Open Access Article
Open Access ArticleThe molecular complexity of terpene biosynthesis in red algae: current state and future perspectives
Wanessa Francesconi Stida
Peixoto
 ab,
Renato Crespo
Pereira
ab,
Renato Crespo
Pereira
 bc,
Esthfanny dos Santos Souza
Azevedo
bc,
Esthfanny dos Santos Souza
Azevedo
 a,
Fernando Martins
dos Santos
Junior
a,
Fernando Martins
dos Santos
Junior
 d,
Ricardo
Coutinho
d,
Ricardo
Coutinho
 ab and
Louisi Souza
de Oliveira
ab and
Louisi Souza
de Oliveira
 *ab
*ab
aDepartment of Marine Biotechnology, Instituto de Estudos do Mar Almirante Paulo Moreira – IEAPM, Arraial do Cabo, 28930-000, RJ, Brazil. E-mail: louisi.oliveira@marinha.mil.br
bMarine Biotecnology Graduate Program, Instituto de Estudos do Mar Almirante Paulo Moreia – IEAPM, Federal Fluminense University – UFF, Brazil
cDepartament of Marine Biology, Biology Institute, Federal Fluminense University – UFF, Niterói, RJ 21941-590, Brazil
dDepartamento of Organic Chemistry, Chemistry Institute, Federal Fluminense University – UFF, Niterói, RJ 24.020-141, Brazil
First published on 24th February 2025
Abstract
Covering the period 1998–2024
Red algae are the largest group of seaweeds and rich sources of bioactive terpenes with broad and significant biotechnological potential. However, the main obstacle to the economic exploitation of these compounds is the difficulty of obtaining them on an industrial and sustainable scale. Genetic engineering and heterologous biosynthesis are promising tools for overcoming this limitation, but little is known about red algal terpene biosynthetic routes. In general, terpene biosynthesis relies on complex mechanisms that produce a wide array of chemically diverse compounds. In this article, we review the main processes that contribute to such chemical diversity of terpenes, which are divided into four biosynthetic steps: (i) biosynthesis of isoprenoid precursors, (ii) linear condensation of precursors to produce polyisoprenyl diphosphate intermediary molecules, (iii) terpene synthase-catalyzed chemical/structural modifications, and (iv) additional chemical/structural modifications on the basic terpene carbon skeleton. Terpene synthase evolution in algae and topics that have only recently been explored, such as terpene synthase catalytic and substrate promiscuity, have also been analyzed in detail. We present a detailed analysis of terpenoid metabolism in red algae, highlighting the mechanisms that generate their chemical diversity and identifying knowledge gaps. Additionally, we provide perspectives to guide future studies, aiming to advance the heterologous biosynthesis of terpenes from red algae for biotechnological development and application.
1. Introduction
Marine organisms produce an enormous diversity of natural products, also known as secondary metabolites, or specialized metabolites, exhibiting a wide range of chemical structures and unique bioactive capabilities. It is estimated that over 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 natural products have already been isolated from marine organisms1 with approximately 1000 new compounds being discovered annually. It is believed that over 70% of these substances occur exclusively in marine organisms.2–5
000 natural products have already been isolated from marine organisms1 with approximately 1000 new compounds being discovered annually. It is believed that over 70% of these substances occur exclusively in marine organisms.2–5
Terpenes represent one of the most important and diverse classes of these chemicals, which exhibit several vital ecological roles, such as intraspecific communication, chemical defense against herbivores, suppression of bacterial and fungal contamination, and inhibition of biofouling, among others.6–9 Terpenes and their precursor isoprenoid biomolecules are generally also primary metabolites, such as chlorophylls, carotenoids, and steroids.10
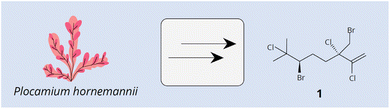
Red algae (Rhodophycota phylum) comprise over 7500 nomenclatural valid and living species, most of them (around 98%) are macroscopic and found in marine environments.11 These algae are notable for producing a wide variety of structurally diverse terpenes, primarily mono-, sesqui-, and diterpenes.1,12,13 The biotechnological importance of these compounds captured the interest of scientists globally due to their diverse potential applications. Terpenes derived from red algae demonstrated antimicrobial, anti-inflammatory, antioxidant, and anticancer properties, among others.14–21 Several studies have highlighted the biotechnological potential of different red algae monoterpene compounds.22–26 For example, acyclic monoterpenes, such as halomon [1] (Fig. 1) from Plocamium hornemannii, have demonstrated high cytotoxic activity against human tumor cell lines, including those from the brain, colon, and kidney.12 Antunes et al.27 also reported significant cytotoxic activity for halogenated monoterpenes from Plocamium suhrii and P. cornutum.
In fact, among red algae, the genus Laurencia is particularly notable for its rich diversity of biologically active terpenes (Fig. 2). For example, the sesquiterpene elatol [2], one of the most common terpenes of the red macroalga Laurencia, stands out for its effectiveness as an antitumor agent.28 Research also highlights its biocidal properties, which could be beneficial for developing antifouling or antimicrobial treatments.29,30 Furthermore, elatol [2] is a promising antiproliferative agent in the treatment of leishmaniasis.31 Similarly, lauremantanones A [3] and B [4], dendroidiol [5], and obtusol [6], also from Laurencia species showed pronounced cytotoxic activity against HeLa and P-388 cell lines. The lauremantanones A [3] and B [4] exhibited high antibacterial activity against Bacillus cereus.32–34 However, despite the broad biotechnological potential of red algae terpenes, their practical and industrial application faces several challenges that limit their use.
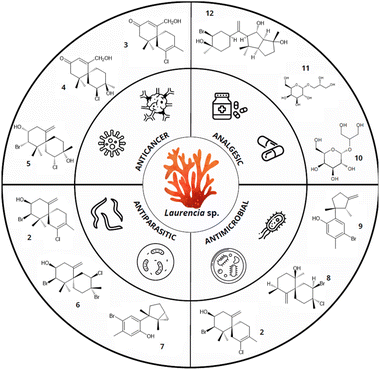 | ||
| Fig. 2 Biotechnological properties of terpenes from Laurencia species. Compounds: elatol [2], lauremantanones A [3] and B [4], dendroidiol [5], obtusol [6], laurinterol [7], panosanol [8], allolaurinterol [9], floridoside [10], D-isofloridoside [11], neorogioltriol [12] (data obtained from Cikoš et al.12 and Machado et al.32). | ||
Indeed, only an extremely limited amount of these marine natural products reach industrial utilization. Some obstacles to the biotechnological application of these compounds include their biosynthesis at low concentrations, the difficulty to adapt algae to cultivation conditions, the reduced production of the metabolites of interest under these conditions, and the structural complexity of these substances rendering their total chemical synthesis economically unfeasible.35,36
The use of genetic engineering techniques and molecular cloning, such as heterologous biosynthesis in recombinant hosts, emerges as an interesting, sustainable, and economically viable approach for the robust large-scale production of specific metabolites of biotechnological interest.37–40 However, the complex mechanisms involved in red algal terpene biosynthesis are still not fully elucidated or understood. Thus, a deeper understanding of how molecular, biochemical, and reactive processes interact to produce terpenes in red algae can offer crucial insights to direct and improve in vitro terpene synthesis for biotechnological uses. This article reviews the biosynthetic processes that contribute to producing the structural and functional diversity of terpenes in red algae, highlights knowledge gaps, and suggests future directions for the heterologous biosynthesis of red algal terpenes.
2. The biosynthetic mechanisms responsible for terpene diversity and structural complexity
With over 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 known compounds, terpenes represent the largest and most diverse class of natural products.41 Such structural and functional diversity depends on a complex network of processes and mechanisms that operate from the assembly of basic precursor units to the stages where structural chemical modifications occur, leading to the final products. According to the number of isoprene units in their structure, terpenes are classified as hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), and triterpenes (C30), among others.42 In red algae, the biosynthesis of basic precursor units of isoprene occurs through one or two different biosynthetic pathways, as detailed in Section 2.1. Further, the cyclization and post-cyclization processes, including conjugation with other molecules, contribute to the structural diversity of these compounds.
000 known compounds, terpenes represent the largest and most diverse class of natural products.41 Such structural and functional diversity depends on a complex network of processes and mechanisms that operate from the assembly of basic precursor units to the stages where structural chemical modifications occur, leading to the final products. According to the number of isoprene units in their structure, terpenes are classified as hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), and triterpenes (C30), among others.42 In red algae, the biosynthesis of basic precursor units of isoprene occurs through one or two different biosynthetic pathways, as detailed in Section 2.1. Further, the cyclization and post-cyclization processes, including conjugation with other molecules, contribute to the structural diversity of these compounds.
In summary, red algal terpene biosynthesis can be divided into four main sequential steps, which will be further explored in the subsequent sections of this review: (i) the biosynthesis of isoprenoid precursors, (ii) the linear condensation of precursors to produce polyisoprenyl diphosphate intermediary molecules, (iii) the terpene synthase-catalyzed chemical modifications, and (iv) the additional chemical modifications of the basic terpene carbon skeleton.
2.1. The biosynthesis of isoprenoid precursors
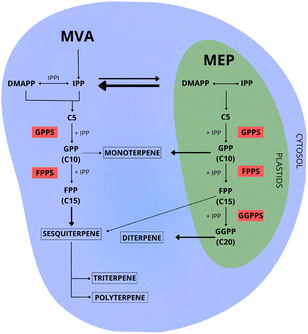
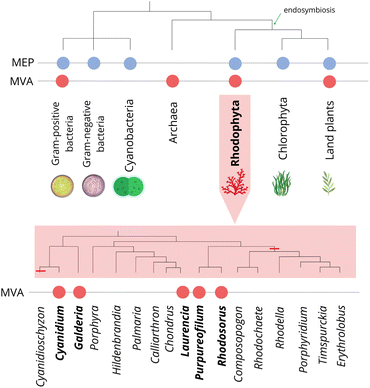
The initial step in terpene biosynthesis typically involves the linear condensation of two types of five-carbon isoprenoid precursors: isopentenyl diphosphate (IPP), and its allylic isomer dimethylallyl diphosphate (DMAPP). These isoprene precursors are biosynthesized through two distinct pathways: the mevalonate (MVA) and the 2-C-methyl-D-erythritol-4-phosphate (MEP)43,44 (Fig. 3).Throughout the evolutionary process, different groups of organisms have acquired the ability to biosynthesize precursors for terpene biosynthesis through their specialization, utilizing one or both biosynthetic pathways (MVA and MEP). Archaea, some Gram-positive bacteria, and most eukaryotes utilize the MVA pathway.45 In contrast, Gram-negative bacteria, cyanobacteria, and green algae rely exclusively on the MEP pathway for the biosynthesis of prenyl substrates.46 Plants and some species of red algae are capable of biosynthesizing precursors through both pathways47 (Fig. 4). Similarly, some bacteria are capable of utilizing both pathways.48
The MVA pathway, predominantly occurring in the cytosol of the cell,49 exclusively generates IPP as its final product. The conversion of IPP to DMAPP requires an additional isomerization process, mediated by the enzyme isopentenyl diphosphate isomerase50–52 (Fig. 3).
