0.1 M NaOH, ΦF = 0.93).
e
Φ
F was calculated in 50 mM Tris buffer (pH 6.8) containing 100 mM KCl at 25 °C. All values are reported for a 1 1 binding model on the fluorimetric titration data was performed using Bindfit, which employed multiple global fitting methods (Nelder–Mead method).
c Not determined (ND) due to the absence of a saturation binding profile.
d Fluorescein is used as the reference standard (0.1 M NaOH, ΦF = 0.93).
e
Φ
F was calculated in 50 mM Tris buffer (pH 6.8) containing 100 mM KCl at 25 °C. All values are reported for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DBI-POE/oligonucleotide ratio. See Table S1 (ESI) for additional data.
f Fluorescein is used as the reference standard (0.1 1 DBI-POE/oligonucleotide ratio. See Table S1 (ESI) for additional data.
f Fluorescein is used as the reference standard (0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M NaOH).
g Determined by direct 1O2 phosphorescence method in EtOH or MeOH using Rose Bengal in MeOH as the reference standard (ΦΔ(RB) = 0.76).
h Determined by ABDA bleaching experiments using Methylene Blue in water as the reference standard (ΦΔ(MB) = 0.52). Experiments are reported for a 1 M NaOH).
g Determined by direct 1O2 phosphorescence method in EtOH or MeOH using Rose Bengal in MeOH as the reference standard (ΦΔ(RB) = 0.76).
h Determined by ABDA bleaching experiments using Methylene Blue in water as the reference standard (ΦΔ(MB) = 0.52). Experiments are reported for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DBI-POE/c-MYC Pu22 ratio in 50 mM Tris buffer (pH 6.8) containing 100 mM KCl at 25 °C. 1 DBI-POE/c-MYC Pu22 ratio in 50 mM Tris buffer (pH 6.8) containing 100 mM KCl at 25 °C.
|
|||||||||
| DBI-POE | — | — | 565 | 4e | 3.4 | 13.0 | 281.5 | 15.8 | 84g/72g |
| c-MYC Pu22 | P | 2.7 × 105 | 566 | 11e | 5.2 | 20.2 | 170.8 | 69.5 | 22h |
| HIF-1α | P | 2.7 × 105 | 568 | 9e | 4.3 | 24.4 | 206.6 | — | — |
| VEGF | P | 1.8 × 105 | 567 | — | — | — | — | — | — |
| Tel-23 | H | 4.6 × 104 | 565 | — | — | — | — | — | — |
| TBA | AP | NDc | 566 | 6e | 3.5 | 16.6 | 268.0 | 30.6 | — |
| ss-DNA | NG4 | NDc | 566 | 5e | 3.3 | 15.4 | 287.0 | ND | — |
| ds-DNA | NG4 | NDc | 566 | 6e | 3.6 | 15.8 | 263.6 | 28.5 | — |
Quantitative analysis using global nonlinear fitting of the emission data supported a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model, yielding an association constant (Ka) of 2.7 × 105 M−1 (Fig. 2B).75 This value aligns with those reported for other G4 fluorescent probes,29,31,52 including our own works on related DBI derivatives.67,71 Job's plot analysis further confirmed the stoichiometry by showing saturation at a mole fraction of approximately 0.5 (Fig. S8, ESI†). This stoichiometry is preferred over higher-order complexes, which can lead to multiple ligands binding to the G4 template and uncontrolled photodamage.48 To probe the specificity of this interaction, we expanded our studies to include a diverse panel of previously characterized G4 structures, including parallel (c-MYC Pu22; vascular endothelial growth factor, VEGF; hypoxia-inducible factor 1-alpha, HIF-1α), hybrid (Telomeric, Tel-23), and antiparallel (thrombin-binding aptamer, TBA) topologies, as well as non-G4 controls such as single-stranded (ss) and double-stranded (ds) DNA (Table S2, ESI†).26,67 These G4 DNA structures were selected for their biological relevance and well-characterized folding topologies, representing the broad structural diversity that G4s can adopt.48,76–79 Strikingly, only parallel G4 structures, and to a lesser extent, the hybrid conformation, induced significant disassembly of DBI-POE aggregates, whereas antiparallel G4, ssDNA, and dsDNA produced minimal or no spectral changes (Fig. 2B, Fig. S9, S10 (ESI†) and Table 1). The absorption spectral changes observed during the titration of DBI-POE with either antiparallel G4 or non-G4 sequences likely result from a reorganization of DBI-POE's supramolecular state, during which the oligonucleotides act as a crowder agent rather than engaging in the specific interactions that lead to complex formation (Fig. S9, ESI†).26,29,30 This selectivity has also been observed for other G4-selective probes operating via DIE and likely arises from the accessible π-stacking surfaces of parallel G4s, which lack lateral or diagonal loops.25,26,29,31,32 This compact architecture facilitates strong interactions with the aromatic core of DBI-POE. Complementary steady-state and time-dependent competition assays using the well-established G4 ligand PhenDC380–83 further confirmed the specific binding of DBI-POE to c-MYC Pu22 (Fig. 2C, Fig. S11 and Scheme S1, ESI†). In these assays, PhenDC3's high affinity for the c-MYC Pu22 G4 directly competes with DBI-POE for the same binding site. As a result, DBI-POE is displaced, leading to a significant drop in its emission intensity. This observation reinforces the notion that DBI-POE interacts specifically with the G4 structure and that its binding is reversible and competitive, as evidenced by its displacement by PhenDC3.
1 binding model, yielding an association constant (Ka) of 2.7 × 105 M−1 (Fig. 2B).75 This value aligns with those reported for other G4 fluorescent probes,29,31,52 including our own works on related DBI derivatives.67,71 Job's plot analysis further confirmed the stoichiometry by showing saturation at a mole fraction of approximately 0.5 (Fig. S8, ESI†). This stoichiometry is preferred over higher-order complexes, which can lead to multiple ligands binding to the G4 template and uncontrolled photodamage.48 To probe the specificity of this interaction, we expanded our studies to include a diverse panel of previously characterized G4 structures, including parallel (c-MYC Pu22; vascular endothelial growth factor, VEGF; hypoxia-inducible factor 1-alpha, HIF-1α), hybrid (Telomeric, Tel-23), and antiparallel (thrombin-binding aptamer, TBA) topologies, as well as non-G4 controls such as single-stranded (ss) and double-stranded (ds) DNA (Table S2, ESI†).26,67 These G4 DNA structures were selected for their biological relevance and well-characterized folding topologies, representing the broad structural diversity that G4s can adopt.48,76–79 Strikingly, only parallel G4 structures, and to a lesser extent, the hybrid conformation, induced significant disassembly of DBI-POE aggregates, whereas antiparallel G4, ssDNA, and dsDNA produced minimal or no spectral changes (Fig. 2B, Fig. S9, S10 (ESI†) and Table 1). The absorption spectral changes observed during the titration of DBI-POE with either antiparallel G4 or non-G4 sequences likely result from a reorganization of DBI-POE's supramolecular state, during which the oligonucleotides act as a crowder agent rather than engaging in the specific interactions that lead to complex formation (Fig. S9, ESI†).26,29,30 This selectivity has also been observed for other G4-selective probes operating via DIE and likely arises from the accessible π-stacking surfaces of parallel G4s, which lack lateral or diagonal loops.25,26,29,31,32 This compact architecture facilitates strong interactions with the aromatic core of DBI-POE. Complementary steady-state and time-dependent competition assays using the well-established G4 ligand PhenDC380–83 further confirmed the specific binding of DBI-POE to c-MYC Pu22 (Fig. 2C, Fig. S11 and Scheme S1, ESI†). In these assays, PhenDC3's high affinity for the c-MYC Pu22 G4 directly competes with DBI-POE for the same binding site. As a result, DBI-POE is displaced, leading to a significant drop in its emission intensity. This observation reinforces the notion that DBI-POE interacts specifically with the G4 structure and that its binding is reversible and competitive, as evidenced by its displacement by PhenDC3.
Time-correlated single-photon counting (TCSPC) measurements provided additional mechanistic insights. They revealed that parallel G4 structures (c-MYC Pu22 or HIF-1α) significantly extended DBI-POE's fluorescence lifetime by reducing non-radiative decay (knr) and enhancing radiative decay (kr) (Fig. S12 and Table 1, Table S1, ESI†). In contrast, antiparallel G4 and non-G4 sequences had minimal effects, indicating that parallel G4 binding effectively suppresses non-radiative pathways and boosts both emission intensity and excited-state lifetime.
Two-photon excited fluorescence (TPEF) studies demonstrated that while unbound DBI-POE exhibited a modest two-photon absorption cross section (σ2 = 15.8 GM at 760 nm), its complexation with c-MYC Pu22 increased this value by a factor of approximately 4.5, thereby enabling effective photosensitization of the dye in the biologically favorable first NIR window (Fig. 2D and Table 1).28 In line with our above mentioned disassembly experiments, this enhancement was much less pronounced (∼two-fold) when DBI-POE was incubated with the antiparallel G4 TBA or ds-DNA (Fig. S13 (ESI†) and Table 1). NIR light (700–1000 nm) offers deeper tissue penetration, reduced scattering and absorption, and minimal photodamage, while its confined excitation minimizes off-target effects, potentially enhancing tumor targeting when DBI-POE binds to c-MYC G4.84 Although promising, two-photon G4-interactive compounds are still in their infancy.28,85,86 Our data indicate that DBI-POE is a promising scaffold for two-photon PDT, as G4-triggered disaggregation significantly enhances its emissive properties and σ2, thereby increasing its two-photon brightness (ΦF × σ2)28,87 and enabling advanced imaging and phototherapeutic strategies targeting oncogene promoter regions.
G4-driven type I-to-type II photophysical conversion
The generation of reactive triplet states is an essential prerequisite for initiating either type I or type II photochemical mechanisms. We have then estimated the intersystem crossing (ISC) probability for DBI-POE, DBI-POE2, and DBI-POE:c-MYC Pu22 complex by computing spin–orbit coupling (SOC) terms at the position of the minimum-energy geometry in the S1 electronic state (S1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min). The DBI-POE:c-MYC Pu22 complex exhibits larger SOCs for the closest triplet in energy from S1 (Table S6, ESI†), being almost double that of DBI-POE. Having established that the triplets can be potentially populated, we systematically evaluated the ROS-generating potential of DBI-POE by NIR 1O2 phosphorescence measurements in MeOH and ethanol (EtOH), solvents in which DBI-POE is highly soluble (Fig. S14, ESI†).67,88 In EtOH, DBI-POE exhibited a high singlet oxygen quantum yield (ΦΔ = 84%), consistent with the almost unitary values found for its lipophilic counterpart S-DBI in chloroform.67 In comparison, a decreased yield is found in MeOH (ΦΔ = 72%). It should be noted that direct detection of 1O2via its phosphorescence band is inherently limited by low sensitivity, particularly in aqueous media. Consequently, DBI-POE alone or in its complex with c-MYC Pu22 in aqueous solution did not yield a sufficiently strong signal, making direct 1O2 detection impractical and necessitating the use of alternative assays.
min). The DBI-POE:c-MYC Pu22 complex exhibits larger SOCs for the closest triplet in energy from S1 (Table S6, ESI†), being almost double that of DBI-POE. Having established that the triplets can be potentially populated, we systematically evaluated the ROS-generating potential of DBI-POE by NIR 1O2 phosphorescence measurements in MeOH and ethanol (EtOH), solvents in which DBI-POE is highly soluble (Fig. S14, ESI†).67,88 In EtOH, DBI-POE exhibited a high singlet oxygen quantum yield (ΦΔ = 84%), consistent with the almost unitary values found for its lipophilic counterpart S-DBI in chloroform.67 In comparison, a decreased yield is found in MeOH (ΦΔ = 72%). It should be noted that direct detection of 1O2via its phosphorescence band is inherently limited by low sensitivity, particularly in aqueous media. Consequently, DBI-POE alone or in its complex with c-MYC Pu22 in aqueous solution did not yield a sufficiently strong signal, making direct 1O2 detection impractical and necessitating the use of alternative assays.
To overcome this limitation, we employed 9,10-anthracenediyl-bis(methylene)dimalonic acid (ABDA) as a specific water-soluble 1O2 sensor.89 ABDA can be readily oxidized to an endoperoxide by 1O2, resulting in a decrease in its absorption that can be easily tracked by UV/vis spectroscopy.89 Under our irradiation protocol, ABDA alone or in the presence of either S-DBI or the S-DBI:c-MYC Pu22 complex exhibited negligible spectral changes (Fig. S15, ESI†). In contrast, exposure to DBI-POE led to progressive ABDA bleaching, indicating effective photoinduced ROS production (Fig. 3A). Notably, the addition of the c-MYC Pu22 G4 sequence resulted in a comparable ABDA bleaching profile, demonstrating that DBI-POE retains good ROS-generating capability even when bound to G4 structures (Fig. 3A). This effect was found to be independent of the c-MYC Pu22 concentration (Fig. S16, ESI†).
Under oxygen-depleted conditions, neither free DBI-POE nor its c-MYC Pu22 complex induced significant ABDA bleaching, confirming the oxygen-dependent nature of the process, and its dependency on ROS generation (Fig. S17, ESI†).
To determine the chemical nature of the generated ROS, we introduced NaN3, a well-established 1O2 quencher.89 To our surprise, NaN3 completely suppressed ABDA bleaching only in samples containing the DBI-POE:c-MYC Pu22 complex, whereas it had no effect on free DBI-POE, even at elevated concentrations, pointing to a predominant type I pathway (likely involving O2˙− or OH˙ species) for the free dye and a type II mechanism (selective for 1O2) upon G4 binding (Fig. 3B and Fig. S18, ESI†). Based on this premise, we calculated a ΦΔ of 22% from ABDA bleaching for the DBI-POE:c-MYC Pu22 complex, a significant value for phototherapeutic applications.68 Substituting ds-DNA for c-MYC Pu22 again led to NaN3-insensitive bleaching, as observed for free DBI-POE, highlighting the critical role of the G4 conformation in eliciting 1O2 production (Fig. S19, ESI†).
To further investigate the generation of 1O2 by DBI-POE when bound to c-MYC Pu22, we conducted ABDA bleaching experiments in a buffered solution containing 85% deuterium oxide (D2O). Since, opposite to NaN3, D2O extends the lifetime of 1O2, we anticipate that it should amplify ABDA bleaching and thus provide direct evidence of 1O2 involvement.89 Indeed, in D2O, the DBI-POE:c-MYC Pu22 complex exhibited a two-fold increase in ABDA bleaching rate, which was completely suppressed by NaN3, whereas free DBI-POE remained robustly insensitive to both D2O and NaN3 (Fig. 3A and B). Together, these observations indicate that complexation with c-MYC Pu22 shifts DBI-POE from a type I to a type II photochemical pathway, which constitutes to the best of our knowledge an unprecedented observation.