In evolutionary terms, the MVA pathway is considered the ancestral isoprenoid metabolic route present in the last common ancestor, and it was vertically transmitted to descendants across all three domains of life.53 Several species of red algae have lost the ability to biosynthesize terpenes through this pathway, possibly due to genome reduction events.45,54,55 However, this loss was not uniform (Fig. 4). Therefore, it is plausible to question the idea that all red algal lineages underwent a widespread loss of the MVA pathway during the evolutionary process. Recent studies suggest that red algal genomes were shaped by at least two distinct episodes of gene loss and genome contraction throughout their evolutionary history.47,56
The loss of the MVA pathway in red algae was attributed to recent genetic events in specific lineages, since this pathway was retained in several species. For example, genetic and biochemical evidences suggest that the red macroalga Laurencia dendroidea57,58 and the unicellular red algae Galdieria sulphuraria,59Rhodosorus marinus,47Purpureofilum apyrenoidigerum,47 and Cyanidium caldarium60 remained capable of biosynthesizing terpenes through both the MVA and MEP pathways (Fig. 4). Genome and transcriptome analysis of the red macroalgae Porphyra purpurea and P. umbilicalis, and the red microalgae Cyanidioschyzon merolae and Porphyridium purpureum revealed the lack of most regulatory genes or the complete absence of the MVA pathway.61–65 Besides, phylogenomic analysis of 15 species of red algae revealed that the mevalonate pathway (MVA) was missing in most of these lineages,47 suggesting that the absence of the MVA pathway in red algae is more extensive than initially estimated.
In that case, a more detailed analysis of the genome context and gene characteristics of the MVA pathway in red algal lineages will be necessary to clarify these issues. However, this in-depth analysis relies on the development of new data. According to Lohr et al.,61 genomic sequencing of species is expected to provide more evidence regarding the distribution of these two pathways, while the lack of high-quality genomes represents the biggest challenge for red algal research.
The MEP pathway can simultaneously produce IPP and DMAPP (Fig. 3). With a stoichiometry of approximately five molecules of IPP for each molecule of DMAPP, this pathway plays a crucial role in maintaining adequate amounts of both precursors for terpene biosynthesis. Among the seven enzymes in the MEP pathway, two of them stand out as potential regulators of carbon flux: 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR).66–68 Therefore, the overexpression or repression of these genes through genetic engineering has a direct impact on terpene biosynthesis.50,69–72
In photosynthetic organisms, the presence of plastids reflects a complex evolutionary heritage, associated with acquiring them through the endosymbiosis process. Recent studies revealed that eukaryotes with plastids, whose MEP pathway genes originate from cyanobacteria and Chlamydiae, show the combined contributions of these groups to the evolution of primary plastids.46 Thus, the MEP pathway originated from an ancestral MEP pathway present in these symbiotic organisms that interacted with early eukaryotes.46,54,73
The subcellular localization of the MEP pathway in red algae was not yet clearly determined.74 According to predictions, in P. purpureum, some of the MEP pathway enzymes would be located in the plastids, while in other species these genes were expected to be found in the cytosol or other organelles.74,75
In plants, several studies indicate that different metabolic pathways involved in terpene biosynthesis lead to the preferential production of specific types of terpenes (Fig. 3). However, few studies investigated the preferential biosynthesis of specific terpene classes in red algae. To investigate the location of red algal prenyltransferases and the preferential biosynthesis of specific terpene classes in particular cell compartments, Deng et al.76 used in vivo analysis in protoplasts of the plant Arabidopsis thaliana. Contradicting what is known for plants, the authors did not find evidence of chloroplast localization for GGPS isolated from the red algae Bangia fuscopurpurea (BfGGPPS). However, they raise the hypothesis that the chloroplasts of the protoplasts failed to recognize the transit peptide from BfGGPPS because it is shorter than those from plants. Also, differently from what is observed in plants, Steele et al.77 did not find an N-terminal signal sequence in monoterpene synthase genes from the red seaweeds Portieria hornemannii and Plocamium pacificum, suggesting a cytosolic monoterpene production in both species.78
In plants, the bidirectional exchange of precursors between the MVA and MEP pathways and the co-expression of specific genes from both pathways, indicates the existence of feedback and metabolic interaction which act as a compensatory mechanism in response to specific environmental and cellular conditions43,78,79 (Fig. 3). Although it is known that there are interactions between both pathways in various plant species, there is still a gap in the understanding of the cooperative metabolic network involved in terpene biosynthesis in red algae. Specifically, how these metabolic pathways collaborate to exchange precursors during terpene biosynthesis in this group of organisms is not well understood. Due to the simultaneous occurrence of the MVA and MEP pathways, raises questions about the regulation of metabolic fluxes between different cellular compartments and the contribution of one or both pathways to the biosynthesis of isoprenoid compounds.
Based on what is known for plants, despite the supposed metabolic cost, maintaining both pathways could bring functional benefits, such as a greater capacity for the evolution of specialized pathways and specific control of isoprenoid pools in each cellular compartment.43,78,80,81 In this scenario, the presence of one or both pathways for isoprene biosynthesis can be seen as the first level of a complex network that influences the diversity of terpenes generated by different lineages of red algae. Furthermore, the exchange of intermediates between these pathways could result in the formation of mixed-origin MVA/MEP isoprenoid compounds, further expanding the terpene diversity.82–84 Therefore, to fill the existing gaps, specific studies are needed to investigate the evolution of isoprenoid biosynthesis and intracellular trafficking between compartments in red algae.
2.2. The condensation process to produce polyisoprenyl diphosphate intermediary compounds
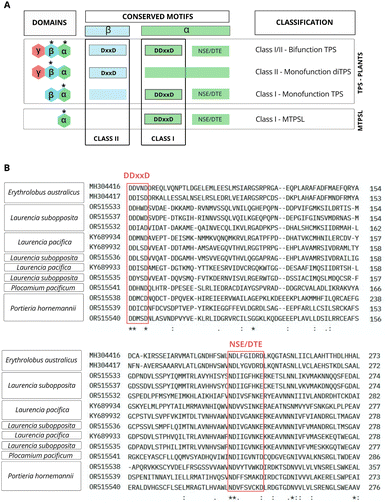
The second step for the biosynthesis of terpenes consists of condensing IPP and DMAPP precursors to biosynthesize linear prenyl diphosphate intermediaries, such as geranyl diphosphate (GPP-C10), farnesyl diphosphate (FPP-C15), geranylgeranyl diphosphate (GGPP-C20), as well as medium-chain (C30–C35) and long-chain (C40–Cn) prenyl substrates. These reactions are catalyzed by isoprenyl diphosphate synthases (IPPS), a class of prenyltransferases. The resulting prenyl diphosphate intermediaries are used as substrates by specific terpene synthases to produce specialized metabolites53,85,86 (Fig. 3). In plants, the distribution of prenyltransferases among cellular compartments creates a regulatory network that coordinates the allocation of isoprenoid precursors for biosynthesizing different terpene classes. These mechanisms are well understood for plants and involve transcriptional co-regulation and the formation of complexes of sequential enzymes. However, the IPPS regulation systems are largely unknown for red algae.The activity of IPPS can determine the carbon chain length and the stereochemical properties of the produced isoprenoid polymers. Based on the stereochemistry of the double bond, the isoprenoid products can be classified as trans (E) or cis (Z). Although they share the same isoprenoid substrates and catalytic mechanisms, trans and cis enzymes exhibit low similarity in their primary and tertiary structure.85,87Trans-IPPS tend to biosynthesize products ranging from short to medium chain lengths (C15 to C50). This enzymatic class typically shares two conserved motifs rich in aspartate (typically, DDx2–4D and DDxxD), positioned in opposite helices to the substrate-binding pocket, both crucial for substrate binding and catalysis, as well as involved in chelation with Mg2+ ions. The second motif, DDxxD, particularly facilitates the binding of isopentenyl diphosphate (IPP). Besides, a third conserved motif containing two sequential arginines (RR) is essential for catalysis and binding of the allylic substrate79,85,88 (Fig. 5).
In contrast, cis-IPPS do not exhibit the conserved DDxxD motif. Initially, it was believed that cis-IPPS were involved only in the biosynthesis of large prenyl diphosphates with more than 50 carbon atoms.86,89 However, cis enzymes capable of producing short-chain compounds have already been identified in plants, indicating that these enzymes present greater metabolic versatility than previously expected.90 The cis-prenyltransferase can be classified into two subfamilies of homodimeric and heteromeric enzymes, and these enzymes were found in a variety of organisms, including bacteria, fungi, plants, and animals.91
Despite being considered the gatekeepers to the diverse branches of terpene biosynthesis, few studies were dedicated to prenyltransferases in algae. Gong et al.92 identified cis-FPP, trans-FPP, and GGP synthases in the green microalga Haematococcus pluvialis. Similarly, FPPS and GGPPS synthases have been identified in the green microalgae Dunaliella salina, Chlamydomonas reinhardtii and H. pluvialis, as well as GPP synthase in D. salina and H. pluvialis.93,94 Besides, Lohr et al.61 searched for trans-IDS candidates in different algal types, including the green microalgae Ostreococcus tauri, Chlamydomonas reinhardtii, and Chlorella variabilis, the diatom Phaeodactylum tricornutum, and the brown macroalga Ectocarpus siliculosus, finding candidate enzymes for trans-GGPPS and trans-FPPS in all of them.
Regarding red algae, studies on prenyltransferases are particularly scarce. As an example, we could mention the identification of candidate sequences for GGPPS and FPPS in the genome of the red microalga C. merolae.61 Moreover, Yang et al.95 cloned the gene encoding GGPS in P. umbilicalis (PuGGPS) and determined that it functions in the biosynthesis of GGPP for carotenoid biosynthesis. Finally, Deng et al.,76 using in vivo and in vitro analyses, identified and confirmed the presence of a synthase capable of producing GGPP in the red macroalga B. fuscopurpurea.
The phylum Rhodophycota is known by the production of a large array of monoterpenes. The GGPPS are present in all plants, whereas GPPS are uncommon outside of gymnosperms. In angiosperms, GGPPS typically utilize type I small subunit proteins (SSUIs) to generate GPP. Song et al.,96 in a recent study, evaluated the functional divergence of GGPPS enzymes across different organisms. The authors observed that homologs of GGPPS and GPPS in algae cannot produce GPP, which is a crucial substrate for monoterpene biosynthesis. Similarly, Deng et al.76 cloned and expressed one GGPPS gene from B. fuscopurpurea and verified that this enzyme produces exclusively GGPP. Steele et al.77 reported the cloning and characterization of TPS from the red macroalgae P. hornemannii and P. pacificum, which acted as selective monoterpene synthases, converting geranyl diphosphate (GPP) to acyclic terpene products.
However, the limited availability of sequencing data and the scarcity of studies characterizing prenyltransferases in red algae hampers the understanding of what kind of enzyme red algae employ to produce GPP, the substrate for monoterpene synthases. Further studies are necessary to clarify the monoterpene biosynthetic pathway in red algae. In particular, effort should be dedicated to the search for GPPS and/or heteromeric GGPPS candidate genes in red algae.
In general, studies involving in vitro enzymatic and mutagenesis assays are necessary for IPPS enzyme classification in red algae. Further, a deeper understanding of the mechanisms of action and regulation of this pathway in red algae and how it interacts with other pathways is essential for advancing the heterologous synthesis of biotechnologically interesting terpenes.