To explore the reversibility of G4 binding and its direct influence on ROS generation, we conducted competitive displacement assays using PhenDC3 (vide supra). Under strongly competitive conditions (PhenDC3 = 25 μM), DBI-POE was displaced from the c-MYC Pu22 template, restoring its self-assembly and NaN3-resistant photosensitization properties, as seen from ABDA bleaching profile (Fig. S20A, ESI†). At lower PhenDC3 concentrations (1 μM), the complex remained largely intact, and NaN3-sensitive bleaching persisted (Fig. S20B, ESI†).
Additionally, we performed scavenger studies using mannitol,90 sodium pyruvate,91 and the superoxide dismutase mimetic manganese(III)tetrakis(4-benzoic acid)porphyrin (MnTBAP),92 which are selective for OH˙ radicals,90 hydrogen peroxide (H2O2),91 and O2˙−,92 respectively. Among the tested scavengers, MnTBAP stood out for its ability to significantly reduce ABDA bleaching induced by free DBI-POE under conditions where NaN3 had no effect, confirming the involvement of O2˙− in its photochemical pathway (Fig. S21, ESI†).
Complementary experiments using Singlet Oxygen Sensor Green (SOSG),93 as an additional 1O2 control probe, revealed that both free DBI-POE and its c-MYC Pu22 complex activated SOSG fluorescence, albeit to different extents (Fig. 3C). Notably, only the fluorescence enhancement in the G4-bound state was suppressed by NaN3, consistent with exclusive 1O2 production (Fig. 3C). Moreover, in agreement with the ABDA studies, MnTBAP significantly reduced the SOSG emission enhancement induced by DBI-POE, supporting the generation of O2˙− radicals (Fig. 3D).
It has been well documented that ABDA and SOSG exhibit high specificity for 1O2 compared to other ROS, with minimal off-target interactions reported.89 However, our findings underscore the importance of employing an extensive array of control experiments when utilizing ROS probes. Such rigorous cross-control is essential to accurately assess the specificity of the generated ROS and to discard false-positive results.
Next, we hypothesized that free DBI-POE and its c-MYC Pu22-bound complex would exhibit contrasting photophysical responses when probed with O2˙−-specific sensors. To test this, we employed dihydrorhodamine-123 (DHR-123), a non-fluorescent compound that is oxidized to the highly fluorescent rhodamine-123 upon exposure to O2˙− radicals.11,94 The DHR-123 assays revealed that free DBI-POE induced a strong fluorescence response, indicative of robust O2˙− generation (Fig. 3E). In contrast, the DBI-POE:c-MYC Pu22 complex elicited minimal DHR-123 activation, suggesting that G4 binding suppresses O2˙− production (Fig. 3E). Control experiments with ds-DNA instead of c-MYC Pu22 produced DHR-123 activation levels comparable to those of free DBI-POE (Fig. 3E). Furthermore, the addition of increasing concentrations of MnTBAP resulted in a dose-dependent decrease in DHR-123 fluorescence, thereby confirming that free DBI-POE generates O2˙− radicals (Fig. 3E).
Further evidence was obtained using the OH˙-selective fluorescent probe hydroxyphenyl fluorescein (HPF), which specifically emits fluorescence upon reaction with OH˙ radicals.11 Free DBI-POE strongly induced HPF fluorescence, indicating efficient OH˙ generation through a type I photochemical pathway. In contrast, the DBI-POE:c-MYC Pu22 complex exhibited minimal HPF fluorescence enhancement, highlighting the shift toward a predominantly 1O2-mediated photochemical mechanism (Fig. 3F and G).
Collectively, these findings establish DBI-POE as the first example of a supramolecular, G4-specific PS capable of toggling between two distinct photophysical pathways, type I and type II, depending on its aggregation state and G4 binding. This G4-induced disassembly not only enables robust 1O2 production, which preferentially oxidizes guanine residues, but also significantly enhances the prospects for targeted PDT that exploit the unique structural and electronic features of G4 motifs.
Light-mediated G4 unfolding and site-specific oxidative guanine damage
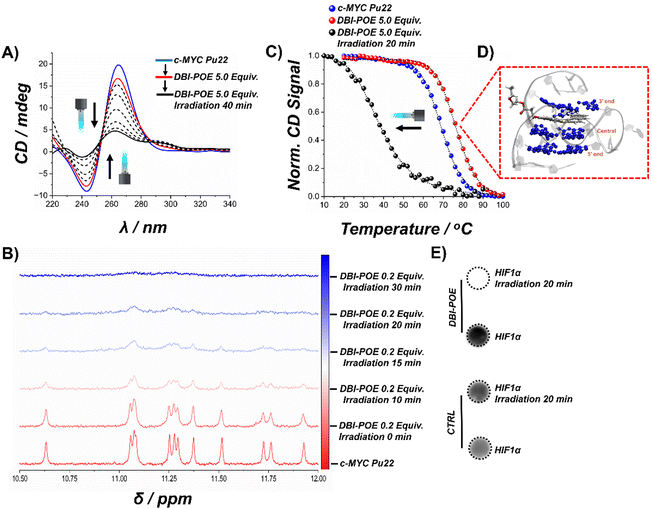
To elucidate the effect of photogenerated 1O2 by DBI-POE on the G4 scaffold, we employed a multi-technique approach that integrates spectroscopic, biochemical, and computational methods.Initially, CD spectroscopy was used to monitor structural changes in the c-MYC Pu22 sequence. In the absence of DBI-POE, the G4 exhibited the expected CD spectral features characterized by a positive peak at ∼260 nm and a negative peak at ∼240 nm (Fig. 4A).48,52,95 When DBI-POE was added in the dark, only minimal spectral alterations were observed, akin to those induced by other uncharged G4-binding compounds.26–28 However, exposure to light produced a dose-dependent decrease in CD band intensity, ultimately yielding a spectrum suggestive of an unfolded G4 (Fig. 4A).96 These results suggest that photoactivation of DBI-POE leads to guanine oxidation and subsequent G4 disruption.97
Complementary 1H NMR spectroscopy further corroborated these findings. The free c-MYC Pu22 oligonucleotide displayed the expected 12 imino proton resonances associated with G4 formation (Fig. 4B).48,51 While dark-incubated DBI-POE caused only slight spectral changes, with a moderate broadening of the peaks, light irradiation resulted in a progressive disappearance of these imino signals, confirming a light-dependent unfolding of the G4 structure (Fig. 4B).
Thermal stability assessments via temperature-dependent CD melting assays provided additional insight on the photodegradation mechanism (Fig. 4C). Under control conditions, c-MYC Pu22 demonstrated high thermal stability (Tm(c-MYC Pu22) = 69 °C). Notably, DBI-POE in the dark increased the melting temperature by ∼8 °C (Tm(DBI-POE:c-MYC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) Pu22) = 77 °C), reflecting the stabilizing influence of S-DBI.67 In stark contrast, light activation of DBI-POE reduced the melting temperature by ∼40 °C (Tm(DBI-POE:c-MYC
Pu22) = 77 °C), reflecting the stabilizing influence of S-DBI.67 In stark contrast, light activation of DBI-POE reduced the melting temperature by ∼40 °C (Tm(DBI-POE:c-MYC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pu22
Pu22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) +
+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Light) = 37 °C) decisively indicating that photooxidative damage, most likely mediated by guanine oxidation, disrupts the tetrad integrity essential for G4 stability.