2.3. Terpene synthase-catalyzed chemical modifications
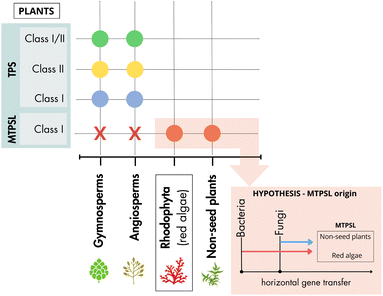
During the third step of the terpenes biosynthesis, the prenyl diphosphate intermediary compounds are converted into cyclic and acyclic terpenes through a cascade of stereospecific carbocation reactions catalyzed by terpene synthase (TPS) enzymes. TPSs facilitate cyclization reactions, rearrangements, and hydride shifts and mediate different termination reactions, such as deprotonation and water addition. During terpene biosynthesis, a single carbocation can undergo multiple cyclizations and hydride shifts before the completion of the reaction, which expands the possibilities of chemical structures for the produced terpenes.97–99 Moreover, the diversity of terpene synthases in a single organism, the ability of some enzymes to accept prenyl diphosphate substrates of different lengths, and their multiproduct nature contribute to the biosynthesis of a wide array of chemically diverse terpenes. Considering the relevance of TPS for the molecular chemical diversity of terpenes in red algae, the evolutionary process and the functional plasticity of these enzymes are detailed in the following subsections.The evolutionary origin of MTPSL genes remains unclear. However, recent studies revealed that these genes are absent in seed plants and green algae, but are widely distributed in red algae and non-seed plants.74,100,102 Also, MTPSL genes have been found in different orders of red algae not phylogenetically related to each other, such as Gigartinales, Plocamiales, Ceramiales, and Phorphyridiales, but are absent in various other groups of Rhodophycota.74,77,101 Since the presence and absence of these genes in red algae has not been fully elucidated, this gap may be attributed to limited sequencing depth, or the low quality of the sequences used. A more comprehensive understanding of the distribution of these genes depends on a broader approach and more complete sequencing of genomes and transcriptomes from different species of red algae (Fig. 6).
Moreover, the time of red algal divergence from the rest of the eukaryotic tree of life is still under debate, with some authors proposing it to have occurred between the late Mesoproterozoic to early Neoproterozoic era (1.3–0.9 billion years ago). Others suggest an earlier appearance in the late Paleoproterozoic (2.5 to 1.6 billion years ago) (reviewed by Borg et al.56). The uncertainty regarding the evolution of Rhodophycota and the scarcity of available genome sequences for this group pose a challenge to understanding MTPSL evolution. Despite clustering with microbial terpene synthase genes, MTPSL genes in Rhodophycota were proved to be of red algal origin, instead of microbial contamination.74 However, the question remains whether red algal MTPSL genes originated from horizontal gene transfer events (either from single or multiple events) or if these genes were present in the Rhodophycota ancestor and secondarily lost in several lineages74 (Fig. 6).
TPS exhibit varied domain architectures, including α, αβ, αβγ, βγ, and β domains, which reflects the chemical diversity of terpenes. The typical plant TPS and MTPSL enzymes have different domain compositions. The first present two domains (αβ) or three domains (αβγ), while MTPSL enzymes exclusively present a single α domain and present a conserved DDxxD motif in aspartic acid (Fig. 7A and B). Additionally, non-canonical motifs like DDxxxD and DDxxx were also found.102
Based on the reaction mechanism, these enzymes, in plants, can be categorized into two main classes based on their formation mechanism of carbocations: ionization metal-dependent (class I) and protonation-dependent (class II) (Fig. 8).103 However, a specific group of bifunctional enzymes possesses characteristics that allow them to perform the functions of both class I and class II enzymes.
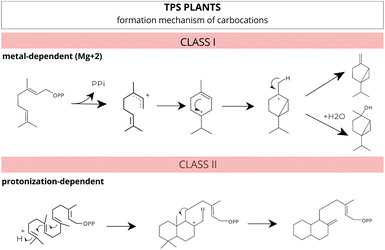 | ||
| Fig. 8 Different mechanisms for carbocation formation. Class I enzymes catalyze ionization-dependent reactions, while class II enzymes catalyze protonation-dependent reactions. Adapted from Huang et al.103 | ||
These class I enzymes catalyze their reactions within the cavity of an alpha-helical bundle protein structure, using one or two conserved metal-binding motifs, DDxxD and NSE/DTE, which coordinate the necessary trio of magnesium (Mg2+) cofactors41,104–106 (Fig. 7A and B). In this reaction mechanism, carbocation formation is induced by metal-dependent ionization, and the stochastic occurrence of rearrangements and chemical often results in multiple products.77,98,104,107
In contrast, class II enzymes host a single active site located in the β domain or at the interface between the β and γ domains. This conserved domain harbours the DxDD motif, rich in aspartate, which is responsible for forming a tertiary carbocation initiated through the protonation mechanism of the terminal C–C double bond in the isoprenoid substrate41,97,108 (Fig. 7A). In a recent study, Pan et al.109 extensively reviewed and compiled information about class II TPS, including functional motifs, catalytic mechanisms, and genetic engineering.
A few TPSs enzymes exhibit all three functional active sites. These enzymes are bifunctional and, along with diterpene synthases, possess an additional domain (γ) which are believed to be involved in binding Mg2+ and diphosphate.108,110 No class II or bifunctional enzymes have been identified in red algae. In plants, all recognized class II terpene synthases are part of the plant TPS family, not the MTPSL (microbial terpene synthase-like) family106 (Fig. 7A).
To date, the complex mechanisms involved in terpene biosynthesis in red algae are still not fully understood. Most of the issues related to establishing pathways for optimizing the development of terpene synthase (TPS) products and enzymes in these organisms remain unresolved. In general, functionally classifying terpene synthase enzymes based solely on primary sequence is not possible due to the lack of sequence–function relationships within the family. TPS amino acid sequence similarities appear more closely associated with taxonomic affinities than with the type of products formed. Consequently, predicting the three-dimensional (3D) structure of these proteins is a challenging task.105,111
Few experiments have been dedicated to elucidating the molecular architecture of the MTPSL enzymes. For example, Yan et al.112 elucidated the catalytically active structure of the class I MTPSL enzyme from the liverwort Jungermannia exsertifolia. The authors demonstrated the substrate binding environment and magnesium coordination configuration using ab initio modeling, molecular docking, and molecular dynamics simulations of the enzyme in complex with the substrate. Moreover, Kersten et al.101 characterized a bourbonane-producing sesquiterpene synthase, the first sesquiterpene synthase to be biochemically characterized in red algae. According to the authors, the combined use of multi-omics techniques, genetic engineering, and approaches for absolute molecular structure determination represents an excellent alternative for biochemical characterization and the discovery of new compounds in red algae.
Therefore, accurate functional characterization of red algal TPS should consider using computational approaches to predict the product through an enzymatic homology model that adjusts cationic intermediates in the active site. Further, the resolution of TPS crystal structures and directed mutagenesis studies for structure–function investigations may provide essential information for understanding how TPS enzyme diversity relates to the huge chemical diversity of red algal terpenes.
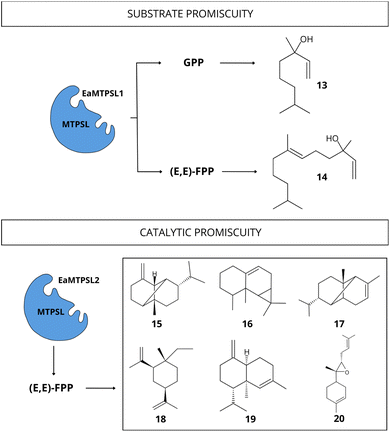 | ||
| Fig. 9 Diversity of terpenoid compounds biosynthesized by promiscuous MTPSL enzyme from E. australicus with the ability to accept different substrates and catalyze the production of multiple compounds. Substrates: (E,E)-FPP and GPP. Compounds: linalool [13], nerolidol [14], β-copaene [15], aristol-9-ene [16], α-ylangene [17], β-elemene [18], α-muurolene [19], (Z)-α-bisabolene epoxide [20] (data obtained from Wei et al.74). | ||
These characteristics highlight TPS structural and catalytic complexity, and the diversity of reactions involved in terpene biosynthesis. Several studies revealed the existence of promiscuous TPS enzymes in plants capable of converting a single substrate into various terpenoid products through conformational flexibility mechanisms of their active sites and a series of structural rearrangements.116 Besides, plant TPS enzymes capable of accepting polyprenyl diphosphates with various chain lengths as substrates have been identified.53,116,117 Indeed, recent studies highlight the importance of the multi-product nature of MTPSL enzymes for terpene biosynthesis in plants,112,117 describing enzymes that can biosynthesize up to 18 compounds from one substrate.118
The substrate promiscuity was also recently demonstrated for the lycophyte Selaginella moellendorffii possessing eight MTPSL enzymes that effectively bind to FPP or GPP, functioning as mono- and sesquiterpene synthases.117 Likewise, several studies have demonstrated the ability of plant monoterpene synthase enzymes to use neryl diphosphate (NPP), FPP, and GGPP as alternative substrates.119–121
To our knowledge, there are few studies demonstrating TPS promiscuity in red algae. Wei et al.74 observed a substrate promiscuous MTPSL enzymes from two the red microalgae: Erythrolobus australicus (EaMTPSL1) and P. purpureum (PpMTPSL). The EaMTPSL1 enzyme could utilize both GPP and (E,E)-FPP as substrates, converting them into linalool [13] and nerolidol [14], respectively (Fig. 9). Meanwhile, the PpMTPSL enzyme was active with both (E,E)-FPP and GGPP.74 In the same study, the catalytic promiscuity was demonstrated for MTPSL enzymes from E. australicus (EaMTPSL2) capable of utilizing (E,E)-FPP to produce a mixture of sesquiterpenes (β-copaene [15], aristol-9-ene [16], α-ylangene [17], β-elemene [18], α-muurolene [19], two unidentified sesquiterpenes, (Z)-α-bisabolene epoxide [20], and an oxygenated sesquiterpene) (Fig. 9), and also a MTPSL enzyme from P. purpureum (PpMTPSL) capable of producing eight distinct sesquiterpenes (β-copaene [15], aristol-9-ene [16], β-elemene [18], germacrene A [21], and four others unidentified sesquiterpenes).74 Further studies are necessary to analyse other red algae lineages and expand the current knowledge regarding TPS promiscuity. Besides, experimental studies should consider increasing the diversity of substrates tested to fully encompass the wide range of activities and products that these enzymes can exhibit, and consequently, demonstrate their total activity.
Moreover, directed mutagenesis can be used to modify specific amino acid residues on TPS active sites to increase the biosynthesis of a specific product enantiomer. Also, several studies with plants induced modifications to one or more residues controlling the active site volume and were able to produce substrate selectivity changes in TPS enzyme variants.122–126 Applying directed mutagenesis tools to red algal TPS should expand the array of substances produced through heterologous biosynthesis and increase the enantio-specificity of the process to biosynthesize biotechnologically valuable red algal terpenes.