Light) = 37 °C) decisively indicating that photooxidative damage, most likely mediated by guanine oxidation, disrupts the tetrad integrity essential for G4 stability.
Indeed, docking followed by MD simulations also support these findings, as DBI-POE stabilizes the G4 structure in its most favorable pose in the dark (Fig. 4D and S28, see ESI† for details), while the oxidized G4 becomes disrupted upon light exposure (Fig. S29, ESI†).
Control experiments, where c-MYC Pu22 was exposed to light irradiation in the absence of DBI-POE, resulted in no spectral alterations.
In line with our observations, previous studies have demonstrated that oxidative lesions, such as 8-oxo-7,8-dihydroguanine (8-oxoG), generally impair the hydrogen-bonding capacity of guanine, often leading to a loss of G4 structure.98 However, notable exceptions exist.99 For example, the Burrows group reported that an additional guanine track in oncogene promoter G4 sequences can serve as a “spare tire,” helping to preserve the G4 fold under oxidative stress.100 Similarly, the Opresko lab showed that telomeric G4 sequences containing a single 8-oxoG mutation retained their hybrid conformation, although the binding affinity of the G4-specific antibody BG433 was reduced depending on the lesion's precise location.101
Therefore, to further validate the impact of oxidative damage mediated by DBI-POE, we performed immuno dot blot assays using BG4 (Fig. 4E). In our hands, the c-MYC Pu22 sequence was poorly recognized by BG4 resulting in a weak signal. Therefore, we employed the HIF-1α G4 sequence, which is effectively targeted by both BG4 and DBI-POE (Fig. 2B).67 Control samples maintained BG4 targeting capability, as evidenced by a robust immunosignal in the dot blot assay, whereas light-activated DBI-POE led to significant disruption of the G4 structure, accompanied by a marked decrease in BG4 binding, a clear hallmark of oxidative damage.
Our experiments clearly demonstrate that DBI-POE's ability to generate 1O2 efficiently damages the G4 structure, resulting in its complete destabilization and unfolding.
To reconstitute in vitro DNA replication across a G4-containing template, we used a primer extension assay that faithfully mimics the natural progression of DNA synthesis.50,102,103 Our assay employs DNA templates, either containing or lacking the G4 sequence, annealed to a fluorescently labeled primer, along with a DNA polymerase, dNTPs, and essential cofactors like Mg2+ (Fig. 5A and D). As synthesis proceeds, the extension of the labeled primer is observed; when the polymerase encounters a blockage, it pauses and ultimately dissociates, resulting in truncated products compared to full-length extensions.50,102,103 These products are then separated via denaturing polyacrylamide gel electrophoresis (PAGE) and visualized.
In the absence of DBI-POE, the polymerase efficiently synthesized full-length products without interruption (Fig. 5B and C). However, when DBI-POE was added in the dark, a prominent pause site emerged at T20, immediately upstream of the first guanine tract, consistent with the structural stabilization observed via CD spectroscopy (Fig. 4C) and previously reported for other G4 ligands (Fig. 5B and C).27,50,102 Under light irradiation, additional pausing occurred at positions T11 and G10, and after 20 minutes, polymerase processivity was almost completely halted (Fig. 5B and C). We speculate that DBI-POE localizes near G9 and G10 within the c-MYC G4 structure, thereby triggering its oxidation.
To further explore the binding site of DBI-POE in c-MYC Pu22, we conducted molecular docking and MD simulations and investigated the intercalation of DBI-POE by opening the 3′-end and central tetrads, where G10 and G9 are located, through a combination of MD and umbrella sampling techniques (see ESI† for details). After equilibrating the final opened G4 structure, docking calculations identified the most populated intercalated poses. As illustrated in Fig. S26 (ESI†), we distinguish two main binding sites depending on the moiety of DBI-POE that stacks between the tetrads: the hydrophobic core (pose A) or the hydrophilic arm (pose B). The MD simulations (Fig. S28, ESI†) showed that pose B is not stable, as it disrupted the tetrad integrity in the G4 (Fig. S28B, ESI†), whereas pose A remained stable (Fig. 4D and Fig. S28A, ESI†). This indicates that pose A is the most probable DBI-POE binding site as, in accordance with experimental studies, it stabilizes the G4 in the dark.
The overall reduction in TET label fluorescence is attributable to light-induced bleaching. To test our hypothesis that these lesions resulted from guanine oxidation, we designed a c-MYC Pu22 template with site-specific 8-oxoG substitutions at positions G10 and G9, which are predicted to be the primary oxidation targets (Fig. 5A). This modified template produced a pausing pattern nearly identical to that observed with light-activated DBI-POE, although with a more intense signal due to the absence of light-mediated TET bleaching, ultimately leading to complete polymerase arrest at the corresponding sites (Fig. 5B). This conclusively demonstrates that DBI-POE induces site-specific oxidative damage via the formation of 8-oxoG.
As control, we used a DNA template lacking any G4 motif (Fig. 5D).102 Under both dark and light conditions, no polymerase pausing was observed (Fig. 5E and F). Similarly to the G4 reaction, light-mediated bleaching of TET label caused an overall decrease of the signal (Fig. 5E and F). These results indicate that DBI-POE's 1O2-induced damage is selective for G4 structures and does not cause nonspecific DNA damage when DBI-POE remains in its unbound state.
We further evaluated the structural impact of 8-oxoG incorporation on the c-MYC Pu22 G4 scaffold using MD simulations. While formation of a single 8-oxoG at G10 preserved the overall G4 conformation, simultaneous substitution at G10 and G9 resulted in significant structural disruption (Fig. 6). These computational insights suggest that multiple oxidative lesions are required to compromise G4 stability and underscore the importance of precise PS positioning for effective phototherapeutic activity.
Collectively, these integrated studies reveal a novel, light-mediated mechanism in which DBI-POE precisely oxidizes and disrupts G4 structures. This strategy holds significant promise for targeted PDT, as G4-targeted PSs can downregulate cancer-associated genes by destabilizing their regulatory G4 elements.62
Conclusions
G4 structures are significantly overrepresented in cancer cells, making them appealing targets for precision therapy.33,104,105 Their enrichment in the promoter regions of difficult-to-treat oncogenes,46 such as MYC and HIF, suggests that small molecule ligands that stabilize G4s can effectively hinder DNA and/or RNA polymerase progression, thereby triggering replication stress, DNA damage, and ultimately cell death.48,74,77,78Despite significant progress in developing light-activated compounds that modulate G4 activity with high spatiotemporal control, several challenges persist.106–108 In particular, the limited bidirectional modulation of molecular photochromes calls for more versatile photoregulation strategies.51,108–110 Moreover, while the susceptibility of G4s to oxidation presents an opportunity, it also poses a challenge, especially since current G4-targeting PSs predominantly rely on highly reactive and poorly selective type I mechanisms that have not adequately addressed this vulnerability.59–63 To overcome these limitations, we introduce DBI-POE, a novel PS that transitions from a type I to a type II mechanism upon binding to G4 structures, thereby combining both structure-specific and site-specific oxidative damage.
Our comprehensive studies revealed that DBI-POE binds selectively to parallel and hybrid G4 conformations, triggering supramolecular disassembly that “turns on” its fluorescence and significantly enhances two-photon absorption in the NIR region. Photophysical analyses showed that while free DBI-POE predominantly generates ROS via a type I pathway (producing O2˙− and OH˙ species), its complexation with G4s reprograms its photochemistry toward a highly selective type II mechanism that yields 1O2. This switching mechanism was rigorously supported by multiple discriminant assays. In-depth biophysical studies confirm that light-activated DBI-POE induces selective oxidative damage and unfolding of the G4 scaffold.
Further validation came from primer extension assays, which demonstrated site-specific stalling at oxidatively modified guanine residues, as well as from MD simulations that highlighted structural destabilization associated with 8-oxoG formation. Importantly, the oxidative damage induced by DBI-POE was confined to the DNA template containing the G4 structure, leaving the non-G4 template unaffected, underscoring its exceptional selectivity.
Collectively, these findings provide unprecedented mechanistic insights into a G4-targeted PS and highlight the potential of DBI-POE as a platform for developing next-generation agents for precision photodynamic anticancer therapy.
Ongoing studies are actively investigating its photoactivation in cells, both as a standalone approach and in combination with delivery systems such as G4 aptamers. These strategies are also being evaluated alongside DNA-damaging agents and/or DNA damage response (DDR) inhibitors, with the aim of further enhancing the overall phototherapeutic efficacy.
Author contributions
Conceptualization (M. Deiana and C. Cabanetos), synthesis and characterization of the compound (D. Puchán Sánchez, A. Kassem), photophysical, biophysical and biochemical studies (K. Saczuk, M. V. Cottini, M. Dudek, L. M. Mazur, C. Monnereau, J. Jamroskovic, M. Deiana), supervision (J. J. Nogueira, L. Martínez-Fernández, J. Jamroskovic, C. Cabanetos, M. Deiana), computational data (L. López-Pacios, J. J. Nogueira, L. Martínez-Fernández), writing original draft (M. Deiana with inputs from all authors), funding acquisition (J. J. Nogueira, L. Martínez-Fernández, J. Jamroskovic, C. Cabanetos, M. Deiana), project administration (M. Deiana), revision of the manuscript (M. Deiana and K. Saczuk). All authors approved the final version of the manuscript.Data availability
All the data that support this study are included in this article and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
M. Deiana would like to acknowledge financial support from project no. 2022/47/P/NZ5/01156, which is co-funded by the National Science Centre and the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 945339. Funding for J. Jamroskovic was provided by the IMPULZ program of the Slovak Academy of Sciences under the Agreement on the Provision of Funds No. IM-2022-62. L. López-Pacios acknowledges the FPU22/02196 grant from the Spanish Ministry of Science, Innovation and Universities (MICINN). L. Martínez-Fernández acknowledges the grant PID2023-151719NA-I00 funded by MICIU/AEI/10.13039/501100011033 and FEDER, UE. J. Nogueira thanks the Spanish Ministry of Science and Innovation for funding support through the project CNS2022-135720 (MCIN/AEI/10.13039/501100011033). This research project was made possible through the access granted by the Galician Supercomputing Center (CESGA) to its supercomputing infrastructure. The authors thank Dr Aeson Chang at Monash University (Australia) for carefully reading the manuscript and providing insightful comments. We acknowledge Protein Production Sweden (PPS) for providing facilities and experimental support. PPS is funded by the Swedish Research Council as a national research infrastructure. We thank the members of Dr. Barak’s laboratory (Slovak Academy of Sciences) for their help with the initial biochemical experiments. We also thank Dr. Barancik (Slovak Academy of Sciences) for providing access to the Typhoon scanner.Notes and references
- P. Kobauri, F. J. Dekker, W. Szymanski and B. L. Feringa, Angew. Chem., Int. Ed., 2023, 62, e202300681 CrossRef CAS PubMed.
- X. Li, J. F. Lovell, J. Yoon and X. Chen, Nat. Rev. Clin. Oncol., 2020, 17, 657–674 CrossRef PubMed.
- B. M. Vickerman, E. M. Zywot, T. K. Tarrant and D. S. Lawrence, Nat. Rev. Chem., 2021, 5, 816–834 CrossRef PubMed.
- W. A. Velema, W. Szymanski and B. L. Feringa, J. Am. Chem. Soc., 2014, 136, 2178–2191 CrossRef CAS PubMed.
- T. C. Pham, V.-N. Nguyen, Y. Choi, S. Lee and J. Yoon, Chem. Rev., 2021, 121, 13454–13619 CrossRef CAS PubMed.
- X. Zhao, J. Liu, J. Fan, H. Chao and X. Peng, Chem. Soc. Rev., 2021, 50, 4185–4219 RSC.
- V.-N. Nguyen, Y. Yan, J. Zhao and J. Yoon, Acc. Chem. Res., 2021, 54, 207–220 CrossRef CAS PubMed.
- B. Lu, L. Wang, H. Tang and D. Cao, J. Mater. Chem. B, 2023, 11, 4600–4618 RSC.
- G. Xu, C. Li, C. Chi, L. Wu, Y. Sun, J. Zhao, X.-H. Xia and S. Gou, Nat. Commun., 2022, 13, 3064 CrossRef CAS PubMed.
- K.-X. Teng, L.-Y. Niu, N. Xie and Q.-Z. Yang, Nat. Commun., 2022, 13, 6179 CrossRef CAS PubMed.
- Y. Tang, Y. Li, B. Li, W. Song, G. Qi, J. Tian, W. Huang, Q. Fan and B. Liu, Nat. Commun., 2024, 15, 2530 CrossRef CAS PubMed.
- Z. Li, Z. Zhang, L. Ma, H. Wen, M. Kang, D. Li, W. Zhang, S. Luo, W. Wang, M. Zhang, D. Wang, H. Li, X. Li and H. Wang, Angew. Chem., Int. Ed., 2024, 63, e202400049 CrossRef CAS PubMed.
- K.-X. Teng, L.-Y. Niu and Q.-Z. Yang, J. Am. Chem. Soc., 2023, 145, 4081–4087 CrossRef CAS PubMed.
- J. Zhuang, B. Wang, H. Chen, K. Zhang, N. Li, N. Zhao and B. Z. Tang, ACS Nano, 2023, 17, 9110–9125 CrossRef CAS PubMed.
- Y. Wang, S. Xu, L. Shi, C. Teh, G. Qi and B. Liu, Angew. Chem., Int. Ed., 2021, 60, 14945–14953 CrossRef CAS PubMed.
- M. Wu, X. Liu, H. Chen, Y. Duan, J. Liu, Y. Pan and B. Liu, Angew. Chem., Int. Ed., 2021, 60, 9093–9098 CrossRef CAS PubMed.