2.4. Chemical diversity and additional modifications to the basic terpene skeleton
As a result of the mentioned complex metabolic network, red algae stand out for their great chemical diversity. Terpene compounds are recognized as unique due to their structural and functional diversity and can be found in nature with acyclic, monocyclic, bicyclic, and polycyclic chemical structures.13,18,127,128 Further, a series of sequential rearrangements mediated by enzymatic carbocation intermediates, results in the formation of a wide variety of terpenes belonging to different classes,129,130 which contribute significantly to the diversity of red algal terpenes (Fig. 10).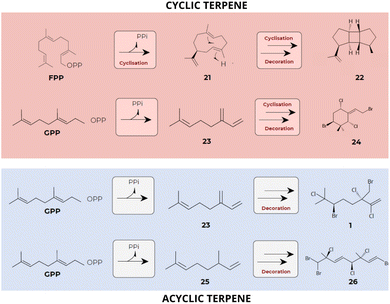 | ||
| Fig. 10 Sequential skeletal rearrangements leading to the synthesis of diverse terpene classes. The initial cyclic or acyclic terpene skeletons produced by terpene synthases (TPS) undergo subsequent chemical modifications, greatly contributing to the diversity of terpenes found in red algae. Substrates: FPP and GPP. Compounds: hamolon [1], germacrene A [21], prespatane [22], myrcene [23], 1-ethylidene-3,3-dimethylcyclohexane [24], omicene [25] and hexabromochlorinated [26] (data obtained from Cikoš et al.12 and Kersten et al.101). | ||
At least 700 compounds with unique structures have been identified in 60 species of Laurencia.131 Among these, over 500 are sesquiterpenes, approximately 100 are diterpenes, 50 are triterpenes, and 50 belong to other classes of metabolites.13,130,131
Laurencia species are recognized as the most appealing source of sesquiterpenes offering a diverse array of structurally distinct compounds. These sesquiterpenes can be categorized based on their unique skeleton types.130 In general, in sesquiterpene biosynthesis, multiple pathways contribute to the production of structurally diverse compounds (Fig. 11). For example, in two of these pathways, terpene biosynthesis occurs without initial isomerization of FPP, resulting from a cyclization process induced at carbons 1,10 for the (E,E)-germacradienyl cation or 1,11 for the (E,E)-humulyl cation formed from the ionization of the farnesyl cation [27]. Alternatively, FPP can undergo isomerization to two enantiomers (3S) [(35)-28] or (3R)-NPP [(3R)-28], an essential process for the common 1,6-cyclization to the bisabolyl cation [29].132
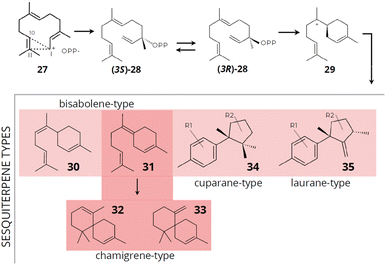 | ||
| Fig. 11 Proposed mechanisms for the initial cyclization of FPP and the synthesis of typical sesquiterpene skeleton types. Compounds: farnesyl cation [27], (3S)-NPP [(3S)-28], (3R)-NPP [(3R)-28], bisabolyl cation [29], α-bisabolane [30], y-bisabolane [31], α-chamigrane [32], β-chamigrane [33], cuparane-type [34] and laurane-type [35] (data obtained from Cikoš et al.130). | ||
The bisabolyl cation [29] can result in the formation of sesquiterpenes of the chamigrane and bisabolane skeleton types (α-bisabolane [30] and y-bisabolane [31]), while the precursor γ-bisabolane can produce derivatives of α- [32] and β-chamigrane [33] through enzymatic cyclization. The chamigrane skeleton can act as an intermediate in forming new carbon skeletons, potentially generating a chemical diversity of produced compounds. Cuparane-type [34] sesquiterpenes are derived through protonation of γ-bisabolane followed by cyclization to cuparane cation, which subsequently yields sesquiterpenes with a cyclopropane ring and can be the precursor for the formation of laurane-type [35] sesquiterpenes by methyl migration. Laurane-type [35] sesquiterpenes are derived from the bisabolyl cation intermediate, while cuparane-type [34] sesquiterpenes are derived through protonation of γ-bisabolane followed by cyclization to cuparane cation, which subsequently yields sesquiterpenes with a cyclopropane ring and can be the precursor for the formation of laurane-type sesquiterpenes by methyl migration.130,133,134
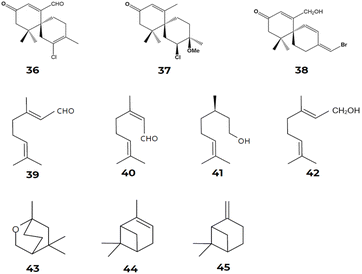
The chamigrane-type sesquiterpenes are the most common in the Laurencia species, presenting skeletons of the α- [32] and β-chamigrane types [33]. Several studies revealed the presence of chamigrane-type sesquiterpenes in Laurencia.135–140 Kamada et al.141 described the presence of α-chamigrane [32] sesquiterpenes in Laurencia majuscula, including 7-aldehyde-laurencenone B [36], 2-chloro-3-methoxy-α-chamigrane-9-one [37], and laurebornone [38]. Similarly, Kamada et al.142 observed the production of lauremantanones A [3] and B [4], derived from the α-chamigrane skeleton [32], and elatol [2], dendroidiol [5] and obtusol [6], derived from the β-chamigrane skeleton [33].
Similarly, Cikoš et al.12 surveyed the biodiversity of red macroalgal monoterpenes and identified 136 different chemical structures of monoterpenes from the genera Plocamium, Portieria, Ochtodes, and Microcladia. The presence of reaction cascades associated with highly reactive intermediates enables the biosynthesis of a wide variety of chemically and structurally distinct chemical compounds, as described earlier. Following a divalent ionization process assisted by cations from the diphosphate group during the biosynthetic process for forming cyclic monoterpenes' carbon skeletons. The cyclic cation α-terpinyl is also formed and undergoes various distinct reaction mechanisms, such as oxidation, reduction, isomerization, or conjugation, resulting in a variety of mono- and bicyclic monoterpene carbon skeletons. According to Cikoš et al.,12 the most common acyclic monoterpenes in red algae are myrcene [23], ocimene [25], geranial [39], neral [40], citronellol [41], and geraniol [42]. On the other hand, 1,8-cineole [43] stands out as the most frequent monocyclic monoterpene, while α-pinene [44] and β-pinene [45] are the most common examples of bicyclic monoterpenes found in these marine macroalgae22–26 (Fig. 12).
Moreover, several enzymes catalyze complex chemical modifications, including glycosylation, methylation, acetylation, halogenation, phosphorylation, and oxidation,143–145 increasing the chemical diversity of red algal terpenes. These reactions play an important role in the broad biological activity of numerous natural products. Among the enzymes that modify terpene chemical structures, cytochrome P450 catalyzes common reactions of hydroxylation and epoxidation, and a variety of “uncommon” reactions involving the formation and cleavage of carbon–carbon (C–C), carbon–nitrogen (C–N), and carbon–sulfur (C–S) bonds, as well as ring modifications. These multiple reactions catalyzed by P450 greatly expand the diversity of the chemical structures and biological activities of natural products.
Comparative genomic and transcriptomic analyses revealed a high diversity of P450 genes in algae.146–148 As the most conserved P450 family, CYP51 is widely distributed across all kingdoms. According to Teng et al.,148 both CYP51 and CYP97 show a close relationship among red algae, green algae, and higher plants. The genome of the red microalgae C. merolae revealed sequences corresponding to CYP51 and CYP710 and contained five putative P450-coding genes: CMD096C, CMJ270C, CMJ284C, CMS319C, and CMR093C.149,150 Yang et al.146 identified and functionally characterized the first cytochrome P450-type carotene hydroxylase (PuCHY1) in the red alga P. umbilicalis. Sequence comparisons showed that PuCHY1 belongs to the CYP97B family. The authors suggest that members of the CYP97B subfamily of cytochrome P450-type carotene hydroxylases emerged early in the evolution of eukaryotic photosynthetic organisms. Recent advances in cultivation and genetic manipulation technologies have expanded the possibilities for studying red algal enzymes. However, research on red alga P450 is still scarce and concentrated on the application of P450 inhibitors to cell cultures. One potential obstacle to the functional characterization of red algal P450 candidate genes is that the heterologous expression of membrane-bound proteins is challenging.147,151
Red seaweeds produce a wide range of halogenated terpene products with biotechnological potential and industrial applications. The biosynthesis of halogenated metabolites in these organisms relies on numerous enzymatic and biosynthetic reactions.152 Haloperoxidase-mediated terpene halogenation is recognized as an important process in the special metabolism of red algae.153–155 Butler and Sandy156 conducted a study on the vanadium haloperoxidases found in marine algae, highlighting the importance of these enzymes in the bromocyclization of terpenes. This reaction is crucial for the biosynthesis of halogenated natural products, which possess significant biological properties. Additionally, Butler et al.157 discussed the reactivity of vanadium bromoperoxidase (V-BPO), examining its role in the production of halogenated compounds.
Together, these studies provide the foundation for understanding the essential roles of marine V-BPO in the bromination and cyclization of terpene products.77,155,156,158 A common feature of V-BPOs found in red algae is their ability to produce prolific brominated compounds from a single precursor, reflecting the versatility of these enzymes.155 V-BPO isolated from Laurencia, for example, catalyzes the cyclization and bromination of the sesquiterpene nerolidol [46], leading to the formation of α- [47], β- [48], and γ-snyderol [49] and (+)-3β-bromo-8-epicaparrapi oxide [50]. Here, γ-snyderol [49] is proposed as an intermediate in the synthesis of other bicyclic compounds (Fig. 13).156
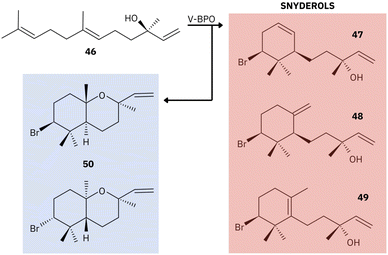 | ||
| Fig. 13 Example of the proposed mechanism of catalysis by red algal V-BPO. Compounds: nerolidol [46], α- [47], β- [48], and γ-snyderol [49] and (+)-3β-bromo-8-epicaparrapi oxide [50]. Adapted from Butler and Sandy.156 | ||
The family Rhodomelaceae (order Ceramiales, class Rhodophyceae, and phylum Rhodophycota) stands out as the largest family of marine red macroalgae and biosynthesizes thousands of compounds, most of them (about 80%) being halogenated metabolites. Notably, most of these halogenated compounds from Rhodomelaceae consist of bromine or chlorine halogenation in a wide variety of carbon distinct skeletons.13 These halogenated terpenes were mainly found in the genus Laurencia, which comprises about 144 known species.11
Red algal terpenes have been extensively isolated and tested in the last decades. Future interdisciplinary studies should use the accumulated knowledge of red algal terpenes' chemical diversity to expand the comprehension of the underlying molecular mechanisms. Further, new biosynthetic methods are needed for the sustainable biosynthesis of these valuable compounds.
3. Heterologous biosynthesis of terpenes in red algae
The biotechnological applicability of red algal terpenes depends on the development of sustainable production methods. Recent advances in synthetic biology and genetic engineering have facilitated the development of new tools for gene construction, control, and insertion, as well as for the optimization and manipulation of metabolic pathways and biosynthesis in heterologous systems. However, few studies have been dedicated to the characterization and expression of genes involved in the biosynthesis of terpenes from red algae in heterologous systems (Table 1).| Species | Substrate | Products | Ref. |
|---|---|---|---|
| L. pacifica | (E,E)-FPP | Prespatane | 77 and 101 |
| (E,E)-FPP | Bicyclogermacrene | ||
| (E,E)-FPP | Sesquiterpene alcohol | ||
| L. subopposite | (E,E)-FPP | (Z,E)-α-Farnesene | 77 |
| (E,E)-FPP | Germacrene D-4-ol | ||
| (E,E)-FPP | Germacrene B | ||
| (E,E)-FPP | Ledene | ||
| (E,E)-FPP | Prepatane | ||
| P. hornemannii | GPP | B-mircene | 77 |
| (E,E)-FPP | Germacrene D | ||
| (E,E)-FPP | Valencene | ||
| P. pacificum | GPP | Trans-β-ocimene | 77 |
| P. purpureum | (E,E)-FPP | β-Elemene, aristol-9-ene, β-copaene, germacrene A, four others unidentified sesquiterpenes | 74 |
| GGPP | 7,11-Dimethyl-3-methylene-1,6,10-dodecatriene, cembrene A | ||
| E. australicus | GPP | Linalool | 74 |
| (E,E)-FPP | Nerolidol | ||
| (E,E)-FPP | β-Copaene, aristol-9-ene, α-ylangene, β-elemene, α-muurolene, two unidentified sesquiterpenes, (Z)-α-bisabolene epoxide, oxygenated sesquiterpene | ||
| P. umbilicalis | GGPP | β-Carotene, lycopene | 95 |
| B. fuscopurpurea | GGPP | Geranylgeraniol | 76 |
Besides, the number of red algal species with available genome sequences is low compared to other major eukaryotic groups.56 Indeed, Hanschen and Starkenburg159 report an overall reduction in algae genome completeness, assembly, and gene annotation quality. The incomplete annotation of important red algal metabolic pathways was also reported by McKinnie et al.152 The authors functionally annotated the genome and transcriptomes of 72 red algal species and demonstrated that the secondary metabolic pathways are especially underrepresented. This hampers the comprehension of the regulation mechanisms involved in terpene biosynthesis and the application of valuable tools such as gene knockout, overexpression, and knockdown.
Indeed, cloning and expression techniques were rarely employed for the functional characterization of red algal genes but brought important insights. As an example, Yang et al.95 used a co-transformation method to characterize a gene coding for a prenyltransferase, and was able to prove that the gene PuGGPS from the red macroalgae Pyropia umbilicalis codes for geranylgeranyl diphosphate synthase (GGPPS), based on the accumulation of β-carotene and lycopene by E. coli transformed cells. The authors combined these tools with gene expression analysis of algae treated with a specific inhibitor of carotenoid biosynthesis, high-performance liquid chromatography, and enzymatic docking analyses. As such, this study characterized the first GGPS gene for the order Bangiales and contributed to the understanding of the carotenoid metabolism in red algae.
Similarly, Deng et al.76 cloned and expressed a candidate GGPPS gene from the red macroalgae B. fuscopurpurea in E. coli and confirmed, through enzymatic assays, that BfGGPPS is capable of biosynthesizing geranylgeraniol as its only product. By using yeast-two-hybrid assays these authors detected the interaction of BfGGPPS with a candidate phytoene synthase and suggested a putative mechanism modulating metabolic flux through protein–protein interaction in red algal carotenoid biosynthesis. The authors also suggested an early divergence of GGPSs from red and green algae lineages during evolution, based on phylogenetic analyses. Further, a recent study from Li et al.160 functionally characterized the first phytoene synthase from a red alga through cloning and expressing a candidate gene from P. yezoensis in E. coli cells. The authors also analyzed the phylogenetic relationship of this gene in different organisms, adding another important piece of information regarding the evolution of carotenoid biosynthesis. Several studies also applied heterologous gene expression to characterize TPS genes and produce valuable terpenes. As an example, Kersten et al.101 identified and characterized a sesquiterpene synthase from Laurencia pacifica through the expression of the LphTPS-A gene in Saccharomyces cerevisiae and E. coli, resulting in the principal production of prespatane [23], a bourbonane-type sesquiterpene. This was the first study to functionally define a bourbonane-producing sesquiterpene synthase from red algae. Also, Wei et al.74 determined the biochemical functions of terpene synthases from the red microalgae P. purpureum (PpMTPSL) and E. australicus (EaMTPSL1 and EaMTPSL2) by cloning and expressing the candidate genes in E. coli cells. Through in vitro assays, the authors demonstrated that PpMTPSL was active with both substrates (E,E)-FPP and (E,E,E)-GGPP and produced sesquiterpenes and diterpenes, respectively. In contrast, EaMTPSL1 was able to accept GPP and (E,E)-FPP as substrates and converted them into specific mono- and sesquiterpenes, respectively, whereas EaMTPSL2 converted (E,E)-FPP into a mixture of sesquiterpenes. This was the first study to demonstrate a promiscuous potential for red algal TPS, although further analyses are necessary to confirm these findings and evaluate the contribution of this mechanism to the chemical diversity of terpenes in Rhodophycota.
Moreover, Steele et al.77 functionally characterized 10 new type I terpene synthase sequences from the red macroalgae species P. hornemannii, P. pacificum, L. pacifica, and L. subopposita through the heterologous gene expression in E. coli. The authors reported the selective activity of monoterpene synthases from P. pacificum and P. hornemannii, and the selective sesquiterpene synthase activity of MTPSL sequences from P. hornemannii, L. subopposita and L. pacifica. This work significantly expanded the knowledge regarding the red algal terpene synthase family.
Finally, Kaneko et al.161 reported the functional characterization and bromination activity of a vanadium-dependent bromoperoxidase (V-BPO) from the red seaweed Laurencia saitoi by cloning the candidate gene in E. coli cells. This is the first report of cloning and expression of a gene involved in the chemical modification of terpenes and represents an important step towards the heterologous synthesis of halogenated red algal compounds. Genetic manipulation for the biosynthesis of natural products from red algae remains an underexplored area. Notably, the few studies in this field focused on E. coli and yeast as hosts, despite the advances on the engineering of cyanobacteria and microalgae for terpene production.162,163 In special, the green microalga C. reinhardtii and the primitive red microalga C. merolae are emerging as platforms for the biosynthesis of bioproducts, including specialized terpenes and carotenoids.150,163 Important technological advances for the broad utilization of microalgae as hosts include new methods for gene delivery, gene editing, promoter engineering and transcription factors-based metabolic engineering.164,165
In general, more studies applying cloning and expression techniques for the functional characterization of red algal genes are needed. Also, the use of long-read sequencing and the adoption of high-standard bioinformatic approaches for assembly and annotation are important measures to increase genomic information for Rhodophycota. Combined with bioinformatics tools, the use of multi-omics techniques, such as genomics, transcriptomics, proteomics, and metabolomics, can integrate heterologous expression platforms, allowing the identification of bottlenecks in the production of commercially valuable compounds.166–168 Through molecular approaches, it is possible to clone and co-express different genes, enabling the biosynthesis of a wide range of compounds of biotechnological interest. Further, protein engineering provides the opportunity to improve the properties of enzymes involved in terpene biosynthesis by modifying specific amino acid residues. Also, gene editing techniques can target enzymes responsible for rate-limiting steps to enhance metabolite production rates.126,169–172 Future advances in these areas may open new perspectives for genetic engineering and the production of commercially valuable natural products derived from red algae.
4. Conclusion and future perspectives
Despite the broad biotechnological potential of red algal terpenes, the complex molecular and chemical processes underlying the biosynthesis of this diversified class of natural products are still poorly understood. In the present review, we categorized the biosynthetic steps of red algal terpenes into four complexity levels: the synthesis of isoprenoid precursors; the linear condensation of precursors to produce polyisoprenyl diphosphate; the chemical/structural modifications catalyzed by terpene synthases; and the additional chemical/structural modifications in the basic terpene skeleton. The studies regarding the contribution of each of these levels to the chemical diversity of terpenes in red algae were carefully discussed, and the knowledge gaps were highlighted. The complete or partial absence of the MVA in several, but not all, red macroalgae was considered in light of the evolutionary trajectory and the availability of transcriptomic data for red algal lineages. The studies characterizing IPS genes in red algae were also analyzed and the need for investigating their occurrence, distribution, and regulatory processes was emphasized. The molecular diversity and the catalytic and substrate promiscuity of TPS were discussed as sources of variations that contribute to the wide array of chemically diverse red algal terpenes. Finally, the chemical modifications of the basic terpene skeleton were analyzed.Future research should expand the genomic and transcriptomic data for understudied red algal species, elucidate the protein crystal structures for these algal TPS, and use computational approaches to predict their products. Co-expression analyses may also reveal the relevant pathways for terpene chemical modifications. Finally, directed mutagenesis studies have the potential to elucidate structure–function relationships and improve the heterologous synthesis of valuable red algal terpenes.
5. Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.6. Author contributions
WFSP participated in conceptualization, investigation, methodology, original draft writing and manuscript review. RCP participated in conceptualization, original draft writing and manuscript review. ESSA participated in original draft writing and manuscript review. FMSJ participated in manuscript review. RC participated in original draft writing. LSO participated in conceptualization, investigation, methodology, original draft writing and manuscript review.7. Conflicts of interest
There are no conflicts to declare.8. Acknowledgements
This work was supported by CAPES, CNPq, and FAPERJ. WFSP thanks FAPERJ for her PhD Scholarship. RC thanks CNPq for his Research Productivity Fellowships. RC and RCP thank FAPERJ for the Scientist of State Grant. LSO thanks FAPERJ for the granting of the Young Researcher and APQ1 projects (grant numbers E-26/210.341/2022 and E-26/2010.997/2024).9. References
- A. R. Carroll, B. R. Copp, T. Grkovic, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2024, 41, 162–207 RSC.
- E. Alves, M. Dias, D. Lopes, A. Almeida, M. d. R. Domingues and F. Rey, Antibiotics, 2020, 9, 441 CrossRef CAS PubMed.
- Y. Hu, J. Chen, G. Hu, J. Yu, X. Zhu, Y. Lin, S. Chen and J. Yuan, Mar. Drugs, 2015, 13, 202–221 CrossRef PubMed.
- P. Kiuru, M. D'Auria, C. Muller, P. Tammela, H. Vuorela and J. Yli-Kauhaluoma, Planta Med., 2014, 80, 1234–1246 CrossRef CAS PubMed.
- R. Montaser and H. Luesch, Future Med. Chem., 2011, 3, 1475–1489 CrossRef CAS PubMed.
- V. Ninkuu, L. Zhang, J. Yan, Z. Fu, T. Yang and H. Zeng, Int. J. Mol. Sci., 2021, 22, 5710 CrossRef CAS PubMed.
- C. Li, W. Zha, W. Li, J. Wang and A. You, Int. J. Mol. Sci., 2023, 24, 11561 CrossRef CAS PubMed.
- D. D. Das, N. Sharma, V. Chawla and P. A. Chawla, Crit. Rev. Anal. Chem., 2023, 1–17 Search PubMed.
- C. D. Amsler, in Algal Chemical Ecology, Springer Berlin Heidelberg, Berlin, Heidelberg, 2008, pp. 297–309 Search PubMed.
- S. D. Tetali, Planta, 2019, 249, 1–8 CrossRef CAS PubMed.
- M. D. Guiry, J. Phycol., 2024, 60, 214–228 CrossRef PubMed.
- A.-M. Cikoš, M. Jurin, R. Čož-Rakovac, S. Jokić and I. Jerković, Mar. Drugs, 2019, 17, 537 CrossRef PubMed.
- B.-G. Wang, J. B. Gloer, N.-Y. Ji and J.-C. Zhao, Chem. Rev., 2013, 113, 3632–3685 CrossRef CAS PubMed.