- H. Wang, Q. Li, P. Alam, H. Bai, V. Bhalla, M. R. Bryce, M. Cao, C. Chen, S. Chen, X. Chen, Y. Chen, Z. Chen, D. Dang, D. Ding, S. Ding, Y. Duo, M. Gao, W. He, X. He, X. Hong, Y. Hong, J.-J. Hu, R. Hu, X. Huang, T. D. James, X. Jiang, G.-I. Konishi, R. T. K. Kwok, J. W. Y. Lam, C. Li, H. Li, K. Li, N. Li, W.-J. Li, Y. Li, X.-J. Liang, Y. Liang, B. Liu, G. Liu, X. Liu, X. Lou, X.-Y. Lou, L. Luo, P. R. McGonigal, Z.-W. Mao, G. Niu, T. C. Owyong, A. Pucci, J. Qian, A. Qin, Z. Qiu, A. L. Rogach, B. Situ, K. Tanaka, Y. Tang, B. Wang, D. Wang, J. Wang, W. Wang, W.-X. Wang, W.-J. Wang, X. Wang, Y.-F. Wang, S. Wu, Y. Wu, Y. Xiong, R. Xu, C. Yan, S. Yan, H.-B. Yang, L.-L. Yang, M. Yang, Y.-W. Yang, J. Yoon, S.-Q. Zang, J. Zhang, P. Zhang, T. Zhang, X. Zhang, N. Zhao, Z. Zhao, J. Zheng, L. Zheng, Z. Zheng, M.-Q. Zhu, W.-H. Zhu, H. Zou and B. Z. Tang, ACS Nano, 2023, 17, 14347–14405 CrossRef CAS PubMed.
- N. Kwon, H. Weng, M. A. Rajora and G. Zheng, Angew. Chem., Int. Ed., 2025, e202423348 CAS.
- L. Zhang, F. Chai, H. Dong, Y. Bao, K. Yan, S. Min, Y. Yao, S. Li, Y. Liu, T. Gao, J. Wang and Y. Liu, J. Phys. Chem. Lett., 2024, 15, 10866–10872 CrossRef CAS PubMed.
- G. Liu, H. Wang, C. Shi, G. Chen, Y. Wang, W. Huang and H. Zhao, Chem. Commun., 2022, 58, 10813–10816 RSC.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed.
- F. Bouhedda, K. T. Fam, M. Collot, A. Autour, S. Marzi, A. Klymchenko and M. Ryckelynck, Nat. Chem. Biol., 2020, 16, 69–76 CrossRef CAS PubMed.
- I. O. Aparin, R. Yan, R. Pelletier, A. A. Choi, D. I. Danylchuk, K. Xu and A. S. Klymchenko, J. Am. Chem. Soc., 2022, 144, 18043–18053 CrossRef CAS PubMed.
- M. Collot, P. Ashokkumar, H. Anton, E. Boutant, O. Faklaris, T. Galli, Y. Mély, L. Danglot and A. S. Klymchenko, Cell Chem. Biol., 2019, 26, 600–614.e607 CrossRef CAS PubMed.
- K. Saczuk, M. Dudek, K. Matczyszyn and M. Deiana, Nanoscale Horiz., 2024, 9, 1390–1416 RSC.
- M. Deiana, K. Chand, J. Jamroskovic, I. Obi, E. Chorell and N. Sabouri, Angew. Chem., Int. Ed., 2020, 59, 896–902 CrossRef CAS PubMed.
- M. Deiana, K. Chand, J. Jamroskovic, R. N. Das, I. Obi, E. Chorell and N. Sabouri, Nanoscale, 2020, 12, 12950–12957 RSC.
- V. Grande, C.-A. Shen, M. Deiana, M. Dudek, J. Olesiak-Banska, K. Matczyszyn and F. Würthner, Chem. Sci., 2018, 9, 8375–8381 RSC.
- M. Deiana, K. Chand, E. Chorell and N. Sabouri, J. Phys. Chem. Lett., 2023, 14, 1862–1869 CrossRef CAS PubMed.
- V. Grande, F. Doria, M. Freccero and F. Würthner, Angew. Chem., Int. Ed., 2017, 56, 7520–7524 CrossRef CAS PubMed.
- M. Zuffo, A. Guédin, E.-D. Leriche, F. Doria, V. Pirota, V. Gabelica, J.-L. Mergny and M. Freccero, Nucleic Acids Res., 2018, 46, e115 CrossRef PubMed.
- A. Pandith, Y. Luo, Y. Jang, J. Bae and Y. Kim, Angew. Chem., Int. Ed., 2023, 62, e202215049 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- G. Biffi, M. Di Antonio, D. Tannahill and S. Balasubramanian, Nat. Chem., 2014, 6, 75–80 CrossRef CAS PubMed.
- P. A. Summers, B. W. Lewis, J. Gonzalez-Garcia, R. M. Porreca, A. H. M. Lim, P. Cadinu, N. Martin-Pintado, D. J. Mann, J. B. Edel, J. B. Vannier, M. K. Kuimova and R. Vilar, Nat. Commun., 2021, 12, 162 CrossRef CAS PubMed.
- M. Di Antonio, A. Ponjavic, A. Radzevičius, R. T. Ranasinghe, M. Catalano, X. Zhang, J. Shen, L.-M. Needham, S. F. Lee, D. Klenerman and S. Balasubramanian, Nat. Chem., 2020, 12, 832–837 CrossRef CAS PubMed.
- K. T. McQuaid, A. Pipier, C. J. Cardin and D. Monchaud, Nucleic Acids Res., 2022, 50, 12636–12656 CrossRef CAS PubMed.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005, 33, 2908–2916 CrossRef CAS PubMed.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. Di Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877–881 CrossRef CAS PubMed.
- R. Hänsel-Hertsch, D. Beraldi, S. V. Lensing, G. Marsico, K. Zyner, A. Parry, M. Di Antonio, J. Pike, H. Kimura, M. Narita, D. Tannahill and S. Balasubramanian, Nat. Genet., 2016, 48, 1267–1272 CrossRef PubMed.
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- N. Kosiol, S. Juranek, P. Brossart, A. Heine and K. Paeschke, Mol. Cancer, 2021, 20, 40 CrossRef CAS PubMed.
- M. Sauer and K. Paeschke, Biochem. Soc. Trans., 2017, 45, 1173–1182 CrossRef CAS PubMed.
- Kevin K.-P. Yan, I. Obi and N. Sabouri, Nucleic Acids Res., 2021, 49, 8339–8354 CrossRef CAS PubMed.
- P. Lejault, J. Mitteaux, F. R. Sperti and D. Monchaud, Cell Chem. Biol., 2021, 28, 436–455 CrossRef CAS PubMed.
- S. Neidle, Nat. Rev. Chem., 2017, 1, 0041 CrossRef CAS.
- S. Neidle, J. Med. Chem., 2016, 59, 5987–6011 CrossRef CAS PubMed.
- D. R. Calabrese, X. Chen, E. C. Leon, S. M. Gaikwad, Z. Phyo, W. M. Hewitt, S. Alden, T. A. Hilimire, F. He, A. M. Michalowski, J. K. Simmons, L. B. Saunders, S. Zhang, D. Connors, K. J. Walters, B. A. Mock and J. S. Schneekloth, Nat. Commun., 2018, 9, 4229 CrossRef PubMed.
- H. Xu, M. Di Antonio, S. McKinney, V. Mathew, B. Ho, N. J. O’Neil, N. D. Santos, J. Silvester, V. Wei, J. Garcia, F. Kabeer, D. Lai, P. Soriano, J. Banáth, D. S. Chiu, D. Yap, D. D. Le, F. B. Ye, A. Zhang, K. Thu, J. Soong, S.-C. Lin, A. H. C. Tsai, T. Osako, T. Algara, D. N. Saunders, J. Wong, J. Xian, M. B. Bally, J. D. Brenton, G. W. Brown, S. P. Shah, D. Cescon, T. W. Mak, C. Caldas, P. C. Stirling, P. Hieter, S. Balasubramanian and S. Aparicio, Nat. Commun., 2017, 8, 14432 CrossRef CAS PubMed.