- C.-Q. Li, Q.-Y. Ma, X.-Z. Gao, X. Wang and B.-L. Zhang, Mar. Drugs, 2021, 19, 572 CrossRef CAS PubMed.
- M. L. Del Prado-Audelo, H. Cortés, I. H. Caballero-Florán, M. González-Torres, L. Escutia-Guadarrama, S. A. Bernal-Chávez, D. M. Giraldo-Gomez, J. J. Magaña and G. Leyva-Gómez, Front. Pharmacol., 2021, 12, 704197 CrossRef CAS PubMed.
- L. A. Minhas, M. Kaleem, H. M. U. Farooqi, F. Kausar, R. Waqar, T. Bhatti, S. Aziz, D. W. Jung and A. S. Mumtaz, Algal Res., 2024, 78, 103396 CrossRef.
- L.-A. Tziveleka, M. A. Tammam, O. Tzakou, V. Roussis and E. Ioannou, Antioxidants, 2021, 10, 1431 CrossRef CAS PubMed.
- P. Nova, A. M. Gomes and A. R. Costa-Pinto, Crit. Rev. Biotechnol., 2023, 1–15 Search PubMed.
- M. M. El-Sheekh, R. A. E. K. El-Shenody, E. A. Bases and S. M. El Shafay, Food Sci. Technol., 2021, 41, 29–40 CrossRef.
- J. R. B. Monteiro, R. P. Rodrigues, A. C. Mazzuco, R. de Cassia Ribeiro Gonçalves, A. F. Bernardino, R. M. Kuster and R. R. Kitagawa, Mar. Drugs, 2023, 21, 318 CrossRef CAS PubMed.
- R. Chatter, R. Ben Othman, S. Rabhi, M. Kladi, S. Tarhouni, C. Vagias, V. Roussis, L. Guizani-Tabbane and R. Kharrat, Mar. Drugs, 2011, 9, 1293–1306 CrossRef CAS PubMed.
- M. G. Knott, J.-A. de la Mare, A. L. Edkins, A. Zhang, M. J. Stillman, J. J. Bolton, E. M. Antunes and D. R. Beukes, Phytochem. Lett., 2019, 29, 182–185 CrossRef CAS.
- L. P. Machado, L. R. Carvalho, M. C. M. Young, E. M. Cardoso-Lopes, D. C. Centeno, L. Zambotti-Villela, P. Colepicolo and N. S. Yokoya, Rev. Bras. Farmacogn., 2015, 25, 657–662 CrossRef CAS.
- C. A. Motti, P. Thomas-Hall, K. A. Hagiwara, C. J. Simmons, R. Willis and A. D. Wright, J. Nat. Prod., 2014, 77, 1193–1200 CrossRef CAS PubMed.
- M. Pérez, E. Falqué and H. Domínguez, Mar. Drugs, 2016, 14, 52 CrossRef PubMed.
- A. J. Shilling, J. L. von Salm, A. R. Sanchez, Y. Kee, C. D. Amsler, J. B. McClintock and B. J. Baker, Mar. Drugs, 2019, 17, 230 CrossRef CAS PubMed.
- E. M. Antunes, A. F. Afolayan, M. T. Chiwakata, J. Fakee, M. G. Knott, C. E. Whibley, D. T. Hendricks, J. J. Bolton and D. R. Beukes, Phytochemistry, 2011, 72, 769–772 CrossRef CAS PubMed.
- A. Campos, C. B. Souza, C. Lhullier, M. Falkenberg, E. P. Schenkel, R. M. Ribeiro-do-Valle and J. M. Siqueira, J. Pharm. Pharmacol., 2012, 64, 1146–1154 CrossRef CAS PubMed.
- B. A. P. Da Gama, R. C. Pereira, A. G. V. Carvalho, R. Coutinho and Y. Yoneshigue-Valentin, Biofouling, 2002, 18, 13–20 CrossRef CAS.
- R. C. Pereira, B. A. P. da Gama, V. L. Teixeira and Y. Yoneshigue-Valentin, Braz. J. Microbiol., 2003, 63, 665–672 CAS.
- A. O. dos Santos, P. Veiga-Santos, T. Ueda-Nakamura, B. P. D. Filho, D. B. Sudatti, É. M. Bianco, R. C. Pereira and C. V. Nakamura, Mar. Drugs, 2010, 8, 2733–2743 CrossRef PubMed.
- F. L. d. S. Machado, C. R. Kaiser, S. S. Costa, L. M. Gestinari and A. R. Soares, Rev. Bras. Farmacogn., 2010, 20, 441–452 CrossRef CAS.
- Z.-B. Hu, X.-Q. Yu, B. Wang, A.-H. Liu, T.-S. Zhao, Y.-W. Guo, H.-L. Huang and S.-C. Mao, Fitoterapia, 2020, 146, 104716 CrossRef CAS PubMed.
- K. R. R. Rengasamy, L. P. Slavětínská, M. G. Kulkarni, W. A. Stirk and J. Van Staden, Algal Res., 2017, 25, 178–183 CrossRef.
- R. C. Pereira and L. V. Costa-Lotufo, Rev. Bras. Farmacogn., 2012, 22, 894–905 CrossRef CAS.
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch and C. T. Supuran, Nat. Rev. Drug Discovery, 2021, 20, 200–216 CrossRef CAS PubMed.
- W. M. Brück, L. Santiago-Vázquez and T. B. Brück, Front. Mar. Sci., 2023, 10, 1188090 CrossRef.
- M. S. Belcher, J. Mahinthakumar and J. D. Keasling, Curr. Opin. Biotechnol., 2020, 65, 88–93 CrossRef CAS PubMed.
- L. Pimentel, E. Carsanba, F. Teixeira, S. Vidigal, M. Pintado, C. Oliveira and L. M. Rodríguez-Alcalá, in Microbial Production of Food Bioactive Compounds, Springer International Publishing, Cham, 2022, pp. 1–38 Search PubMed.
- R. Chen, M. Wang, J. D. Keasling, T. Hu and X. Yin, Trends Biotechnol., 2024, 42(6), 699–713 CrossRef CAS PubMed.
- D. W. Christianson, Chem. Rev., 2017, 117, 11570–11648 CrossRef CAS PubMed.
- C. Ignea, M. H. Raadam, A. Koutsaviti, Y. Zhao, Y.-T. Duan, M. Harizani, K. Miettinen, P. Georgantea, M. Rosenfeldt, S. E. Viejo-Ledesma, M. A. Petersen, W. L. P. Bredie, D. Staerk, V. Roussis, E. Ioannou and S. C. Kampranis, Nat. Commun., 2022, 13, 5188 CrossRef CAS PubMed.
- E. Vranová, D. Coman and W. Gruissem, Annu. Rev. Plant Biol., 2013, 64, 665–700 CrossRef PubMed.
- A. O. Chatzivasileiou, V. Ward, S. M. Edgar and G. Stephanopoulos, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 506–511 CrossRef CAS PubMed.
- J. Lombard and D. Moreira, Mol. Biol. Evol., 2011, 28, 87–99 CrossRef CAS PubMed.
- L. Zeng and K. Dehesh, BMC Genomics, 2021, 22, 137 CrossRef CAS PubMed.
- H. Qiu, H. S. Yoon and D. Bhattacharya, PLOS Curr, 2016, 8, 1 Search PubMed.
- T. Kuzuyama and H. Seto, Proc. Jpn. Acad., Ser. B, 2012, 88, 41–52 CrossRef CAS PubMed.
- W. C. Paradas, T. M. Crespo, L. T. Salgado, L. R. de Andrade, A. R. Soares, C. Hellio, R. R. Paranhos, L. J. Hill, G. M. de Souza, A. G. A. C. Kelecom, B. A. P. Da Gama, R. C. Pereira and G. M. Amado-Filho, J. Phycol., 2015, 51, 225–235 CrossRef CAS PubMed.
- F. Abbas, Y. Ke, R. Yu, Y. Yue, S. Amanullah, M. M. Jahangir and Y. Fan, Planta, 2017, 246, 803–816 CrossRef CAS PubMed.
- D. Tholl, Curr. Opin. Plant Biol., 2006, 9, 297–304 CrossRef CAS PubMed.
- D. Tholl and S. Lee, Arabidopsis Book, 2011, vol. 9, p. e0143 Search PubMed.
- P. S. Karunanithi and P. Zerbe, Front. Plant Sci., 2019, 10, 1066 CrossRef PubMed.
- F. Q. Y. Goh, J. Jeyakani, P. Tipthara, A. Cazenave-Gassiot, R. Ghosh, N. Bogard, Z. Yeo, G. K.-S. Wong, M. Melkonian, M. R. Wenk and N. D. Clarke, Sci. Rep., 2019, 9, 10482 CrossRef PubMed.
- H. Qiu, D. C. Price, E. C. Yang, H. S. Yoon and D. Bhattacharya, J. Phycol., 2015, 51, 624–636 CrossRef CAS PubMed.
- M. Borg, S. A. Krueger-Hadfield, C. Destombe, J. Collén, A. Lipinska and S. M. Coelho, New Phytol., 2023, 240, 471–488 CrossRef PubMed.
- L. de Oliveira, D. Tschoeke, A. de Oliveira, L. Hill, W. Paradas, L. Salgado, C. Thompson, R. Pereira and F. Thompson, Mar. Drugs, 2015, 13, 879–902 CrossRef PubMed.
- L. S. de Oliveira, G. B. Gregoracci, G. G. Z. Silva, L. T. Salgado, G. A. Filho, M. Alves-Ferreira, R. C. Pereira and F. L. Thompson, BMC Genomics, 2012, 13, 487 CrossRef PubMed.
- C. Grauvogel and J. Petersen, Gene, 2007, 396, 125–133 CrossRef CAS PubMed.
- A. Disch, J. Schwender, C. Muller, H. K. Lichtenthaler and M. Rohmer, Biochem. J., 1998, 333, 381–388 CrossRef CAS PubMed.
- M. Lohr, J. Schwender and J. E. W. Polle, Plant Sci., 2012, 185–186, 9–22 CrossRef CAS PubMed.
- M. Matsuzaki, O. Misumi, T. Shin-I, S. Maruyama, M. Takahara, S.-Y. Miyagishima, T. Mori, K. Nishida, F. Yagisawa, K. Nishida, Y. Yoshida, Y. Nishimura, S. Nakao, T. Kobayashi, Y. Momoyama, T. Higashiyama, A. Minoda, M. Sano, H. Nomoto, K. Oishi, H. Hayashi, F. Ohta, S. Nishizaka, S. Haga, S. Miura, T. Morishita, Y. Kabeya, K. Terasawa, Y. Suzuki, Y. Ishii, S. Asakawa, H. Takano, N. Ohta, H. Kuroiwa, K. Tanaka, N. Shimizu, S. Sugano, N. Sato, H. Nozaki, N. Ogasawara, Y. Kohara and T. Kuroiwa, Nature, 2004, 482, 653–657 CrossRef PubMed.
- S. H. Brawley, N. A. Blouin, E. Ficko-Blean, G. L. Wheeler, M. Lohr, H. V. Goodson, J. W. Jenkins, C. E. Blaby-Haas, K. E. Helliwell, C. X. Chan, T. N. Marriage, D. Bhattacharya, A. S. Klein, Y. Badis, J. Brodie, Y. Cao, J. Collén, S. M. Dittami, C. M. M. Gachon, B. R. Green, S. J. Karpowicz, J. W. Kim, U. J. Kudahl, S. Lin, G. Michel, M. Mittag, B. J. S. C. Olson, J. L. Pangilinan, Y. Peng, H. Qiu, S. Shu, J. T. Singer, A. G. Smith, B. N. Sprecher, V. Wagner, W. Wang, Z.-Y. Wang, J. Yan, C. Yarish, S. Zäuner-Riek, Y. Zhuang, Y. Zou, E. A. Lindquist, J. Grimwood, K. W. Barry, D. S. Rokhsar, J. Schmutz, J. W. Stiller, A. R. Grossman and S. E. Prochnik, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6361–6370 CrossRef PubMed.
- C. X. Chan, N. A. Blouin, Y. Zhuang, S. Zäuner, S. E. Prochnik, E. Lindquist, S. Lin, C. Benning, M. Lohr, C. Yarish, E. Gantt, A. R. Grossman, S. Lu, K. Müller, J. W. Stiller, S. H. Brawley and D. Bhattacharya, J. Phycol., 2012, 48, 1328–1342 CrossRef CAS PubMed.
- D. Bhattacharya, H. Qiu, J. M. Lee, H. Su Yoon, A. P. M. Weber and D. C. Price, CRC Crit. Rev. Plant Sci., 2018, 37, 81–99 CrossRef.
- P. Pulido, C. Perello and M. Rodriguez-Concepcion, Mol. Plant, 2012, 5, 964–967 CrossRef CAS PubMed.
- M. E. Bergman, B. Davis and M. A. Phillips, Molecules, 2019, 24, 3961 CrossRef CAS PubMed.
- Q. Wang, S. Quan and H. Xiao, Bioresour. Bioprocess., 2019, 6, 6 CrossRef.
- L. Carretero-Paulet, A. Cairó, P. Botella-Pavía, O. Besumbes, N. Campos, A. Boronat and M. Rodríguez-Concepción, Plant Mol. Biol., 2006, 62, 683–695 CrossRef CAS PubMed.
- Y. Du, J. Guan, R. Xu, X. Liu, W. Shen, Y. Ma, Y. He and S. Shen, Algae, 2017, 32, 359–377 CrossRef CAS.
- J. Liu, Y. Xu, L. Liang and J. Wei, J. Genet., 2015, 94, 239–249 CrossRef CAS PubMed.
- A. Rautela and S. Kumar, Plant Cell Rep., 2022, 41, 1791–1803 CrossRef CAS PubMed.
- J. F. H. Strassert, I. Irisarri, T. A. Williams and F. Burki, Nat. Commun., 2021, 12, 1879 CrossRef CAS PubMed.
- G. Wei, Q. Jia, X. Chen, T. G. Köllner, D. Bhattacharya, G. K.-S. Wong, J. Gershenzon and F. Chen, Plant Physiol., 2019, 179, 382–390 CrossRef CAS PubMed.
- D. Bhattacharya, D. C. Price, C. X. Chan, H. Qiu, N. Rose, S. Ball, A. P. M. Weber, M. Cecilia Arias, B. Henrissat, P. M. Coutinho, A. Krishnan, S. Zäuner, S. Morath, F. Hilliou, A. Egizi, M.-M. Perrineau and H. S. Yoon, Nat. Commun., 2013, 4, 1941 CrossRef PubMed.
- Y.-Y. Deng, Q. Wang, T.-J. Cao, H. Zheng, Z.-H. Ge, L.-E. Yang and S. Lu, Algal Res., 2020, 48, 101935 CrossRef.
- T. S. Steele, I. Burkhardt, M. L. Moore, T. de Rond, H. K. Bone, K. Barry, V. M. Bunting, J. Grimwood, L. H. Handley, S. Rajasekar, J. Talag, T. P. Michael and B. S. Moore, ACS Chem. Biol., 2023, 19, 185–192 CrossRef PubMed.
- D. Manzano, P. Andrade, D. Caudepón, T. Altabella, M. Arró and A. Ferrer, Plant Physiol., 2016, 172, 93–117 CrossRef CAS PubMed.
- D. Kopcsayová and E. Vranová, Molecules, 2019, 24, 4556 CrossRef PubMed.
- A. Hemmerlin, Plant Sci., 2013, 203–204, 41–54 CrossRef CAS PubMed.
- K. Okada, H. Kasahara, S. Yamaguchi, H. Kawaide, Y. Kamiya, H. Nojiri and H. Yamane, Plant Cell Physiol., 2008, 49, 604–616 CrossRef CAS PubMed.
- A. Lipko, C. Pączkowski, L. Perez-Fons, P. D. Fraser, M. Kania, M. Hoffman-Sommer, W. Danikiewicz, M. Rohmer, J. Poznanski and E. Swiezewska, Biochem. J., 2023, 480, 495–520 CrossRef CAS PubMed.
- S. Opitz, W. D. Nes and J. Gershenzon, Phytochemistry, 2014, 98, 110–119 CrossRef CAS PubMed.
- K. Skorupinska-Tudek, J. Poznanski, J. Wojcik, T. Bienkowski, I. Szostkiewicz, M. Zelman-Femiak, A. Bajda, T. Chojnacki, O. Olszowska, J. Grunler, O. Meyer, M. Rohmer, W. Danikiewicz and E. Swiezewska, J. Biol. Chem., 2008, 283, 21024–21035 CrossRef CAS PubMed.
- H. Chang, T. Cheng and A. H. J. Wang, IUBMB Life, 2021, 73, 40–63 CrossRef CAS PubMed.
- S. Takahashi and T. Koyama, Chem. Rec., 2006, 6, 194–205 CrossRef CAS PubMed.
- P. Liang, T. Ko and A. H. J. Wang, Eur. J. Biochem., 2002, 269, 3339–3354 CrossRef CAS PubMed.
- A. Satta, L. Esquirol, B. E. Ebert, J. Newman, T. S. Peat, M. Plan, G. Schenk and C. E. Vickers, FEBS J., 2022, 289, 6672–6693 CrossRef CAS PubMed.
- L. Surmacz and E. Swiezewska, Biochem. Biophys. Res. Commun., 2011, 407, 627–632 CrossRef CAS PubMed.
- C. Sallaud, D. Rontein, S. Onillon, F. Jabès, P. Duffé, C. Giacalone, S. Thoraval, C. Escoffier, G. Herbette, N. Leonhardt, M. Causse and A. Tissier, Plant Cell, 2009, 21, 301–317 CrossRef CAS PubMed.
- K. A. Grabińska, E. J. Park and W. C. Sessa, J. Biol. Chem., 2016, 291, 18582–18590 CrossRef PubMed.
- F. Gong, C. Zhang, L. Zhang and J. Liu, Aquacult. Res., 2020, 51, 770–778 CrossRef CAS.
- Q. Luo, C. Bian, M. Tao, Y. Huang, Y. Zheng, Y. Lv, J. Li, C. Wang, X. You, B. Jia, J. Xu, J. Li, Z. Li, Q. Shi and Z. Hu, Genome Biol. Evol., 2019, 11, 166–173 CrossRef CAS PubMed.
- J. E. W. Polle, S. Calhoun, Z. McKie-Krisberg, S. Prochnik, P. Neofotis, W. C. Yim, L. T. Hathwaik, J. Jenkins, H. Molina, J. Bunkenborg, I. V. Grigoriev, K. Barry, J. Schmutz, E. Jin, J. C. Cushman and J. K. Magnusson, Algal Res., 2020, 50, 101990 CrossRef.
- L.-E. Yang, X.-Q. Huang, Q.-Q. Lu, J.-Y. Zhu and S. Lu, J. Appl. Phycol., 2016, 28, 671–678 CrossRef CAS.
- S. Song, R. Jin, Y. Chen, S. He, K. Li, Q. Tang, Q. Wang, L. Wang, M. Kong, N. Dudareva, B. J. Smith, F. Zhou and S. Lu, Plant Cell, 2023, 35, 2293–2315 CrossRef PubMed.
- F. Chen, D. Tholl, J. Bohlmann and E. Pichersky, Plant J., 2011, 66, 212–229 CrossRef CAS PubMed.
- J. Degenhardt, T. G. Köllner and J. Gershenzon, Phytochemistry, 2009, 70, 1621–1637 CrossRef CAS PubMed.
- F. Zhou and E. Pichersky, Curr. Opin. Plant Biol., 2020, 55, 1–10 CrossRef CAS PubMed.
- Q. Jia, G. Li, T. G. Köllner, J. Fu, X. Chen, W. Xiong, B. J. Crandall-Stotler, J. L. Bowman, D. J. Weston, Y. Zhang, L. Chen, Y. Xie, F. W. Li, C. J. Rothfels, A. Larsson, S. W. Graham, D. W. Stevenson, G. K. S. Wong, J. Gershenzon and F. Chen, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, 12328–12333 CrossRef CAS PubMed.
- R. D. Kersten, S. Lee, D. Fujita, T. Pluskal, S. Kram, J. E. Smith, T. Iwai, J. P. Noel, M. Fujita and J. K. Weng, J. Am. Chem. Soc., 2017, 139, 16838–16844 CrossRef CAS PubMed.
- Q. Jia, T. G. Köllner, J. Gershenzon and F. Chen, Trends Plant Sci., 2018, 23, 121–128 CrossRef CAS PubMed.
- Z.-Y. Huang, R.-Y. Ye, H.-L. Yu, A.-T. Li and J.-H. Xu, Bioresour. Bioprocess., 2021, 8, 66 CrossRef PubMed.
- D. W. Christianson, Chem. Rev., 2006, 106, 3412–3442 CrossRef CAS PubMed.
- J. L. Faylo, T. A. Ronnebaum and D. W. Christianson, Acc. Chem. Res., 2021, 54, 3780–3791 CrossRef CAS PubMed.
- Q. Jia, R. Brown, T. G. Köllner, J. Fu, X. Chen, G. Ka-Shu Wong, J. Gershenzon, R. J. Peters and F. Chen, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2100361119 CrossRef PubMed.
- F. Zhou and E. Pichersky, New Phytol., 2020, 226, 1341–1360 CrossRef CAS PubMed.
- R. Cao, Y. Zhang, F. M. Mann, C. Huang, D. Mukkamala, M. P. Hudock, M. E. Mead, S. Prisic, K. Wang, F. Lin, T. Chang, R. J. Peters and E. Oldfield, Proteins: Struct., Funct., Bioinf., 2010, 78, 2417–2432 CrossRef CAS PubMed.
- X. Pan, J. D. Rudolf and L.-B. Dong, Nat. Prod. Rep., 2024, 41, 402–433 RSC.
- J. Bohlmann, G. Meyer-Gauen and R. Croteau, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 4126–4133 CrossRef CAS PubMed.
- K. Schriever, P. Saenz-Mendez, R. S. Rudraraju, N. M. Hendrikse, E. P. Hudson, A. Biundo, R. Schnell and P.-O. Syrén, J. Am. Chem. Soc., 2021, 143, 3794–3807 CrossRef CAS PubMed.
- X. Yan, J. Zhou, J. Ge, W. Li, D. Liang, W. Singh, G. Black, S. Nie, J. Liu, M. Sun, J. Qiao and M. Huang, ACS Catal., 2022, 12, 4037–4045 CrossRef CAS.
- M. Loizzi, D. J. Miller and R. K. Allemann, Org. Biomol. Chem., 2019, 17, 1206–1214 RSC.