- J. Jamroskovic, M. Doimo, K. Chand, I. Obi, R. Kumar, K. Brännström, M. Hedenström, R. Nath Das, A. Akhunzianov, M. Deiana, K. Kasho, S. Sulis Sato, P. L. Pourbozorgi, J. E. Mason, P. Medini, D. Öhlund, S. Wanrooij, E. Chorell and N. Sabouri, J. Am. Chem. Soc., 2020, 142, 2876–2888 CrossRef CAS PubMed.
- M. Dudek, L. López-Pacios, N. Sabouri, J. J. Nogueira, L. Martinez-Fernandez and M. Deiana, Angew. Chem., Int. Ed., 2025, 64, e202413000 CrossRef CAS PubMed.
- M. Deiana, I. Obi, M. Andreasson, S. Tamilselvi, K. Chand, E. Chorell and N. Sabouri, ACS Chem. Biol., 2021, 16, 1365–1376 CrossRef CAS PubMed.
- A. M. Fleming and C. J. Burrows, J. Am. Chem. Soc., 2020, 142, 1115–1136 CrossRef CAS PubMed.
- A. M. Fleming, B. L. Guerra Castañaza Jenkins, B. A. Buck and C. J. Burrows, J. Am. Chem. Soc., 2024, 146, 11364–11370 CAS.
- A. M. Fleming and C. J. Burrows, NAR Cancer, 2021, 3, zcab038 CrossRef PubMed.
- Y. Ding, A. M. Fleming and C. J. Burrows, J. Am. Chem. Soc., 2017, 139, 2569–2572 CrossRef CAS PubMed.
- P. Di Mascio, G. R. Martinez, S. Miyamoto, G. E. Ronsein, M. H. G. Medeiros and J. Cadet, Chem. Rev., 2019, 119, 2043–2086 CrossRef CAS PubMed.
- J. Cadet, K. J. A. Davies, M. H. G. Medeiros, P. Di Mascio and J. R. Wagner, Free Radicals Biol. Med., 2017, 107, 13–34 CrossRef CAS PubMed.
- W. Chen, Y. Zhang, H.-B. Yi, F. Wang, X. Chu and J.-H. Jiang, Angew. Chem., Int. Ed., 2023, 62, e202300162 CrossRef CAS PubMed.
- X. Zhang, J. Wang and M.-H. Hu, ACS Pharmacol. Transl. Sci., 2024, 7, 2174–2184 CrossRef CAS PubMed.
- X. Zhang and M.-H. Hu, Sens. Actuators, B, 2025, 425, 136952 CrossRef CAS.
- K. Kawauchi, W. Sugimoto, T. Yasui, K. Murata, K. Itoh, K. Takagi, T. Tsuruoka, K. Akamatsu, H. Tateishi-Karimata, N. Sugimoto and D. Miyoshi, Nat. Commun., 2018, 9, 2271 CrossRef PubMed.
- L. Holden, R. C. Curley, G. Avella, C. Long and T. E. Keyes, Angew. Chem., Int. Ed., 2024, 63, e202408581 CrossRef CAS PubMed.
- M. Bartlett, J. Burke, P. Sherin, M. K. Kuimova, M. Barahona and R. Vilar, Chem. – Eur. J., 2024, 30, e202402465 CrossRef CAS PubMed.
- B. Halliwell, A. Adhikary, M. Dingfelder and M. Dizdaroglu, Chem. Soc. Rev., 2021, 50, 8355–8360 RSC.
- M. Deiana, P. Josse, C. Dalinot, A. Osmolovskyi, P. S. Marqués, J. M. A. Castán, L. Abad Galán, M. Allain, L. Khrouz, O. Maury, T. Le Bahers, P. Blanchard, S. Dabos-Seignon, C. Monnereau, N. Sabouri and C. Cabanetos, Commun. Chem., 2022, 5, 142 CrossRef CAS PubMed.
- M. Deiana, J. M. Andrés Castán, P. Josse, A. Kahsay, D. P. Sánchez, K. Morice, N. Gillet, R. Ravindranath, A. K. Patel, P. Sengupta, I. Obi, E. Rodriguez-Marquez, L. Khrouz, E. Dumont, L. Abad Galán, M. Allain, B. Walker, H. S. Ahn, O. Maury, P. Blanchard, T. Le Bahers, D. Öhlund, J. von Hofsten, C. Monnereau, C. Cabanetos and N. Sabouri, Nucleic Acids Res., 2023, 51, 6264–6285 CrossRef CAS PubMed.
- D. P. Sánchez, K. Morice, M. G. Mutovska, L. Khrouz, P. Josse, M. Allain, F. Gohier, P. Blanchard, C. Monnereau, T. Le Bahers, N. Sabouri, Y. Zagranyarski, C. Cabanetos and M. Deiana, J. Mater. Chem. B, 2024, 12, 8107–8121 RSC.
- A. Merabti, D. P. Sánchez, A. Nocentini, L. M. A. Ali, C. Nguyen, D. Durand, K. Hamon, T. Ghanem, P. Arnoux, P. Josse, C. Frochot, R. Zalubovskis, S. Richeter, M. Gary-Bobo, C. T. Supuran, C. Cabanetos, J.-Y. Winum and S. Clément, Mater. Adv., 2024, 5, 4172–4177 RSC.
- A. Nsubuga, K. Morice, N. Fayad, F. Pini, V. Josserand, X. Le Guével, A. Alhabi, M. Henry, D. Puchán Sánchez, N. Plassais, P. Josse, J. Boixel, P. Blanchard, C. Cabanetos and N. Hildebrandt, Adv. Funct. Mater., 2024, 2410077 Search PubMed.
- K. Saczuk, A. Kassem, M. Dudek, D. P. Sánchez, L. Khrouz, M. Allain, G. C. Welch, N. Sabouri, C. Monnereau, P. Josse, C. Cabanetos and M. Deiana, J. Phys. Chem. Lett., 2025, 16, 2273–2282 CrossRef CAS PubMed.
- E. Rodriguez-Marquez, H. Nord, D. Puchán Sánchez, A. Kassem, J. M. Andrés Castán, M. Deiana, C. Cabanetos, N. Sabouri and J. von Hofsten, ACS Pharmacol. Transl. Sci., 2025 DOI:10.1021/acsptsci.5c00061.
- V. Grande, B. Soberats, S. Herbst, V. Stepanenko and F. Würthner, Chem. Sci., 2018, 9, 6904–6911 RSC.
- A. Berner, R. N. Das, N. Bhuma, J. Golebiewska, A. Abrahamsson, M. Andréasson, N. Chaudhari, M. Doimo, P. P. Bose, K. Chand, R. Strömberg, S. Wanrooij and E. Chorell, J. Am. Chem. Soc., 2024, 146, 6926–6935 CrossRef CAS PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- B.-C. Zhu, J. He, X.-Y. Xia, J. Jiang, W. Liu, L.-Y. Liu, B.-B. Liang, H.-G. Yao, Z. Ke, W. Xia and Z.-W. Mao, Chem. Sci., 2022, 13, 8371–8379 RSC.
- S. J. Welsh, A. G. Dale, C. M. Lombardo, H. Valentine, M. de la Fuente, A. Schatzlein and S. Neidle, Sci. Rep., 2013, 3, 2799 CrossRef PubMed.
- H. Chen, H. Long, X. Cui, J. Zhou, M. Xu and G. Yuan, J. Am. Chem. Soc., 2014, 136, 2583–2591 CrossRef CAS PubMed.