- Y. Tong, T. Hu, L. Tu, K. Chen, T. Liu, P. Su, Y. Song, Y. Liu, L. Huang and W. Gao, Int. J. Biol. Macromol., 2021, 185, 949–958 CrossRef CAS PubMed.
- J. N. Whitehead, N. G. H. Leferink, L. O. Johannissen, S. Hay and N. S. Scrutton, ACS Catal., 2023, 13, 12774–12802 CrossRef CAS PubMed.
- L. Pazouki and Ü. Niinemets, Front. Plant Sci., 2016, 7, 1–16 Search PubMed.
- Y. Zhao, T. Hu, R. Liu, Z. Hao, G. Liang and G. Li, Int. J. Mol. Sci., 2021, 22, 605 CrossRef CAS PubMed.
- W. Xiong, J. Fu, T. G. Köllner, X. Chen, Q. Jia, H. Guo, P. Qian, H. Guo, G. Wu and F. Chen, Phytochemistry, 2018, 149, 116–122 CrossRef CAS PubMed.
- J. Hattan, K. Shindo, T. Sasaki and N. Misawa, J. Oleo Sci., 2018, 67, 1235–1246 CrossRef CAS PubMed.
- J.-X. Ruan, J.-X. Li, X. Fang, L.-J. Wang, W.-L. Hu, X.-Y. Chen and C.-Q. Yang, Front. Plant Sci., 2016, 7, 638 Search PubMed.
- M. Zhang, J. Liu, K. Li and D. Yu, PLoS One, 2013, 8, e75972 CrossRef CAS PubMed.
- A. Di Girolamo, J. Durairaj, A. van Houwelingen, F. Verstappen, D. Bosch, K. Cankar, H. Bouwmeester, D. de Ridder, A. D. J. van Dijk and J. Beekwilder, Arch. Biochem. Biophys., 2020, 695, 108647 CrossRef CAS PubMed.
- C. Ignea, M. Pontini, M. E. Maffei, A. M. Makris and S. C. Kampranis, ACS Synth. Biol., 2014, 3, 298–306 CrossRef CAS PubMed.
- D.-S. Ker, K. G. Chan, R. Othman, M. Hassan and C. L. Ng, Phytochemistry, 2020, 173, 112286 CrossRef CAS PubMed.
- J.-X. Li, X. Fang, Q. Zhao, J.-X. Ruan, C.-Q. Yang, L.-J. Wang, D. J. Miller, J. A. Faraldos, R. K. Allemann, X.-Y. Chen and P. Zhang, Biochem. J., 2013, 451, 417–426 CrossRef CAS PubMed.
- H. Xu and J. S. Dickschat, Synthesis, 2022, 54, 1551–1565 CrossRef CAS.
- D. H. Lamparelli, M. Winnacker and C. Capacchione, Chempluschem, 2022, 87, e202100366 CrossRef CAS PubMed.
- M. Plaza, S. Santoyo, L. Jaime, G. García-Blairsy Reina, M. Herrero, F. J. Señoráns and E. Ibáñez, J. Pharm. Biomed. Anal., 2010, 51, 450–455 CrossRef CAS PubMed.
- M. Ashour, M. Wink and J. Gershenzon, in Biochemistry of Plant Secondary Metabolism, Wiley-Blackwell, Oxford, UK, pp. , pp. 258–303 Search PubMed.
- A.-M. Cikoš, M. Jurin, R. Čož-Rakovac, D. Gašo-Sokač, S. Jokić and I. Jerković, Algal Res., 2021, 56, 102330 CrossRef.
- M. Harizani, E. Ioannou and V. Roussis, in Progress in the Chemistry of Organic Natural Products, 2016, vol. 102, pp. 91–252 Search PubMed.
- J. S. Dickschat, Nat. Prod. Rep., 2011, 28, 1917 RSC.
- L. R. de Carvalho, M. T. Fujii, N. F. Roque, M. J. Kato and J. H. G. Lago, Tetrahedron Lett., 2003, 44, 2637–2640 CrossRef CAS.
- X.-Q. Yu, W.-F. He, D.-Q. Liu, M.-T. Feng, Y. Fang, B. Wang, L.-H. Feng, Y.-W. Guo and S.-C. Mao, Phytochemistry, 2014, 103, 162–170 CrossRef CAS PubMed.
- F. da Silva Machado, W. Pacienza-Lima, B. Rossi-Bergmann, L. de Souza Gestinari, M. Fujii, J. Campos de Paula, S. Costa, N. Lopes, C. Kaiser and A. Soares, Planta Med., 2011, 77, 733–735 CrossRef PubMed.
- D. Davyt, R. Fernandez, L. Suescun, A. W. Mombrú, J. Saldaña, L. Domínguez, J. Coll, M. T. Fujii and E. Manta, J. Nat. Prod., 2001, 64, 1552–1555 CrossRef CAS PubMed.
- A. R. Díaz-Marrero, I. Brito, J. M. de la Rosa, L. D'Croz, O. Fabelo, C. Ruiz-Pérez, J. Darias and M. Cueto, Eur. J. Org. Chem., 2009, 2009, 1407–1411 CrossRef.
- E. Dorta, A. R. Díaz-Marrero, M. Cueto, L. D'Croz, J. L. Maté and J. Darias, Tetrahedron Lett., 2004, 45, 7065–7068 CrossRef CAS.
- X. Li, W. Ding, F. Miao and N. Ji, Magn. Reson. Chem., 2012, 50, 174–177 CrossRef CAS PubMed.
- D. E. White, I. C. Stewart, R. H. Grubbs and B. M. Stoltz, J. Am. Chem. Soc., 2008, 130, 810–811 CrossRef CAS PubMed.
- T. Kamada, C.-S. Phan, V. S.-T. Sien and C. S. Vairappan, J. Appl. Phycol., 2018, 30, 3373–3378 CrossRef CAS.
- T. Kamada, C.-S. Phan, T. Okino and C. S. Vairappan, Planta Med. Int. Open, 2019, 6, e36–e40 CrossRef.
- S. Cai and H. Xiao, in Microbial Cell Factories Engineering for Production of Biomolecules, Elsevier, 2021, pp. 37–49 Search PubMed.
- Z. Chang, Y. Li, Y. Lu and H. Xiao, Enzyme Microb. Technol., 2023, 162, 110148 CrossRef CAS PubMed.
- R. D. Kersten, J. K. Diedrich, J. R. Yates and J. P. Noel, ACS Chem. Biol., 2015, 10, 2501–2511 CrossRef CAS PubMed.
- L. Yang, X. Huang, Y. Hang, Y. Deng, Q. Lu, S. Lu and J. Integr, Plant Biol., 2014, 56, 902–915 CAS.
- S. Zheng, J. Guo, F. Cheng, Z. Gao, L. Du, C. Meng, S. Li and X. Zhang, Acta Pharm. Sin. B., 2022, 12, 2832–2844 CrossRef CAS PubMed.
- L. Teng, X. Fan, D. R. Nelson, W. Han, X. Zhang, D. Xu, H. Renault, G. V. Markov and N. Ye, Planta, 2019, 249, 647–661 CrossRef CAS PubMed.
- F. X. Cunningham, H. Lee and E. Gantt, Eukaryotic Cell, 2007, 6, 533–545 CrossRef CAS PubMed.
- M. Seger, F. Mammadova, M. Villegas-Valencia, B. Bastos de Freitas, C. Chang, I. Isachsen, H. Hemstreet, F. Abualsaud, M. Boring, P. J. Lammers and K. J. Lauersen, Metab. Eng. Commun., 2023, 17, e00226 CrossRef CAS PubMed.
- J. Hausjell, H. Halbwirth and O. Spadiut, Biosci. Rep., 2018, 38(2), BSR20171290 CrossRef CAS PubMed.
- L. McKinnie, S. Cummins and M. Zhao, Mar. Drugs, 2023, 22, 3 CrossRef PubMed.
- J. N. Carter-Franklin and A. Butler, J. Am. Chem. Soc., 2004, 126, 15060–15066 CrossRef CAS PubMed.
- C. Paul and G. Pohnert, Nat. Prod. Rep., 2011, 28, 186–195 RSC.
- T. Ishikawa, K. Washio, K. Kaneko, X. R. Tang, M. Morikawa and T. Okino, Appl. Phycol., 2022, 3, 120–131 CrossRef.
- A. Butler and M. Sandy, Nature, 2009, 460, 848–854 CrossRef CAS PubMed.
- A. Butler and J. N. Carter-Franklin, Nat. Prod. Rep., 2004, 21, 180 RSC.
- H. Esselin, S. Sutour, A. Bighelli and F. Tomi, Biochem. Syst. Ecol., 2019, 82, 24–26 CrossRef CAS.
- E. R. Hanschen and S. R. Starkenburg, Algal Res., 2020, 50, 101968 CrossRef.
- C.-L. Li, J.-Q. Pu, W. Zhou, C.-M. Hu, Y.-Y. Deng, Y.-Y. Sun and L.-E. Yang, Mar. Drugs, 2024, 22, 257 CrossRef CAS PubMed.
- K. Kaneko, D. Kobayashi, S. Masaki, K. Washio, M. Morikawa and T. Okino, J. Appl. Phycol., 2023, 35, 1443–1452 CrossRef CAS.
- P.-C. Lin and H. B. Pakrasi, Planta, 2019, 249, 145–154 CrossRef CAS PubMed.
- A. Einhaus, T. Baier, M. Rosenstengel, R. A. Freudenberg and O. Kruse, ACS Synth. Biol., 2021, 10, 847–856 CrossRef CAS PubMed.
- S. Gutiérrez and K. J. Lauersen, Biology, 2021, 10, 265 CrossRef PubMed.
- A. Gupta, K. Kang, R. Pathania, L. Saxton, B. Saucedo, A. Malik, Y. Torres-Tiji, C. J. Diaz, J. V. Dutra Molino and S. P. Mayfield, Front. Bioeng. Biotechnol., 12, 1350722 CrossRef PubMed.
- J. J. Zhang, X. Tang and B. S. Moore, Nat. Prod. Rep., 2019, 36, 1313–1332 RSC.
- Y. Luo, B.-Z. Li, D. Liu, L. Zhang, Y. Chen, B. Jia, B.-X. Zeng, H. Zhao and Y.-J. Yuan, Chem. Soc. Rev., 2015, 44, 5265–5290 RSC.
- M. B. Quin and C. Schmidt-Dannert, Curr. Opin. Biotechnol., 2014, 29, 55–61 CrossRef CAS PubMed.
- X. Chen, C. Gao, L. Guo, G. Hu, Q. Luo, J. Liu, J. Nielsen, J. Chen and L. Liu, Chem. Rev., 2018, 118, 4–72 CrossRef CAS PubMed.
- E. Fordjour, E. O. Mensah, Y. Hao, Y. Yang, X. Liu, Y. Li, C.-L. Liu and Z. Bai, Bioresour. Bioprocess., 2022, 9, 6 CrossRef PubMed.
- C. Li, R. Zhang, J. Wang, L. M. Wilson and Y. Yan, Trends Biotechnol., 2020, 38, 729–744 CrossRef CAS PubMed.
- M. J. R. Segura, B. E. Jackson and S. P. T. Matsuda, Nat. Prod. Rep., 2003, 20, 304–317 RSC.
| This journal is © The Royal Society of Chemistry 2025 |