- P. Agrawal, E. Hatzakis, K. Guo, M. Carver and D. Yang, Nucleic Acids Res., 2013, 41, 10584–10592 CrossRef CAS PubMed.
- I. Obi, M. Rentoft, V. Singh, J. Jamroskovic, K. Chand, E. Chorell, F. Westerlund and N. Sabouri, Nucleic Acids Res., 2020, 48, 10998–11015 CrossRef CAS PubMed.
- M. Deiana, J. Jamroskovic, I. Obi and N. Sabouri, Chem. Commun., 2020, 56, 14251–14254 RSC.
- A. Ghosh, M. Trajkovski, M.-P. Teulade-Fichou, V. Gabelica and J. Plavec, Angew. Chem., Int. Ed., 2022, 61, e202207384 CrossRef CAS PubMed.
- B. Prasad, M. Doimo, M. Andréasson, V. L’Hôte, E. Chorell and S. Wanrooij, Chem. Sci., 2022, 13, 2347–2354 RSC.
- A. Soleimany, D. K. Aghmiouni, M. Amirikhah, M. A. Shokrgozar, S. Khoee and B. Sarmento, Adv. Funct. Mater., 2024, 2408594 CrossRef CAS.
- M. Deiana, B. Mettra, L. Martinez-Fernandez, L. M. Mazur, K. Pawlik, C. Andraud, M. Samoc, R. Improta, C. Monnereau and K. Matczyszyn, J. Phys. Chem. Lett., 2017, 8, 5915–5920 CrossRef CAS PubMed.
- S. Liu, L. Bu, Y. Zhang, J. Yan, L. Li, G. Li, Z. Song and J. Huang, Anal. Chem., 2021, 93, 5267–5276 CrossRef CAS PubMed.
- T.-C. Lin, W. Chien, L. M. Mazur, Y.-Y. Liu, K. Jakubowski, K. Matczyszyn, M. Samoc and R. W. Amini, J. Mater. Chem. C, 2017, 5, 8219–8232 RSC.
- L. M. Mazur, T. Roland, S. Leroy-Lhez, V. Sol, M. Samoc, I. D. W. Samuel and K. Matczyszyn, J. Phys. Chem. B, 2019, 123, 4271–4277 CrossRef CAS PubMed.
- T. Entradas, S. Waldron and M. Volk, J. Photochem. Photobiol., B, 2020, 204, 111787 CrossRef CAS PubMed.
- Y.-L. Zeng, L.-Y. Liu, T.-Z. Ma, Y. Liu, B. Liu, W. Liu, Q.-H. Shen, C. Wu and Z.-W. Mao, Angew. Chem., Int. Ed., 2024, 63, e202410803 CrossRef CAS PubMed.
- F. J. Ballester, E. Ortega, D. Bautista, M. D. Santana and J. Ruiz, Chem. Commun., 2020, 56, 10301–10304 RSC.
- J. Bonelli, E. Ortega-Forte, G. Vigueras, J. Follana-Berná, P. Ashoo, D. Abad-Montero, N. Isidro, M. López-Corrales, A. Hernández, J. Ortiz, E. Izquierdo-García, M. Bosch, J. Rocas, Á. Sastre-Santos, J. Ruiz and V. Marchán, ACS Appl. Mater. Interfaces, 2024, 16, 38916–38930 CrossRef CAS PubMed.
- M. Bregnhøj, F. Thorning and P. R. Ogilby, Chem. Rev., 2024, 124, 9949–10051 CrossRef PubMed.
- J. Zhuang, S. Liu, B. Li, Z. Li, C. Wu, D. Xu, W. Pan, Z. Li, X. Liu and B. Liu, Angew. Chem., Int. Ed., 2024, e202420643 Search PubMed.
- R. del Villar-Guerra, J. O. Trent and J. B. Chaires, Angew. Chem., Int. Ed., 2018, 57, 7171–7175 CrossRef CAS PubMed.
- B. G. Kim, H. M. Evans, D. N. Dubins and T. V. Chalikian, Biochemistry, 2015, 54, 3420–3430 CrossRef CAS PubMed.
- L. De Paepe, A. Madder and E. Cadoni, Small Methods, 2024, 8, 2301570 CrossRef CAS PubMed.
- S. Bielskutė, J. Plavec and P. Podbevšek, J. Am. Chem. Soc., 2019, 141, 2594–2603 CrossRef PubMed.
- S. Bielskutė, J. Plavec and P. Podbevšek, Nucleic Acids Res., 2021, 49, 2346–2356 CrossRef PubMed.
- A. M. Fleming, J. Zhou, S. S. Wallace and C. J. Burrows, ACS Cent. Sci., 2015, 1, 226–233 CrossRef CAS PubMed.
- S. A. Johnson, T. Paul, S. L. Sanford, B. L. Schnable, A. C. Detwiler, S. A. Thosar, B. Van Houten, S. Myong and P. L. Opresko, Nucleic Acids Res., 2024, 52, 1763–1778 CrossRef CAS PubMed.
- J. Jamroskovic, M. Deiana and N. Sabouri, Biochimie, 2022, 199, 81–91 CrossRef CAS PubMed.
- M. Deiana, M. Mosser, T. Le Bahers, E. Dumont, M. Dudek, S. Denis-Quanquin, N. Sabouri, C. Andraud, K. Matczyszyn, C. Monnereau and L. Guy, Nanoscale, 2021, 13, 13795–13808 RSC.
- G. Biffi, D. Tannahill, J. Miller, W. J. Howat and S. Balasubramanian, PLoS One, 2014, 9, e102711 CrossRef PubMed.
- R. Hänsel-Hertsch, A. Simeone, A. Shea, W. W. I. Hui, K. G. Zyner, G. Marsico, O. M. Rueda, A. Bruna, A. Martin, X. Zhang, S. Adhikari, D. Tannahill, C. Caldas and S. Balasubramanian, Nat. Genet., 2020, 52, 878–883 CrossRef PubMed.
- M. P. O’Hagan, J. Ramos-Soriano, S. Haldar, S. Sheikh, J. C. Morales, A. J. Mulholland and M. C. Galan, Chem. Commun., 2020, 56, 5186–5189 RSC.
- J. Ramos-Soriano and M. C. Galan, JACS Au, 2021, 1, 1516–1526 CrossRef CAS PubMed.
- M. P. O'Hagan, S. Haldar, M. Duchi, T. A. A. Oliver, A. J. Mulholland, J. C. Morales and M. C. Galan, Angew. Chem., Int. Ed., 2019, 58, 4334–4338 CrossRef PubMed.
- M. Dudek, L. López-Pacios, N. Sabouri, J. J. Nogueira, L. Martinez-Fernandez and M. Deiana, J. Phys. Chem. Lett., 2024, 15, 9757–9765 CrossRef CAS PubMed.
- A. Matusiak, M. Drąg, M. Deiana, M. J. Janicki and M. Dudek, Chem. – Eur. J., 2025, e202404365 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, experimental procedures, synthesis, characterization, photostability and aggregation studies, G4-binding interactions, absorption, emission, time-resolved fluorescence (TCSPC), two-photon absorption, singlet oxygen spectra, ABDA bleaching experiments, molecular docking, quantum mechanical calculations, and molecular dynamics simulations. See DOI: https://doi.org/10.1039/d5nh00237k |
| This journal is © The Royal Society of Chemistry 2025 |